Abstract
Surveys for conserved noncoding sequences (CNS) among genes from monocot cereal species were conducted to assess the general properties of CNS in grass genomes and their correlation with known promoter regulatory elements. Initial comparisons of 11 orthologous maize-rice gene pairs found that previously defined regulatory motifs could be identified within short CNS but could not be distinguished reliably from random sequence matches. Among the different phylogenetic footprinting algorithms tested, the VISTA tool yielded the most informative alignments of noncoding sequence. VISTA was used to survey for CNS among all publicly available genomic sequences from maize, rice, wheat, barley, and sorghum, representing >300 gene comparisons. Comparisons of orthologous maize-rice and maize-sorghum gene pairs identified 20 bp as a minimal length criterion for a significant CNS among grass genes, with few such CNS found to be conserved across rice, maize, sorghum, and barley. The frequency and length of cereal CNS as well as nucleotide substitution rates within CNS were consistent with the known phylogenetic distances among the species compared. The implications of these findings for the evolution of cereal gene promoter sequences and the utility of using the nearly completed rice genome sequence to predict candidate regulatory elements in other cereal genes by phylogenetic footprinting are discussed.
INTRODUCTION
Comparative analyses of noncoding DNA sequences, also known as phylogenetic footprinting (Tagle et al., 1988), is becoming an increasingly important tool to identify candidate gene regulatory elements, based on the premise that regulatory elements have been conserved during evolution as a result of functional constraints. Surveys for conserved noncoding sequences (CNS) among orthologous human and mouse genes have found that CNS are enriched significantly in regulatory sequence elements (Levy et al., 2001). Phylogenetic footprinting, followed by experimental verification, has been shown to be an efficient method for the identification of transcriptional regulatory regions in genes from humans (Loots et al., 2000), Caenorhabditis elegans (Thacker et al., 1999), Drosophila (Bergman and Kreitman, 2001), and yeast (Cliften et al., 2001). CNS also were identified readily in comparisons between genomic sequences from Arabidopsis and related Brassica species (Koch et al., 2001; Quiros et al., 2001; Colinas et al., 2002) and among different grass genomes (Kaplinsky et al., 2002; Morishige et al., 2002). However, the functional significance of plant CNS is not yet known, and surveys for plant CNS have been limited to <15 genes in each case.
The power of phylogenetic footprinting is enhanced significantly when genomic sequences are available from a number of related species that have diverged sufficiently so that conserved functional elements can be distinguished from nonconserved sequences (Dubchak et al., 2000; Cliften et al., 2001; Koch et al., 2001; Boffelli et al., 2003). Cereal genomes are uniquely poised among plant genomes for phylogenetic footprinting. Cereal genomes exhibit a high degree of synteny (Bennetzen, 2000; Devos and Gale, 2000) and sequence identity within coding sequences, facilitating the identification of orthologous gene sequences. However, noncoding sequences show little overall conservation, suggesting that there has been sufficient time for the accumulation of nonselected sequence changes. The major cereal crop species maize, rice, wheat, barley, and sorghum diverged from a common ancestor ∼50 million years ago (Kellogg, 2001), which is less than the evolutionary distance between the human and mouse genomes (80 million years) (Li and Graur, 1991). The phylogenetic relationships among these cereal species are known (Kellogg, 2001), with at least one representative from each of the grass subfamilies Pooideae (wheat and barley), Erhartoideae (rice), and Panicoideae (maize and sorghum). Within the Pooideae, wheat and barley diverged ∼10 to 14 million years ago (Wolfe et al., 1989), whereas within the Panicoideae, maize and sorghum diverged ∼16.5 million years ago (Gaut and Doebley, 1997), which allows for comparisons that can track the directionality of CNS evolution. Importantly, two different draft rice genome sequences are available (Goff et al., 2002; Yu et al., 2002), and a large number of maize genomic sequences have been characterized previously without the benefit of a dedicated sequencing project. Characterizations of the sorghum, barley, and wheat genomes also are progressing (Ramakrishna et al., 2002a). In addition, ongoing structural and functional genomics efforts are rapidly increasing our knowledge about expressed genes and their functions within cereal genomes, particularly maize and rice (Chandler and Wessler, 2001; Shimamoto and Kyozuka, 2002).
Despite the relatively low overall sequence identity among promoter sequences from cereal genes compared with their coding regions, numerous expression analyses and transgenic experiments involving promoter-reporter gene constructs have demonstrated that orthologous genes from different cereal species often share highly regulated patterns of gene expression. Thus, these conserved patterns of gene expression might be programmed by regulatory sequences associated with CNS.
We tested this hypothesis by surveying for CNS among orthologous genes from maize, rice, barely, wheat, and sorghum. The genomic sequences from 652 genes of these species were gleaned from public databases, annotated, and compared with orthologous gene sets. Using this data set, we performed analyses to determine the general properties of CNS among cereal genes, particularly their frequency, length, and distribution within genes and across cereal taxa. We found that CNS can be identified in the vast majority of comparisons involving orthologous cereal genes, but their frequency and length are less compared with CNS in other species. For the 31 known regulatory motifs examined, 28 were identified within CNS by one or more of the phylogenetic footprinting algorithms tested, but only under relaxed definitions for CNS length that did not reliably distinguish regulatory elements above background noise. Thus, although phylogenetic footprinting comparisons can predict functional regulatory elements within cereal promoter sequences, their relatively short length limits the utility of this approach to identify new unknown motifs, particularly when sequences from only two species are compared. Together, these analyses suggest that the promoter sequences among cereal genomes are evolving rapidly and show little sequence conservation even among orthologous genes. We also discuss the prospects for phylogenetic footprinting approaches with the rice genome sequence to help predict promoter elements that define regulatory networks of gene expression in maize and other grass species.
RESULTS
Compilation of Genomic Sequences from Orthologous Cereal Genes
A prerequisite for evaluating phylogenetic footprinting between maize and other grasses on a genome scale is the availability of large numbers of genomic sequences from grass species. Rice genome sequencing projects have generated large amounts of sequence available for comparison, but promoter sequences from other grass genomes are limiting. Among other cereals, searches of GenBank identified 652 annotated genomic sequences from maize, barley, wheat, and sorghum that contained at least 300 bp of 5′ upstream noncoding sequences, representing 2.165 Mbp. For gene families with many highly related members, such as those that encode seed storage proteins, only one representative sequence was included in the data set. The list of annotated maize genomic sequences can be found in the supplemental data at http://www.cropsci.uiuc.edu/faculty/moose/PromoterComparisons.htm and is summarized in Table 1.
Table 1.
Summary Data of Annotated Genomic Sequences Used for Identification and Analyses of CNS in This Study
| Total Genomic Sequence |
5′ Upstream Sequence (kb) |
Promoter Sequence
|
|||
|---|---|---|---|---|---|
| Species | No. | kb | No. | kb | |
| Maize | 288 | 1123 | 420 | 120 | 199 |
| Barley | 123 | 450 | 141 | 49 | 56 |
| Wheat | 89 | 312 | 132 | 31 | 36 |
| Sorghum | 59 | 280 | 95 | 1 | 1.5 |
All GenBank sequences from the four species listed that contained at least 300 bp of 5′ upstream sequences (as defined by the translation start codon) were included in the data set. Promoter sequence was defined as sequences upstream of experimentally determined transcription start sites, as indicated in the annotation for the GenBank entry.
The largest group of genomic sequences was from maize and represented 1.12 Mbp. Approximately 40% of the maize, barley, and wheat sequences were annotated previously for promoter regions, which totaled 291 kb from 288 genes. The vast majority of sorghum genomic sequences were obtained from recently sequenced BAC clones and thus were not annotated for transcription start sites. Within BAC clones, the 5′ boundary of upstream sequences was defined as either 3000 bp 5′ to the start codon or the 3′ end of the closest upstream gene or repetitive DNA sequence (e.g., retrotransposons). The entire set of 652 genes contained on average 3958 bp of genomic sequence and 1441 bp of 5′ upstream sequence. For those genes with annotated promoters, the average promoter length was 1450 bp.
The maize genomic sequences gleaned from GenBank entries were used in sequence similarity searches to identify putative orthologous genes from publicly available rice genome sequences (see Methods). From these searches, 78 likely pairs of orthologous maize and rice genes were identified based on high nucleotide identity (>80%) throughout their coding regions and additional criteria, such as expression and proposed gene function based on mutant analyses or biochemical assays. A complete listing of these likely orthologous gene sets is available as supplemental data at http://www.cropsci.uiuc.edu/faculty/moose/PromoterComparisons.htm.
The set of orthologous gene pairs is representative of most classes of gene functions, including housekeeping proteins, structural genes, enzymes that participate in specific metabolic or physiological processes such as starch synthesis, and genes that encode photosynthetic proteins. The average nucleotide identity within coding regions was 90.1% between maize and rice orthologous gene pairs. The total sequence space in the orthologous gene pair data set was 103 kb for maize and 150 kb for rice, with 73.1 and 98.8 kb representing 5′ upstream sequences from maize and rice, respectively.
Evaluation of Phylogenetic Footprinting for Promoter Element Identification in Maize and Rice
Regulatory cis-acting sequence elements within promoter DNA often are short and orientation independent and contain frequent gaps of variable size. Thus, typical local (e.g., BLAST [Basic Local Alignment Search Tool]) or global (e.g., CLUSTAL W) sequence alignment tools, which were designed for protein-coding sequences, perform poorly with promoter DNA sequences (Jareborg et al., 1999). Phylogenetic footprinting has been used to identify CNS that contain previously defined regulatory sequence elements within human (Loots et al., 2000), yeast (Cliften et al., 2001), and Arabidopsis (Koch et al., 2001) promoters. Although the parameters for the BL2SEQ pair-wise alignment algorithm can be adjusted to identify CNS among orthologous gene sequences from grass genomes (Kaplinsky et al., 2002), a number of other tools have been developed recently specifically for the purpose of identifying CNS. These tools include PipMaker (Schwartz et al., 2000), DNA Block Aligner (DBA: www.ebi.ac.uk/Wise2/dbaform.html), Bayes Block Aligner (BBA; Zhu et al., 1998), DIALIGN (Morgenstern, 1999), and the VISualization Tool for Alignments (VISTA; Mayor et al., 2000).
Each of these algorithms is designed to rapidly align long genomic sequences, but via different approaches (Frazer et al., 2003). PipMaker represents a local alignment strategy that produces optimal similarity scores between subregions of the sequences and is advantageous when long sequences containing multiple genes are being compared. Each of the other programs uses global alignments that optimize similarity over the entire length of the compared sequences, which is more appropriate for comparing individual gene sequences. The utility of these phylogenetic footprinting tools to identify CNS containing candidate regulatory sequences was evaluated initially using a set of 10 maize-rice gene pairs that contain 31 experimentally defined regulatory elements within their promoters. These elements are listed in Table 2 and are supported in most cases by both functional assays of promoter activity using promoter-reporter constructs and sequence-specific biochemical interactions with DNA binding proteins (either in vivo or in vitro). The elements tested function in the regulation of gene expression in response to the cell cycle (histone H3 octamer; Brignon and Chaubet, 1993), sugars (Sbe1; Kim and Guiltinan, 1999), the growth regulator abscisic acid (ABRE; Busk et al., 1999), iron (Fer1; Petit et al., 2001), light (rbcS; Martínez-Hernández et al., 2002), and drought or salt stress (DRE1; Kizis and Pages, 2002). For each of the genes examined, the functions and expression patterns of the maize and rice genes are similar, suggesting that the defined promoter regulatory elements also may be conserved.
Table 2.
Tests of Different Phylogenetic Footprinting Tools for Identifying Known Regulatory Motifs in Maize and Rice Orthologous Gene Pairs
| Gene Name | Element | Element Length | VISTA | DIALIGN | BL2SEQ | BBA |
|---|---|---|---|---|---|---|
| Histone H3 | Octamer | 8 | 8 | 8 | 8 | 8 |
| Nonamer | 10 | 5 | 3 | 0 | 0 | |
| Starch-branching enzyme1 (sbe1) | −314 … −295 | 19 | 13 | 0 | 7 | 5 |
| −284 … −255 | 29 | 25 | 0 | 22 | 8 | |
| G-box | 10 | 0 | 0 | 7 | 10 | |
| Ferritin1 | IDRS | 18 | 18 | 18 | 18 | 18 |
| G-box | 7 | 7 | 7 | 7 | 7 | |
| Block II | 10 | 10 | 10 | 10 | 10 | |
| Ribulose bisphosphate carboxylase (rbcS) | GC-rich region | 13 | 10 | 0 | 11 | 9 |
| Monocot rbcS | 19 | 12 | 12 | 0 | 5 | |
| TATA-box | 18 | 6 | 9 | 0 | 5 | |
| rab17 | DRE1 | 7 | 0 | 7 | 0 | 0 |
| DRE2 | 6 | 0 | 3 | 6 | 0 | |
| ABRE1 | 6 | 0 | 6 | 5 | 0 | |
| ABRE2 | 6 | 4 | 6 | 6 | 0 | |
| ABRE4 | 6 | 5 | 6 | 6 | 0 | |
| GRA | 12 | 0 | 0 | 7 | 0 | |
| Sph | 8 | 8 | 8 | 0 | 0 | |
| rab28 | ABRE A | 10 | 7 | 0 | 0 | 7 |
| ABRE B | 10 | 6 | 9 | 0 | 9 | |
| CE3 | 12 | 0 | 0 | 0 | 7 | |
| GRA | 9 | 7 | 0 | 0 | 9 | |
| Catalase1 | ABRE1 | 8 | 8 | 0 | 0 | 0 |
| ABRE2 | 8 | 8 | 0 | 5 | 0 | |
| ARE | 11 | 8 | 0 | 1 | 0 | |
| CE1-like | 5 | 4 | 0 | 4 | 0 | |
| Alcohol dehydrogenase2 (Adh2) | −160 | 25 | 0 | 4 | 24 | 4 |
| Anthocyaninless1 (a1) | ccggtagtt | 9 | 0 | 0 | 0 | 0 |
| acctaccaacc | 11 | 0 | 0 | 0 | 0 | |
| Anthocyaninless2 (a2) | −99 | 8 | 0 | 8 | 0 | 0 |
| −91 | 9 | 0 | 1 | 8 | 8 | |
| Elements identified successfully | 19 | 15 | 16 | 11 | ||
| Background nucleotide identity | 12.34% | 36.41% | 51.80% | 14.74% |
The number of nucleotides aligned successfully between the maize and rice promoter sequences are indicated for comparisons using the VISTA, DIALIGN, BL2SEQ, and BBA tools. An element was considered to be identified successfully if at least 50% of the nucleotides within the elements were aligned. The background nucleotide identity refers to the total fraction of the nucleotides compared that were contained within CNS using criteria of 70% identity in a 10-bp window. References for regulatory elements in maize genes are as follows: histone H3 (Brignon and Chaubet, 1993); sbe1 (Kim and Guiltinan, 1999); ferritin1 (Petit et al., 2001); rbcS (Martínez-Hernández et al., 2002); rab17 (Kizis and Pages, 2002); rab28 (Busk et al., 1999); catalase1 (Guan et al., 2000); adh2 (Paul and Ferl, 1994); a1 (Sainz et al., 1997); and a2 (Lesnick and Chandler, 1998).
The orthologous maize and rice gene pairs were compared for CNS using each of the BL2SEQ, PipMaker, DBA, DIALIGN, BBA, and VISTA algorithms. Similar to the findings reported by Kaplinsky et al. (2002) for a set of seven orthologous maize-rice gene pairs using BL2SEQ, we did not identify any CNS that fit the definition of CNS for mammalian gene pairs (i.e., >100 bp in length and >70% identity). Because our goal was to detect short (<10-bp) sequence elements corresponding to cis-acting regulatory sequences, we searched for CNS using reduced length criteria while keeping identity within a window at >70% to allow for degeneracy within regulatory elements. In evaluating different length criteria for the identification of promoter elements within CNS, we used as a benchmark the presence of the octamer motif within the CNS found for the maize and rice histone H3 genes. The histone H3 genes are highly conserved in sequence, protein function, and expression pattern among all eukaryotes, and the octamer motif has been demonstrated to be a functional regulatory motif in many plant histone promoters, including Arabidopsis (Brignon and Chaubet, 1993), that span a greater evolutionary distance than that from maize to rice. CNS contained the histone octamer motif when the length of CNS was reduced to 10 bp but not at 25 bp (Figure 1). Similar results were observed for many of the other candidate regulatory elements (Table 1), in which the elements often were found in CNS defined as >70% identity within a 10-bp, but not a 25-bp, window.
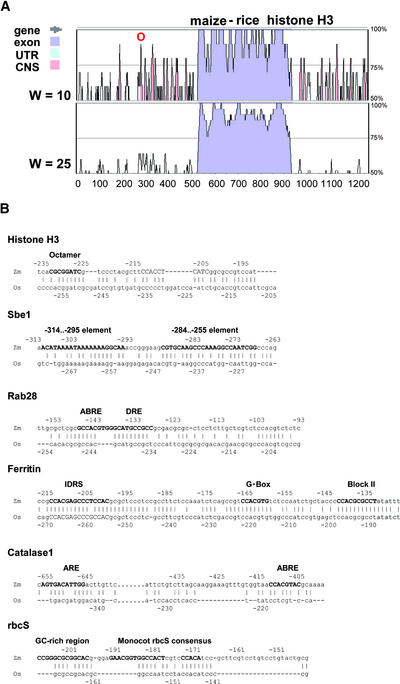
Figure 1.
Examples of Regulatory Elements Contained within Maize and Rice CNS Identified Using VISTA.
(A) Graphic output for comparisons of maize and rice histone H3 genes using VISTA and parameters of at least 70% identity within a window (W) of either 10 or 25 bp. The CNS denoted “O” represents the highly conserved octamer motif. UTR, untranslated region.
(B) Nucleotide alignments of CNS identified in comparisons of the maize and rice histone H3, Sbe1, Rab28, ferritin, catalase1, and rbcS genes. The previously defined sequences of each element (based on biochemical or functional assays) are shown in boldface. Nucleotide positions are given relative to the experimentally defined transcription start site.
The six phylogenetic footprinting algorithms tested performed differently in their ability to predict known promoter regulatory motifs. PipMaker and DBA detected <10 of the 31 elements under any of the evaluated length parameters (data not shown). Under the relaxed criteria to define CNS, approximately half of the known promoter motifs were found within CNS identified by BL2SEQ, DIALIGN, or BBA, but these tools also produced a relatively high background in which >36% of all nucleotides within the promoter sequences were defined as CNS. The CNS identified by VISTA contained 19 of the 30 promoter elements, but only 12% of the total promoter sequences were contained within CNS. Thus, VISTA appeared to offer the best combination of predictive success for known regulatory elements (63.3%) within the smallest fraction of promoter sequence defined as CNS.
Figure 1B shows the nucleotide alignments of the CNS identified by VISTA for six maize-rice orthologous gene pairs in the regions surrounding 11 of the known regulatory sequence elements. What is striking in these alignments is that the CNS often localize over the regulatory sequence elements, with the flanking DNA sequences diverging sufficiently to fail to be defined as CNS. This is clearly evident for CNS observed in the maize Sbe1 promoter, in which each of the three positive regulatory sequence elements defined by Kim and Guiltinan (1999) also are identified as CNS, but their intervening sequences are not. Similarly, the octamer and nonamer elements in the histone H3 promoter, the ABRE and anaerobic response element (ARE) binding sites in the catalase1 promoter, and the ABRE site in the rab28 promoter are contained within short CNS, but their flanking sequences are not. The three distinct iron-responsive elements are highly conserved between maize and rice Fer promoters, but their intervening sequences also still fit the definition of CNS in this study, albeit with less nucleotide identity than the elements themselves. These findings demonstrate that when CNS are minimally defined as 10 bp of >70% identity, VISTA often identifies CNS between maize and rice gene pairs that contain known functional regulatory elements. DIALIGN, BL2SEQ, and BBA also were capable of localizing known regulatory elements within CNS, but often with less precision as a result of the higher overall frequency of nucleotides aligned with these methods.
Surveys for CNS in Orthologous Maize and Rice Genes
Rice and maize currently possess the greatest amount of functional information for gene regulation and available genomic sequence among cereal crop species. Thus, maize and rice comparisons are likely to be the primary basis to assess the utility of CNS in predicting regulatory elements of gene expression for cereal genes on a genome scale. We surveyed 78 maize and rice orthologous gene pairs for CNS using VISTA to gain some understanding of the properties of CNS on a wider spectrum of genes than has been studied previously (Kaplinsky et al., 2002; Morishige et al., 2002). Because our previous comparisons of maize and rice genes (Table 2) indicated that CNS between maize and rice genes can identify functional regulatory elements with criteria of >70% identity for a minimum of 10 bp, these criteria were used to compare maize and rice genes (Table 3).
Table 3.
Summary of Properties for CNS Identified in Comparisons of 75 Maize-Rice Orthologous Gene Pairs
| Property | Mean ± SD | Range |
|---|---|---|
| Number of blocks per kilobase pair of noncoding sequence |
11.98 ± 6.44 | 1.1 to 31.4 |
| CNS block length | 11.86 ± 6.02 | 7 to 88 |
| Percentage of total noncoding sequence within CNS |
15.2 ± 8.3 | 1.1 to 33.8 |
| Percentage of promoter sequence contained within CNS |
8.7 ± 5.6 | 1.1 to 24.8 |
| Percentage of 5′ untranslated leader sequence contained within CNS |
28.6 ± 30.74 | 0 to 100 |
| Percentage of intron sequence contained within CNS |
23.1 ± 13.53 | 5.2 to 45 |
| Percentage of 3′ untranslated region sequence contained within CNS |
14.5 ± 9.25 | 0 to 35.4 |
All comparisons used VISTA with parameters of >70% identity in a 10-bp window.
The frequency of CNS blocks among the 78 gene pairs ranged from 1 to 31 per 1000 bp of noncoding sequence compared, with the mean being 11.98 blocks per 1000 bp. The total fraction of noncoding sequence contained within CNS ranged from 1 to 34%, with a mean of 15.2%. The mean block length for CNS identified between maize and rice promoter sequences was 11.9 bp, with 88 bp being the largest CNS in this data set. The vast majority (67%) of maize-rice CNS are between 8 and 11 bp, which is consistent with the observation that most of these CNS are not identified with larger window sizes.
Among the 78 gene pairs compared, the five genes with the highest proportions of CNS (>30%) relative to the entire data set (ferritin1, alcohol dehydrogenase2, hsp70, proliferating cell nuclear antigen, and sbe1) tended to represent genes that perform housekeeping or metabolic functions and whose expression patterns might be expected to be more highly conserved between maize and rice. Conversely, five gene pairs showed <3% CNS (nitrate reductase1, rf2 and rf2B aldehyde dehydrogenases, sugary1, and Mha1 proton-transporting ATPase). These genes could represent gene pairs whose noncoding sequences have diverged more rapidly than those of most other genes. Alternatively, they may represent paralogous rather than orthologous gene pairs whose noncoding sequences have diverged.
CNS were not distributed equally among different genic regions (Table 3). CNS at least 10 bp in length were most prevalent in the 5′ untranslated leader sequences of mRNAs upstream of the start codon, followed by transcribed sequences downstream of stop codons and then intron sequences. CNS were least frequent (8.7%) in promoter sequences upstream of defined or predicted transcription start sites. Within each of these noncoding sequences, CNS tend to be found more often closest to coding sequences. This distribution may reflect the strong selection pressure on exonic coding sequences, which indirectly preserves adjacent untranslated sequences, or could indicate functional conservation of sequences surrounding transcription start sites, exon-intron splice junctions, or polyadenylation signals. The frequency of CNS within promoter sequences decreases as distance 5′ to the transcription start site or start codon increases. Very few CNS are identified >1000 bp upstream of transcription start sites. This distribution is consistent with the compact nature of plant gene promoters, in which <1000 bp of promoter sequence often is sufficient to drive proper regulated patterns of transcription and regulatory elements tend to be clustered near the transcription start site.
Maize-Rice CNS Are Not Significantly Enriched for Known Regulatory Elements
The most striking feature of CNS identified between maize-rice orthologs is their short length. Although the mean length of maize-rice CNS (∼12 bp; Table 3) is longer than that of most previously identified promoter regulatory elements, the utility of these CNS in predicting true regulatory elements over background noise remained unclear. This issue is particularly important in assessing the value of phylogenetic footprinting to identify candidate regulatory elements among genes that have not been characterized previously by functional assays for promoter activity.
One test for the regulatory significance of short CNS identified in maize-rice gene comparisons is to determine whether known functional promoter regulatory elements appear at a higher frequency in CNS compared with the entire maize and rice promoter sequence data set, which has been observed in comparisons of orthologous human and mouse genes (Levy et al., 2001). We performed such a test by first calculating the frequency of occurrence for all combinations of heptad sequences in two sets of sequences: the 277.2 kb of annotated promoter sequences from the 78 maize-rice orthologous gene pairs, and the entire collection of CNS (8.5 kb) identified in the VISTA comparisons of these same promoter sequences (Figure 2; see also supplemental data at http://www.cropsci.uiuc.edu/faculty/moose/PromoterComparisons.htm). We tested the occurrence of heptad sequences because this represents the lower boundary for the identification of CNS using criteria of 70% identity in a 10-bp window.
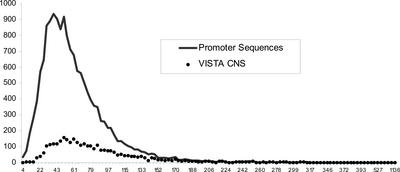
Figure 2.
Heptad Sequence Frequency Distributions in Promoter and CNS Data Sets.
Frequencies of occurrence per megabase pair are given for all possible combinations of heptad sequences in either the sequences upstream of the transcription start site for 78 pairs of orthologous maize and rice genes (promoter sequences; solid line) or the CNS identified by VISTA comparisons of these same maize-rice gene pairs (VISTA CNS; dotted line).
The heptad sequences within maize and rice gene promoters showed a relatively normal distribution, with an observed mean identical to that expected (1/47 × 106 bp = 61 per Mbp). However, this distribution was biased by a small group of ∼300 elements that appeared with very high frequencies, which shifted the median peak occurrence frequency in the cereal promoter sequence data set to ∼51 per Mbp. Only 3283 of the possible 16,384 heptad sequences appeared in the smaller VISTA CNS data set; thus, the occurrence frequencies for individual heptad repeats were inflated relative to the total cereal promoter sequence data set, preventing a direct comparison of these two distributions. However, it was possible to compare whether the distribution of the 3283 heptad sequences common to the two data sets differed from that of the total cereal promoter sequence data set. The log likelihood goodness-of-fit test (G = 865.11, P < 0.001) showed that the distribution of heptad sequences identified by VISTA as CNS was not the same as that of the total data set, with the most common heptad sequences from the total promoter data set occurring at even higher frequencies within CNS (Figure 2). The 5′-AAAAAAA-3′ heptad and its complement were the two most frequent heptads in both data sets, with similar AT-rich and CT-rich heptads also being very common. The high occurrence frequencies of AT-rich sequences in maize and rice promoters and their enrichment in maize-rice CNS are consistent with the recent observation that human-mouse CNS also are enriched significantly in AT-rich sequences associated with nuclear matrix attachment regions (Glazko et al., 2003).
The occurrence frequencies for each of the 19 elements identified by VISTA in Table 2 were compared between the total cereal promoter and VISTA CNS data sets. For elements >7 bp, each 7-bp window within the total element was compared, and the results were averaged to derive a mean frequency for the whole element. Only 5 of the 19 elements (Sbe1 −284 to −255, rab28 ABRE, rab28 DRE, Fer1 IDRS, and Fer1 G-box) showed a greater than twofold increase in their occurrences per megabase pair for the CNS data set compared with the total promoter sequence data set. Thus, although the distribution of heptad sequences differed between CNS and the total promoter data set, these differences do not appear to be attributable to an enrichment of the 19 known regulatory elements considered here.
Twenty Base Pairs Is the Minimum Length for a Significant CNS among Orthologous Cereal Genes
Another question raised by the short CNS identified in comparisons of maize-rice orthologs is whether these blocks are truly under functional constraint or do they instead represent sequences that are identified by chance using global alignment algorithms such as VISTA. As observed for Drosophila (Bergman and Kreitman, 2001), the frequency distribution of maize-rice CNS fits a log-normal distribution that is biased toward shorter blocks (Figure 3A), which is consistent with a pure mutation-drift model (Clark, 2001). If CNS defined in this study are under functional constraints, then the properties of the CNS identified for orthologous genes would be expected to deviate from those of sequence blocks identified by the same algorithm using a random set of sequences. One strategy to calibrate the significance of output by algorithms such as VISTA is to perform runs on multiple permutations of random sequences (McCue et al., 2002). However, the critical question in this study concerned the criteria by which CNS in promoter sequences from orthologous cereal genes can be shown to be significantly different from those of nonorthologous cereal genes. This stricter definition of baseline sequence conservation is important considering the short nature of both promoter regulatory elements and CNS identified between maize and rice genes.
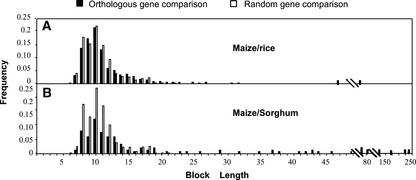
Figure 3.
Frequency Distributions of CNS Identified in Promoter Sequences of Maize-Rice and Maize-Sorghum Orthologous Gene Pairs Relative to Random Promoter Sequence Comparisons.
The distributions of orthologous promoter CNS (black bars) are based on comparisons of 78 maize-rice and 10 maize-sorghum gene pairs. Random distributions (white bars) are based on comparisons in which all promoter sequences first were fused to orthologous coding sequences and then each “hybrid” test sequence was compared with all other hybrid sequences (see Methods). All comparisons were performed using VISTA with parameters of >70% identity in a window of 10 bp.
To assess whether or not the distribution of CNS block lengths identified in comparisons of orthologous maize-rice gene pairs was significantly different from that of sequence blocks observed in random comparisons, a sequence data set was constructed in which the promoter sequences from the 50 maize genes were fused to the maize sbe1 coding region and the 50 rice gene promoters were fused to the rice sbe1 coding region. These gene constructs then were compared in all possible pair-wise combinations using VISTA, and all noncoding sequence blocks that fit the same criteria as CNS (>70% identity in a 10-bp window) were included in the random comparison data set. The distribution of the sequence block lengths from the random maize-rice gene comparisons (Figure 3A, white bars) was essentially the same as that observed for the orthologous gene comparisons, except that the random comparisons did not contain any sequence blocks of >20 bp. Thus, for maize-rice gene comparisons using VISTA, only those CNS of >20 bp showed frequencies that were higher than would be expected at random.
The frequency distribution of CNS lengths observed between maize and rice genes was only slightly different from what might be expected at random, suggesting that the evolutionary distance between maize and rice is on average too great to identify CNS. Thus, sorghum was investigated as an alternative to compare with maize sequences because it diverged from maize ∼15 to 20 million years ago, which is more recent than the 50 million years that has been estimated for the separation of maize and rice. Among the 56 sorghum genomic sequences, 10 were orthologous with maize genes also present in the maize-rice orthologous gene set. These 10 maize-sorghum orthologous gene pairs were compared for CNS (>70% identity in a 10-bp window) using VISTA as well as all pair-wise comparisons in which the maize and sorghum promoter sequences were fused to the same orthologous coding region (sbe1). The frequency distribution of the CNS identified in the orthologous and random maize-sorghum gene comparisons is shown in Figure 3B. It is evident that the frequency distribution of CNS obtained from orthologous maize-sorghum comparisons is different from that of conserved blocks observed in the random maize-sorghum comparisons. Maize-sorghum CNS tend to be longer, with 30% being >20 bp. The distribution of blocks from random maize-sorghum comparisons is similar to that observed for the random maize-rice comparisons, with no blocks of >21 bp. Thus, as observed for maize-rice comparisons, maize-sorghum CNS of >20 bp occur at a higher frequency than might be expected at random. The greater frequency of these longer CNS in maize-sorghum orthologous gene pairs compared with maize-rice pairs is consistent with the known phylogenetic relationships between maize, sorghum, and rice.
CNS in Genes from Three or More Cereal Species
The results from the collective comparisons of maize-rice and maize-sorghum gene pairs suggested that the frequency of CNS of >20 bp follows expected phylogenetic relationships; hence, they are likely to be relevant to the evolution of cereal genes. Further evidence for this hypothesis was obtained by evaluating the proportion of CNS among a set of 130 genes from maize, rice, sorghum, barley, and wheat in which orthologous gene sequences were available for at least three different cereal species (Table 4). Based on the frequency distributions for CNS shown in Figure 3, reasonable criteria for significant CNS across cereal genes appeared to be >70% identity in a 20-bp window. The highest proportion of CNS was observed for the maize-sorghum (32.2%) and barley-wheat (34.9%) comparisons, consistent with the fact that maize and sorghum are both members of the Panicoideae subfamily and wheat and barley are both members of the Pooideae subfamily, whereas rice is a member of the distinct Ehrhartoideae subfamily. Interestingly, despite the relatively high nucleotide identity (>80%) within the coding regions among the 18 orthologous gene sets compared across these five cereals, the proportion of CNS was not >10% except for in the maize-sorghum and wheat-barley gene pairs. Maize-rice (5.7%) and sorghum-rice (9.4%) gene pairs showed higher proportions of CNS relative to either wheat or barley compared with the other cereals (all <4%). Although the number of genes compared here is relatively small, the proportions of CNS generally are consistent with the known phylogenetic relationships among these five cereal crop species.
Table 4.
Summary of Multiple-Species Comparisons to Identify CNS among Orthologous Cereal Genes
| Maize
|
Rice
|
Sorghum
|
Barley
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | No. of Genes |
Coding Identity (%) |
Percent CNS |
No. of Genes |
Coding Identity (%) |
Percent CNS |
No. of Genes |
Coding Identity (%) |
Percent CNS |
No. of Genes |
Coding Identity (%) |
Percent CNS |
| Rice | 15 | 82.7 | 5.7 | |||||||||
| Sorghum | 7 | 89.4 | 32.2 | 7 | 81.0 | 9.4 | ||||||
| Barley | 8 | 85.7 | 2.9 | 8 | 88.6 | 4.1 | 2 | 92.4 | 3.2 | |||
| Wheat | 6 | 85.8 | 0.4 | 6 | 85.5 | 0.5 | 1 | 81.1 | 0.0 | 7 | 92.3 | 34.9 |
Sixty-seven genes representing 18 orthologous gene sets that contained sequences from at least three cereal species (see Methods) were used in multiple-species sequence comparisons with VISTA (>70% identity, 20-bp window size).
The inclusion of promoter sequences from additional species at intermediate levels of evolutionary divergence often improves the identification of functionally important sites by phylogenetic footprinting (Boffelli et al., 2003). Observing a candidate CNS among three or more species provides greater evidence that the CNS is likely to be significant. VISTA was used to compare the orthologous Adh1 genes from maize, rice, barley, and sorghum (Figure 4A); it is the only gene for which a four-way species comparison was possible. CNS (defined as >70% identity in a window of 20 bp) were identified for each pair-wise species comparison, with the frequency and length of CNS being much greater for the maize-sorghum comparison than for any of the other comparisons. Many of the CNS found in pair-wise comparisons also were coincident with CNS from at least one other species. The Adh1 CNS are found in 5′ upstream sequences, introns, and 3′ untranslated regions, with the promoter CNS occurring at a higher frequency near the transcription start site. Within introns, CNS tend to be found more frequently near exon-intron boundaries than in internal sequences, but Adh1 intron 6 represents an exception to this generalization.
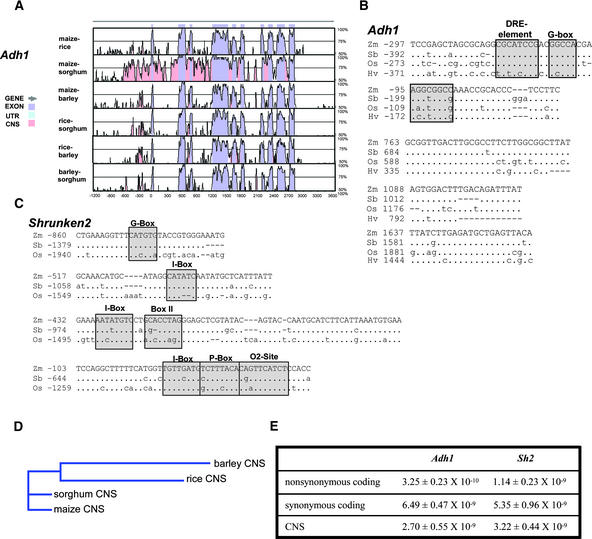
Figure 4.
CNS Evolution in Cereal Adh1 and Sh2 Genes.
(A) Graphic VISTA output obtained after comparison of the maize, rice, sorghum, and barley Adh1 genes (criteria of >70% identity with a window size of 20 bp). UTR, untranslated region.
(B) Sequence alignments and positions within the Adh1 gene for the five CNS conserved among Adh1 genes from all four species (Hv, barley; Os, rice; Sb, sorghum; Zm, maize). Dots indicate nucleotide identity, nucleotide substitutions are given in lowercase letters, and dashes show gaps introduced by the alignment algorithm. Boxed and shaded regions indicate sequences similar to known promoter regulatory elements as predicted by the Search for CARE tool at http://oberon.rug.ac.be:8080/PlantCARE/index.html (Lescot et al., 2002), with the shaded sequence from −201 to −194 of the maize Adh1 promoter corresponding to the sequences that show differential biochemical footprints when Adh1 expression is induced by hypoxia (Paul and Ferl, 1991).
(C) Sequence alignments and positions for the four CNS conserved among the orthologous Sh2 genes from maize, rice, and sorghum. Details and symbols are as in (B).
(D) Predicted phylogenetic tree using the combined Adh1 and Sh2 CNS alignments from (B) and (C).
(E) Estimates of nucleotide substitution rates (substitutions per site per year) for coding sequences and CNS from the Adh1 and Sh2 genes compared in (A) to (C).
Five Adh1 CNS were conserved across all four of the cereal species (Figure 4B). Two of these CNS reside in the promoters of these genes, the most distal of which contains sequences similar to the DRE element and G-box motifs known to interact with DNA binding proteins (Menkens et al., 1995; Kizis and Pages, 2002). Both the DRE and G-box elements have been implicated in mediating transcriptional activation in response to environmental stresses, which is consistent with Adh1 gene function (Freeling and Bennett, 1985). The distal CNS also is contained within a region of the maize Adh1 gene that has been shown to have nuclear matrix binding activity (Avramova and Bennetzen, 1993). The CNS beginning at position −201 relative to the maize Adh1 transcription start site also is relevant in this context, because it localizes over a sequence that shows enhanced sensitivity to dimethyl sulfate by in vivo footprinting, but only when Adh1 transcription is induced by hypoxia (Ferl and Nick, 1987; Paul and Ferl, 1991). Surprisingly, no CNS identified by these criteria were found to colocalize with the extensively studied ARE between −140 and −99 of the maize Adh1 gene (Walker et al., 1987; Olive et al., 1991), which also has been demonstrated to be important for proper anaerobically induced expression from the maize Adh1 promoter in transgenic rice (Kyozuka et al., 1994). However, as observed previously for other known regulatory motifs (Table 2), the important sequence elements within the Adh1 ARE were found as CNS when the window size was reduced to 10 bp (data not shown). The other three CNS contained within introns are of unknown function. The conservation of these sequences across each of the four cereal species suggests that they may contribute to common properties of Adh1 gene function in these grasses.
Inspection of the outputs from the 17 other orthologous gene sets for which three-way species comparisons were possible (Table 4) identified few CNS in the promoter regions of genes from all three species. However, Figure 4C shows that four CNS of 68, 50, 41, and 32 bp were found for the maize, rice, and sorghum genes encoding the large subunit of ADP-glucose pyrophosphorylase (maize Shrunken2 [Sh2]). Each of these promoter CNS contains sequences with similarity to at least one known promoter regulatory element. Maize Sh2 and its rice ortholog are expressed in seed endosperm tissue (Bhave et al., 1990; Anderson et al., 1991), and maize Sh2 functions as the rate-limiting step for endosperm starch biosynthesis. Thus, the Sh2 promoter CNS may be important for the abundant endosperm expression of these genes. Of particular interest in this regard is the presence of adjacent P-box and O2-site elements in the Sh2 promoter CNS that lies closest to the transcription start site for these genes. This tandem arrangement of binding sites for the Dof (P-box) and basic domain/Leu zipper (O2-site) classes of plant transcription factors, also known as the endosperm motif, is found in the promoters of many cereal storage protein genes that are highly expressed in endosperm tissue (Forde et al., 1985; Vicente-Carbajosa et al., 1997).
It is evident from visual inspection of the alignments that the maize and sorghum sequences show the highest degree of conservation, with rice and barley being somewhat divergent from each other as well as from the maize and sorghum sequences. These relationships are consistent with the known phylogeny of these grasses. Figure 4D provides estimates of nucleotide substitution rates based on the Adh1 and Sh2 CNS alignments. The estimates for nonsynonymous and synonymous substitution rates within the Adh1 coding sequence are very close to the mean estimates of Gaut et al. (1996) for grass Adh1genes (3.21 × 10−10 nonsynonymous, 7.00 × 10−9 synonymous). The nucleotide substitution rate estimates for the Sh2 coding region are similar to those for Adh1, with a slightly faster rate of nonsynonymous substitutions. The estimated substitution rates for both the Adh1 and Sh2 CNS were similar and in the range of nonsynonymous substitution rates for the Adh1 coding region. Thus, the CNS identified in at least the Adh1 and Sh2 genes, and probably many other cereal genes as well, show evolutionary patterns that reflect known phylogenetic relationships and accumulate substitutions at rates comparable to nonsynonymous substitutions within coding sequences.
DISCUSSION
We have surveyed for CNS from a large number of genes from the major cereal crop species to determine the general properties and distribution of CNS among grass genomes. Our findings have practical implications for comparative genomics within this important plant family, suggest strategies and limitations for the identification of promoter regulatory elements in cereal genes by phylogenetic footprinting, and provide insights into the evolution of regulatory sequences in the grasses.
An Annotated Database for Cereal Gene Promoter Sequences
Databases of annotated promoter sequences, such as the Eukaryotic Promoter Database (EPD; Praz et al., 2002), have been used widely as a resource to characterize eukaryotic transcription control elements and to develop eukaryotic promoter prediction tools. The utility of EPD for maize and rice is limited because it contains only 21 maize and 7 rice promoter sequences, with 11 of the maize promoters derived from the closely related α-zein class of seed storage proteins. The nearly 600 annotated promoter sequences from maize, rice, sorghum, wheat, and barley collected in this study significantly expands the number of cereal promoter sequences that are available in a central repository, which may be used as a “training set” for the evaluation of existing and newly developed algorithms for promoter sequence analysis. The number of promoter sequences also is sufficiently large to characterize the general properties of promoter sequences in grass species. For example, our analysis of the frequency distribution of heptad repeat sequences (Figure 2) reveals a very strong bias against guanine nucleotides in the frequently occurring heptads, resulting in a very low GC content (3.1% among the top-50 heptads). Conversely, the 37 rarest heptads, which occurred only once in the cereal promoter data set, are composed of 62.9% GC (data not shown). These dramatic differences in GC content lead to the complete exclusion of CG or CXG plant DNA methylation targets (Gruenbaum et al., 1981) on either strand of the 50 most frequent heptads, in contrast to the rare heptads in which 33 of 37 sequences contained a CG or CXG methylation target. This type of information, which will only be improved with the continued sequencing of grass genomes, will be valuable in the computational prediction of promoter regulatory elements.
Strong candidate orthologous rice genes were identified for only 78 of the 288 maize genes tested (27.1%), which is much less than the estimated coverage of phase-2 rice genome sequences in GenBank (currently ∼80%) (Buell, 2002). The ability to define best candidates for orthologous gene pairs was improved only slightly when nearly complete draft genome rice sequences (Yu et al., 2002) were searched. This finding largely reflects difficulties in distinguishing among different members of gene families or between duplicated genes. The best evidence for orthology is obtained by sequencing and comparative analysis of orthologous BAC clones from multiple grass species; however, few such comparisons have been performed to date (Chen et al., 1998; Tikhonov et al., 1999; Tarchini et al., 2000; Morishige et al., 2002; Ramakrishna et al., 2002a, 2002b; Song et al., 2002). As demonstrated in our surveys for conserved sequence blocks among both maize-rice and maize-sorghum gene pairs, blocks of >70% identity over a window of 20 bp are likely to be found only among orthologous genes. Thus, just as greater sequence similarity within protein-coding sequences provides greater evidence for orthology, CNS also may prove useful as another criterion to help distinguish orthologous genes from paralogs among duplicated genes.
Among the available sequence comparisons cited above, two examples exist in which CNS provides supporting evidence for orthology. Chen et al. (1998) found that sorghum contains two tandemly duplicated homologs (A1-a and A1-b) of the maize a1 gene. Nucleotide similarities within exons were very similar (90 to 91%) when the maize a1 homolog was compared with either of the sorghum A1 genes. However, we found using VISTA that maize a1 showed more CNS with sorghum A1-b (7.6% of total noncoding sequence) compared with sorghum A1-a (6.1%), particularly in the 5′ untranslated mRNA leader and first intron (data not shown). This observation suggests that A1-b is more likely to be orthologous with the maize a1 gene than with sorghum A1-a. Similarly, VISTA comparisons of the maize Rp1-D gene with the different duplicated sorghum rph1 genes characterized by Ramakrishna et al. (2002b) found that maize Rp1-D showed CNS only with the sorghum rph1-1 gene promoter region (data not shown), despite the fact that the rph1-1 coding region is truncated. This finding suggests that maize Rp1-D and sorghum rph1-1 may be orthologous genes that subsequently underwent independent duplication and diversification in the maize and sorghum lineages.
Utility of Phylogenetic Footprinting to Predict Regulatory Motifs in Cereal Gene Promoters
The principle of phylogenetic footprinting has been applied previously to the analysis of many plant gene promoters, with one of the earliest examples being the identification of the highly conserved prolamin box sequence in cereal seed storage protein genes (Forde et al., 1985). In this study, we directly tested the ability of recently developed tools for phylogenetic footprinting to identify known regulatory sequence elements in a set of 11 orthologous maize-rice gene pairs. Collectively, the four best tools (VISTA, DIALIGN, BL2SEQ, and BBA) identified 28 of the 31 known elements, with 18 being detected by two or more algorithms. Thus, the global alignment tools evaluated in this study were successful in identifying short CNS that colocalized with known regulatory elements, although comparisons with multiple alignment tools are likely to enhance the efficacy of promoter regulatory element prediction.
VISTA was clearly superior to the other alignment methods used, at least with this set of genes. The three other algorithms all share the property of discarding short blocks, which constitute the vast majority of the CNS contained in this data set. VISTA also exhibits biases, because it tends to omit small blocks that flank longer, strongly conserved blocks or to omit small blocks that lie between two larger blocks. The net result of these biases is to subdivide larger CNS blocks into multiple smaller blocks, which is one contributing factor to the high frequency of small CNS blocks identified by VISTA. It is important to note that each of the regulatory elements identified by the different tools evaluated here (Figure 1) were parts of regulatory “modules” containing more than one adjacent transcription factor binding site. Levy et al. (2001) also found that CNS contain transcription factor binding sites that frequently are clustered. These observations suggest that the phylogenetic footprinting approach may be most effective at identifying regulatory modules rather than isolated sequence blocks containing single transcription factor binding sites, because the presence of adjacent regulatory elements creates larger blocks of functionally constrained sequence that are easier to detect as a significant CNS.
Our primary goal in surveying for CNS among a large number of cereal genes was to develop criteria with which to assess their biological significance, particularly for comparisons with the nearly completed rice genome. We found for maize-rice comparisons using VISTA that the majority of CNS are between 8 and 15 bp (Figure 3), which offers potential advantages in precisely defining candidate regulatory sequences. However, our analyses of the occurrence frequencies for all possible combinations of heptad sequences in the CNS compared with total promoter sequence data sets did not provide any evidence that CNS are enriched significantly in known regulatory elements. Thus, the short CNS identified among cereal gene promoters are unlikely to be useful in identifying functionally significant promoter regulatory sequences.
CNS among grass genomes are much smaller than those observed in mammals (Kaplinsky et al., 2002; this study). The distribution frequency for CNS block lengths for maize-rice and maize-sorghum comparisons (Figure 3) was similar to that observed for comparisons of 38 orthologous Drosophila melanogaster and Drosophila virilis gene pairs (Bergman and Kreitman, 2001). In contrast to Drosophila, in which promoter and intron sequences showed similar proportions of CNS (22 to 26%), we found that the frequency of CNS varied with different functional regions of cereal genes (Table 3), being highest in 5′ untranslated leader sequences and introns (means of 28.6 and 23.1%, respectively) but lowest in promoters (mean of 8.7%). This result suggests that promoters of genes from cereal species are evolving at a much faster rate than other noncoding sequences from the same gene.
The small size of the CNS identified in the maize-rice comparisons raises the possibility that they represent background noise generated by the alignment tool. Indeed, we found that the frequency distributions of CNS for orthologous maize-rice gene comparisons did not deviate significantly from those obtained in the random comparisons (Figure 3), whereas those from the orthologous maize-sorghum comparisons did differ from random. In both sets, only blocks of >20 bp were found at frequencies greater than those observed in the random gene comparison data set; hence, 20 bp would seem to be a lower boundary by which to define CNS among grass genomes. In their comparisons of two promoter sequences from 22 cruciferous species spanning 45 million years of evolution, Koch et al. (2001) found nine CNS blocks that averaged 25 bp in length. Thus, 20 bp may be a reasonable minimum length criterion for CNS in both dicot and monocot plant species. However, it is evident from our searches for CNS among genes with experimentally determined regulatory motifs (Table 2) that blocks of <20 bp do contain important regulatory sequences.
A second criterion that we used to assess the significance of CNS was to determine if they could be identified in comparisons of orthologous genes from three or more cereal species. This analysis could be performed for only 18 gene sets, but the results demonstrate that this strategy greatly reduced the number of CNS identified. Of course, such a criterion is biased for regulatory elements that contribute to highly conserved patterns of gene expression among the grasses. Interestingly, maize, sorghum, barley, and wheat each showed <10% CNS compared with rice, suggesting that the problem of background noise found in the maize-rice comparisons also applies when using the rice genome for comparisons with other economically important cereals. However, the Adh1 and Sh2 examples (Figure 4) as well as the results of Kaplinsky et al. (2002) demonstrate that such comparisons can yield CNS across a number of distantly related grass species.
Together, our results suggest that stringent criteria for defining CNS in the grasses would be a length of at least 20 bp and the presence of the CNS in at least one additional grass species with an intermediate level of divergence, such as rice-sorghum-maize. Although additional comparisons need to be made to better assess the utility of phylogenetic footprinting for a larger set of cereal genes, the low frequencies of CNS observed here between the rice, barley-wheat, and sorghum-maize clades (Table 4) suggest that shorter evolutionary distances may be required for effective promoter comparisons. Thus, as has been demonstrated recently for primate genomes (Boffelli et al., 2003), the sequencing of orthologous regions from other grass species with intermediate levels of divergence between the other major cereals would greatly enhance the power of comparative genomics for the analysis of transcriptional gene regulation. Cliften et al. (2001) found that Saccharomyces species that showed ∼80% identity in coding regions and 40% identity in noncoding sequences were the most effective in predicting regulatory sequences. Sorghum exhibits these levels of divergence with maize and thus serves as a good bridge species between maize and other cereals. Perhaps the diploid forage grass species Festuca pratensis and Lolium perenne (perennial ryegrass) would serve similar purposes for barley and wheat. Additional species, such as the diploid Eleusine indica (goosegrass) from the Chloroidoideae subfamily, which also has a relatively small genome size (0.73 pg of DNA per 1C nucleus), would aid in filling taxonomic gaps among other grass species. Although a full genome sequence for these grasses is not a realistic short-term goal, the targeted sequencing of desired orthologous regions (Morishige et al., 2002) is feasible and will be an important tool for future investigations of cereal promoter evolution.
CNS and the Evolution of Cereal Gene Promoters
The frequency and size of CNS among cereal genes follow known phylogenetic relationships (Table 4, Figure 4), indicating that they may be used to gain insights into features of grass genome evolution. Estimates of nucleotide substitution rates (Figure 4) demonstrate that CNS in cereal genes, particularly those of >20 bp, are subject to evolutionary forces that indicate functional constraints. The rate of evolution within CNS, at least for the Adh1 and Sh2 genes, appears to be similar to the nonsynonymous substitution rate within coding regions, suggesting that CNS are subject to the same degree of selective pressure as amino acid sequences.
Song et al. (2002) have proposed that the combined effects of recent transposon movement and amplification, polyploidization, segmental chromosomal duplications, and gene amplification contribute to a much greater rate of evolution in the grasses than would be predicted by comparing coding sequence substitution rates. The properties of CNS from the current survey (Figure 3) support this view. Cereal CNS are smaller and less frequent than CNS observed in comparisons of mammalian (Frazer et al., 2003) or Drosophila (Bergman and Kreitman, 2001) genes, despite the fact that the degree of nucleotide similarity among cereal gene coding regions (80 to 85%) and divergence times (∼50 million years ago) are approximately equal to those for mammalian (human-mouse; 80 million years ago) and D. melanogaster–D. virilis (40 million years ago) gene comparisons. The rapid evolution of noncoding sequences does not appear to be unique to the grasses. Colinas et al. (2002) used VISTA to compare 13 orthologous gene pairs from cauliflower and Arabidopsis, which diverged 15 to 20 million years ago (Quiros et al., 2001). Twenty-four Arabidopsis-cauliflower CNS were identified that averaged 49 bp and covered 28% of the total 5′ noncoding sequence. These features are similar to those observed for maize and sorghum comparisons (Figure 3, Table 4), which diverged 16.5 million years ago.
Our surveys for CNS among orthologous cereal genes generally found low proportions of CNS within promoter regions. A simple interpretation of this observation is that expression patterns have diverged significantly among apparently orthologous genes in the grasses. Certainly, this will be the case for some genes, especially those that have been subjected to one or more rounds of gene duplication. However, studies that have analyzed endogenous expression patterns and characterized promoter-reporter constructs in transgenic cereals have found that regulated patterns of gene expression often are highly conserved across different cereal species. Therefore, the few CNS that are identified among cereal gene promoters may be the primary sequence determinants for conserved patterns of gene expression. For example, a short region of the maize Adh1 promoter confers proper gene expression in transgenic rice (Kyozuka et al., 1994), and this region contained two of the prominent CNS within the maize and rice Adh1 promoters (Figure 4). Alternatively, the low proportions of CNS suggest that for many orthologous cereal genes, stabilizing selection for expression pattern may not be reflected in the conservation of promoter nucleotide sequences. A particularly striking example of this possibility is the waxy1 gene, which encodes granule-bound starch synthase. The maize waxy1 gene is expressed specifically in endosperm and pollen (Shure et al., 1983), as are its rice, sorghum, and wheat orthologs (Okagaki and Wessler, 1988). However, only two 10-bp blocks were identified as CNS in comparisons of the waxy1 gene, and neither of these was conserved across more than two species (see supplemental data online).
Certainly, the regulation of gene expression in the grasses and other plant species is complex. The use of comparative genomics approaches that reveal patterns of noncoding sequence evolution, particularly in promoter regions, offers important insights into this problem. Promoter CNS often are likely to correspond to transcription factor binding sites, but they also could mediate sites of interaction with chromatin complexes that influence gene expression. The strategies described here for identifying CNS among grass genes can be applied readily to the analysis of other plant genes, with CNS helping guide laboratory experiments to determine their functional roles in the regulation of gene expression. We expect that as the amount of plant genomic sequence information expands to include more species and taxa, appropriate comparisons of noncoding sequences that consider the significance tests presented here will become an increasingly powerful approach to elucidating mechanisms of plant gene regulation.
METHODS
Compilation of Genomic Sequences from Orthologous Cereal Genes
The Entrez nucleotide database from GenBank (http://www.ncbi.nlm.nih.gov) was searched for all maize (Zea mays) sequence entries by setting the following limits parameters: genomic DNA/RNA sequences, exclude ESTs, exclude sequence tagged sites, exclude genomic survey sequences, but include patents. Approximately 5000 sequence entries were returned that were inspected manually for the presence and annotation of genomic DNA sequences with features such as transcription start sites, the TATA-box promoter element, coding sequence, intron-exon boundaries, and poly(A) signals. Among sequences contained in patent applications, genomic annotations frequently were absent but often could be ascertained by comparisons of genomic and cDNA sequences for the same gene when available. Because many sequences contained annotation only of the start codon, only those sequences that contained at least 200 bp of 5′ upstream sequence were included in the final database. This process was repeated in a search for all sorghum (Sorghum bicolor), wheat (Triticum aestivum), and barley (Hordeum vulgare) genomic sequence entries that contained at least 200 bp of 5′ upstream sequence. When multiple sequences from the same gene were available, only the longest sequence was included, and preference also was given to sequences from wild-type functional alleles rather than mutant alleles. All entries that represented unique gene sequences were downloaded as annotated sequence files into a local database within the Vector NTI Suite 7.0 (Informax, Bethesda, MD) software. The final database, containing 652 annotated entries, then was imported into a Microsoft Access (Redmond, WA) relational database, including html links to National Center for Biotechnology Information GenBank entries, which can be viewed as supplemental data at http://www.cropsci.uiuc.edu/faculty/moose/PromoterComparisons.htm.
The sequence entries in this database were used to extract putative orthologous gene sequences as follows. First, GenBank nonredundant and high-throughput genomic sequence databases were searched with coding region sequences using TBLASTX. The top maize, rice (Oryza sativa), barley, sorghum, or wheat hits for each sequence from these searches then were compared with the genomic DNA sequences using BLASTN. All searches were performed on a local server (Keck Center for Comparative and Functional Genomics, University of Illinois at Urbana-Champaign). Only those genomic sequences that showed probability scores of >1 × 10−5 and >80% nucleotide identity within target coding regions were considered to be candidate orthologous genes. Members of known multigene families that showed similarity only in highly conserved protein domains were excluded to minimize the chance of comparing paralogs rather than orthologs. Only those gene pairs that contained at least 200 bp of 5′ upstream sequence were included in the final data set. The majority of rice and sorghum genomic sequences were identified within BAC or P1 artificial chromosome clones; thus, the genomic sequences containing conserved coding sequences as well as 3000 bp of 5′ flanking sequence and 1000 bp of 3′ flanking sequence were selected manually from the larger sequence and saved as annotated files in a Vector NTI Explorer local database. A listing of the orthologous genes identified in this manner is included as a table in the relational database housed at http://www.cropsci.uiuc.edu/faculty/moose/PromoterComparisons.htm.
Phylogenetic Footprinting Comparisons
The six alignment tools used in this study were VISTA (Mayor et al., 2000; www-gsd.lbl.gov/vista/VistaInput.html), DIALIGN (Morgenstern, 1999; bibiserv.techfak.uni-bielefeld.de/cgi-bin/dialign_submit), BL2SEQ (Tatusova and Madden, 1999; www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html), Bayesian Block Aligner (Zhu et al., 1998; http://bayesweb.wadsworth.org/cgi-bin/bayes_align12.pl), PipMaker (Schwartz et al., 2000; http://bio.cse.psu.edu/pipmaker/), and DBA (www.ebi.ac.uk/Wise2/dbaform.html). Each of these tools was evaluated initially on a set of 11 maize-rice gene pairs containing known regulatory sequence elements using a number of different parameters that varied the minimum length and percent nucleotide identity required for definition as conserved noncoding sequences (CNS). These initial experiments indicated that requiring 70% identity within a 10-bp window returned the greatest number of regulatory elements with acceptable specificity. Based on the performance of these six tools in identifying CNS during these evaluations (Figure 2), VISTA was selected as the tool for further genome-scale analyses. All subsequent comparisons with VISTA used the minimum percent identity threshold of 70% and window size of 10, 20, or 25 bp. Annotation files were included in all comparisons that defined promoter, 5′ untranslated region, exon, intron, and 3′ untranslated region portions of the genes. Outputs from these and all other comparisons described here are available upon request.
The surveys for CNS among multiple-species comparisons involved orthologous genes from maize, rice, sorghum, barley, and wheat. The gene sequences compared and their accession numbers are listed at the end of Methods.
Estimation of Heptad Frequency Distributions in Sequence Data Sets
A file was created that contained as entries each of the possible 16,384 combinations of heptad sequences (e.g., AAAAAAA through to TTTTTTT). A PERL program (www.perl.com) was written to use the heptad sequences as an input file to screen for the number of occurrences of each heptad sequence in a sequence data set. The PERL program was run against the 277.2 kb of promoter sequences (all sequences upstream of defined transcription start sites) contained in the 78 pairs of orthologous maize and rice genes (see supplemental data online), and the sequences were defined as CNS from VISTA outputs comparing these same 78 maize-rice gene pairs. The number of heptad sequence occurrences was normalized to their frequency per megabase pair by dividing the number of occurrences by the total number of kilobases in the data set and then multiplying by 1000. These normalized values then were used to plot a distribution of the number of elements exhibiting a given normalized frequency per megabase pair using the X-Y Scatter Graph function in Microsoft Excel.
Analyses of CNS
PERL scripts were written to parse VISTA output. The information on block length and identity from the VISTA “regions” output files was used to calculate the number of CNS blocks per kilobase pair of total noncoding sequence and the proportion of noncoding sequences defined as CNS (% CNS) for the total sequence. The inclusion of gene annotation files allowed parsing the CNS into promoter, intron, 5′ leader, and 3′ untranslated region sequences. The values obtained for each of these parameters in the 78 individual maize-rice gene comparisons then were used to generate the data shown in Table 3. The values obtained for the multiple-species comparisons were used to generate the data shown in Table 4. Using the information about the relative positions of CNS contained in the VISTA regions output files, PERL scripts extracted the CNS from the gene sequences that were downloaded from GenBank in FASTA format. The conserved sequence blocks then were used to analyze the frequency of CNS. Each of the CNS sequences from the 78 maize-rice comparisons was placed into a database annotated by the source gene and the position of the CNS within the gene sequence.
The frequency distribution of block lengths from comparisons of orthologous maize-rice or maize-sorghum gene pairs (Figure 2) was determined using the Frequency function in Microsoft Excel. To generate a data set that assessed the frequency with which sequences that met the criteria for CNS (70% identity in a 10-bp window) occurred in random comparisons of noncoding sequences, each of the 50 maize upstream noncoding sequences was fused to the maize sbe1 coding region and each of the 50 rice upstream noncoding sequences was fused to the rice sbe1 coding region. Thus, each of these gene constructs contained the same pair of orthologous coding regions, which facilitated proper alignment of the entire sequence by VISTA. Each of the 50 maize constructs then was compared with the 49 nonorthologous rice constructs using VISTA and criteria of 70% identity in a 10-bp window. The VISTA output then was used to generate the frequency distribution for random gene comparisons, as described above for orthologous gene comparisons.
Estimation of Nucleotide Substitution Rates
The entire coding sequences from the maize, sorghum, rice, and barley Adh1 genes were aligned using the CLUSTAL W algorithm within the Vector NTI Suite 7.0 software package. A total of 1140 contiguously aligned nucleotides from each of the four sequences then were used as inputs into the codonml program within the PAML version 3.13 program package (Yang, 1997). Synonymous and nonsynonymous nucleotide distance measures were estimated using an empirical codon model and estimated κ and no-clock parameters. All other parameters were set as defaults. Synonymous and nonsynonymous nucleotide distance measures were estimated similarly for 1548 aligned nucleotides from the maize, sorghum, and rice Sh2 coding sequences.
The alignments of CNS from Adh1 and Sh2 (Figure 3) were pooled into single alignments of 137 bp (Adh1) and 192 bp (Sh2) and used as inputs into the baseml program within PAML. Nucleotide distance measures were estimated under the model of Tamura and Nei (1993) with no-clock and estimated κ parameters. All other parameters were set as defaults.
Nucleotide substitution rates were estimated using these computed distance measures and the formula described by Gaut et al. (1996). For simplicity, divergence times between maize and rice, sorghum and rice, maize and barley, sorghum and barley, and barley and rice were estimated at 50 million years ago, a reasonable estimate for the radiation of the cereals into the Panicoideae, Pooideae, and Ehrhartoideae subfamilies (Kellogg, 2001).
Upon request, all novel materials described in this article will be made available in timely manner for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the sequences described in this article are as follows: maize Adh1 (X04049), sorghum Adh1 (AF124045), rice Adh1 (AF172282), barley Adh1 (AF253472), maize Sh2 (M81603), sorghum Sh2 (AF010283), and rice Sh2 (AF101045). Accession numbers for the gene sequences compared are as follows: ADP-glucose pyrophosphorylase (Sh2), M81603 (maize), AF101045 (rice), and AF010283 (sorghum); alcohol dehydrogenase1, X04049 (maize), AF172282 (rice), X04049 (sorghum), and AF253472 (barley); alcohol dehydrogenase2, X02915 (maize), AF172282 (rice), and X12733 (barley); chalcone synthase, X60204 (maize), AP003380 (rice), and X58339 (barley); chlorophyll a/b binding protein, X14794 (maize), AF162665 (rice), X12735 (barley), and M10144 (wheat); gene X transcription factor, AF434193 (maize), AF101045 (rice), and AF010283 (sorghum); histone H3, M13379 (maize), AB026295 (rice), and X00937 (wheat); hydroxy-rich glycoprotein, AJ131535 (rice), X61280.1 (sorghum), and X56010 (barley); NADPH-dependent reductase (maize A1), X05068 (maize); U70541 (rice), AF010283 (sorghum), S69616 (barley), and AF434703 (wheat); phosphoenolpyruvate carboxylase (PEPC), E17154 (maize), AP003052 (rice), and X63756 (sorghum); starch-branching enzyme1, AF072724 (maize), D10838 (rice), and AJ237897 (barley); starch synthase1, AF036891 (maize), D38221 (rice), AF234163 (barley), and AF091802 (wheat); high-affinity sulfate transporter, AF075720 (barley) and AJ238244 (wheat); teosinte branched1, AF415063 (maize), AY043215 (rice), and AF466204 (barley); type-A α-amylase, X16509 (rice), M15208 (barley), and X13576 (wheat); granule-bound starch synthase (waxy1), X03935 (maize), AF141954 (rice), X07931 (barley), and AY050175 (wheat); α-amylase (Amy32b), X16509 (rice), X05166 (barley), and X13577 (wheat); and β-amylase, AF068119 (maize), L10345 (rice), and AJ301645 (barley).
Supplementary Material
Acknowledgments
We thank the Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign for access to its batch BLAST server as well as for server space to perform phylogenetic footprinting comparisons. We thank Nick Lauter and an anonymous reviewer for helpful suggestions regarding the statistical evaluation of CNS. We also acknowledge access to draft rice genome sequences from Monsanto (rice-research.org) and Syngenta's Torrey Mesa Research Institute, which allowed confirmation of orthologous maize-rice gene sequences. Funding for this research was provided by the College of Agriculture and Consumer Economics at the University of Illinois and by grants to S.P.M. from the University of Illinois Research Board and the U.S. Department of Agriculture (Award 2002-35301-12157).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010181.
Footnotes
Online version contains Web-only data.
References
- Anderson, J.M., Larsen, R., Laudencia, D., Kim, W.T., Morrow, D., Okita, T.W., and Preiss, J. (1991). Molecular characterization of the gene encoding a rice endosperm-specific ADP-glucose pyrophosphorylase subunit and its developmental pattern of transcription. Gene 97, 199–205. [DOI] [PubMed] [Google Scholar]
- Avramova, Z., and Bennetzen, J.L. (1993). Isolation of matrices from maize leaf nuclei: Identification of a matrix-binding site adjacent to the Adh1 gene. Plant Mol. Biol. 22, 1135–1143. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L. (2000). Comparative sequence analysis of plant nuclear genomes: Microcolinearity and its many exceptions. Plant Cell 12, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman, C., and Kreitman, M. (2001). Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res. 11, 1335–1345. [DOI] [PubMed] [Google Scholar]
- Bhave, M.R., Lawrence, S., Barton, C., and Hannah, L.C. (1990). Identification and molecular characterization of shrunken-2 cDNA clones of maize. Plant Cell 2, 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. (2003). Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299, 1391–1394. [DOI] [PubMed] [Google Scholar]
- Brignon, P., and Chaubet, N. (1993). Constitutive and cell-division-inducible protein-DNA interactions in two maize histone gene promoters. Plant J. 4, 445–457. [DOI] [PubMed] [Google Scholar]
- Buell, C.R. (2002). Obtaining the sequence of the rice genome and lessons learned along the way. Trends Plant Sci. 7, 521–565. [DOI] [PubMed] [Google Scholar]
- Busk, P.K., Pujal, J., Jessop, A., Lumbreras, V., and Pages, M. (1999). Constitutive protein-DNA interactions on the abscisic acid-responsive element before and after developmental activation of the rab28 gene. Plant Mol. Biol. 41, 529–536. [DOI] [PubMed] [Google Scholar]
- Chandler, V.L., and Wessler, S. (2001). Grasses: A collective model genetic system. Plant Physiol. 125, 1155–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., SanMiguel, P., and Bennetzen, J.L. (1998). Sequence organization and conservation in sh2/a1-homologous regions of sorghum and rice. Genetics 148, 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A.G. (2001). The search for meaning in noncoding DNA. Genome Res. 11, 1319–1320. [DOI] [PubMed] [Google Scholar]
- Cliften, P.F., Hillier, L.W., Fulton, L., Graves, T., Miner, T., Gish, W.R., Waterston, R.H., and Johnston, M. (2001). Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 11, 1175–1186. [DOI] [PubMed] [Google Scholar]
- Colinas, J., Birnbaum, K., and Benfey, P.N. (2002). Using cauliflower to find conserved non-coding regions in Arabidopsis. Plant Physiol. 129, 451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K.M., and Gale, M.D. (2000). Genome relationships: The grass model in current research. Plant Cell 12, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak, I., Brudno, M., Loots, G.G., Mayor, C., Pachter, L., Rubin, E.M., and Frazer, K.A. (2000). Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 10, 1304–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl, R.J., and Nick, H.S. (1987). In vivo detection of regulatory factor binding sites in the 5′ flanking region of maize Adh1. J. Biol. Chem. 262, 7947–7950. [PubMed] [Google Scholar]
- Forde, B.G., Heyworth, A., Pywell, J., and Kreis, M. (1985). Nucleotide sequence of a B1 hordein gene and the identification of possible upstream regulatory elements in endosperm storage protein genes from barley, wheat, and maize. Nucleic Acids Res. 13, 7327–7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer, K.A., Elnitski, L., Church, D.M., Dubchak, I., and Hardison, R.C. (2003). Cross-species sequence comparisons: A review of methods and available resources. Genome Res. 13, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling, M., and Bennett, D.C. (1985). Maize Adh1. Annu. Rev. Genet. 19, 297–323. [DOI] [PubMed] [Google Scholar]
- Gaut, B.S., and Doebley, J.F. (1997). DNA sequence evidence for the segmental allotretraploid origin of maize. Proc. Natl. Acad. Sci. USA 94, 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., Morton, B.R., McCaig, B.C., and Clegg, M.T. (1996). Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear Adh1 gene parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93, 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko, G.V., Konin, E.V., Rogozin, I.B., and Shabalina, S.A. (2003). A significant fraction of conserved noncoding DNA in human and mouse consists of predicted matrix attachment regions. Trends Genet. 19, 119–124. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Gruenbaum, Y., Naveh-Many, T., Cedar, H., and Razin, A. (1981). Sequence specificity of ethylation in higher plant DNA. Nature 292, 860–862. [DOI] [PubMed] [Google Scholar]
- Guan, L.M., Zhao, J., and Scandalios, G. (2000). Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signaling molecule for the response. Plant J. 22, 87–95. [DOI] [PubMed] [Google Scholar]
- Jareborg, N., Birney, E., and Durbin, R. (1999). Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 9, 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplinsky, N.J., Braun, D.M., Penterman, J., Goff, S.A., and Freeling, M. (2002). Utility and distribution of conserved noncoding sequences in the grasses. Proc. Natl. Acad. Sci. USA 99, 6147–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, E.A. (2001). Evolutionary history of the grasses. Plant Physiol. 125, 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.N., and Guiltinan, M.J. (1999). Identification of cis-acting elements important for expression of the starch-branching enzyme I gene in maize endosperm. Plant Physiol. 121, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizis, D., and Pages, M. (2002). Maize DRE binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J. 30, 679–689. [DOI] [PubMed] [Google Scholar]
- Koch, M.A., Weishaar, B., Kroyman, J., Haubold, B., and Mitchell-Olds, T. (2001). Comparative genomics and regulatory evolution: Conservation and function of the Chs and Apetala3 promoters. Mol. Biol. Evol. 18, 1882–1891. [DOI] [PubMed] [Google Scholar]
- Kyozuka, J., Olive, M., Peacock, W.J., Dennis, E.S., and Shimamoto, K. (1994). Promoter elements required for developmental expression of the maize Adh1 gene in transgenic rice. Plant Cell 6, 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., Rouzé, P., and Rombauts, S. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnick, M.L., and Chandler, V.L. (1998). Activation of the maize anthocyanin gene a2 is mediated by an element conserved in many anthocyanin promoters. Plant Physiol. 117, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, S., Hannennalli, S., and Workman, C. (2001). Enrichment of regulatory signals in conserved non-coding genomic sequence. Bioinformatics 17, 871–877. [DOI] [PubMed] [Google Scholar]
- Li, W.-H., and Graur, D. (1991). Fundamentals of Molecular Evolution. (Sunderland, MA: Sinauer Associates).
- Loots, G.G., Locksley, R.M., Blankenspoor, C.M., Wang, Z.E., Miller, W., Rubin, E.M., and Frazer, K.A. (2000). Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288, 136–140. [DOI] [PubMed] [Google Scholar]
- Martínez-Hernández, A., López-Ochoa, L., Argüello-Astorga, G., and Herrera-Estrella, L. (2002). Functional properties and regulatory complexity of a minimal RBCS light-responsive unit activated by phytochrome, cryptochrome, and plastid signals. Plant Physiol. 128, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor, C., Brudno, M., Schwartz, J.R., Poliakov, A., Rubin, E.M., Frazer, K.A., Pachter, L.S., and Dubchak, I. (2000). VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047. [DOI] [PubMed] [Google Scholar]
- McCue, L.A., Thompson, W., Carmack, C.S., and Lawrence, C.E. (2002). Factors influencing the identification of transcription factor binding sites by cross-species comparison. Genome Res. 12, 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens, A.E., Schindler, U., and Cashmore, A.R. (1995). The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20, 506–510. [DOI] [PubMed] [Google Scholar]
- Morgenstern, B. (1999). DIALIGN 2: Improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15, 211–218. [DOI] [PubMed] [Google Scholar]
- Morishige, D.T., Childs, K.L., Moore, L.D., and Mullet, J.E. (2002). Targeted analysis of orthologous phytochrome A regions of the sorghum, maize, and rice genomes using comparative gene-island sequencing. Plant Physiol. 130, 1614–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki, R.J., and Wessler, S.R. (1988). Comparison of mutant and non-mutant waxy genes in rice and maize. Genetics 120, 1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive, M.R., Peacock, W.J., and Dennis, E.S. (1991). The anaerobic responsive element contains two GC-rich sequences essential for binding a nuclear protein and hypoxic activation of the maize Adh1 promoter. Nucleic Acids Res. 19, 7053–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, A.L., and Ferl, R.J. (1991). In vivo footprinting reveals unique cis-elements and different modes of hypoxic induction in maize Adh1 and Adh2. Plant Cell 3, 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, A.L., and Ferl, R.J. (1994). In vivo footprinting identifies an activating element of the maize Adh2 promoter specific for root and vascular tissues. Plant J. 5, 523–533. [DOI] [PubMed] [Google Scholar]
- Petit, J.-M., van Wuytswinkel, O., Briat, J.-F., and Lobreaux, S. (2001). Characterization of an iron-dependent regulatory sequence involved in the transcriptional control of AtFer1 and ZmFer1 plant ferritin genes by iron. J. Biol. Chem. 276, 5584–5590. [DOI] [PubMed] [Google Scholar]
- Praz, V., Perier, R., Bonnard, C., and Bucher, P. (2002). The Eukaryotic Promoter Database, EPD: New entry types and links to gene expression data. Nucleic Acids Res. 30, 322–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros, C.F., Grellet, F., Sadowski, J., Suzuki, T., Li, G., and Wroblewski, T. (2001). Arabidopsis and Brassica comparative genomics: Sequence, structure and gene content in the ABI1-Rps2-Ck1 chromosomal segment and related regions. Genetics 157, 1321–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, W., Dubcovsky, J., Park, Y.-J., Busso, C., Emberton, J., SanMiguel, P., and Bennetzen, J.L. (2002. a). Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics 162, 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, W., Emberton, J., SanMiguel, P., Ogden, M., Llaca, V., Messing, J., and Bennetzen, J.L. (2002. b). Comparative sequence analysis of the sorghum Rph region and the maize Rp1 resistance gene complex. Plant Physiol. 130, 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz, M.B., Grotewold, E., and Chandler, V.L. (1997). Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9, 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, A., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. (2000). PipMaker: A web server for aligning two genomic DNA sequences. Genome Res. 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto, K., and Kyozuka, J. (2002). Rice as a model for comparative genomics of plants. Annu. Rev. Plant Biol. 53, 399–419. [DOI] [PubMed] [Google Scholar]
- Shure, M., Wessler, S., and Fedoroff, N. (1983). Molecular identification and isolation of the waxy locus in maize. Cell 35, 225–235. [DOI] [PubMed] [Google Scholar]
- Song, R., Llaca, V., and Messing, J. (2002). Mosaic organization of orthologous sequences in grass genomes. Genome Res. 12, 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagle, D.A., Koop, B.F., Goodman, M., Slightom, J.L., Hess, D.L., and Jones, R.T. (1988). Embryonic epsilon and gamma globin genes of a prosimian primate (Galago crassicaudatus): Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol. 203, 439–455. [DOI] [PubMed] [Google Scholar]
- Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. [DOI] [PubMed] [Google Scholar]
- Tarchini, R., Biddle, P., Wineland, R., Tingey, S., and Rafalski, A. (2000). The complete sequence of the 340 kb of DNA around the maize Adh1-Adh2 region reveals interrupted colinearity with maize chromosome 4. Plant Cell 12, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova, T.A., and Madden, T.L. (1999). BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174, 247–250. [DOI] [PubMed] [Google Scholar]
- Thacker, C., Marra, M.A., Jones, A., Baillie, D.L., and Rose, A.M. (1999). Functional genomics in Caenorhabditis elegans: An approach involving comparisons of sequences from related nematodes. Genome Res. 9, 348–359. [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A.P., SanMiguel, P.J., Nakajima, Y., Gorenstein, N.M., Bennetzen, J.L., and Avramova, Z. (1999). Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96, 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa, J., Moose, S.P., Parson, R.L., and Schmidt, R.J. (1997). A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc. Natl. Acad. Sci. USA 94, 7685–7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J.C., Howard, E.A., Dennis, E.S., and Peacock, W.J. (1987). DNA sequences required for anaerobic expression of the maize alcohol dehydrogenase1 gene. Proc. Natl. Acad. Sci. USA 84, 6624–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K.H., Gouy, M., Yang, Y.-W., Sharp, P.M., and Li, W.-H. (1989). Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 86, 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (1997). PAML: A program package for phylogenetic analysis by maximum likelihood. CABIOS 13, 555–556. [DOI] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
- Zhu, J., Liu, J.S., and Lawrence, C.E. (1998). Bayesian adaptive sequence alignment algorithms. Bioinformatics 14, 25–39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.