Abstract
Lignin quantity and reactivity [which is associated with its syringyl/guaiacyl (S/G) constituent ratio] are two major barriers to wood-pulp production. To verify our contention that these traits are regulated by distinct monolignol biosynthesis genes, encoding 4-coumarate–CoA ligase (4CL) and coniferaldehyde 5-hydroxylase (CAld5H), we used Agrobacterium to cotransfer antisense 4CL and sense CAld5H genes into aspen (Populus tremuloides). Trees expressing each one and both of the transgenes were produced with high efficiency. Lignin reduction by as much as 40% with 14% cellulose augmentation was achieved in antisense 4CL plants; S/G-ratio increases as much as 3-fold were observed without lignin quantity change in sense CAld5H plants. Consistent with our contention, these effects were independent but additive, with plants expressing both transgenes having up to 52% less lignin, a 64% higher S/G ratio, and 30% more cellulose. An S/G-ratio increase also accelerated cell maturation in stem secondary xylem, pointing to a role for syringyl lignin moieties in coordinating xylem secondary wall biosynthesis. The results suggest that this multigene cotransfer system should be broadly useful for plant genetic engineering and functional genomics.
Tremendous efforts have been devoted to developing genetically engineered trees, with the emphasis on reducing lignin quantity, to improve wood-pulp production efficiency (1–5). However, lignin chemical reactivity also is a critical barrier to wood-pulp production, because lignin removal from wood is either initiated by chemical degradations or, in most cases, accomplished entirely through chemical reactions. Thus, the current tree biotechnology emphasis on low lignin quantity must be expanded to include greater lignin reactivity and, ultimately, a combination of low and reactive lignin traits.
Lignin in angiosperm trees is polymerized from the guaiacyl and syringyl monolignols (Fig. 1) with a syringyl/guaiacyl (S/G) ratio of ≈2–2.5 (6–10). Wood-pulping kinetics further revealed that every unit increase in the lignin S/G ratio would roughly double the rate of lignin removal (8), supporting the idea that combinations of S/G-ratio augmentations and lignin reductions may offer far-reaching opportunities for maximizing wood-pulp production efficiency. However, in trees genetic reduction of lignin, which has been achieved through the suppression of the monolignol pathway gene encoding either 4-coumarate–CoA ligase (4CL) (2) or caffeoyl CoA O-methyltransferase (3), had no significant effect on the S/G ratio. Attempts to modify the S/G ratio in trees could not succeed in lignin reduction (11, 12). These results argue that lignin quantity and the S/G ratio are regulated independently during lignin biosynthesis in trees.
Figure 1.
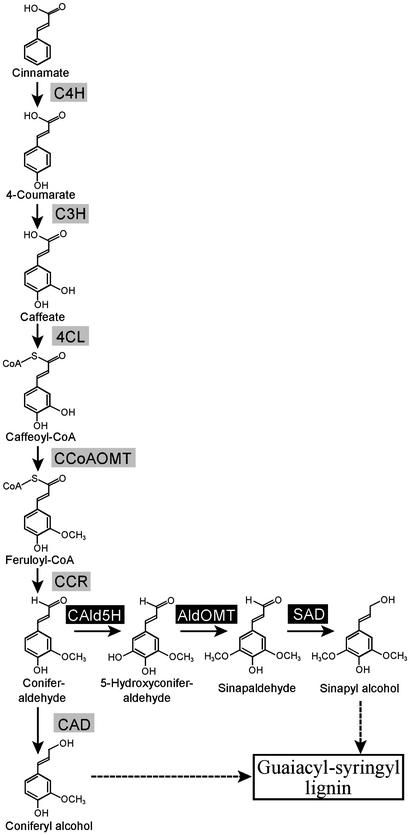
Proposed principal biosynthetic pathway for the formation of monolignols in woody angiosperms. C4H, cinnamate 4-hydroxylase; C3H, 4-coumarate 3-hydroxylase; CCoAOMT, caffeoyl CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; AldOMT, 5-hydroxyconiferaldehyde O-methyltransferase; SAD, sinapyl alcohol dehydrogenase; CAD, cinnamyl alcohol dehydrogenase.
Considerable evidence is now available that shows that in angiosperm trees, the syringyl monolignol pathway (Fig. 1) branches out from the guaiacyl pathway through coniferaldehyde and is regulated in sequence by three genes encoding coniferaldehyde 5-hydroxylase (CAld5H) (13), 5-hydroxyconiferaldehyde O-methyltransferase (14), and sinapyl alcohol dehydrogenase (15). Enzyme kinetics further demonstrated that CAld5H has a 6- to 50-times-slower turnover rate than the other two syringyl pathway enzymes (13–15), pointing to a key role for CAld5H in limiting syringyl monolignol biosynthesis and, therefore, lignin S/G ratio. Thus, simultaneously up-regulating CAld5H and down-regulating 4CL gene expression may lead to a concurrent lignin-reactivity augmentation and quantity reduction in plants. Although multigene manipulation in plants could be achieved by repetitive gene insertion or crosspollination (refs. 16–18; see ref. 19 for review), these approaches are impractical for trees, which have long life cycles. Transformation with a single multigene-encompassing construct is another possibility, which, if successful, may lead to transgenics integrating only a given set of the transgenes (20). But the central question from a practical viewpoint relating to the perspective of maximizing the efficiency of various wood-conversion processes is how genetic integrations involving individual transgenes as well as combinations of these genes can be achieved concurrently to yield distinct transgenic tree clones with low lignin, a high S/G ratio, and a combination of the two.
Here we report the modification of multiple lignin traits in a tree species using an Agrobacterium-mediated cotransformation system. Two different genetic constructs harboring aspen xylem-specific promoter (Pt4CL1P)-driven aspen antisense Pt4CL (2, 21) and sweetgum (Liquidambar styraciflua) sense LsCAld5H (13) genes were cotransferred into aspen, which led to the production of a variety of phenotypically normal transgenics expressing each one and both of the transgenes. The transgenics expressing a single transgene showed that antisense down-regulation of 4CL gene expression selectively mediated lignin reduction, whereas overexpression of the CAld5H gene specifically induced S/G-ratio augmentation. These independent effects became additive, with transgenic trees simultaneously expressing two transgenes exhibiting strong lignin reductions and drastic lignin S/G-ratio augmentations.
Materials and Methods
Transformation of Aspen with Multiple Genes.
Aspen xylem-specific promoter (Pt4CL1P)-GUS binary plasmid DNA (22) was used as a module for preparing the genetic constructs. The GUS fragment was replaced with either an antisense Pt4CL1 or a sense LsCAld5H cDNA with respect to the Pt4CL1P promoter. Each construct was mobilized into disarmed Agrobacterium tumefaciens C58 strain and used to coinoculate aspen leaf discs for the production of transgenic trees as described (2). Wild-type aspen plants derived from in vitro micropropagation were used as the control.
PCR, Protein Blot, and Enzyme-Activity Analyses.
PCR on aspen genomic DNA was used to verify transgene integration. Pt4CL1P promoter-specific sense primer (5′-CAGGAATGCTCTGCACTCTG-3′) coupled with Pt4CL1 5′-end sense primer (5′-ATGAATCCACAAGAATTCAT-3′) was used for amplifying the antisense Pt4CL1 transgene and with LsCAld5H 3′-end antisense primer (5′-ATAGAGAGGACAGAGAAGGCG-3′) for amplifying the LsCAld5H sense transgene. Crude protein was extracted from developing xylem of 10-month-old greenhouse-grown trees, used in protein gel-blot analysis probed with anti-4CL1 (21) or anti-CAld5H (13) antibodies, and used for assaying 4CL and CAld5H activities with caffeate and coniferaldehyde as substrates, respectively, as described (15).
Lignin Histochemical Analysis and Protein Immunolocalization.
Fresh hand-cut sections from stem internodes of transgenics used for the protein extraction described above were used for lignin histochemical localization by using the Cross–Bevan method as described (15). Thin sections (3 μm thick) from the same stem segments used for histochemical analysis were made for immunolocalizing 4CL and CAld5H proteins by using corresponding antibodies as we did before (15).
Lignin Analysis.
After stem developing xylem was collected for protein extraction, the woody tissue was ball-milled and used for lignin-content determination by using the Klason method (2) and for lignin S/G-ratio analysis by thioacidolysis (2). The rest of the ball-milled woody tissue was used to isolate lignin (23) for NMR analysis. The 1D 13C and 2D heteronuclear multiple-bond correlation (HMBC) NMR spectra were taken on a Bruker DRX-360 instrument fitted with a 5-mm 1H/broadband gradient probe with inverse geometry (proton coils closest to the sample). The conditions used for all samples were ≈100 mg of acetylated lignin in 0.4 ml of acetone-d6, with the central solvent peak as internal reference (δH 2.04, δC 29.80). Experiments used were standard Bruker implementations of gradient-selected versions of inverse (1H-detected) HMBC experiments (80-ms long-range coupling delay) as described (24).
Carbohydrate Analysis.
The stem woody tissue used for lignin analysis was used also for the determination of cellulose and hemicellulose contents as described (2).
Results
Establishment of Agrobacterium-Mediated Cotransformation System.
To establish a multigene cotransformation system, four pBI101-based binary vectors, each containing a cauliflower mosaic virus 35S-driven monolignol pathway cDNA sequence, were mobilized individually into independent A. tumefaciens C58 strains for cotransforming tobacco. Forty-eight independent kanamycin-resistant transgenic plants were regenerated after cocultivating leaf tissues with a mixture of these four independent Agrobacterium strains. Of these transgenic plants, 35%, 27%, 19%, and 19% contained one, two, three, and four transgene constructs, respectively, based on PCR (data not shown), validating the multigene cotransformation system. We then applied this system to manipulate the lignin content and S/G ratio in aspen.
Characterization of Transgenic Aspen.
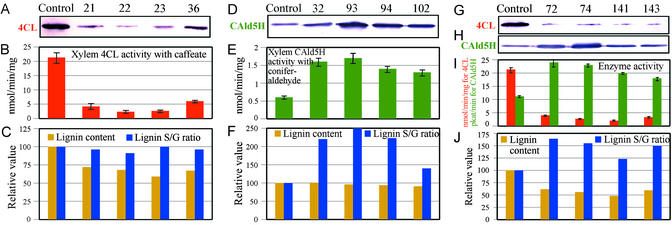
Heterologous LsCAld5H was used to avoid possible sequence homology-based gene silencing (25, 26). Forty phenotypically normal transgenic aspen lines were obtained, of which 37%, 40%, and 23% harbored antisense Pt4CL, sense LsCAld5H, and antisense Pt4CL + sense LsCAld5H gene constructs, respectively, as confirmed by genomic PCR. From each of these three transgenic groups grown in a greenhouse, several trees were selected randomly (Fig. 2) and harvested at the age of 10 months during the growing season for various characterizations. The effects of transgene expression are shown in Fig. 3. 4CL protein levels were reduced drastically in lines (Fig. 3A) harboring only antisense Pt4CL transgene (Fig. 2), leading to a 70–90% reduction in xylem 4CL enzyme activity (Fig. 3B) and a 30–40% reduction in stem lignin (Fig. 3C and Table 1). No significant effect on the lignin S/G ratio was found (Fig. 3C and Table 1). Overexpressing the LsCAld5H gene alone (Fig. 2) drastically elevated the xylem CAld5H protein levels (Fig. 3D), giving rise to a 2.2- to 2.8-fold increase in xylem CAld5H enzyme activity (Fig. 3E). As a result, these transgenics exhibited up to a remarkable 2.5-fold increase in the S/G ratio as compared with the control (Fig. 3F and Table 1). The single CAld5H gene effect had no influence on total lignin accumulation in transgenic trees (Fig. 3F and Table 1). However, the single-gene effects became additive in transgenics harboring both antisense Pt4CL and sense LsCAld5H genes. Alterations of 4CL and CAld5H protein levels in these trees were consistent with changes of the corresponding enzyme activities: 80–90% reduction in 4CL and 60–110% increase in CAld5H (Fig. 3 G, H, and I). This combinatorial gene manipulation led to a 38–52% reduction in stem lignin and 22–64% increase in the lignin S/G ratio (Fig. 3J and Table 1).
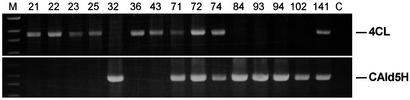
Figure 2.
PCR analysis of the integration of antisense Pt4CL1 and sense LsCAld5H transgenes in various transgenic aspen (lane numbers represent different transgenic lines). A 1-kb DNA ladder was used for both panels (lane M). The PCR fragments seen are 1.66 and 1.61 kb in size, encompassing a portion of the aspen xylem-specific promoter and the full-length antisense Pt4CL and sense LsCAld5H coding sequences, respectively. Such transgene fragments were absent from the control (lane C).
Figure 3.
The effects of down-regulation of 4CL and up-regulation of CAld5H on 4CL and CAld5H enzyme activities and lignin accumulation and S/G ratio. (A, D, G, and H) Protein gel-blot (10 μg of protein extracts per lane) analysis of xylem 4CL and CAld5H protein levels by using anti-4CL (A and G) and anti-CAld5H (D and H) antibody probes, respectively. (B, E, and I) 4CL and CAld5H enzyme activities in stem developing xylem tissue. Crude protein (20–40 μg) was assayed for 4CL (B and I) and CAld5H (E and I) activities with caffeate and coniferaldehyde, respectively, by using HPLC/MS. Error bars represent standard deviation values of three replicates. (C, F, and J) The levels of lignin reduction and lignin S/G ratio increase in stem wood of transgenic lines as compared with the control.
Table 1.
Chemical compositions in stem wood of control and transgenic aspen
| Plant | Control | 21 | 22 | 23 | 25 | 36 | 32 | 84 | 93 | 94 | 102 | 72 | 74 | 141 | 143 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene integrated | |||||||||||||||
| −4CL | −4CL | −4CL | −4CL | −4CL | −4CL | −4CL | −4CL | −4CL | −4CL | ||||||
| +CAId5H | +CAId5H | +CAId5H | +CAId5H | +CAId5H | +CAId5H | +CAId5H | +CAId5H | +CAId5H | +CAId5H | ||||||
| Lignin content, % | 22.2 ± 0.8 | 16.0 ± 0.6 | 15.3 ± 0.4 | 14.4 ± 0.5 | 13.1 ± 0.3 | 14.9 ± 0.2 | 22.4 ± 0.5 | 21.6 ± 0.4 | 21.1 ± 0.4 | 20.7 ± 0.6 | 19.7 ± 0.4 | 13.7 ± 0.4 | 12.4 ± 0.5 | 10.7 ± 0.4 | 13.2 ± 0.3 |
| Lignin S/G ratio | 2.2 | 2.1 | 2.0 | 2.2 | 2.3 | 2.1 | 4.8 | 4.0 | 5.5 | 4.9 | 3.0 | 3.6 | 3.4 | 2.7 | 3.3 |
| Syringyl content, μmol/g lignin | 1190 ± 94 | 1067 ± 87 | 924 ± 72 | 1027 ± 88 | 864 ± 71 | 838 ± 69 | 1815 ± 98 | 1441 ± 103 | 1735 ± 121 | 1502 ± 120 | 1567 ± 110 | 1109 ± 86 | 1145 ± 82 | 986 ± 71 | 1241 ± 79 |
| Guaiacyl content, μmol/g lignin | 546 ± 38 | 504 ± 39 | 455 ± 40 | 469 ± 42 | 370 ± 27 | 394 ± 32 | 377 ± 30 | 362 ± 28 | 318 ± 25 | 305 ± 24 | 519 ± 38 | 307 ± 20 | 340 ± 22 | 373 ± 36 | 371 ± 31 |
| Syringyl + guaiacyl, μmol/g lignin | 1736 | 1571 | 1379 | 1496 | 1234 | 1232 | 2192 | 1803 | 2053 | 1807 | 2086 | 1416 | 1485 | 1359 | 1612 |
| Cellulose content, % | 41.4 ± 0.4 | 43.1 ± 0.3 | ND | 44.8 ± 0.4 | 47.3 ± 0.5 | ND | 40.0 ± 0.3 | 42.6 ± 0.2 | 44.7 ± 0.1 | 43.4 ± 0.2 | 44.3 ± 0.1 | 49.2 ± 0.3 | 50.9 ± 0.1 | 53.3 ± 0.2 | ND |
| Xylan content, % | 15.8 ± 0.2 | 16.8 ± 0.3 | ND | 16.1 ± 0.3 | 16.9 ± 0.4 | ND | 16.5 ± 0.4 | 15.3 ± 0.1 | 15.7 ± 0.5 | 15.6 ± 0.5 | 15.2 ± 0.1 | 15.3 ± 0.3 | 14.6 ± 0.2 | 15.4 ± 0.4 | ND |
| Cellulose/lignin ratio | 1.9 | 2.7 | ND | 3.1 | 3.6 | ND | 1.8 | 1.9 | 2.1 | 2.0 | 2.2 | 3.6 | 4.1 | 5.0 | ND |
Values are means ± SE of two to three assays of different samples from each line. −4CL and +CAId5H, antisense 4CL and sense CAId5H transgenes, respectively. Lignin, cellulose, and xylan contents are percentages of dry wood weight. ND, not determined.
Immunochemical and Histochemical Characterization of Transgenic Aspen.
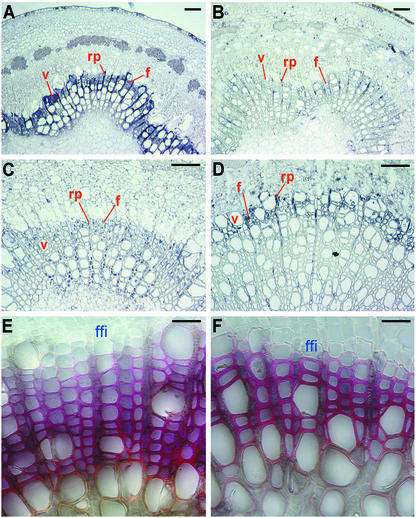
We next examined the indigenous levels of stem 4CL and CAld5H proteins in control and transgenic plants. In control plants, 4CL protein signals were most conspicuous in the stem secondary developing xylem (Fig. 4A). Confirming the gene-suppression effects, these 4CL signals were diminished drastically in stem xylem elements of the lignin-reduced antisense Pt4CL transgenics as shown in Fig. 4B for plant 23. In sense LsCAld5H transgenics such as plant 93 (Fig. 4D), the xylem-specific overexpression of the transgene was evidenced by the explicit elevation of CAld5H protein signals in a few layers of the developing xylem elements, particularly the fiber cells, located at the periphery of the secondary xylem. But in control plants, the indigenous CAld5H was distributed in multiple cell layers in stem xylem (Fig. 4C). The absence of CAld5H signals from the interior of the secondary xylem in the sense LsCAld5H transgenic may suggest completed lignification in these xylem elements. Furthermore, in sense LsCAld5H transgenics, secondary cell-wall thickening occurred rapidly and apparently completed in elements adjacent to the periphery of the secondary xylem (Fig. 4F, plant 93) as revealed by the Cross–Bevan cell-wall staining. Because syringyl lignin deposition marks the concluding phase of lignification (27–30), the intense syringyl-specific Cross–Bevan staining of the peripheral xylem elements in sense LsCAld5H transgenics exemplified an extensive lignification occurring already in the newly fusiform initial-derived xylem elements. These phenomena were observed for the transgenics expressing the sense LsCAld5H gene alone or together with antisense Pt4CL transgene (data not shown). In contrast, in control plants the secondary cell-wall thickening and lignification in xylem elements proceeded gradually along the centripetal course of cell differentiation (thin-walled, pink cells) and reached completion (thick-walled, dark-red cells) near the interior of the secondary xylem (Fig. 4E). These results suggest that the sense overexpression of CAld5H triggering an accelerated syringyl lignin deposition induces a rapid maturation/lignification of stem secondary xylem cells.
Figure 4.
Immunodection of 4CL and CAld5H protein levels and cell-wall histochemical staining in transgenic and control plants. Light micrographs of stem transverse sections showing protein localization (A–D) and cell-wall lignin staining (E and F) are shown. (A and B) In the fifth internode, strong 4CL protein signals were present in vessel (v), ray parenchyma (rp), and fiber (f) cells of developing xylem of control (A) but were reduced greatly in these cells/elements in 4CL down-regulated transgenic plants such as plant 23 (B) having a 35% lignin reduction (Table 1). (C and D) In the ninth internode, the CAld5H protein signals were distributed widely in secondary xylem of control (C), whereas in transgenic lines having an augmented lignin S/G ratio, such as plant 93, strong CAld5H protein signals were localized exclusively to a few layers of the developing xylem fiber and ray parenchyma cells (D) where syringyl lignin deposits. (E and F) Cross–Bevan cell-wall staining shows the normal centripetal course of cell maturation starting from the fusiform initials (ffi) in control (E) and accelerated cell-wall thickening in CAld5H up-regulated transgenic plants such as plant 93 (F). (Bars: A–D, 50 μm; E and F, 30 μm.)
Structural Analysis of Lignin.
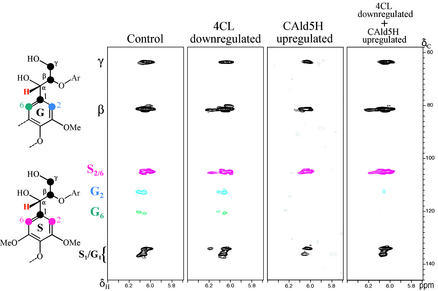
Fig. 5 shows long-range 13C 1H correlations, from gradient HMBC (31), involving the α-protons of the major β-aryl ether units in control and transgenics (plants 23, 93, and 72) lignins. The equivalent 2- and 6-carbons of the symmetrical syringyl units resonate at ≈105 ppm and are easily distinguished from the corresponding guaiacyl carbons (for which the 2- and 6-carbons resonate differentially at ≈113 and ≈121 ppm). It is evident that the lignins from the control and the 4CL-down-regulated transgenic exhibited a similar S/G ratio (Fig. 5). However, the syringyl content was enriched considerably in lignin from the CAld5H-up-regulated transgenic; only trace guaiacyl correlations appear at the base-plane noise level (Fig. 5). Similarly, the lignin from the 4CL-down-regulated + CAld5H-up-regulated transgenic also had a very low guaiacyl component compared with wild type; guaiacyl unit correlations are seen at lower contour levels approaching the base-plane noise level (Fig. 5). The guaiacyl/syringyl compositional shifts observed in β-ether units among the control and various transgenics are in line with the gross compositional shifts evident in the 1D 13C NMR spectra (data not shown) and with the chemical S/G-ratio analyses (Table 1).
Figure 5.
NMR spectra of milled wood lignin from control and transgenic aspen. Gradient-selected 2D HMBC subspectra showing α-proton correlations in β-aryl ether units revealing the S/G compositional differences among the control and the transgenics (4CL-down-regulated plant 23, CAld5H-up-regulated plant 93, and 4CL-down-regulated + CAld5H-up-regulated plant 72). Structures show the five carbons that are two or three bonds from the proton H-α (red), revealed in HMBC correlations. Symmetrical syringyl units, with 2/6-carbon (rose) correlations at ≈105 ppm, are readily distinguished from asymmetric guaiacyl units with 2- and 6-carbons (blue and green, respectively) at ≈113 and ≈121 ppm. Note that the spectra are not calibrated to an absolute concentration; relative levels of syringyl and guaiacyl units can be discerned within individual spectra but not between them.
To test whether up-regulating CAld5H may divert its reaction product 5-hydroxyconiferaldehdye into the lignin sink, we examined lignin HMBC spectra for the benzodioxan (4-O-β/5-O-α) units demonstrated to derive from 5-hydroxyconiferyl alcohol (32). However, such units in all four lignins (control, 23, 93, and 72) were barely detectable. 1D and 2D NMR also did not reveal any sinapaldehyde incorporation in these lignins. Thus, when CAld5H is up-regulated, the CAld5H/5-hydroxyconiferaldehyde O-methyltransferase/sinapyl alcohol dehydrogenase pathway (Fig. 1) is exclusive for the biosynthesis of sinapyl alcohol, the syringyl monolignol.
Carbohydrate Analysis.
Transgenic trees with reduced lignin exhibited an increase in cellulose content, and up to a remarkable 30% increase was observed in antisense-Pt4CL/sense-LsCAld5H transgenic line 141 due to a 52% lignin reduction (Table 1). Consistent with the observation reported by Hu et al. (2), the increased cellulose content together with reduced lignin quantity resulted in a cellulose/lignin ratio of 3 to 5 in the transgenic lines as opposed to 2.2 in the control (Table 1). The relative abundance of the major hemicellulose component xylan was essentially unaffected in all transgenic lines, confirming our previous results (2).
Discussion
We reported here an Agrobacterium system for transforming tobacco and aspen with multiple genes to develop transgenics that would incorporate various combinations of the transgenes. No significant differences were observed for either tobacco or aspen transgenics in the frequency of transgene incorporation; each transgene had an ≈50–60% chance of insertion. These results suggest that this system could be broadly useful for dissecting complex biosynthetic pathways and engineering polygenic agronomic traits in plants.
Using this system we produced tree clones with diverse transgenic wood properties such as low lignin, a high S/G ratio, a high cellulose/lignin ratio, and various combinations of these traits. However, the growth enhancement noted in the previous transgenics having 4CL constitutively down-regulated was not found in this study. It is likely that the use of a xylem-specific promoter obviating the constitutive promoter-induced pleiotropic effects (2) might account for the normal phenotype of the transgenics reported in this study. The compensatory deposition of lignin and cellulose (Table 1) observed in this and previous (2) studies would then suggest intrinsic crosstalk between these two major cell-wall components and is consistent with the well known fact that trees naturally regulate the deposition of lignin and cellulose during wood formation (33–35). Thus, cellulose augmentation observed in transgenic trees could be a tree-specific adaptation to sustain mechanical strength in lignin-deficient xylem cells.
The accelerated maturation (wall thickening and lignification; Fig. 4F) of secondary xylem cells in CAld5H-up-regulated transgenics was a striking observation for any transgenics with altered lignin biosynthesis. Cell maturation involves a series of events in which secondary cell-wall thickening and lignification are the two overlapping terminal processes (36). In angiosperm trees, the deposition of syringyl lignin in secondary xylem elements further signifies the end of lignification (15, 28–30). Therefore, in the context of these cytodifferentiation events, the accelerated cell maturation in transgenics as a result of CAld5H up-regulation-mediated early deposition of syringyl lignin suggests that the formation of syringyl oligomers may be an important part of signaling mechanisms for secondary wall thickening. Although considerable evidence is available to show that complex oligosaccharides have signaling and growth-regulating properties during primary wall formation (37, 38), our transgenic results need further verification as a model for investigating possible roles for oligomeric lignin moieties in coordinating secondary wall biosynthesis in woody xylem cells.
The S/G-ratio increase as well as lignin content-reduction efficiencies were well correlated with the activity levels of the enzymes involved and were independent of the numbers of the integrated transgenes (Fig. 3 and Table 1). However, the possibility may also exist that the less-prominent S/G-ratio increases seen in certain transgenics (Fig. 3 J vs. F) might be caused by transgene-induced abnormal branch pathways diverting metabolites away from the CAld5H/5-hydroxyconiferaldehyde O-methyltransferase/sinapyl alcohol dehydrogenase flux, thereby attenuating the biosynthesis of sinapyl alcohol. However, this possibility was eliminated, because neither benzodioxans, which could originate from 5-hydroxyconiferaldehyde (32), nor sinapaldehyde were found incorporated in transgenic or control lignins as demonstrated by 1D and 2D NMR. In sharp contrast, benzodioxanes were one of the major subunit types (>10% of the total lignin subunits) found in transgenic Arabidopsis, in which ferulate 5-hydroxylase was up-regulated (39), suggesting distinct demands on lignin structural units between Arabidopsis and trees. In fact, normal angiosperm trees having a lignin S/G ratio nearing 10 times that of wild-type Arabidopsis (0.28) (39) suggests a considerably higher demand placed by trees on syringyl lignin, a xylem cell-wall-strengthening component (15, 28, 29), than herbaceous species.
In trees, down-regulation of 5-hydroxyconiferaldehyde O-methyltransferase has been shown to result in S/G-ratio reductions, with, however, concomitantly increased incorporation of coniferaldehyde and 5-hydroxyconiferyl alcohol derived directly from 5-hydroxyconiferaldehyde, producing cyclic benzodioxan 5-hydroxyguaiacyl moieties (32, 39). This suggests a certain metabolic flexibility during monolignol biosynthesis. Down-regulating cinnamyl alcohol dehydrogenase also induced a shunt of its substrate coniferaldehyde into lignin, diminishing the conversion of coniferaldehyde into both syringyl and guaiacyl monolignols to result in lignins with conserved S/G ratios (4, 40). Importantly, regulation in trees of these and CAld5H enzymes, which all are downstream of coniferaldehyde, has not resulted in lignin quantity change. Rather, it affects the overall lignin structure. Thus, these results would suggest a model that monolignol metabolic flexibility is operative downstream of coniferaldehyde, the major branch point in the monolignol biosynthetic pathway (Fig. 1).
It is becoming increasingly evident that interactive functions (13–15) of pathway enzymes and the resulting “substrate-channeling” (41) operations may be critical in regulating metabolic flexibility for such a physiologically important process as monolignol biosynthesis. The multigene cotransformation approach described here represents a promising tool to allow the elucidation of these interactive enzyme functions in vivo to add mechanistic insights to the knowledge of lignin biosynthesis that cannot be acquired effectively by the traditional single-gene approach. Without an efficient multiple-gene manipulation technology, functional genomics and genome-wide expression profiling intended for developing economically important wood-quality traits in trees might still remain as distant reality.
Acknowledgments
We acknowledge Wen-Jing Hu and Cheng-Chung Tsao for cloning aspen 4CL and sweetgum CAld5H genes, respectively, and Fachuang Lu for confirming S/G ratios for selected transgenic trees using the derivatization followed by reductive cleavage method. This study was supported by U.S. Department of Energy Division of Energy Biosciences Grants DE-FG02-01ER15179 (to V.L.C.) and DE-AI02-00ER15067 (to J.R.) and U.S. Department of Agriculture (Agricultural Plant Biochemistry, National Research Initiative Competitive Grants Program) Grant 2001-35318-11268 (to L.L.).
Abbreviations
- S/G
syringyl/guaiacyl
- 4CL
4-coumarate–CoA ligase
- CAld5H
coniferaldehyde 5-hydroxylase
- HMBC
heteronuclear multiple-bond correlation
References
- 1.Whetten R W, MacKay J J, Sederoff R R. Plant Mol Biol. 1998;49:585–609. doi: 10.1146/annurev.arplant.49.1.585. [DOI] [PubMed] [Google Scholar]
- 2.Hu W-J, Lung J, Harding S A, Popko J L, Ralph J, Stokke D D, Tsai C-J, Chiang V L. Nat Biotechnol. 1999;17:808–812. doi: 10.1038/11758. [DOI] [PubMed] [Google Scholar]
- 3.Zhong R, Morrison W H, Himmelsbach D S, Poole F L, Ye Z H. Plant Physiol. 2000;124:563–577. doi: 10.1104/pp.124.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pilate G, Guieny E, Holt K, Petit-Conil M, Lapierre C, Leple J-C, Pollet B, Mila I, Webster E A, Marstorp G G, et al. Nat Biotechnol. 2002;20:607–612. doi: 10.1038/nbt0602-607. [DOI] [PubMed] [Google Scholar]
- 5.Chiang V L. Nat Biotechnol. 2002;20:557–558. doi: 10.1038/nbt0602-557. [DOI] [PubMed] [Google Scholar]
- 6.Tower G H N, Gibbs R D. Nature. 1953;172:25–26. doi: 10.1038/172025a0. [DOI] [PubMed] [Google Scholar]
- 7.Sarkanen K V. In: Lignins: Occurrence, Formation, Structure and Reaction. Sarkanen K V, Ludwig C H, editors. New York: Wiley Interscience; 1971. pp. 19–42. [Google Scholar]
- 8.Chang H M, Sarkanen K V. Techn Assoc Pulp Pap Ind. 1973;56:132–143. [Google Scholar]
- 9.Trotter P C. Techn Assoc Pulp Pap Ind. 1986;69:22–28. [Google Scholar]
- 10.Chiang V L, Funaoka M. Holzforschung. 1990;44:147–156. [Google Scholar]
- 11.Van Doorsselaere J, Baucher M, Chognot E, Chabbert B, Tollier M-T, Petit-Conil M, Leple J-C, Pilate G, Cornu D, Monties B, et al. Plant J. 1996;8:855–864. doi: 10.1104/pp.112.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franke R, McMichael C M, Meyer K, Shirley A M, Cusumano J C, Chapple C. Plant J. 2000;22:223–234. doi: 10.1046/j.1365-313x.2000.00727.x. [DOI] [PubMed] [Google Scholar]
- 13.Osakabe K, Tsao C C, Li L, Popko J L, Umezawa T, Carraway D T, Smeltzer R H, Joshi C P, Chiang V L. Proc Natl Acad Sci USA. 1999;96:8955–8960. doi: 10.1073/pnas.96.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Popko J L, Umezawa T, Chiang V L. J Biol Chem. 2000;275:6537–6545. doi: 10.1074/jbc.275.9.6537. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Cheng X F, Leshkevich J, Umezawa T, Harding S A, Chiang V L. Plant Cell. 2001;13:1567–1585. doi: 10.1105/TPC.010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pincon G, Chabannes M, Lapierre C, Pollet B, Ruel K, Joseleau J-P, Boudet A M, Legrand M. Plant Physiol. 2001;126:145–155. doi: 10.1104/pp.126.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chabannes M, Barakate A, Lapieere C, Marita J M, Ralph J, Pean M, Danoun S, Halpin C, Grima-Pettenati J, Boudet A M. Plant J. 2001;28:257–270. doi: 10.1046/j.1365-313x.2001.01140.x. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Marmey P, Taylor N J, Brizard J-P, Espinoza C, D'Cruz P, Huet H, Zhang S, de Kochko A, Beachy R N, et al. Nat Biotechnol. 1998;16:1060–1064. doi: 10.1038/3511. [DOI] [PubMed] [Google Scholar]
- 19.Halpin C, Barakate A, Askari B M, Abbott J C, Ryan E D. Plant Mol Biol. 2001;47:295–310. [PubMed] [Google Scholar]
- 20.Tricoli D M, Carney K J, Russell P F, McMaster J R, Groffi D W, Hadden K C, Himmel P T, Hubbard J P, Boeshore M L, Quemada H D. Bio/Technology. 1995;13:1458–1473. [Google Scholar]
- 21.Hu W-J, Kawaoka A, Tsai C J, Lung J, Osakabe K, Ebinuma H, Chiang V L. Proc Natl Acad Sci USA. 1998;95:5407–5412. doi: 10.1073/pnas.95.9.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harding S A, Leshkevich J, Chiang V L, Tsai C J. Plant Physiol. 2002;128:428–438. doi: 10.1104/pp.010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marita J, Ralph J, Hatfield R D, Chapple C. Proc Natl Acad Sci USA. 1999;96:12328–12332. doi: 10.1073/pnas.96.22.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ralph J, Marita J M, Ralph S A, Hatfield R D, Lu F, Ede R M, Peng J, Quideau S, Helm R F, Grabber J H, et al. In: Advances in Lignocellulosics Characterization. Argyropoulos D S, Rials T, editors. Atlanta: Tech. Assoc. Pulp Pap. Ind.; 1999. pp. 55–108. [Google Scholar]
- 25.Jorgensen R A, Cluster P D, English J, Que Q, Napoli C A. Plant Mol Biol. 1996;31:957–973. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- 26.Tsai C-J, Popko J L, Mielke M R, Hu W J, Podila G K, Chiang V L. Plant Physiol. 1998;117:101–112. doi: 10.1104/pp.117.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musha Y, Goring D A I. Wood Sci Technol. 1975;9:45–58. [Google Scholar]
- 28.Saka S, Goring D A I. Holzforschung. 1988;42:149–153. [Google Scholar]
- 29.Terashima N, Fukushima K, Takabe K. Holzforschung. 1986;42:101–105. [Google Scholar]
- 30.Wardrop A B. In: Xylem Cell Development. Barnett J R, editor. Tunbridge Wells, U.K.: Castle House; 1981. pp. 115–152. [Google Scholar]
- 31.Ruiz-Cabello J, Vuister G W, Moonen C T W, Van Gelderen P, Cohen J S, Van Zijl P C M. J Magn Reson. 1992;100:282–302. doi: 10.1016/j.jmr.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Ralph J, Lapierre C, Lu F, Marita J M, Pilate G, Van Doorsselaere J, Boerjan W, Jouanin L. J Agric Food Chem. 2001;49:86–91. doi: 10.1021/jf001042+. [DOI] [PubMed] [Google Scholar]
- 33.Timell T E. Compression Wood in Gymnosperms. New York: Springer; 1986. pp. 289–408. [Google Scholar]
- 34.Wardrop A B, Davies G W. Aust J Bot. 1964;12:24–38. [Google Scholar]
- 35.Scurfield G. Science. 1973;179:647–655. doi: 10.1126/science.179.4074.647. [DOI] [PubMed] [Google Scholar]
- 36.Esau K. Plant Anatomy. 2nd Ed. New York: Wiley; 1965. [Google Scholar]
- 37.Cosgrove D J. Plant Cell. 1997;9:1031–1041. doi: 10.1105/tpc.9.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Creelman R A, Mullet J E. Plant Cell. 1997;9:1211–1223. doi: 10.1105/tpc.9.7.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ralph J, Lapierre C, Marita J, Kim H, Lu F, Hatfield R D, Ralph S A, Chapple C, Franke R, Hemm M R, et al. Phytochemistry. 2001;57:993–1003. doi: 10.1016/s0031-9422(01)00109-1. [DOI] [PubMed] [Google Scholar]
- 40.Halpin C, Knight M E, Foxon G A, Campbell M M, Boudet A M, Boon J J, Chabbert B, Tollier M T, Schuch W. Plant J. 1994;6:339–350. [Google Scholar]
- 41.Rasmussen S, Dixon R A. Plant Cell. 1999;11:1537–1551. doi: 10.1105/tpc.11.8.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]