Abstract
Human acute T-cell lymphoblastic leukemias and lymphomas (T-ALL) are commonly associated with gain-of-function mutations in Notch1 that contribute to T-ALL induction and maintenance. Starting from an expression-profiling screen, we identified c-myc as a direct target of Notch1 in Notch-dependent T-ALL cell lines, in which Notch accounts for the majority of c-myc expression. In functional assays, inhibitors of c-myc interfere with the progrowth effects of activated Notch1, and enforced expression of c-myc rescues multiple Notch1-dependent T-ALL cell lines from Notch withdrawal. The existence of a Notch1–c-myc signaling axis was bolstered further by experiments using c-myc-dependent murine T-ALL cells, which are rescued from withdrawal of c-myc by retroviral transduction of activated Notch1. This Notch1-mediated rescue is associated with the up-regulation of endogenous murine c-myc and its downstream transcriptional targets, and the acquisition of sensitivity to Notch pathway inhibitors. Additionally, we show that primary murine thymocytes at the DN3 stage of development depend on ligand-induced Notch signaling to maintain c-myc expression. Together, these data implicate c-myc as a developmentally regulated direct downstream target of Notch1 that contributes to the growth of T-ALL cells.
Keywords: Notch, Myc, leukemia, T cell, transformation
Notch receptors participate in a conserved signaling pathway that regulates the development of diverse cell and tissue types in metazoans. Outcomes resulting from Notch signals are highly pleiotropic, depending on dose and context (Artavanis-Tsakonas et al. 1999). Within the hematolymphoid compartment, Notch signaling affects lineage commitment at multiple developmental stages (for review, see Radtke et al. 2004; Maillard et al. 2005). Notch influences the self-renewal of hematopoietic stem cells (HSCs) (Varnum-Finney et al. 2000; Calvi et al. 2003; Duncan et al. 2005), T-cell specification from a multipotent precursor (Pui et al. 1999; Radtke et al. 1999; Sambandam et al. 2005; Tan et al. 2005), maturation of double-negative (DN) thymocytes, especially at the β-selection checkpoint (Wolfer et al. 2002; Tanigaki et al. 2004; Ciofani and Zuniga-Pflucker 2005; Taghon et al. 2006), and the differentiation of CD4+ T cells along either TH1 or TH2 pathways (Amsen et al. 2004; Tanigaki et al. 2004; Minter et al. 2005; Tu et al. 2005). Since the discovery of Notch1 through the analysis of a rare (7;9) chromosomal translocation in human T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) (Ellisen et al. 1991), abundant evidence has also accumulated implicating Notch1 in the pathogenesis of this aggressive cancer (Pear et al. 1996; Aster et al. 2000; Bellavia et al. 2000). Recently, activating mutations in Notch1 were discovered in 55%–60% of human T-ALLs (Weng et al. 2004), and emerging data indicate that similar types of Notch1 mutations occur frequently in many different murine T-ALL models as secondary events (Dumortier et al. 2006; Lin et al. 2006; O’Neil et al. 2006).
Normal Notch signaling is initiated by the binding of ligands of the Delta/Serrate/Lag-2 (DSL) family to the Notch ectodomain, which result in cleavage at a site just external to the transmembrane domain by ADAM metalloproteases (Brou et al. 2000; Mumm et al. 2000). This event creates a short-lived Notch intermediate (NTM*) that is recognized by nicastrin (Shah et al. 2005), a component of γ-secretase, which in turn, cleaves NTM* within its transmembrane domain (Schroeter et al. 1998; De Strooper et al. 1999; Kimberly et al. 2003). This final cleavage releases the intracellular domain of Notch (ICN) from the membrane, allowing it to translocate to the nucleus and form a transcriptional activation complex with a DNA-binding protein termed CSL (for CBF1, Suppressor of Hairless, Lag-1) (Jarriault et al. 1995; Kopan et al. 1996; Struhl and Greenwald 1999; Ye et al. 1999) and transcriptional coactivators of the Mastermind-like (MAML) family (Petcherski and Kimble 2000a, b; Wu et al. 2000).
Although signals mediated through Notch receptors have diverse outcomes, only a fairly limited set of Notch target genes have been identified in various cellular/developmental contexts. The hairy/enhancer of split (Hes) genes are highly conserved target genes that are regulated by Notch in multiple cell types (Preiss et al. 1988; Jarriault et al. 1995). On the other hand, investigators studying Notch1 function in immature T cells identified several likely T-cell-specific target genes, including CD25 and pre-Tα (Deftos et al. 2000; Reizis and Leder 2002). Other putative context-specific target genes that may promote cell growth include cyclinD1, which was identified as a transcriptional target in RKE cells (Ronchini and Capobianco 2001), and c-myc (Satoh et al. 2004), which was identified as a possible Notch target in hematopoietic stem cells. In this latter study, Notch responsiveness was linked to an ∼200-base-pair (bp) element lying immediately 5′ of the c-myc transcriptional start site, but neither direct association of Notch with this site nor its functional importance was demonstrated. Thus, the identities of the genes downstream of Notch1 that maintain the growth of T-ALL cells have yet to be determined.
To address this uncertainty, we used expression profiling to identify genes that are down-regulated by Notch pathway inhibitors in T6E, a murine T-ALL cell line whose growth depends on a membrane-tethered form of Notch1 resembling NTM (Weng et al. 2003). Among the potential target genes identified was c-myc, which has been shown to induce T-ALL in animal models when overexpressed (Girard et al. 1996; Felsher and Bishop 1999; Langenau et al. 2003). This insight led to a series of functional studies, which showed that c-myc is an important target of Notch not only in T-ALL cells, but also at a critical stage of normal pre-T-cell development.
Results
Identification of c-myc as a putative Notch1 target gene
To identify potential Notch1 target genes, we performed expression profiling on a set of RNAs obtained from T6E T-ALL cells in which Notch was turned “off” (γ-secretase inhibitor [GSI]-treated or DN-MAML1 transduced) or left “on” (untreated and mock GSI-treated cells, sorted GFP+ cells transduced with empty MigRI virus, and sorted GFP− cells from MigRI-DN-MAML1 cultures). RNAs were hybridized to Affymetrix U74Av2 GeneChip arrays (∼12,000 genes). Raw data were analyzed with dChip software (Li and Hung Wong 2001) by applying standard normalization and modeling routines, and by filtering gene lists based on “presence” call, expression level, and variation criteria. Hierarchical clustering performed using filtered gene sets resulted in unsupervised discovery of the Notch “on” and “off” sample groups. Supervised analysis was then applied to a filtered list of ∼600 genes to identify genes for which the expression levels were most highly correlated with Notch activation status.
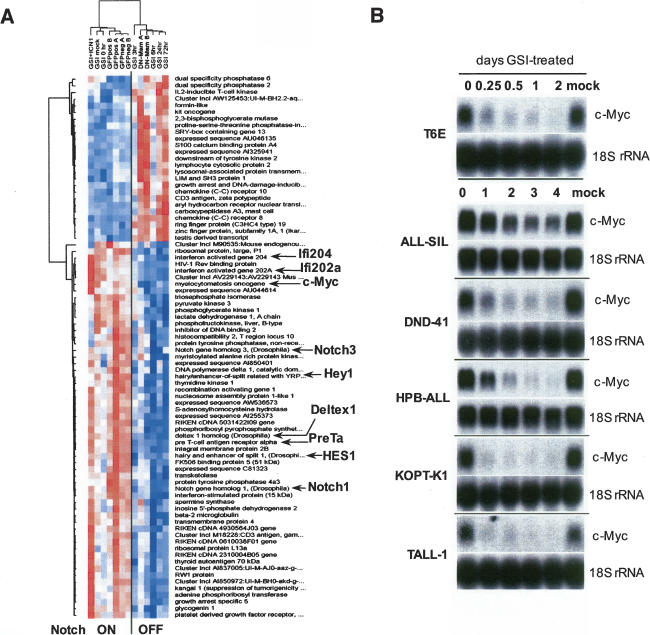
A total of 83 genes demonstrated multiple-comparison-adjusted p-values of <0.05 (Fig. 1A; see Supplementary Table S1 for the full list of genes, fold changes, and p-values). Genes down-regulated in the Notch “off” group included the majority of known/previously described Notch targets in immature T cells (hes1, hey1, pre-Tα, deltex1, and several interferon-induced genes), and c-myc. We also performed an independent set of expression profiling experiments in which we sought to identify pathways that were perturbed by GSI treatment of T6E. When analyzed with Ingenuity software, the pathway most highly down-regulated by Notch inhibition was that involving c-myc (data not shown).
Figure 1.
Withdrawal of Notch1 signals down-regulates c-myc expression in T-ALL cells. (A) Identification of Notch1-sensitive genes in T6E cells. Columns represent experimental samples, while rows represent genes. Each colored box indicates relative expression level (normalized for each gene), where red indicates high and blue indicates low. The seven columns on the left are samples from T6E cells with active Notch signaling, and the six columns on the right are samples from T6E cells in which Notch signaling was inhibited. Expression data from 82 genes significantly correlated with the Notch “on” (left) versus “off” (right) distinction (p < 0.05) are depicted; expression of the 25 genes in the upper cluster increased on inhibition of Notch signaling, expression of the 57 in the lower cluster decreased. Genes implicated in the literature as related to Notch signaling as well as the novel gene c-myc are highlighted. Sample label key: (GSI [1 μM compound E] + ICN1) sorted ICN1 transduced cells treated with GSI; (GSI mock) DMSO vehicle-treated cells; (GSI 0 h) untreated cells; (GFPposA/B) sorted GFP-only cells; (GFPnegA/B) sorted untransduced cells from the same cultures as transduced cells; (GSI) cells treated with GSI for the indicated number of hours; (DN-MamA/B) sorted dominant-negative MAML1 transduced cells. (B) Northern blot analysis demonstrating the down-regulation of c-myc following treatment with GSI (1 μM compound E). Notch-dependent T6E cells and human T-ALL cell lines were treated with carrier (DMSO) or GSI for the indicated time periods. Blots were hybridized with probes indicated in the right margin.
The identification of c-myc as a potential Notch1 target in studies using different inhibitors and several independent analytical tools provided the impetus for more focused studies. As an initial test, we assessed the effects of γ-secretase blockade on c-myc mRNA levels in five human T-ALL cell lines that require Notch signals for growth (Weng et al. 2004). Notch pathway blockade with a GSI led to the down-regulation of c-myc in all five of the human T-ALL cell lines that require Notch signals, as well as murine T6E cells (Fig. 1B).
c-Myc is a direct target of Notch1
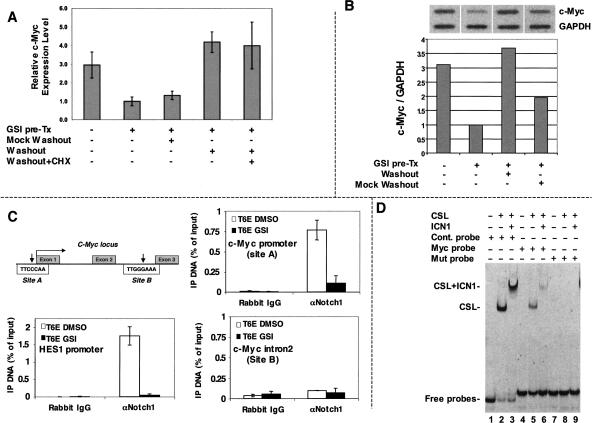
Forms of Notch1 bearing activating mutations within the extracellular domain are susceptible to ligand-independent cleavage by metalloproteases at site S2 (Sanchez-Irizarry et al. 2004). The product of this cleavage, NTM*, is normally rapidly recruited to the γ-secretase complex and further processed into ICN1 (Shah et al. 2005). However, we noted previously that NTM* in T-ALL cells is stabilized and accumulates in the presence of GSI (Weng et al. 2003), creating a pool of NTM* that can be rapidly converted to ICN1 upon GSI washout.
We used this maneuver to determine if the transcriptional up-regulation of c-myc by Notch1 requires protein synthesis. These experiments used the human T-ALL cell line KOPT-K1, which bears a mutation in the extracellular heterodimerization domain of Notch1 that results in ligand-independent S2 cleavage (Weng et al. 2004; Malecki et al. 2006). Washout of GSI in KOPT-K1 cells led to the rapid up-regulation of c-myc transcripts even in the presence of cycloheximide, an inhibitor of protein synthesis (Fig. 2A). The up-regulation of c-myc upon GSI washout was observed over a wide range of cycloheximide doses (5–40 μg/mL), all of which were sufficient to suppress the growth of KOPT-K1 cells (data not shown).
Figure 2.
c-Myc is a direct target of Notch1. (A) c-Myc up-regulation by Notch does not require de novo protein synthesis. KOPT-K1 cells were treated with GSI (1 μM compound E) for 48 h to permit accumulation of the γ-secretase substrate NTM*. Cells were then washed and refed medium containing GSI (mock washout), or medium lacking GSI (washout) with or without 20 μM cycloheximide (CHX). c-Myc RNA levels were determined after 4 h of additional culture by qPCR. Each sample was assayed in triplicate; error bars correspond to standard deviations. Similar results were obtained in four independent experiments. (B) Notch1 stimulates c-myc transcription. Nuclei were prepared from KOPT-K1 cells treated with vehicle (0.01% DMSO) for 48 h; treated with GSI (1 μM compound E) for 48 h; treated with GSI for 48 h, then washed thrice and cultured 2 h in fresh medium without GSI (washout); or treated with GSI for 48 h, then washed three times and cultured 2 h in fresh medium with GSI (mock washout). RNAs isolated from runoff reactions were hybridized to slot-blots containing probes specific for c-myc and GAPDH. Bound radioactivity was quantified using a PhosphorImager. (Top panel) Phosphorimages. (Bottom panel) Calculated c-myc/GAPDH transcript ratios. Results of a representative experiment are shown. (C) Notch1 binds to the c-myc promoter through a region containing a conserved CSL consensus sequence. Chromatin immunoprecipitates were performed on cross-linked fragmented DNAs prepared from T6E cells treated with DMSO or 1 μM compound E (GSI) for 24 h. Eluted DNAs were then analyzed by qPCR performed with primers flanking putative CSL-binding sites A and B. The amount of DNA amplified from immunoprecipitated DNAs was normalized to that amplified from input DNA. (D) CSL binds to the putative target sequence in the c-myc promoter. Oligonucleotides labeled with carboxyfluorescein (FAM) were mixed with buffer alone or buffer containing purified CSL and Notch1 polypeptides. Following electrophoresis in 10% native gels, fluorescently labeled probes were detected with an 860 Storm FluorImager (Amersham Pharmacia Biotech).
To determine if Notch stimulates the synthesis of c-myc transcripts, nuclear runoff experiments were performed with KOPT-K1 nuclei obtained from cells treated with DMSO or GSI (Fig. 2B). GSI treatment diminished c-myc transcription, whereas washout of GSI for as little as 2 h produced a rebound in c-myc transcription comparable to that observed in the experiments conducted with cycloheximide. Importantly, the increase in c-myc transcription was largely abrogated when cells were refed fresh medium containing GSI (mock washout). Thus, Notch1 increases c-myc RNA in T-ALL cells by stimulating transcription.
Scanning of c-myc genomic sequences revealed two potential CSL-binding sites that are conserved between humans and mice: (1) site A (TTCCCAA), which lies within the putative Notch-responsive element identified by Satoh et al. (2004); and (2) site B (TTGGGAAA), which is within intron 2 of c-myc (Fig. 2C). Chromatin immunoprecipitates prepared from T6E cells revealed that ICN1 associated with DNA fragments containing site A, and that GSI treatment depleted ICN1 from this site (Fig. 2C). In contrast, ICN1 did not associate with site B. As anticipated, GSI treatment also depleted ICN1 from CSL-binding sites in the hes1 promoter, a well-characterized Notch1 target (Jarriault et al. 1995). Electrophoretic mobility shift assays (EMSA) showed that CSL and CSL/ICN1 complexes bind to an oligonucleotide that contains site A (Fig. 2D), albeit with an affinity that is about fivefold lower than that of CSL for a consensus binding site (Supplementary Fig. S1). Taken together, these data suggest that activated Notch1 up-regulates c-myc directly.
c-Myc is necessary and sufficient to rescue T6E cells from Notch1 withdrawal
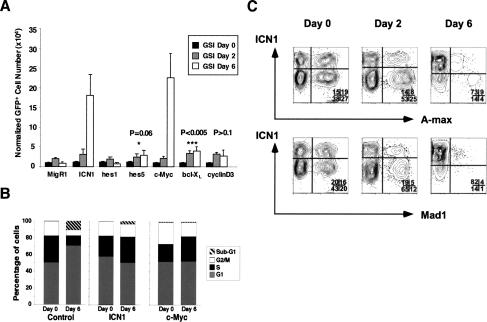
Treatment of T6E cells with GSI leads to a G0/G1 cell cycle arrest that is prevented by transduction of ICN1 (Weng et al. 2003). Given the potent progrowth effects of c-myc (for recent review, see Oskarsson and Trumpp 2005), we investigated whether enforced expression of c-myc mimicked the effects of ICN1. T6E cells were transduced with either empty MigRI, or with c-myc, ICN1, other Notch1 targets (hes1 and hes5), the antiapoptotic gene bcl-xL, or cyclinD3, a gene implicated in the growth and proliferation of normal T-cell progenitors (Sicinska et al. 2003). Only ICN1 and c-myc fully rescued T6E cells from GSI-induced growth arrest (Fig. 3A); a small but significant survival advantage was imparted by bcl-xL (data not shown), whereas hes1, hes5, and cyclinD3 did not have effects significantly different than empty virus. Like ICN1, c-myc also maintained the cell cycle progression of T6E cells in the presence of GSI (Fig. 3B).
Figure 3.
c-Myc is necessary and sufficient to rescue T6E cells from withdrawal of Notch1 signals. (A) c-Myc rescues growth. T6E cells, which express a membrane-tethered form of Notch1 that requires cleavage by γ-secretase for activation, were transduced with MigRI retroviruses expressing GFP alone (MigRI) or GFP and the indicated polypeptides, and then treated with GSI (1 μM compound E) for up to 6 d. The numbers of GFP+ cells at each time point are shown. (B) c-Myc restores cell cycle kinetics. The DNA content of T6E cells transduced with the indicated MigRI retroviruses was measured by flow cytometry at baseline and after 6 d of treatment with GSI. (C) Inhibitors of c-myc function prevent ICN1 from rescuing T6E cells treated with GSI. T6E cells were cotransduced with retroviruses expressing ICN1 and either A-max or mad1. Cells were then cultured in the presence of 1 μM compound E for 6 d. Cells transduced with ICN1 alone (top left quadrants) are enriched over the course of the experiment, while cells expressing either A-Max or Mad1 alone (bottom right quadrants) or one of these two c-myc inhibitors and ICN1 (top right quadrant) are selected against.
To further explore the significance of c-myc as a target, T6E cells were simultaneously transduced with ICN1 and either mad1 or dominant-negative (DN) max (A-max) (Krylov et al. 1997) to antagonize the action of c-myc (Fig. 3C). Upon GSI treatment, the substantial growth advantage of ICN1 transduced cells (compared with nontransduced cells) was abrogated by mad1 and DN max, indicating that c-myc function is necessary for ICN1 to rescue T6E cells (Fig. 3C).
c-Myc rescues a subset of human T-ALL cell lines from Notch1 withdrawal
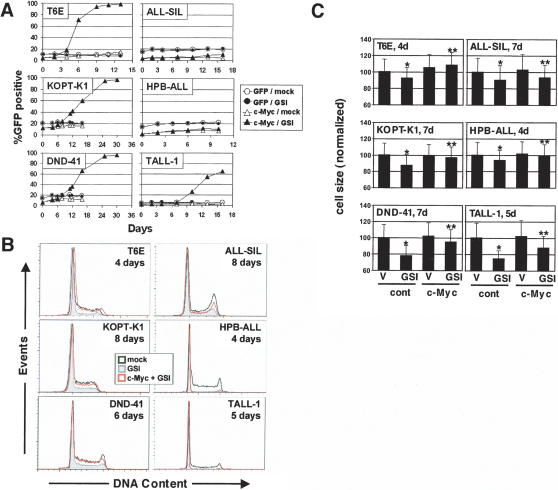
To investigate whether c-myc is generally sufficient to rescue T-ALL cell lines from Notch withdrawal, we transduced five human Notch-dependent T-ALL cell lines with c-myc. Like ICN1 (Weng et al. 2004), c-myc provided a partial or complete rescue of three of these cell lines (KOPT-K1, DND-41, TALL-1) from GSI treatment, as judged by proliferation (Fig. 4A) and cell cycle parameters (Fig. 4B), whereas two cell lines (HPB-ALL and ALL-SIL) were not growth-rescued by c-myc. All of the GSI-treated Notch1-dependent cell lines also showed a significant decrease in cell size that was partially or completely abrogated by c-myc (Fig. 4C). Thus, although c-myc appears to be a consistent target of activated Notch1, the degree to which Notch-dependent T-ALL cell lines are rescued by c-myc alone is variable.
Figure 4.
Transduction of c-myc leads to variable rescue of human Notch-sensitive T-ALL cell lines from withdrawal of NOTCH signals. Rescue of growth (A), cell cycle progression (B), and cell size (C) were assessed in control T-ALL cell lines or the same lines transduced with c-myc and a GFP marker (c-myc) or GFP only (GFP). Cells were treated with DMSO vehicle (mock) or 1 μM compound E (GSI) for the indicated time periods. (A) The fraction of live GFP+ cells was monitored by flow cytometry. An increasing GFP+ fraction indicates a growth/survival advantage conferred by the retrovirus over untransduced GFP− cells. (B) The DNA content of control cell lines was compared with that of sorted populations of cells transduced with c-myc by staining with Hoechst 33342 followed by flow cytometry. The DNA histograms shown were derived from >20,000-gated events. (C) Cell size in c-myc transduced cells and untransduced control cells treated with GSI (compound E, 1 μM) or DMSO vehicle (V) for the indicated time was determined by forward scatter of live cells. The relative cell sizes for each population (normalized to a value of 100 for vehicle-treated, untransduced control cells ± 1 SD) are depicted. The decreases in size upon addition of GSI in untransduced cells (*), and the increases in size upon transduction with c-myc in GSI-treated cultures (**, as compared with untransduced GSI-treated cells), were both significant (P < 0.01) in all cell lines tested, as measured by the Bonferroni post-test after two-way ANOVA (unweighted means) analysis.
Activated Notch1 can rescue T-ALL cells that depend on transgenic c-myc
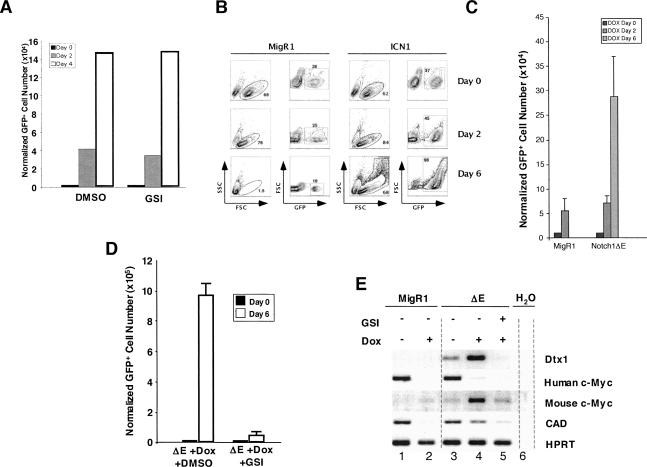
If Notch1 is a major upstream regulator of c-myc in T-ALL cells, activated Notch1 should replace functions provided by trangenic c-myc. This idea was tested using the cell line 8946, which is derived from a murine T-ALL induced with a doxycycline-repressible human c-myc transgene (Felsher and Bishop 1999). 8946 cells undergo growth arrest and apoptosis when treated with doxycycline, but are unaffected by treatment with GSI (Fig. 5A). Transduction of 8946 cells with ICN1 or ΔE (a membrane-tethered form of activated Notch1 resembling NTM*) rescued these cells from c-myc withdrawal, whereas empty virus did not (Fig. 5B,C). Significantly, the rescue by Notch1 was associated with the up-regulation of endogenous murine c-myc (Fig. 5E). Further, when rescued with ΔE, 8946 cells became GSI-sensitive (Fig. 5D) and up-regulated the expression of both Notch1 (dtx1, c-myc) and c-myc (cad) target genes (Fig. 5E). Taken together, these data are consistent with a model in which c-myc acts downstream of Notch to promote the growth of T-ALL cells.
Figure 5.
Activated Notch1 rescues the T-ALL cell line 8946 from withdrawal of transgenic c-myc and conveys sensitivity to Notch pathway inhibitors. (A) Basal 8946 cell growth is not affected by treatment with GSI (1 μM compound E). (B,C) Transduction of 8946 cells with retroviruses encoding two different forms of activated Notch1, ICN1, or ΔE retroviruses, rescue the cells from the effects of doxycycline (20 ng/mL), whereas empty MigRI does not. 8946 cell rescue was judged variously by forward and side scatter (B, which shows the rescue by ICN1), or by the number of GFP positive cells on day 6 post-treatment (C, which shows the rescue by ΔE). (D) When rescued with ΔE from doxycycline treatment, 8946 cells become newly sensitized to GSI. 8946 cells transduced with ΔE were treated for 6 d with doxycline (Dox, 20 ng/mL) ± 1 μM compound E (GSI). Effects of GSI treatment were assessed by flow cytometry. (E) 8946 cells rescued from doxycycline (Dox, 20 ng/mL) by ΔE up-regulate endogenous c-myc and other Notch and c-myc target genes. 8946 cells transduced with MigRI or MigRI-ΔE were treated with doxycline (20 ng/mL) ± GSI (1 μM compound E) for 24 h Expression of deltex1 (a gene that is a up-regulated by Notch1), transgenic human c-myc, endogenous murine c-myc, and the c-myc target cad were monitored by RT–PCR.
Notch regulates c-myc in normal thymocytes
In addition to its role in T-ALL, Notch1 is required for induction of T cell differentiation and subsequent normal maturation of thymocytes (for review, see Radtke et al. 2004). The earliest steps of intrathymic T-cell development are characterized by four hierarchical developmental stages, termed DN1–4. Notch1 signaling peaks in DN3 thymocytes (defined as CD3−, CD4−, CD8−, c-kit−, CD44−, CD25+), and then declines as cells progress to the DN4 stage (CD3−, CD4−, CD8−, c-kit−, CD44−, CD25−). Recently, Rothenberg’s group noted that DN3 thymocytes could be subdivided into DN3a and DN3b cells (Taghon et al. 2006). These cell types are positioned respectively just prior to or following β-selection, which is characterized by the onset of signaling through the pre-TCR receptor and increased cell division. Of note, the expression of Notch1 and downstream Notch target genes such as hes1 and deltex1 falls dramatically as cells progress from the DN3a to DN3b, and in keeping with this, DN3a cells are dependent on Notch signals for growth and survival, whereas DN3b cells are not (Taghon et al. 2006).
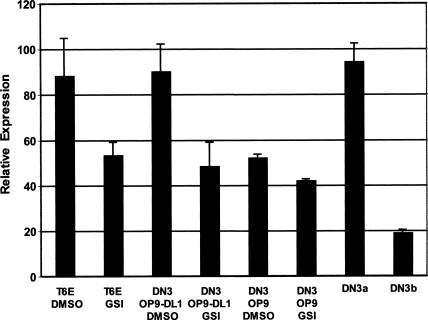
Based on these observations, we performed experiments to determine if Notch signaling affects c-myc expression in DN3 thymocytes. Sorted DN3 cells were cocultured on OP9 cells or OP9 cells expressing the Notch ligand dll1, which are capable of directing T-cell development from progenitors ex vivo (Schmitt et al. 2004). DN3 cells cultivated on OP9-DLL1 cells had higher expression of c-myc than DN3 cells grown on control OP9 cells, and this difference was abrogated by a GSI (Fig. 6). Further, freshly isolated DN3a cells had significantly higher expression of c-myc than did DN3b cells. Thus, Notch signaling correlates with c-myc expression at a critical stage of normal αβ T-cell development.
Figure 6.
Notch signaling up-regulates c-myc in normal thymocytes at the DN3 stage. Sorted DN3 thymocytes from three to five mice were incubated for 16 h on OP9 or OP9-DLL1 feeder cells in the presence of the GSI compound E (1 μM) or DMSO vehicle. Murine T6E T-ALL cells cultivated in the presence or absence of GSI for 24 h served as positive and negative controls. c-Myc transcript levels were determined by qPCR in these cells and in freshly sorted DN3a and DN3b thymocytes. Expression of c-myc was determined in three independent experiments. Mean expression levels ± 1 SD are shown.
Discussion
c-Myc is a Notch1 target in T-ALL cells
Although the Notch signaling pathway is best known for its ability to influence differentiation, inappropriate gains in Notch function can have profound effects on growth in specific contexts (Berry et al. 1997; Weng et al. 2003, 2004). Using genomic, biochemical, and functional approaches, we identified c-myc as an important mediator of Notch’s progrowth effects in T-ALL cells. Importantly, the ability of Notch1 to stimulate c-myc transcription in the presence of the protein synthesis inhibitor cycloheximide is persuasive evidence that it regulates c-myc directly, rather than through a Notch1-dependent intermediate.
c-Myc is a particularly attractive target to explain the progrowth effects of Notch1 in transformed T-cell progenitors. c-Myc drives cell cycle progression and regulates the expression of key enzymes that control cellular metabolism (for review, see Grandori et al. 2000; Levens 2003), and stimulates ribosome biogenesis and protein synthesis through interactions with RNA polymerase III (Felton-Edkins et al. 2003) and RNA polymerase I (Arabi et al. 2005; Grandori et al. 2005; Grewal et al. 2005). c-Myc also controls cell size (Johnston et al. 1999; Schuhmacher et al. 1999), presumably through its ability to stimulate protein synthesis (Oskarsson and Trumpp 2005). These functional attributes of c-myc are consistent with the growth arrest and decrease in cell size that are observed upon withdrawal of Notch signals from T-ALL cells.
Implications for prior studies linking Notch and c-myc
Retroviral mutagenesis studies conducted with c-myc (MMTVD/myc) transgenic mice showed that a high frequency of T-ALLs occurring with shortened latencies had proviral integrations into Notch1 (Girard et al. 1996; Hoemann et al. 2000). The simplest explanation for this observation (that c-myc and Notch1 act through independent, complementary pathways to promote pre-T-cell transformation) is at odds with our functional data, which places c-myc downstream of Notch1.
How might these seemingly conflicting data be brought into register? One explanation lies in the different contexts in which the experiments were performed. In vivo models permit detection of oncogenic complementation at different stages of tumor development and progression, whereas our experiments are focused only on growth maintenance in vitro. In mice, retroviral insertions into Notch1 in multipotent bone marrow progenitors are predicted to expand the pool of pre-T cells and thereby increase the likelihood that some cell within this pool will acquire other rate-limiting oncogenic aberrations. This ability to expand the pool of “at-risk” cells may underlie Notch1’s remarkable capacity to complement many (if not all) of the other genetic lesions implicated in human (Weng et al. 2004) and murine T-ALL (Girard et al. 1996; Feldman et al. 2000; Dumortier et al. 2006; Lin et al. 2006; O’Neil et al. 2006). Much less commonly, retroviral insertions into c-myc occur in T-ALLs arising in lck-activated-Notch1 (lck-Nic) transgenic mice (Beverly and Capobianco 2003). Although c-myc should already be up-regulated in lck-Nic mice, supraphysiologic expression of c-myc due to proviral insertion may confer an additional selective advantage. This idea is in line with studies suggesting that tumor latency is shortened in homozygous transgenic c-myc mice relative to heterozygous animals (Sidman et al. 1993). Finally, the failure of c-myc to rescue all Notch-dependent cell lines clearly points to the existence of other Notch1 target genes that contribute to growth and survival, and which also likely provide a selective advantage to tumors in MMTVD/myc transgenic mice bearing Notch1-proviral insertions. It will be important to define the molecular basis for the variation in c-myc dependency among cell lines, and to identify other genes downstream of Notch1 that contribute to T-ALL cell growth and survival.
Importance of Notch/myc interaction in normaland neoplastic pre-T cells
T-ALLs appear to resemble various stages of pre-T-cell development (Ferrando et al. 2002; Asnafi et al. 2003), during which Notch signaling outcomes may differ dramatically. Notch1 is required for both T-lineage specification from multipotent progenitors (Pui et al. 1999; Radtke et al. 1999; Sambandam et al. 2005) and subsequent pre-T-cell development up to and including the β-selection checkpoint (Wolfer et al. 2002; Tanigaki et al. 2004; Ciofani and Zuniga-Pflucker 2005), which occurs during the maturation of “Notch-high” DN3a to “Notch-low” DN3b cells (Taghon et al. 2006). Of note, normal β-selection is accompanied by an expansion of T-cell progenitors that requires both Notch1 and c-myc (Douglas et al. 2001; Iritani et al. 2002). Our ex vivo studies show that Notch signals contribute to c-myc expression in DN3a cells, and that there is a precipitous drop in c-myc expression in post-β-selection DN3b thymocytes, which also down-regulate pre-Tα expression. Of interest, Notch-associated T-ALLs are often associated with persistent expression of pre-Tα (see Fig. 1A; Bellavia et al. 2000), which has been taken as evidence that these cells are arrested at a stage resembling normal β-selection. Together, these findings suggest that the Notch/c-myc signaling axis found in Notch-dependent T-ALL cells reflects the persistence of a stage-specific relationship that is involved in the development of normal T cells.
How are Notch1 targets restricted to specific contexts?
The outcome of Notch signaling varies dramatically from context to context. One probable basis for these different outcomes is the existence of context-specific target genes, such as pre-Tα (Deftos et al. 2000; Reizis and Leder 2002). Since physiologic Notch signals do not generally lead to increased growth, it seems likely that c-myc will also prove to be a context-specific target.
How might this be achieved? Our data suggest that c-myc is targeted by Notch1 in T-ALL cells via a conserved site (TTGGGAA) located within the 5′ c-myc promoter region immediately upstream of the TATA box that was shown to contain a CSL-responsive element previously (Satoh et al. 2004). This sequence differs from the canonical CSL recognition sequence YGTGRGAA (Tun et al. 1994; Nellesen et al. 1999), and binds CSL relatively weakly, which may have a role in restricting CSL occupancy to particular contexts. Other factors may further modulate occupancy positively and negatively. In this regard, the putative CSL-binding site in the c-myc promoter is a high similarity match to the consensus binding site for Ikaros (Molnar and Georgopoulos 1994), a transcriptional repressor that regulates pre-T-cell development (Georgopoulos et al. 1994; Winandy et al. 1999). Ikaros loss-of-function is associated with T-ALL development (Winandy et al. 1995), and it has been hypothesized that Ikaros and CSL compete for access to the promoter sequences of genes that determine T-ALL development in mouse models (Beverly and Capobianco 2003); c-myc could be one such gene. Conversely, unknown cis-acting factors could enhance site-occupancy by CSL and/or transcriptional activation by the CSL/ICN1/MAML ternary complex. Recent work has identified cis-acting elements that contribute to the activation of a number of Notch-responsive elements (Cave et al. 2005; Ong et al. 2006), providing support for this model.
Summary and implications
The existence of a direct link between Notch and c-myc in T-ALL cell lines and normal thymocytes has therapeutic as well as basic implications. Most mutated Notch1 receptors found in T-ALL cells depend on γ-secretase to transduce signals (Weng et al. 2004; Malecki et al. 2006), and the withdrawal of c-myc transgene expression “cures” ∼50% of mice with T-ALL (Felsher and Bishop 1999). Recognition that the primary effect of Notch1 signals in T-ALL cells appears to be on proliferation and metabolism, rather than differentiation or survival, point to rational combinations of Notch pathway inhibitors and other therapeutic agents. This could take the form of agents that also impinge on protein synthesis through independent mechanisms, or drugs that target parallel pathways, such as those that regulate cell survival directly.
Materials and methods
Cell lines
All lymphoid lines were grown in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone), 2 mM L-glutamine, 1 mM sodium pyruvate, and antibiotics. 293T cells were maintained in Dulbecco’s modified Eagle medium (DMEM) (Invitrogen) with the same supplements except sodium pyruvate. Cells were grown at 37°C under 5% CO2. The T6E murine cell line has been described previously (Pear et al. 1996). 8946 is a murine T-ALL cell line derived from a tumor created with a tetracycline-dependent human c-myc transgene (Felsher and Bishop 1999).
Plasmids and probes
cDNAs encoding murine and human c-myc, murine Mad1, HA-tagged A-Max (kind gift of Dr. Charles Vinson, NCI, Bethesda, MD), and Flag-tagged cyclin D3 (kind gift of Dr. Alan Diehl, University of Pennsylvania, Philadelphia, PA) were subcloned into MigR1, a retroviral vector containing an internal ribosomal entry site (IRES) and GFP marker (Pui et al. 1999). The retroviral constructs MigRI–ICN1 and MigRI–MAML1(13–74)-GFP have been described (Weng et al. 2003). All constructs were sequenced and tested for expression and followed by detection on Western blots stained with either anti-Mad1 (#4682, Cell Signaling), anti-HA (clone 6E2, Cell Signaling) to detect the A-Max construct, or anti-Flag (#2368, Cell Signaling) to detect the cyclin D3 construct (#2368, Cell Signaling). For the experiments in which ICN was cotransduced with either Mig A-Max or Mig Mad1, ICN1 was expressed from a MSCV-based retroviral vector that coexpresses the truncated nerve growth factor receptor (tNGFR) as a surrogate marker (Tu et al. 2005). This approach allows detection of GFP in the FL1 channel and tNGFR in the FL2 channel using a NGFR antibody (Pharmingen). The c-myc and 18S rRNA probes used for Northern blot hybridization were generated by random hexamer priming of a ∼2.2-kb human c-myc cDNA (MGC-5183) and a 644-bp PCR product spanning residues 548–1191 of GenBank accession M10098, respectively.
Retroviral transduction and GSI rescue
Production of high-titer ecotropic (Pear et al. 1996) and pseudotyped (Weng et al. 2003) retroviruses and transduction of target cells have been described. For the GSI rescue experiments, 5 × 105 to 7.5 × 105 cells were centrifuged with the appropriate amount of viral supernatant and 4 μg/mL hexadimethidrine bromide (Sigma) for 50 min at 2500 rpm (day −2). Transduction efficiency was measured 48 h post-infection (day 0) by GFP fluorescence on a FACScan flow cytometer (Becton Dickinson). After assessing GFP percentage, cells were immediately counted and prepared for treatment with GSI (1 μM compound E, which was synthesized in the laboratory of M.S. Wolfe). Changes in the percentage of GFP+ cells and other cellular parameters were assessed with a FACScan flow cytometer and total cell number was obtained using a hemocytometer. The number of GFP+ cells was normalized to the number of GFP+ cells at day 0 to correct for differences in the initial retroviral transduction. Average cell counts and standard deviations were calculated for each experiment (carried out in triplicate), and each experiment was carried out at least twice. Where appropriate, a Student’s t-test was used to assess statistical significance.
Expression profiling/analysis
Total RNA prepared using Trizol (Invitrogen) was used to generate labeled cRNA as described (Shipp et al. 2002). cRNAs were hybridized/scanned on AffyMetrix U74Av2 and/or Expression Set 430 A/B, chips, and raw fluorescence data were analyzed using dChip software. GSI-treated cells were cultured in the presence of 1.0 μM compound E for 3, 6, 24, or 72 h. Mock-treated cells were cultured in the presence of DMSO vehicle (0.01% final) for 120 h. DN-MAML1 transduced cells were sorted at 72 h following exposure to retrovirus to purities of 87% GFP+ (DN-MamA) and 98% GFP+ (DN-MamB); GFP-only controls were sorted to purities of 95% GFP+ (GFPposA) and 98% GFP+ (GFPposB); GFP-negative controls were sorted from cells exposed to, but not transduced by, DN-Mam retroviruses to purities of >98% GFP− (GFPnegA) and >99% GFP− (GFPnegB). Sorted cells (2 × 106 to 4 × 106) were harvested for RNA immediately following sorting. Cells transduced with ICN were cultured in the presence of 0.5 μM compound E continuously for several weeks prior to RNA harvest (>99% GFP+).
Northern blots
Total RNAs (10 μg per sample) were run on 1.2% agarose/2.2 M formaldehyde gels and transferred to Hybond-XL nylon membranes (Amersham Biosciences) with UV cross-linking (Stratalinker, Stratagene). Blots were hybridized with radiolabeled 32P-random-primed probes at 65°C and washed under stringent conditions, then exposed to Kodak BioMax MS autoradiography film with intensifying screens.
Quantitative RT–PCR (qRT–PCR)
Oligo-d(T)-primed total RNAs (2 μg per cell line sample and 0.5 μg for thymocytes) were reverse-transcribed with SuperScript II (Invitrogen). For cell line samples, an appropriate dilution of cDNA and gene-specific primers were combined with iQ SYBR Green Supermix (Bio-Rad) and amplified in an iCycler iQ real-time PCR machine (Bio-Rad). All qPCR reactions were performed in triplicate. Ct (threshold cycle number) and expression values with standard deviations were calculated using the Gene Expression Macro for Excel (Bio-Rad). Primer sequences for real-time PCRs were as follows: c-myc forward, 5′-CTTCTCTCC GTCCTCGGATTCT-3′; c-myc reverse, 5′-GAAGGTGATCC AGACTCTGACCTT-3′; β-actin forward, 5′-CGCGAGAAGA TGACCCAGAT-3′; β-actin reverse, 5′-GATAGCACAGCCT GGATAGCAAC-3′. c-Myc and β-actin primers were used at 0.25 μM final and exhibited PCR efficiencies of 95.7% and 92.9%, respectively. Real-time amplification was performed with initial denaturation at 95°C for 3 min, followed by 40 cycles of two-step amplification (95°C for 15 sec, 65°C for 1 min). For thymocyte samples, real-time RT–PCR was performed with TaqMan universal PCR Master Mix (Applied Biosystems) and analyzed on the ABI Prism 7900 (Applied Biosystems). Notch1, c-myc, and deltex1 Taqman primers were obtained from Applied Biosystems.
DNA content/flow cytometric analysis
Live or fixed cells were stained for DNA content with Hoechst 33342 (4 μM, Sigma B2261) or propidium iodide (40 μg/mL), respectively. Flow cytometry was performed using an Influx Analyzer (Cytopeia) equipped with 488 nm and UV lasers to measure GFP and Hoechst 33342 in live cells, while FACSCalibur cytometers (BD Biosciences) were used to measure propidium iodide in fixed cells or GFP in live cells. Data analysis was performed using FlowJo software (Treestar).
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed using ChIP assay kits (Upstate Biotechnology). T6E cells were treated with DMSO or GSI (1 μM Compound E) for 24 h prior to fixation. Cells were fixed with 1% paraformaldehyde at room temperature for 10 min, washed, and lysed with SDS lysis buffer (50 mM Tris-HCl, 1% SDS, 10 mM EDTA, protease inhibitor cocktail, Sigma). The lysates were sonicated to reduce DNA lengths to between 200 and 600 bp. The soluble fraction was diluted, precleared with salmon sperm DNA/protein A-agarose, then divided into two tubes and incubated with 5 μL of antiserum specific for Notch1 TAD domain (Aster et al. 2000) or normal rabbit IgG. The immune complexes were then precipitated with protein A-agarose and eluted with an elution buffer (0.1 M NaHCO3 containing 1% SDS). The eluted material was reverse-cross-linked and treated with proteinase K (20 μg/mL). DNA was purified using a PCR purification kit (Qiagen) and eluted by water (5 × 106 cell equivalents/50 μL). Quantitative PCR (qPCR) was performed using the SYBR Green system on the ABI 7900HT Sequence Detection System (Applied Biosystems) with the following primers: (1) c-myc promoter-forward, 5′-TGAGGCTCCTCCTCCTCTTTC-3′; (2) c-myc promoter reverse, 5′-GCAGACCCCCGGAATATAAA-3′; (3) c-myc intron2 forward, 5′-CACGGGACCTGAAAG GTTCT-3′; (4) c-myc intron2 reverse, 5′-GGGTTAGGGCAC AGGTGAGA-3′; (5) hes1 promoter forward, 5′-CGTGTCT CTTCCTCCCATTG-3′; (6) hes1 promoter reverse, 5′-CCAG GACCAAGGAGAGAGGT-3′. Primers for the hes1 promoter sequence flank two CSL-binding sites that lie just 5′ of the TATA box (Jarriault et al. 1995). Each sample was independently prepared at least two times and run in duplicate. The relative DNA amount was calculated using the standard curve method as described in the ABI 7900HT Sequence Detection System manual. The input DNA was defined as an aliquot of sheared chromatin prior to immunoprecipitation, and was used to normalize the sample to the amount of chromatin added to each ChIP.
EMSA
Oligonucleotides fluorescently 5′-labeled with carboxyfluorescein (FAM) and unlabeled complementary oligonucleotides were obtained from Integrated DNA Technologies. Sequences of the FAM-labeled oligonucleotides were as follows: c-myc promoter, 5′-FAM-CCCCTCCCGGGTTCCCAAAGCAGAGGG CGT-3′; mutated c-myc promoter, 5′-FAM-CCCCTCCCGGG TTCAAAAAGCAGAGGGCGT-3′; consensus CSL-binding site, 5′-FAM-TCCAAATTTTTTCCCACGGCGTGT-3′. CSL and the RAM-ANK domains of Notch1 were prepared as described (Nam et al. 2003). Nonradioisotopic EMSAs were performed by incubating 2 pmol of probes for 30 min at 30°C in binding buffer (10% glycerol, 20 mM HEPES at pH 7.9, 60 mM KCl, 10mM DTT, 5 mM MgCl2, 250 ng dGdC, 0.2 mg/mL bovine serum albumin) in the presence or absence of 10 μg of CSL and RAM-ANK. Gel electrophoresis was performed in 10% native gels at 4°C and 180 V. Following electrophoresis, gels were immediately analyzed by blue-excitation fluorescence scanning with a Storm 860 FluorImager (Amersham Pharmacia Biotech). To compare the affinity of CSL binding to sequences of interest, oligonucleotides with 5′ overhangs were incubated with 32P-α-dCTP (Perkin-Elmer) and the Klenow fragment of Escherichia coli DNA polymerase I (New England Biolabs). 32P-labeled probes were incubated with 0–4000 ng of recombinant CSL as described above. Following electrophoresis, gels were exposed to PhosphorImager screens and analyzed with a Storm 860 PhosphorImager (Amersham Pharmacia Biotech).
Nuclear runoff assays
Nuclei were prepared by hypotonic detergent lysis and centrifugation (300 × g) from 50 × 106 cells/sample as described (Greenberg and Bender 1997). Runoff reactions were performed with α-32P UTP (Perkin-Elmer), followed by DNase I (500 U/sample, Invitrogen) and proteinase K digestion. RNA was isolated using RNeasy Mini columns (Qiagen). A Minifold II appartus (Schleicher & Schuell BioScience) was used to transfer 5 μg of linearized, NaOH-denatured plasmid DNA per slot onto positively charged nylon membranes (Hybond-XL, Amersham Biosciences). Following UV cross-linking (70,000 μJ/cm2; UV Stratalinker 2400, Stratagene), 5 × 106 cpm of each runoff RNA sample was hybridized to slot blots at 65°C overnight. After washing to high stringency (0.1× SSC/0.1% SDS, 65°C), bound radioactivity was quantified using a PhosphorImager (Molecular Dynamics Storm 860) and ImageQuant 5.0 software using “object average” background correction.
Isolation of DN3 thymocytes, OP9/OP9-DL1 culture, and GSI treatment
CD4−CD8− DN thymocytes were negatively selected with anti-CD4 and anti-CD8 MACs beads according to the manufacturer’s recommendations (Miltenyi Biotec). Following staining with antibodies against lineage markers (TCRβ, TCRγ, CD3ε, CD4, NK1.1, CD19, Gr-1, CD11b), CD44, and CD25 antigen (Pharmingen), DN3 thymocytes (CD44−/lo CD25hi Lin−) were purified by cell sorting on a FACS Moflo (Cytomation). DN3a (CD44−/lo CD25hi Lin− CD27lo) and DN3b (CD44−/lo CD25hi Lin− CD27lo) cells were purified by sorting after staining with anti-CD27 antibody (eBioscience) as described (Taghon et al. 2006). Cells were either used for RNA extraction directly, or were cocultured with OP9 or OP9-DLL1 stromal cells in the presence or absence of 1 μM compound E for 16 h as described (Schmitt et al. 2004). RNA was isolated from thymocytes using the RNEasy kit according to the manufacturer’s recommendations (Qiagen).
Statistical analysis
ANOVA and t-test analyses were performed using the Prism 4.03 software package (GraphPad Software).
Acknowledgments
We thank Alan Diehl and Charles Vinson for providing reagents, and Gerd Blobel, Tom Kadesch, David Levens, Steve McMahon, Gary Koretzky, Craig Thompson, and members of the Pear and Aster laboratories for helpful suggestions. J.M. was supported by NIH Training Grant (T32 CA 09140-31-35). C.D.B. is supported by a Long-Term Fellowship from the Human Frontier Science Program Organization, and M.L.A. is supported by the Foundation pour la Recherche Medicale (FRM). A.P.W., D.W.F., S.C.B., W.S.P., and J.C.A. are supported by grants from the NIH.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1450406
References
- Amsen D., Blander J.M., Lee G.R., Tanigaki K., Honjo T., Flavell R.A. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- Arabi A., Wu S., Ridderstrale K., Bierhoff H., Shiue C., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Asnafi V., Beldjord K., Boulanger E., Comba B., Le Tutour P., Estienne M.H., Davi F., Landman-Parker J., Quartier P., Buzyn A., et al. Analysis of TCR, pT α, and RAG-1 in T-acute lymphoblastic leukemias improves understanding of early human T-lymphoid lineage commitment. Blood. 2003;101:2693–2703. doi: 10.1182/blood-2002-08-2438. [DOI] [PubMed] [Google Scholar]
- Aster J.C., Xu L., Karnell F.G., Patriub V., Pui J.C., Pear W.S. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol. Cell. Biol. 2000;20:7505–7515. doi: 10.1128/mcb.20.20.7505-7515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia D., Campese A.F., Alesse E., Vacca A., Felli M.P., Balestri A., Stoppacciaro A., Tiveron C., Tatangelo L., Giovarelli M., et al. Constitutive activation of NF-κB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 2000;19:3337–3348. doi: 10.1093/emboj/19.13.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry L.W., Westlund B., Schedl T. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development. 1997;124:925–936. doi: 10.1242/dev.124.4.925. [DOI] [PubMed] [Google Scholar]
- Beverly L.J., Capobianco A.J. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell. 2003;3:551–564. doi: 10.1016/s1535-6108(03)00137-5. [DOI] [PubMed] [Google Scholar]
- Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J.R., Cumano A., Roux P., Black R.A., Israel A. A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin–metalloprotease TACE. Mol. Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Calvi L.M., Adams G.B., Weibrecht K.W., Weber J.M., Olson D.P., Knight M.C., Martin R.P., Schipani E., Divieti P., Bringhurst F.R., et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- Cave J.W., Loh F., Surpris J.W., Xia L., Caudy M.A. A DNA transcription code for cell-specific gene activation by notch signaling. Curr. Biol. 2005;15:94–104. doi: 10.1016/j.cub.2004.12.070. [DOI] [PubMed] [Google Scholar]
- Ciofani M., Zuniga-Pflucker J.C. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- De Strooper B., Annaert W., Cupers P., Saftig P., Craessaerts K., Mumm J.S., Schroeter E.H., Schrijvers V., Wolfe M.S., Ray W.J., et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Deftos M.L., Huang E., Ojala E.W., Forbush K.A., Bevan M.J. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas N.C., Jacobs H., Bothwell A.L., Hayday A.C. Defining the specific physiological requirements for c-Myc in T cell development. Nat. Immunol. 2001;2:307–315. doi: 10.1038/86308. [DOI] [PubMed] [Google Scholar]
- Dumortier A., Jeannet R., Kirstetter P., Kleinmann E., Sellars M., Dos Santos N.R., Thibault C., Barths J., Ghysdael J., Punt J.A., et al. Notch activation is an early and critical event during T-cell leukemogenesis in ikaros-deficient mice. Mol. Cell. Biol. 2006;26:209–220. doi: 10.1128/MCB.26.1.209-220.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A.W., Rattis F.M., Dimascio L.N., Congdon K.L., Pazianos G., Zhao C., Yoon K., Cook J.M., Willert K., Gaiano N., et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat. Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- Ellisen L.W., Bird J., West D.C., Soreng A.L., Reynolds T.C., Smith S.D., Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- Feldman B.J., Hampton T., Cleary M.L. A carboxy-terminal deletion mutant of Notch1 accelerates lymphoid oncogenesis in E2A-PBX1 transgenic mice. Blood. 2000;96:1906–1913. [PubMed] [Google Scholar]
- Felsher D.W., Bishop J.M. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- Felton-Edkins Z.A., Kenneth N.S., Brown T.R., Daly N.L., Gomez-Roman N., Grandori C., Eisenman R.N., White R.J. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle. 2003;2:181–184. [PubMed] [Google Scholar]
- Ferrando A.A., Neuberg D.S., Staunton J., Loh M.L., Huard C., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K., Bigby M., Wang J.H., Molnar A., Wu P., Winandy S., Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- Girard L., Hanna Z., Beaulieu N., Hoemann C.D., Simard C., Kozak C.A., Jolicoeur P. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes & Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- Grandori C., Cowley S.M., James L.P., Eisenman R.N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Grandori C., Gomez-Roman N., Felton-Edkins Z.A., Ngouenet C., Galloway D.A., Eisenman R.N., White R.J. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Greenberg M.E., Bender T.P. Ausubel F.M., et al. Current protocols in molecular biology. John Wiley & Sons. Inc.; Hoboken, NJ: 1997. Identification of newly transcribed RNA. Unit 4.10. [DOI] [PubMed] [Google Scholar]
- Grewal S.S., Li L., Orian A., Eisenman R.N., Edgar B.A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- Hoemann C.D., Beaulieu N., Girard L., Rebai N., Jolicoeur P. Two distinct Notch1 mutant alleles are involved in the induction of T-cell leukemia in c-myc transgenic mice. Mol. Cell. Biol. 2000;20:3831–3842. doi: 10.1128/mcb.20.11.3831-3842.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iritani B.M., Delrow J., Grandori C., Gomez I., Klacking M., Carlos L.S., Eisenman R.N. Modulation of T-lymphocyte development, growth and cell size by the Myc antagonist and transcriptional repressor Mad1. EMBO J. 2002;21:4820–4830. doi: 10.1093/emboj/cdf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S., Brou C., Logeat F., Schroeter E.H., Kopan R., Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Johnston L.A., Prober D.A., Edgar B.A., Eisenman R.N., Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberly W.T., Esler W.P., Ye W., Ostaszewski B.L., Gao J., Diehl T., Selkoe D.J., Wolfe M.S. Notch and the amyloid precursor protein are cleaved by similar γ-secretase(s). Biochemistry. 2003;42:137–144. doi: 10.1021/bi026888g. [DOI] [PubMed] [Google Scholar]
- Kopan R., Schroeter E.H., Weintraub H., Nye J.S. Signal transduction by activated mNotch: Importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov D., Kasai K., Echlin D.R., Taparowsky E.J., Arnheiter H., Vinson C. A general method to design dominant negatives to B-HLHZip proteins that abolish DNA binding. Proc. Natl. Acad. Sci. 1997;94:12274–12279. doi: 10.1073/pnas.94.23.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau D.M., Traver D., Ferrando A.A., Kutok J.L., Aster J.C., Kanki J.P., Lin S., Prochownik E., Trede N.S., Zon L.I., et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- Levens D.L. Reconstructing MYC. Genes & Dev. 2003;17:1071–1077. doi: 10.1101/gad.1095203. [DOI] [PubMed] [Google Scholar]
- Li C., Hung Wong W. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. 2001;1:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.W., Nichols R.A., Letterio J.J., Aplan P.D. Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood. 2006;107:2540–2543. doi: 10.1182/blood-2005-07-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard I., Fang T., Pear W.S. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- Malecki M.J., Sanchez-Irizarry C., Mitchell J.L., Histen G., Xu M.L., Aster J.C., Blacklow S.C. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol. Cell Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minter L.M., Turley D.M., Das P., Shin H.M., Joshi I., Lawlor R.G., Cho O.H., Palaga T., Gottipati S., Telfer J.C., et al. Inhibitors of γ-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat. Immunol. 2005;6:680–688. [PubMed] [Google Scholar]
- Molnar A., Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell. Biol. 1994;14:8304–8314. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm J.S., Schroeter E.H., Saxena M.T., Griesemer A., Tian X., Pan D.J., Ray W.J., Kopan R. A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- Nam Y., Weng A.P., Aster J.C., Blacklow S.C. Structural requirements for assembly of the CSL·intracellular Notch1·Mastermind-like 1 transcriptional activation complex. J. Biol. Chem. 2003;278:21232–21239. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- Nellesen D.T., Lai E.C., Posakony J.W. Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev. Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- O’Neil J., Calvo J., McKenna K., Krishnamoorthy V., Aster J.C., Bassing C.H., Alt F.W., Kelliher M., Look A.T. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;107:781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C.T., Cheng H.T., Chang L.W., Ohtsuka T., Kageyama R., Stormo G.D., Kopan R. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J. Biol. Chem. 2006;281:5106–5119. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- Oskarsson T., Trumpp A. The Myc trilogy: Lord of RNA polymerases. Nat. Cell Biol. 2005;7:215–217. doi: 10.1038/ncb0305-215. [DOI] [PubMed] [Google Scholar]
- Pear W.S., Aster J.C., Scott M.L., Hasserjian R.P., Soffer B., Sklar J., Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petcherski A.G., Kimble J. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature. 2000a;405:364–368. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- Petcherski A.G., Kimble J. Mastermind is a putative activator for Notch. Curr. Biol. 2000b;10:R471–R473. doi: 10.1016/s0960-9822(00)00577-7. [DOI] [PubMed] [Google Scholar]
- Preiss A., Hartley D.A., Artavanis-Tsakonas S. The molecular genetics of Enhancer of split, a gene required for embryonic neural development in Drosophila. EMBO J. 1988;7:3917–3927. doi: 10.1002/j.1460-2075.1988.tb03278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui J.C., Allman D., Xu L., DeRocco S., Karnell F.G., Bakkour S., Lee J.Y., Kadesch T., Hardy R.R., Aster J.C., et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H.R., Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Radtke F., Wilson A., Mancini S.J., MacDonald H.R. Notch regulation of lymphocyte development and function. Nat. Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- Reizis B., Leder P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes & Dev. 2002;16:295–300. doi: 10.1101/gad.960702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchini C., Capobianco A.J. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): Implication for cell cycle disruption in transformation by Notch(ic). Mol. Cell. Biol. 2001;21:5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam A., Maillard I., Zediak V.P., Xu L., Gerstein R.M., Aster J.C., Pear W.S., Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat. Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- Sanchez-Irizarry C., Carpenter A.C., Weng A.P., Pear W.S., Aster J.C., Blacklow S.C. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol. Cell. Biol. 2004;24:9265–9273. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh Y., Matsumura I., Tanaka H., Ezoe S., Sugahara H., Mizuki M., Shibayama H., Ishiko E., Ishiko J., Nakajima K., et al. Roles for c-Myc in self-renewal of hematopoietic stem cells. J. Biol. Chem. 2004;279:24986–24993. doi: 10.1074/jbc.M400407200. [DOI] [PubMed] [Google Scholar]
- Schmitt T.M., de Pooter R.F., Gronski M.A., Cho S.K., Ohashi P.S., Zuniga-Pflucker J.C. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat. Immunol. 2004;5:410–417. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- Schroeter E.H., Kisslinger J.A., Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Schuhmacher M., Staege M.S., Pajic A., Polack A., Weidle U.H., Bornkamm G.W., Eick D., Kohlhuber F. Control of cell growth by c-Myc in the absence of cell division. Curr. Biol. 1999;9:1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- Shah S., Lee S.F., Tabuchi K., Hao Y.H., Yu C., LaPlant Q., Ball H., Dann C.E., III, Sudhof T., Yu G. Nicastrin functions as a γ-secretase-substrate receptor. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Shipp M.A., Ross K.N., Tamayo P., Weng A.P., Kutok J.L., Aguiar R.C., Gaasenbeek M., Angelo M., Reich M., Pinkus G.S., et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat. Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- Sicinska E., Aifantis I., Le Cam L., Swat W., Borowski C., Yu Q., Ferrando A.A., Levin S.D., Geng Y., von Boehmer H., et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4:451–461. doi: 10.1016/s1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- Sidman C.L., Denial T.M., Marshall J.D., Roths J.B. Multiple mechanisms of tumorigenesis in E μ-myc transgenic mice. Cancer Res. 1993;53:1665–1669. [PubMed] [Google Scholar]
- Struhl G., Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- Taghon T., Yui M.A., Pant R., Diamond R.A., Rothenberg E.V. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Tan J.B., Visan I., Yuan J.S., Guidos C.J. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat. Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- Tanigaki K., Tsuji M., Yamamoto N., Han H., Tsukada J., Inoue H., Kubo M., Honjo T. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- Tu L., Fang T.C., Artis D., Shestova O., Pross S.E., Maillard I., Pear W.S. Notch signaling is an important regulator of type 2 immunity. J. Exp. Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun T., Hamaguchi Y., Matsunami N., Furukawa T., Honjo T., Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-J κ. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum-Finney B., Xu L., Brashem-Stein C., Nourigat C., Flowers D., Bakkour S., Pear W.S., Bernstein I.D. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat. Med. 2000;6:1278–1281. doi: 10.1038/81390. [DOI] [PubMed] [Google Scholar]
- Weng A.P., Nam Y., Wolfe M.S., Pear W.S., Griffin J.D., Blacklow S.C., Aster J.C. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell. Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng A.P., Ferrando A.A., Lee W., Morris J.P., Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Winandy S., Wu P., Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83:289–299. doi: 10.1016/0092-8674(95)90170-1. [DOI] [PubMed] [Google Scholar]
- Winandy S., Wu L., Wang J.H., Georgopoulos K. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J. Exp. Med. 1999;190:1039–1048. doi: 10.1084/jem.190.8.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer A., Wilson A., Nemir M., MacDonald H.R., Radtke F. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early α β Lineage Thymocytes. Immunity. 2002;16:869–879. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- Wu L., Aster J.C., Blacklow S.C., Lake R., Artavanis-Tsakonas S., Griffin J.D. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- Ye Y., Lukinova N., Fortini M.E. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature. 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]