Abstract
Viral replication and latently infected cellular reservoirs persist in HIV-infected patients achieving undetectable plasma virus levels with potent antiretroviral therapy. We exploited a predictable drug resistance mutation in the HIV reverse transcriptase to label and track cells infected during defined intervals of treatment and to identify cells replenished by ongoing replication. Decay rates of subsets of latently HIV-infected cells paradoxically decreased with time since establishment, reflecting heterogeneous lymphocyte activation and clearance. Residual low-level replication can replenish cellular reservoirs; however, it does not account for prolonged clearance rates in patients without detectable viremia. In patients receiving potent antiretroviral therapy, the latent pool has a heterogeneous and dynamic composition that comprises a progressively increasing proportion of stable lymphocytes. Eradication will not be achieved with complete inhibition of viral replication alone.
The vast majority of HIV particles in the blood of infected persons is released by productively infected lymphocytes. Viral kinetics in plasma and lymphoid tissues during highly active antiretroviral therapy (HAART) demonstrate rapid turnover of this virus population (1–3). Infected macrophages and virus trapped on the follicular dendritic network (FDC) of lymphoid tissues are secondary sources (1–3). These virus sources might be eradicated with a few years of treatment (3, 4), although the long-term clearance kinetics of FDC-associated virus is unknown (5, 6). Stable, latently infected cells, harboring replication competent virus have been found to persist after many years of suppression of viremia to <50 copies per ml (7–9). These cells are a barrier to treatment eradication and rekindle productive viral infection when treatment is interrupted (10, 11).
Ascertaining the clearance characteristics of these cellular reservoirs is important for formulating future treatment and eradication strategies but is complicated by the simultaneous persistence of very low levels of viral replication that may replenish these reservoirs, even in patients who achieve suppression of plasma viral load below the level of detection (<50 copies per ml) (9, 12–18). Whether the stability of latent reservoirs is a direct consequence of replenishment by residual replication during HAART is the subject of much debate (12, 19–22). If stability of the latent reservoir results principally from reseeding, then more effective inhibition of HIV replication might alone be enough to permit elimination of the reservoir within 8 to 10 years of treatment (12, 22). Alternatively, the biological need for memory lymphocytes to persist argues that they should be inherently long-lived (19, 20, 23). An intrinsically slow reservoir decay rate (19) would mean that this reservoir, like immune memory, will not be extinguished within the lifetime of patients, even with complete inhibition of viral replication by more potent drugs.
The factors that maintain or clear the latent reservoir in patients on suppressive therapy are not well understood. Elimination of latently infected lymphocytes presumably results either from cell senescence or from cellular activation by encounter with cognate antigen leading to viral gene expression and cell death (Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org). Recent studies have arrived at divergent half-life estimates for the reservoir of latently infected cells ranging from 6 to 44 months (12, 19). These estimates assume that long-lived, infected cells are a homogeneous population exhibiting exponential decay characteristics, and the assays used do not distinguish latently infected cells established before therapy from those produced by residual replication during HAART. However, there is evidence for both subexponential clearance kinetics of viral reservoirs (17, 24) and variable degrees of replenishment caused by residual replication (12, 22).
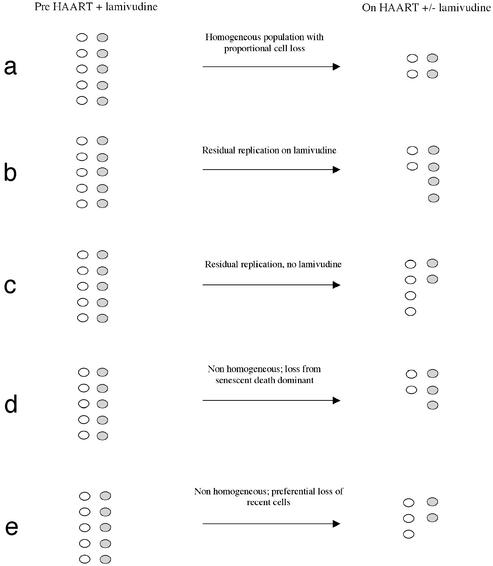
We exploited the predictability of a mutation within the viral reverse transcriptase active site, a methionine to valine replacement at codon 184 (25), to track the fate of infected cell subpopulations during suppressive antiviral therapy. This technique allowed us to quantify both the intrinsic turnover rate of cellular reservoirs and the extent of their replenishment by residual replication. The 184 codon also served as a genetic marker for cells infected before or during defined periods of therapy and thus permitted an analysis of elimination rates based on time of establishment (Fig. 1).
Figure 1.
Fate of infected lymphocytes with M184V mutant and 184M wild-type viruses based on different models of cell clearance and under different treatment conditions. Cells harboring 184M virus are shown as open circles, and cells harboring 184V virus are shaded circles. The expected results shown are theoretical outcomes in isolation from all other processes. (a) Uniform decay predicted by current models which assume a homogeneous population and in the absence of effects due to disproportionate reseeding. (b) Replenishment of reservoir on HAART with lamivudine. (c) Replenishment of reservoir on HAART without lamivudine. (d) Cell loss caused by senescent death. (e) Preferential loss of latently infected cells that were more recently established.
Methods
Patients.
Studies were performed with local Internal Review Board approval and appropriate patient informed consent. Criteria for selection for the present study were previous treatment with regimens containing lamivudine that did not result in suppression of viral replication and hence permitted the emergence of the M184V mutation. In all cases, patients had received multiple nucleoside reverse transcriptase inhibitors (NRTI) as treatment for periods ranging from 7 to 15 months, with continuously detectable viral loads, followed by suppressive combination ART, defined as therapy enabling reduction of plasma viremia to below 50 RNA copies per ml (Fig. 2). Suppressive regimens for four patients included the continuation of lamivudine. Peripheral blood mononuclear cells (PBMC) from multiple time points (median 4.5, range 3 to 7) during nonsuppressive therapy were available from six patients while PBMC from multiple time points (median 7, range 6–14) on suppressive therapy were available from all nine patients.
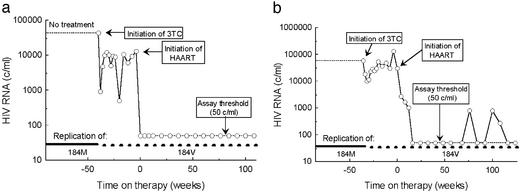
Figure 2.
Plasma RNA response in representative patients, M2 and M1, during suppressive and nonsuppressive therapy. Interval of replication exclusively of 184M virus occurred before therapy (solid bar, lower portion of graph). Interval of preferential replication of 184V genomes began during nonsuppressive treatment and continued during HAART (broken bar, lower portion of graph). (a) Patient M2 experienced rapid and sustained suppression of plasma viremia. (b) Patient M1 received similar treatment but experienced intermittently detectable viremia during HAART.
PBMC Isolation, Nucleic Acid Extraction, and HIV DNA Quantitation.
Buffy coat and cell-free plasma were separated by centrifugation of acid-citrate-dextrose anticoagulated whole blood. Plasma was stored at −80°C. Total PBMC was isolated from the buffy coat by ficoll centrifugation and frozen as pellets of 2 × 106 cells at −80°C or cyropreserved and stored at −150°C until use. HIV RNA was extracted from cell free plasma by using the Qiamp Viral RNA kit (Qiagen, Chatsworth, CA). Cryopreserved cells were thawed and washed in PBS before DNA extraction. DNA was isolated from cell pellets according to published protocols for DNA quantitation (26). In some cases, duplicate aliquots were re-extracted by using Qiamp DNA mini extraction kit (Qiagen) and total DNA was requantified by fluorometry using Hoescht dye 33258 (Sigma). Quantitation of HIV DNA was performed on PBMC DNA extracts according to published protocols (26) with a lower quantitation limit of ≈5–10 HIV DNA copies per microgram of total PBMC DNA.
PCR Amplification for Sequencing Studies.
For sequencing of HIV DNA, ten μl of DNA extract containing 5 to 676 (median 90) HIV DNA templates were subjected to PCR amplification. When fewer than 10 template copies were present in 10 μl of DNA extract, two or three replicate PCRs were performed, and the products were pooled. RNA was extracted from thawed aliquots of plasma by using the Qiagen Rneasy kit (Qiagen), and one-tenth the volume of eluted RNA was amplified. First step PCR or RT-PCR was performed with outer primers RTF1c and RTB1c (Table 6, which is published as supporting information on the PNAS web site). RT-PCR was performed with the Finnzyme Robust RT-PCR kit (MJ Research, Waltham, MA). Second step PCR or RT-PCR was performed by using two μl of template and primers M13 forward-5RT and M13 reverse-3RT (Table 6). Products were purified by using Qiaquick spin columns (Qiagen).
Estimates of Proportions of 184V and 184M by Dye Primer Sequencing.
Dye primer sequencing in both forward and reverse directions by using Prism Big Dye Primer version 3.0 kits was performed according to manufacturer's protocols (Applied Biosystems, Foster City, CA). The proportions of M and V were computed from the relative peak heights, determined from ABI trace files by using Vector NTI 5.0 (Informax, Bethesda, MD). In 14 replicate dye primer assays by using defined mixes of control plasmids, quantification of 184V proportions was consistent, with coefficients of variation ranging from 0.04 to 0.19 (Fig. 5, which is published as supporting information on the PNAS web site).
Virus Isolation by Coculture Near Endpoint Dilution.
PBMC were isolated from 50–150 ml of fresh whole blood anticoagulated with acid citrate dextrose. CD4 enrichment of patient cells and CD8 depletion of donor cells were accomplished by negative selection with either the VarioMacs (Miltenyi Biotec, Auburn, CA) or the Rosette Sep (Stem Cell Technologies, Vancouver) procedures according to manufacturer's directions. Purity of CD4 fractions was confirmed by fluorescence-activated cell sorter (FACS) analysis and was typically >85% CD4+ with fewer than 2% CD8+ cells, natural killer (NK) cells, or monocytes. Patient cells were activated with anti-CD3 antibody (PharMingen) conjugated to Dynal goat anti-mouse M 450 magnetic beads (Dynal, Lake Success, NY) and rIL2 (Chiron, Emeryville, CA) at a concentration of 20 units/ml. Pooled CD8 depleted donor cells were activated with phytohemagglutinin (PHA) (Sigma). Microcultures were performed in 24–100 replicates, each with 105 patient cells and 106 activated donor cells pooled from at least two healthy donors. Half the culture media volume was exchanged twice weekly, and cells were split and replaced with fresh PHA/IL2 stimulated donor cells weekly. Propagating infection was determined by detection of p24 antigen by ELISA (Coulter, Hialeah, FL) at days 7, 14, or 21.
Statistics.
Comparisons of decay rates were made by using nonparametric methods (paired or unpaired Wilcoxon tests). All results reported as significant satisfied P < 0.05 by both the Wilcoxon test and the corresponding two-tailed Student's T test.
Results and Discussion
Nine patients with chronic HIV infection who had received nonsuppressive treatment with nucleoside analogues including lamivudine before initiating suppressive combination antiretroviral treatment as a part of three clinical trials (27, 28, ††) (Table 1 and Fig. 2) were retrospectively identified and studied. All patients remained viremic on regimens of two or three nucleoside analogues. After initiation of HAART, either with or without lamivudine, plasma viral load became undetectable. The patient M2, representative of eight of the nine patients, did not exhibit intermittent viremia (blips) on HAART (Fig. 2a), whereas patient M1 experienced several episodes of detectable viremia between 50 and 700 copies per ml, suggesting a higher level of residual viral replication (12, 16, 18) (Fig. 2b). This patient also required a longer interval of treatment (36 weeks) to achieve a viral load <50 copies per ml.
Table 1.
Baseline patient characteristics
| Patient | Baseline CD4, cells per mm3 | Baseline plasma HIV RNA, copies per ml | Pre-HAART plasma RNA, copies per ml | Mos. on nonsuppressive 3TC regimen | Years on HAART | Pre-HAART regimen | HAART regimen | Blip on HAART |
|---|---|---|---|---|---|---|---|---|
| M1 | 312 | 57,619 | 46,946 | 10 | 4.5 | ZDV/3TC | ZDV/3TC/IDV | Yes |
| M2 | 144 | 43,091 | 12,920 | 10 | 4.5 | ZDV/3TC | ZDV/3TC/IDV | No |
| M3 | 63 | 22,604 | 6,444 | 12 | 4 | ZDV/3TC | ZDV/3TC/IDV | No |
| S1 | 385 | 168,486 | 25,027 | 7 | 3 | ZDV/3TC | ddI/D4T/RIT | No |
| S2 | 588 | 268,765 | 27,317 | 7 | 2.5 | ZDV/3TC | ddI/3TC/IDV | No |
| S3 | 595 | 42,793 | 2,202 | 9 | 2.5 | ZDV/3TC | ddI/D4T/NFV | No |
| D1 | 243* | NA | 118,733 | 9 | 4 | ZDV/3TC/d4T | EFV/IDV | No |
| D2 | 349* | NA | 68,603 | 15 | 3.5 | ZDV/3TC/d4T | EFV/IDV | No |
| D3 | 422* | NA | 21,419 | 10 | 4 | ZDV/3TC/ddC | EFV/IDV | No |
NA, not available; ZDV, zidovudine; 3TC, lamivudine; IDV, indinavir; ddI, didanosine; d4T, stavudine; NFV, nelfinavir; RIT, ritonavir; EFV, efavirenz.
Pre-HAART CD4.
In the patients with available plasma and PBMC samples during nonsuppressive therapy (n = 6), dye primer sequencing, and analysis of relative peak heights (1, 29) demonstrated emergence of the lamivudine-resistant 184V variant within weeks of initiating treatment that included lamivudine. Consistent with previous reports, mutant virus rapidly replaced the wild-type plasma virus population, reflecting the strong selective advantage for the 184V mutant in the presence of drug (25, 30) (Fig. 3a). This fitness advantage predicts that most of the residual viral replication would occur with the 184V mutant in patients that subsequently received suppressive HAART containing lamivudine. Similarly, the wild-type 184M codon is favored in the absence of lamivudine therapy (31) and residual replication of virus with 184M would occur in patients who received suppressive HAART not containing lamivudine.
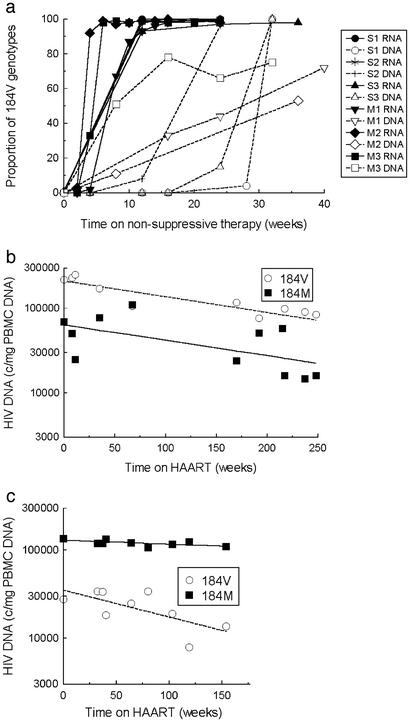
Figure 3.
Rise of M184V mutants in plasma RNA and PBMC DNA. (a) In all six patients, more than 90% of plasma RNA genotypes contained the 184V codon after 12 weeks of treatment with ZDV/3TC. Although the accumulation of 184V in PBMC DNA was slower, the mutant accounted for a majority of genotypes before or at the time HAART was initiated. (b and c) Decay of HIV DNA in representative patients receiving HAART with lamivudine. (b) Patient M1. Viral DNA with the 184V or 184M markers both decay very slowly in this patient, who experienced episodic viremia up to 700 copies per ml during HAART. (c) Patient M2. Viral DNA containing 184M is relatively stable, whereas 184V DNA decays rapidly.
As previously described for other compounds (1, 32), emergence of the drug resistant variant during nonsuppressive therapy was delayed in PBMC DNA (Fig. 3a) indicating that only a small minority of PBMC are productively infected, despite high levels of viral replication in lymphoid tissue. Nevertheless, after this delay, the 184V codon became dominant, reaching maximum proportions of 50–90%. This finding demonstrates that HIV DNA in PBMC, despite the presence of replication impaired and defective forms (33, 34), is maintained dynamically rather than statically.
The intermediate lamivudine resistance mutation 184I was not seen in plasma virus, perhaps because of limited sampling frequency relative to the rapid replacement of the 184I variant by the 184V (30) with greater replication fitness. In two patients, small minority populations with the 184I codon were detected in PBMC DNA at times when plasma virus contained 100% 184V virus (data not shown), verifying the temporary appearance of M184I mutants in vivo (30).
Next, we studied the independent decay characteristics of HIV DNA containing 184M and 184V in PBMC during suppressive therapy including (n = 4) or not including (n = 5) lamivudine. The products of the proportions of 184V and 184M from dye primer sequencing and the total HIV DNA copy numbers from a validated PCR-based assay (26) were used to calculate the size of the subpopulations of cells harboring the 184M and 184V codons (Fig. 3 b and c). The assays used do not distinguish between the different full-length molecular forms of HIV DNA that are present in PBMC. Episomal circular forms are stable, but comprise too small a fraction of the total HIV DNA to affect our estimates (35–37). Linear unintegrated DNA is abundant in PBMC from untreated patients (35) but has a half-life of only ≈6 days (38, 39). Our decay estimates were calculated starting with time points 4–12 weeks after the initiation of HAART to allow for clearance of these labile forms and the few productively infected cells that might be present. Furthermore, although a minority of HIV genomes present in PBMC is replication competent (33–35), decay dynamics appear to be similar for replication-competent and replication-defective genomes, because cell-associated infectivity and total PBMC HIV DNA exhibit comparable decay characteristics after the first several weeks of HAART (3, 13, §§). This similarity in kinetics is consistent with natural history studies showing constant absolute concentrations of HIV DNA in blood cells over the course of infection, rather than rising levels of HIV DNA caused by accumulating defective forms (40). These observations support the measurement of viral DNA clearance to understand turnover of latent reservoirs of replication competent virus in peripheral blood.
Reservoirs of HIV DNA in PBMC behaved as a heterogeneous population with nonuniform decay characteristics (Table 2, Fig. 1 b–e). Cells containing 184M and 184V HIV DNA decayed at significantly different rates (median T1/2 = 151 months vs. 9 months, P = 0.009). Cells labeled with the more recently established 184V codon decayed faster than those harboring 184M in all patients, indicating that clearance of cellular reservoirs of HIV infection is not dominated by the effects of cellular senescence (Fig. 1e). Results were unaffected by censoring of all dye primer results where the minority variant was <10% (data not shown).
Table 2.
Clearance kinetics during HAART of cells containing 184V and 184M DNA
| Patient | HAART with 3TC | Clearance rate on HAART, months−1
|
Half-life or doubling time on HAART, months
|
Months on HAART | Months on 3TC prior to HAART | ||||
|---|---|---|---|---|---|---|---|---|---|
| 184V | 184M | Total DNA | 184V | 184M | Total | ||||
| S1 | No | −0.19 ± 0.18 | 0.114 ± 0.038 | −0.039 ± 0.023 | 3.6 | −6.1 | 18.0 | 32 | 7 |
| S3 | No | −0.28 ± 0.14 | 0.078 ± 0.16 | −0.069 ± 0.027 | 2.5 | −8.8 | 10.1 | 25 | 9 |
| D1 | No | −0.058 ± 0.011 | −0.022 ± 0.010 | −0.033 ± 0.012 | 11.9 | 32.0 | 20.9 | 42 | 9 |
| D2 | No | −0.044 ± 0.021 | 0.016 ± 0.022 | −0.011 ± 0.009 | 15.7 | −43.8 | 61.1 | 39 | 15 |
| D3 | No | −0.197 ± 0.051 | 0.092 ± 0.052 | −0.025 ± 0.018 | 3.5 | −7.5 | 27.8 | 25 | 10 |
| Median | −0.19† | 0.078* | −0.033 | 3.6† | −7.5* | 20.9 | 32 | 9 | |
| S2 | Yes | −0.12 ± 0.054 | −0.0049 ± 0.076 | −0.098 ± 0.022 | 5.9 | 14.3 | 7.1 | 39 | 7 |
| M1 | Yes | −0.020 ± 0.003 | −0.016 ± 0.009 | −0.018 ± 0.004 | 35.3 | 43.6 | 37.8 | 68 | 10 |
| M2 | Yes | −0.030 ± 0.012 | −0.0046 ± 0.002 | −0.008 ± 0.015 | 22.8 | 151.4 | 86.9 | 54 | 10 |
| M3 | Yes | −0.075 ± 0.043 | −0.018 ± 0.054 | −0.016 ± 0.015 | 9.3 | 39.6 | 44.7 | 72 | 12 |
| Median | −0.052† | −0.017* | −0.017 | 16.0† | 41.6* | 41.2 | 39 | 10 | |
Clearance rates indicated best estimate ± standard error. Negative half-lives are given when the best exponential fit indicated growth rather than decay. The numerical value then indicates the doubling time of this growth, so that large negative values reflect near stasis.
P = 0.02.
P = 0.10.
Relative decay of cells infected with HIV containing 184M and 184V during HAART was affected by whether or not patients continued lamivudine. There was a trend toward slower decay of HIV DNA with 184V in patients who continued lamivudine than in those who did not (P = 0.10) (Table 2). Similarly, clearance of HIV DNA with 184M was slower in patients who did not continue lamivudine than in those who did (P = 0.02). These differential decay patterns are consistent with the hypothesis that residual replication replenishes the cellular reservoir. This interpretation is supported by data for patient M1, who experienced frequent episodes of intermittent viremia while continuing lamivudine during HAART. In this patient, the half-life of the cells labeled with 184V was significantly longer (35.3 months) than in any other patient (9.4 ± 7.1 months in others) and approached the half-life of cells labeled with 184M (Fig. 3 b and c and Table 2). To provide a better estimate of the whole-body HIV reservoir size, results were alternatively normalized to blood volume by using available lymphocyte and monocyte counts. Finally, DNA copy numbers per million CD4 cells were calculated to compare the values determined for HIV DNA with clearance of replication competent virus expressed as infectious units per million CD4 cells (12, 13, 19). These alternative normalizations did not significantly affect the results of the analyses (data not shown). Together, these results demonstrate the replenishment of long-lived cellular reservoirs in proportion to the extent of residual replication, as described (12).
The extent of replenishment of cellular reservoirs of HIV by residual replication demonstrated here is surprising because levels of replication on HAART are at least 2 to 3 orders of magnitude lower than in the untreated state. This would require a much more efficient conversion of activated, productively infected cells to a quiescent state than has been estimated based on lymphocyte mitosis (41) or from the ratio of the latent reservoir size to the number of infection events in untreated patients (2, 42). However, a substantial reversion to latency is consistent with the rise of 184V we observed in PBMC DNA during nonsuppressive therapy. Furthermore, latency conversion rates might increase substantially after the normalization of immune activation during successful antiretroviral therapy (43).
If residual replication does reseed the latent reservoir, is this the principal determinant of latent reservoir stability? Notably, cells containing viral DNA bearing the 184V codon were cleared more rapidly than cells containing the wild-type codon, reflecting the faster elimination of cells recently infected compared with those infected earlier for all patients, whether or not patients continued treatment with lamivudine (Table 2). This finding suggests that the decay rates of remotely infected cells are inherently slow and that the persistence of this reservoir was not dependent on residual virus replication.
To analyze the intrinsic clearance characteristics of the latent reservoir (independent of production due to residual replication), we categorized populations of infected cells into those replenished by residual replication and those not replenished by using the 184 codon as a genetic marker. We assumed that the 184M and 184V populations were activated and eliminated with first-order kinetics at rates −δM and −δV, respectively (Tables 3 and 4). As a first approximation, we assumed that all replicating genomes contained the 184V codon (in patients continuing lamivudine) or the 184M codon (in patients not continuing lamivudine). The effect of this reseeding on replenishment of cellular reservoirs was also modeled as a first-order process, with rates πV and πM (Tables 3 and 4). The 184V population decayed more rapidly than did the 184M population (δV > δM, P < 0.01), demonstrating heterogeneity of long-lived cellular reservoirs of HIV. Because it is possible that residual replication of virus with both 184M and 184V codons likely occurs to some extent in patients who continued lamivudine and in those that did not, these values overestimate πM and πV and underestimate δM and δV. After relaxation of assumptions so that only 80% of replicating virus is of the more fit type, the decay of 184V remained significantly more rapid than that of 184M (δV > δM). The significance of the conclusion that the 184M pool was differentially reseeded in patients who did not continue lamivudine was also unaffected by this less stringent model. Normalization of DNA copy numbers to CD4 count or blood volume yielded comparable decay half-life estimates.
Table 3.
Parameters definitions describing differential clearance kinetics of lymphocytes containing 184M and 184V
| Cell population | HAART with lamivudine | HAART without lamivudine |
|---|---|---|
| 184M | −δM | πM − δM |
| 184V | πV − δV | −δV |
Table 4.
Parameters values describing differential clearance kinetics of lymphocytes containing 184M and 184V
| Parameter | Decay rate, months−1 | Half-life, months |
|---|---|---|
| δM | −0.0216 ± 0.0188 | 32 |
| δV | −0.153 ± 0.099 | 4.5 |
| πM | 0.077 ± 0.060 | −9.0* |
| πV | 0.092 ± 0.109 | −7.5* |
Negative half-lives indicate the estimated doubling time of HIV DNA due to ongoing replication.
To determine whether prolonged survival of the cells with 184M was caused by the preferential retention of archived, replication-defective wild-type virus, we performed replicate endpoint dilution microculture of CD4 lymphocytes obtained after 40–58 months of suppressive treatment. Virus was recovered from five of six patients; fresh cells from the three remaining patients were unavailable. As in previous studies, we observed a high ratio of cells with nonreplication competent HIV DNA compared with replication competent HIV (as defined by cultivation of virus in vitro) (3, 35). The replication competent, biological clones did not disproportionately comprise 184V virus (Table 5). In only one patient, D2, from whom relatively few clones were recovered, did the majority of the viral isolates contain 184V. In all other cases, the majority of the replication competent virus carried the 184M codon. The fraction of replication competent virus with 184V or M was not significantly different from estimates extrapolated from dye primer sequencing of HIV DNA in PBMC (Table 5). In one patient, M1, the proportion of clonal viral isolates that had 184M exceeded the estimates from dye primer assays with borderline significance (P = 0.06). Overall, the proportion of 184M HIV DNA that was replication competent was comparable to that for the more recently established 184V HIV DNA, suggesting that the greater stability of the 184M forms reflected a slower clearance rate of archived viral DNA, consistent with recent observations that virus isolated from latently infected cells frequently exhibit ancestral genotypes (13, 18, 21).
Table 5.
Comparison of proportions of replication-competent clones containing 184V with dye primer estimates
| Patient | Mos. on HAART at time of virus isolation | Positive wells/total wells at approximate endpoint dilution | Biological clones with 184V/total positive clones (95% confidence interval for %184V) | % 184V by dye primer sequences from PBMC, %* |
|---|---|---|---|---|
| M1 | 58 | 29/70 | 12/29 (28–57%) | 81 ± 22 |
| M2 | 40 | 18/100 | 3/18 (8–36%) | 5 ± 8 |
| M3 | 58 | 5/50 | 2/5 (15–79%) | 40 ± 27 |
| D1 | 51 | 3/24 | 0/3 (0–53%) | 13 ± 14 |
| D2 | 51 | 5/24 | 4/5 (42–94%) | 37 ± 17 |
| D3 | 56 | 0/24 | Undetected | <3 |
With the exception of M1, fractions of culture positive wells indicate >80% likelihood that viral isolates were clonal. 184M and 184V proportions were not statistically different between cocultured isolates of replication-competent virus and estimates from dye primer sequencing of HIV DNA in PBMC.
Extrapolated to time of coculture from 184V proportions measured after year 1 on HAART.
If cells infected earlier were not disproportionately defective, why were they preferentially retained? Perhaps memory lymphocytes, which recognize a spectrum of antigens, activate and clear at a continuum of rates according to antigen specificities and the abundance of cognate antigens (44, 45). The selective enrichment of HIV-infected cells recognizing rarely encountered antigens (Fig. 6, which is published as supporting information on the PNAS web site) could also explain why some estimates of the reservoir decay half-life (19), including the present estimates, exceed previous measurements of the intermitotic half-life of memory lymphocytes (41). The greater stability of cells infected at earlier times might also reflect different dynamics of phenotypically distinct subpopulations of latently infected cells, such as CD8+ lymphocytes (46), monocytes (47), natural killer cells (48), memory (49) and naïve (50) CD4+ lymphocytes, or effector and central memory lymphocytes (51, 52). Alternatively, the stability of 184M-labeled cells might have resulted from residual replication of wild-type virus regardless of treatment regimen. However, this seems unlikely because lamivudine penetrates well into putative anatomic sanctuaries (53, 54) and the fitness difference between 184M and 184V virus in the presence of lamivudine is large (25). Division of latently infected memory cells without viral production, rather than residual replication, could also result in a net gain of some HIV infected cells (42), but this would not explain the regimen dependence of decay rates. Thus, the biological basis of the observed reservoir heterogeneity deserves future study.
Discrepancies in estimated decay kinetics of cellular reservoirs of HIV have been explained by interpatient differences in viral suppression, as measured by the frequency of intermittent viremia and the stage of disease when treatment is begun (12, 13, 19). Our data support the recent suggestion that the interval of study after initiation of HAART may also impact estimates of reservoir dynamics (24). We observed a median clearance T1/2 of 9 months for infected cells that dominated decay during the early period of HAART and 151 months for cells that dominated later. These results span the range of those previously reported (12, 13, 19).
In summary, our data demonstrate that long-lived cellular reservoirs of HIV in patients on HAART reflect a heterogeneous population of cells with nonuniform decay characteristics and that reservoir stability is not dependent on replenishment by residual viral replication. The hypothesis that antigen specificity and corresponding antigen frequency dictate clearance rate predicts that the population of cells latently infected with HIV will gradually shift toward cells specific for increasingly rare antigens, assuring the life-long persistence of this viral reservoir, in accord with the durability of immunologic memory. Treatments with greater potency, although needed, will not be sufficient to achieve eradication (12, 55). Interventions designed to mobilize these cellular reservoirs or to promote recognition and clearance of cells expressing low levels of viral antigens must be pursued (56–58).
Supplementary Material
Acknowledgments
We thank the participating patients; S. Wilcox, D. Smith, and R. Sysyn for administrative support; L. Meixner, K. Nuffer, C. Grube, and G. Dyak for study coordination; S. Wolinsky, T. Leitner, J. Guatelli, and A. McLean for helpful comments; J. Condra and W. Schleif for sharing patient samples; C. Christopherson, S. Kwok, K. Young, and Roche Molecular Diagnostics for reagents used in DNA quantitation. This work was supported by La Jolla Interfaces in Science Fellowship and National Institutes of Health Medical Scientist Training Program (M.C.S.); Swiss National Science Foundation Grants 3345-062041 and 3345-65168 and Swiss HIV Cohort Study (H.F.G., M.F., and M.O.); National Institutes of Health Grants AI51982 (D.V.H.), AI27670, AI38858, AI29164, and AI36214 (University of California at San Diego Center for AIDS Research), and the San Diego Veterans Administration Research Center for AIDS and HIV Infection (D.D.R.); and National Institutes of Health Grant AI43752 (J.K.W.) and Department of Veteran Affairs Research Merit Awards (J.K.W. and C.A.S.).
Abbreviations
- HAART
highly active antiretroviral therapy
- FDC
follicular dendritic network
- PBMC
peripheral blood mononuclear cells
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Ruiz, N. & Ridler, S. (1997) 4th International Conference on Retrovirus and Opportunistic Infections, Jan. 22–26, Washington, DC LB1, 206 (abstr.).
Strain, M., Little, S., Daar, E., Günthard, H., Spina, C., Lam, R. Y., Daly, O. A., Ignacio, C., Macaranas, T., Kwok, S., et al. (2002) 9th International Conference on Retrovirus and Opportunistic Infections, February 24–28, Seattle, WA, 97 (abstr.).
References
- 1.Wei X, Ghosh S K, Taylor M E, Jonson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, et al. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 2.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 3.Perelson A S, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D D. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 4.Cavert W, Notermans D W, Staskus K, Wietgrefe S, Zupancic M, Gebhard K, Henry K, Zhang Z Q, Mills R, McDade H, et al. Science. 1997;276:960–964. doi: 10.1126/science.276.5314.960. [DOI] [PubMed] [Google Scholar]
- 5.Hlavacek W S, Wofsy C, Perelson A S. Proc Natl Acad Sci USA. 1999;96:14681–14686. doi: 10.1073/pnas.96.26.14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hlavacek W S, Stilianakis N I, Notermans D W, Danner S A, Perelson A S. Proc Natl Acad Sci USA. 2000;97:10966–10971. doi: 10.1073/pnas.190065897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong J K, Hezareh M, Günthard H F, Havlir D, Ignacio C C, Spina C, Richman D D. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 8.Finzi D, Hermankova M, Pierson T, Carruth L, Buck C, Chaisson R, Quinn T, Chadwick K, Margolick J, Brookmeyer R, et al. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 9.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J M, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neumann A U, Tubiana R, Calvez V, Robert C, Li T S, Agut H, Autran B, Katlama C. AIDS. 1999;13:677–683. doi: 10.1097/00002030-199904160-00008. [DOI] [PubMed] [Google Scholar]
- 11.Davey R T, Jr, Bhat N, Yoder C, Chun T W, Metcalf J A, Dewar R, Natarajan V, Lempicki R A, Adelsberger J W, Miller K D, et al. Proc Natl Acad Sci USA. 1999;96:15109–15114. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramratnam B, Mittler J E, Zhang L, Boden D, Hurley A, Fang F, Macken C, Perelson A S, Markowitz M, Ho D D. Nat Med. 2000;6:82–85. doi: 10.1038/71577. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Ramratnam B, Tenner-Racz K, He Y, Vesanen M, Lewin S, Talal A, Racz P, Perelson A S, Korber B T, et al. N Engl J Med. 1999;340:1605–1613. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
- 14.Dornadula G, Zhang H, VanUitert B, Stern J, Livornese L, Jr, Ingerman M J, Witek J, Kedanis R J, Natkin J, DeSimone J, et al. J Am Med Assoc. 1999;282:1627–1632. doi: 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 15.Schockmel G A, Yerly S, Perrin L. J Acquired Immune Defic Syndr Hum Retrovirol. 1997;14:179–183. doi: 10.1097/00042560-199702010-00013. [DOI] [PubMed] [Google Scholar]
- 16.Havlir D V, Bassett R, Levitan D, Gilbert P, Tebas P, Collier A C, Hirsch M S, Ignacio C, Condra J, Günthard H F, et al. J Am Med Assoc. 2001;286:171–179. doi: 10.1001/jama.286.2.171. [DOI] [PubMed] [Google Scholar]
- 17.Furtado M R, Callaway D S, Phair J P, Kunstman K J, Stanton J L, Macken C A, Perelson A S, Wolinsky S M. N Engl J Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 18.Günthard H F, Frost S D, Leigh-Brown A J, Ignacio C C, Kee K, Perelson A S, Spina C A, Havlir D V, Hezareh M, Looney D J, et al. J Virol. 1999;73:9404–9412. doi: 10.1128/jvi.73.11.9404-9412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finzi D, Blankson J, Siliciano J D, Margolick J B, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 20.Blankson J N, Persaud D, Siliciano R F. Annu Rev Med. 2002;53:557–593. doi: 10.1146/annurev.med.53.082901.104024. [DOI] [PubMed] [Google Scholar]
- 21.Hermankova M, Ray S C, Ruff C, Powell-Davis M, Ingersoll R, D'Aquila R T, Quinn T C, Siliciano J D, Siliciano R F, Persaud D. J Am Med Assoc. 2001;286:196–207. doi: 10.1001/jama.286.2.196. [DOI] [PubMed] [Google Scholar]
- 22. Ramratnam, B., Ribeiro, R., He, T., Chung, C., Simon, V., Vanderhoeven, J., Hurley, A., Zhang, L., Perelson, A. S., Ho, D., et al. (2003) Arch Int. Med., in press.
- 23.Antia R, Pilyugin S S, Ahmed R. Proc Natl Acad Sci USA. 1998;95:14926–14931. doi: 10.1073/pnas.95.25.14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holte, S., Melvin, A., Mullins, J. & Frenkel, L. M. (2002) J. Acquired Immune Defic. Syndr., in press. [DOI] [PubMed]
- 25.Boucher C A B, Cammack N, Schipper P, Schuurman R, Rouse P, Wainberg M A, Cameron J M. Antimicrob Agents Chemother. 1993;37:2231–2234. doi: 10.1128/aac.37.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christopherson C, Kidane Y, Conway B, Krowka J, Sheppard H, Kwok S. J Clin Microbiol. 2000;38:630–634. doi: 10.1128/jcm.38.2.630-634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Deutsch P, Meibohm A, et al. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 28.Opravil M, Cone R W, Fischer M, Vernazza P L, Bassetti S, Lorenzi P, Bisset L R, Ott P, Huber W, Knuchel M C, et al. J Acquired Immune Defic Syndr. 2000;23:17–25. doi: 10.1097/00126334-200001010-00003. [DOI] [PubMed] [Google Scholar]
- 29.Leitner T, Halapi E, Scarlatti G, Rossi P, Albert J, Fenyo E M, Uhlen M. Biotechniques. 1993;15:120–127. [PubMed] [Google Scholar]
- 30.Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, Collis P, Danner S, Mulder J, Loveday C, Christopherson C, Kwok S, et al. J Infect Dis. 1995;171:1431–1437. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 31.Back N K T, Nijhuis M, Keulen W, Boucher C A B, Essink B O, van Kullenburg A B P, van Gennip A H, Berkhout B. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 32.Havlir D V, Gamst A, Eastman S, Richman D D. J Virol. 1996;70:7894–7899. doi: 10.1128/jvi.70.11.7894-7899.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez G, Xu X, Chermann J C, Hirsch I. J Virol. 1997;71:2233–2240. doi: 10.1128/jvi.71.3.2233-2240.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Kappes J C, Conway J A, Price R W, Shaw G M, Hahn B H. J Virol. 1991;65:3973–3985. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chun T W, Carruth L, Finzi D, Shen X, DiGiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, et al. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 36.Fischer M, Trkola A, Joos B, Hafner R, Joller H, Muesing M A, Kaufman D R, Berli E, Hirschel B, Weber R, Günthard H F for the Swiss HIV-1 Cohort Study. Antiviral Ther. 2003;8:65–72. [PubMed] [Google Scholar]
- 37.Pierson T C, Kieffer T L, Ruff C T, Buck C, Gange S J, Siliciano R F. J Virol. 2002;76:4138–4144. doi: 10.1128/JVI.76.8.4138-4144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 39.Blankson J N, Finzi D, Pierson T C, Sabundayo B P, Chadwick K, Margolick J B, Quinn T C, Siliciano R F. J Infect Dis. 2000;182:1636–1642. doi: 10.1086/317615. [DOI] [PubMed] [Google Scholar]
- 40.Cone R W, Gowland P, Opravil M, Grob P, Ledergerber B. AIDS. 1998;12:2253–2260. doi: 10.1097/00002030-199817000-00005. [DOI] [PubMed] [Google Scholar]
- 41.McLean A R, Michie C A. Proc Natl Acad Sci USA. 1995;92:3707–3711. doi: 10.1073/pnas.92.9.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierson T, McArthur J C, Siliciano R F. Annu Rev Immunol. 2000;18:665–708. doi: 10.1146/annurev.immunol.18.1.665. [DOI] [PubMed] [Google Scholar]
- 43.Autran B, Carcelain G, Li T S, Blanc C, Mathez D, Tubiana R, Katlama C, Debre P, Leibowitch J. Science. 1997;277:112–116. doi: 10.1126/science.277.5322.112. [DOI] [PubMed] [Google Scholar]
- 44.Grossman Z, Polis M, Feinberg M B, Levi I, Jankelevich S, Yarchoan R, Boon J, de Wolf F, Lange J M, Goudsmit J, et al. Nat Med. 1999;5:1099–1104. doi: 10.1038/13410. [DOI] [PubMed] [Google Scholar]
- 45.Müller V, Vigueras-Gomez J F, Bonhoeffer S. J Virol. 2002;76:8963–8965. doi: 10.1128/JVI.76.17.8963-8965.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McBreen S, Imlach S, Shirafuji T, Scott G R, Leen C, Bell J E, Simmonds P. J Virol. 2001;75:4091–4102. doi: 10.1128/JVI.75.9.4091-4102.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonza S, Mutimer H P, Oelrichs R, Jardine D, Harvey K, Dunne A, Purcell D F, Birch C, Crowe S M. AIDS. 2001;15:17–22. doi: 10.1097/00002030-200101050-00005. [DOI] [PubMed] [Google Scholar]
- 48.Valentin A, Rosati M, Patenaude D J, Hatzakis A, Kostrikis L G, Lazanas M, Wyvill K M, Yarchoan R, Pavlakis G N. Proc Natl Acad Sci USA. 2002;99:7015–7020. doi: 10.1073/pnas.102672999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pierson T, Hoffman T L, Blankson J, Finzi D, Chadwick K, Margolick J B, Buck C, Siliciano J D, Doms R W, Siliciano R F. J Virol. 2000;74:7824–7833. doi: 10.1128/jvi.74.17.7824-7833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brooks D G, Kitchen S G, Kitchen C M, Scripture-Adams D D, Zack J A. Nat Med. 2001;7:459–464. doi: 10.1038/86531. [DOI] [PubMed] [Google Scholar]
- 51.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Nature. 1999;401:708–712. [PubMed] [Google Scholar]
- 52.Pitcher C J, Hagen S I, Walker J M, Lum R, Mitchell B L, Maino V C, Axthelm M K, Picker L J. J Immunol. 2002;168:29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- 53.Foudraine N A, Hoetelmans R M, Lange J M, De Wolf F, van Benthem B H, Maas J J, Keet I P, Portegies P. Lancet. 1998;351:1547–1551. doi: 10.1016/S0140-6736(98)07333-4. [DOI] [PubMed] [Google Scholar]
- 54.Taylor S, van Heeswijk R P, Hoetelmans R M, Workman J, Drake S M, White D J, Pillay D. AIDS. 2000;14:1979–1984. doi: 10.1097/00002030-200009080-00014. [DOI] [PubMed] [Google Scholar]
- 55.Weverling G J, Lange J M A, Jurriaans S, Prins J M, Lukashov V V, Notermans D W, Roos M, Schuitemaker H, Hoetelmans R M W, Danner S, et al. AIDS. 1998;12:F117–F122. doi: 10.1097/00002030-199811000-00003. [DOI] [PubMed] [Google Scholar]
- 56.Prins J M, Jurriaans S, van Praag R M, Blaak H, van Rij R, Schellekens P T, ten Berge I J, Yong S L, Fox C H, Roos M T, et al. AIDS. 1999;13:2405–2410. doi: 10.1097/00002030-199912030-00012. [DOI] [PubMed] [Google Scholar]
- 57.Kulkosky J, Culnan D M, Roman J, Dornadula G, Schnell M, Boyd M R, Pomerantz R J. Blood. 2001;98:3006–3015. doi: 10.1182/blood.v98.10.3006. [DOI] [PubMed] [Google Scholar]
- 58.Chun T W, Engel D, Mizell S B, Ehler L A, Fauci A S. J Exp Med. 1998;188:83–91. doi: 10.1084/jem.188.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.