Abstract
Cytosine bases can be deaminated spontaneously to uracil, causing DNA damage. Uracil-DNA glycosylase (UDG), a ubiquitous uracil-excising enzyme found in bacteria and eukaryotes, is one of the enzymes that repair this kind of DNA damage. To date, no UDG-coding gene has been identified in Methanococcus jannaschii, although its entire genome was deciphered. Here, we have identified and characterized a novel UDG from M.jannaschii designated as MjUDG. It efficiently removed uracil from both single- and double-stranded DNA. MjUDG also catalyzes the excision of 8-oxoguanine from DNA. MjUDG has a helix–hairpin–helix motif and a [4Fe–4S]-binding cluster that is considered to be important for the DNA binding and catalytic activity. Although MjUDG shares these features with other structural families such as endonuclease III and mismatch-specific DNA glycosylase (MIG), unique conserved amino acids and substrate specificity distinguish MjUDG from other families. Also, a homologous member of MjUDG was identified in Aquifex aeolicus. We report that MjUDG belongs to a novel UDG family that has not been described to date.
INTRODUCTION
Damaged DNA lesions occur when DNA is modified by UV light, ionizing radiation, reactive oxygen species or chemical mutagens. Uracil residues can also be produced in DNA via the spontaneous deamination of existing cytosines or the misincorporation of dUMP during DNA synthesis. Uracil is biologically important because cytosine to thymine transition is induced at the site of cytosine deamination in DNA (1). However, most organisms have repair enzymes that are specific for this deaminated base in DNA. DNA glycosylases catalyze the major repair process, the base excision repair (BER) pathway, which is initiated by removal of the damaged base (2,3). Among those DNA glycosylases, uracil-DNA glycosylase (UDG) was the first to be discovered (4), and it acts as a major repair enzyme that protects DNA from mutational damages caused by the misincorporation of uracil due to a polymerase error or deamination of cytosine. This repair enzyme hydrolyzed the N-glycosidic bond. As a result, uracil base is released from the DNA backbone, leaving an abasic site behind. This uracil-removing repair mechanism seems to be one of the essential and common DNA repair systems, since UDG-homologous genes and their corresponding enzymes can be found in a variety of organisms (2,3).
The genes that encode uracil-excising enzyme have been found in bacteria (4), viruses (5) and eukaryotes, including human (6,7). They can be generally classified into six families [five UDG families plus the mismatch-specific DNA glycosylase (MIG) family] according to their differences in substrate recognition and amino acid sequence (8–10). The six families are the Ung family (family I) (4,11,12), the MUG/TDG family (family II) (13,14), the sMUG family (family III) (15), the thermostable UDG family (family IV, TmUDG) (16–18), the UDG-B family (family V) (9,19) and the MIG family. However, these five different UDG families show limited sequence similarity, although they share two active site motifs (motifs A and B) (8,9).
Genes analogous to UDG also appear to be present in a variety of thermophilic eubacteria and archaea. Several thermostable UDGs (thermostable UDG family, family IV) have been characterized from genomes of thermophiles such as Thermotoga maritima (16), Archaeoglobus fulgidus (17) and Pyrobaculum aerophilum (18) since UDG activities were detected in the hyperthermophile extracts (20). These proteins are capable of removing uracil from uracil-mismatched duplex substrates and are also active on single-stranded DNA containing uracil. Motif A of this family does not contain aspartic acid or asparagine at the corresponding position of the catalytic residue in motif A of other families. However, motif A (-GEAPG-) of this family has glutamic acid, which may participate in the activation of the catalytic water molecule (8). Relatively recently, another hyperthermophilic UDG family, family V (UDG-B), has been identified in P.aerophilum (9) and Thermus thermophilus (19). The proteins belonging to this family have a prominent structural feature that is the absence of any polar amino acid residues within motif A (-GLAPA/G-).
One of the members of the hyperthermophilic archaea is Methanococcus jannaschii that grows optimally at pressures of up to >200 atm and 85°C. Although the entire genome sequence has now been determined, making it the first archael organism to be sequenced completely (21), the UDG-encoding gene of M.jannaschii has not yet been identified. Hyper thermophilic organisms living in a high temperature environment are at especially high risk of DNA damage by cytosine deamination because high temperature can significantly promote base deamination (22). Thus it is plausible that they may have more effective DNA damage repair systems compared with other organisms. In this study, we identified and characterized, to our knowledge for the first time, a UDG from M.jannaschii (MjUDG). MjUDG was cloned from the open reading frame (ORF) MJ1434 encoding a protein of 220 amino acids with a mol. wt of 26 kDa. Our results show that the MjUDG catalyzes the removal of uracil from both single- and double-stranded DNA. In addition, they also revealed that MjUDG has an 8-oxoguanine (8-oxoG)-excising function that has not been found in other UDG family enzymes. Based on these observations and the result of extended sequence analysis of known DNA glycosylases, we conclude that MjUDG belongs to a novel family that has not been identified to date. Therefore, we named it the MjUDG family (family VI).
MATERIALS AND METHODS
Bacterial strains and reagents
The expression strain BL21 (DE3) and plasmid pET28a with a polyhistidine tag were obtained from Novagen (WI). The hyperthermophilic archaeon M.jannaschii (DSM 2661) was purchased from the Deutsche Sammlung von Mikroorganism und Zellkultren GmbH (DSM, Braunschweig, Germany). Methanococcus jannaschii was cultivated under conditions as reported previously (23). Genomic DNA was prepared using a Qiagen Genomic DNA Midi Kit (Qiagen GmbH, Hilden, Germany). Restriction endonuclease, T4 DNA ligase, Escherichia coli endonuclease III (EndoIII) and UDG were purchased from New England Biolabs (MA). All other reagents were of analytical grade purity.
Construction of recombinant plasmid pET28a-MjUDG
The M.jannaschii MJ1434 ORF encoding the MjUDG gene was amplified by PCR. The PCR was carried out using M.jannaschii genomic DNA as a template, and forward (5′-CGTTCACATATGAAAGAGAACAAA-3′) and reverse (5′-CATGTCAAGCTTTTACTTTGAGAGGCAGAA-3′) primers. The forward and reverse primers contained NdeI and HindIII restriction sites (underlined), respectively, for direct cloning. The PCR product was purified using the QIAquick PCR purification kit (Qiagen). The PCR product and pET28a vector were digested with restriction enzymes NdeI and HindIII. Recombinant plasmid pET28a-MjUDG was generated by insertion of the PCR product into the cloning site (NdeI and HindIII) of pET28a in order to express MjUDG protein with a polyhistidine tag. The nucleotide sequence was determined by the dideoxy chain termination method as described in the instruction manual of the Perkin-Elmer cycle sequencing kit (Perkin-Elmer, MA). Gene-specific primers of MJ1434 were used to confirm the correct insertion. Briefly, the sequencing reaction products were separated by 6% acrylamide gel electrophoresis, and the sequence was determined using an ABI 373 automatic DNA sequencer (Applied Biosystems, CA).
Protein purification
The recombinant plasmid pET28a-MjUDG was introduced into E.coli strain BL21 (DE3). Then the E.coli BL21 (DE3) harboring pET28a-MjUDG were inoculated into LB medium (1% tryptone, 0.5% yeast extract and 0.5% NaCl) containing 50 µg/ml of kanamycin to a density of A600 1.0 at 37°C. Recombinant proteins were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C for 6 h. The cells were centrifuged at 6000 r.p.m. for 20 min, and then cell pellets were resuspended in buffer A (50 mM NaH2PO4, 300 mM NaCl pH 8.0). Cells were disrupted by ultrasonication and the lysates were centrifuged at 15 000 r.p.m. for 30 min. The supernatant was applied to an Ni-NTA–agarose resin column (Qiagen) pre-equilibrated in buffer A at a flow rate of 1 ml/min. The flow-through was discarded and proteins were washed 10 times with additional column volumes of buffer A. MjUDG was eluted from the column with buffer B (50 mM NaH2PO4, 300 mM NaCl, 300 mM imidazole pH 8.0). The protein fractions were identified by 15% SDS–PAGE. The volume of sample fractions was reduced to 20 ml by concentration with an Amicon YM10 membrane (Millipore, MA), then cleaved using thrombin protease (10 U/mg of fusion protein). The cleavage mixture of fusion protein was dialyzed in buffer A and applied to a Superdex-75 gel filtration FPLC column. The fractions containing homogeneous MjUDG were collected using a fraction collector and concentrated by ultrafiltration using an Amicon YM10 membrane. The pure MjUDG protein obtained was stored at –80°C at a concentration of 1 mg/ml.
DNA substrates
The oligonucleotide sequences used in this work were 32mer strands (5′-GGATCCTCTAGAGTCXACCTGCAGGCATGCAA-3′) or 39mer strands (5′-GGATCCTCTAGAGTCYACCTGCAGGCATGCAAGCTTGAG-3′), where X is the position for adenine (A), thymine (T), cytosine (C), guanine (G), uracil (U), hypoxanthine (HX), 3-methyladenine (3-mA) or 7-methylguanine (7-mG), and Y is the position for 8-oxoG or apurinic/apyrimidinic (AP) site substitution. These oligonucleotides containing a single modified base at position 16 were purchased from Bio-Synthesis, Inc. (TX). The oligonucleotide sequences with a complementary base (A, T, G or C) opposite the X (or Y) position were used as complementary strands. The oligonucleotide duplexes containing thymine glycol (Tg) residues were prepared by treating 32mer oligonucleotide containing a single T residue (5′-AGGAAGAGGAAGGAGTGAAGGGAGAGAGGAGA-3′) with OsO4 before annealing with complementary strands, as described previously (24). The oligonucleotides containing X, Y and Tg were 5′-end labeled by T4 polynucleotide kinase (Takara, Shiga, Japan) in the presence of [γ-32P]ATP (Dupont NEN, MA). Unincorporated [γ-32P]ATP was removed with the QIAquick Nucleotide Removal Kit (Qiagen) following purification of the oligonucleotide. The duplex oligonucleotides were obtained by annealing with an unlabeled complementary strand at a 1.5-fold molar excess in buffer [20 mM Tris–HCl pH 7.4, 100 mM NaCl, 1 mM dithiothreitol (DTT), 1 mM EDTA, 3% glycerol]. The reaction mixture was heated to 75°C for 5 min and then cooled down to room temperature. Annealed DNA was eluted by ethanol precipitation then dried and resuspended in double-distilled water. The protein concentration was determined by the Bradford method (25) with bovine serum albumin as a standard.
DNA glycosylase assay
The glycosylase reactions were performed with 5 pmol of MjUDG protein in a 20 µl reaction mixture containing 20 mM MES pH 6.0, 80 mM NaCl, 1 mM DTT, 1 mM EDTA and 3% glycerol at 55°C for 20 min. The reaction mixture was subjected to hot alkaline treatment by adding 1 M NaOH (to a final concentration of 100 mM) with heating to 95°C for 10 min. The reaction mixture was then neutralized by adding 1 M Tris (to a final concentration of 30 mM), and then an equal volume of formamide loading buffer (containing 0.05% bromophenol blue and 0.05% xylene cyanol) was added to the mixture. The sample solutions were then boiled at 95°C for 5 min and cooled on ice. The samples were analyzed by gel electrophoresis (15% polyacrylamide gel, 7 M urea, 1× TBE running buffer). The gel was then dried and placed on an imaging plate, and the amount of DNA products was quantified using a BAS 2500 image analyzer (Fuji, Tokyo, Japan).
Phylogenetic analysis
Identification of MjUDG homologs in bacterial, archaeal and eukaryotic genomes was carried out using a basic local alignment search tool (BLASTP) search of the National Center for Biotechnology Information (NCBI) database. The sequences retrieved from the BLASTP search were analyzed by multiple sequence alignment using the Clustal W program (26). Distance analysis of the phylogenetic tree was performed using the program TreeView (Win32) 1.5 (27).
RESULTS
Identification and purification of MjUDG
The entire genomic sequence of a hyperthermophilic archaeon M.jannaschii has been published (21), yet the function of a significant fraction of the coding genes and their products remains to be elucidated. Among those genes, the ORF MJ1434 (Q58829, 220 amino acids) has been annotated as a putative EndoIII under the category ‘DNA replication and repair’ in the genome database (TIGR Microbial Database: http://www.tigr.org/tdb/mdb/mdbcomplete.html) until now. Nevertheless, the amino acid sequence of MJ1434 is only 21% identical to that of E.coli EndoIII and even less identical to the DNA repair proteins, such as the DNA glycosylases family, of other organisms (data not shown).
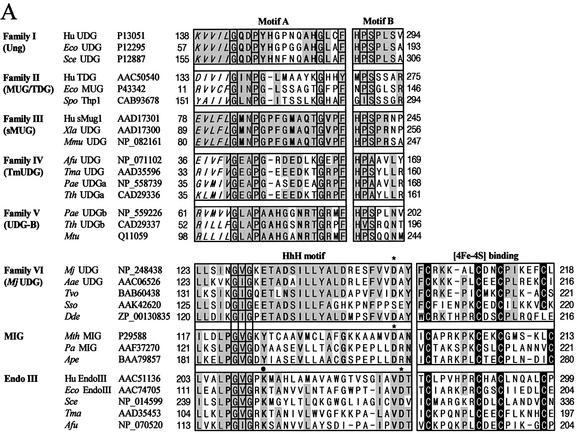
The search using the GenBank database identified several homologous members of MJ1434 in other organisms such as Aquifex aeolicus (AAC06526), Thermoplasma volcanium (BAB60438) and Sulfolobus solfataricus (AAK42620) (Fig. 1). These members fall into the categories of conserved hypothetical protein or putative EndoIII. The amino acid sequence of MJ1434 showed 34–52% identity with these homologous proteins and, interestingly, they share a highly conserved helix–hairpin–helix (HhH) motif and a [4Fe–4S] cluster found most notably in the EndoIII and MutY DNA glycosylase family (Fig. 1). However, these proteins lack the lysine residue within the HhH motif critical for the catalytic reaction of AP lyase, and some amino acid sequences in the motif are different from those of the EndoIII family. Therefore, we investigated the practical activity of MJ1434 protein in order to determine its biochemical function.
Figure 1.
Sequence alignment of MjUDG and its homologs using the program Clustal W. The homologs of MjUDG are: A.aeolicus (AAC06526), T.volcanium (BAB60438) and S.solfataricus (AAK42620). The shadowed boxes represent conserved amino acid residues. The cysteine residues for binding of the [4Fe–4S] cluster are shown in black boxes. The arrow represents the highly conserved helix–hairpin–helix motif.
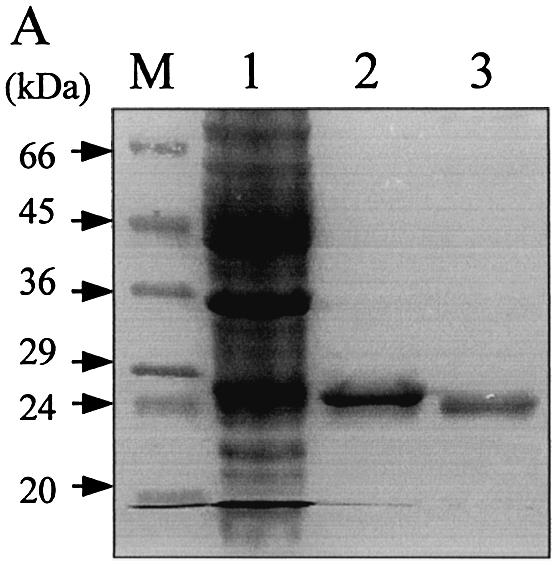
The protein encoding MJ1434 was expressed in E.coli BL21 (DE3) as a hexa-histidine-tagged recombinant protein. The MJ1434 gene was amplified from genomic DNA by PCR using two oligonucleotide primers at the N- and C-terminal ends of the ORF, and the PCR product was cloned in an expression vector, pET28a, which places a hexa-histidine tag at the 5′ end of the gene. The recombinant plasmid was introduced into E.coli BL21 (DE3) for protein expression. The E.coli BL21 (DE3) cells harboring pET28a-Mj1434 were induced to express fusion proteins by the addition of IPTG to the growth medium. The His-tagged MJ1434 protein was eluted from the Ni-NTA affinity column with imidazole, while the other proteins flowed through. After cleavage of the N-terminus of His-tagged MJ1434 protein with thrombin, the MJ1434 protein was purified with Superdex-75 gel filtration FPLC. After the induction with IPTG and purification through the Ni-NTA column, the overexpressed protein (∼26 kDa) was detected on the gel while a control (E.coli harboring a plasmid without the MJ1434 insert) did not display this band (Fig. 2). The resultant purified MJ1434 protein had an additional three amino acids (Gly–Ser–His) at its N-terminus after thrombin cleavage. The final product had a single detectable band at 24 kDa, which is compatible with the predicted molecular mass of MJ1434 protein by SDS–PAGE with >95% purity (Fig. 2).
Figure 2.
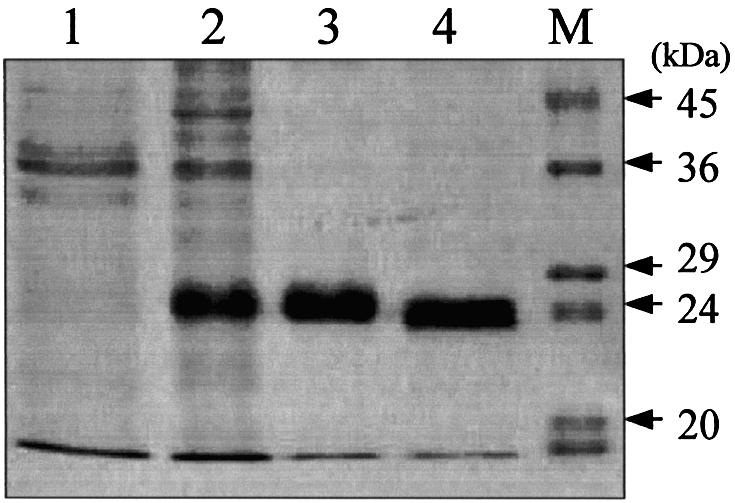
Purification steps of recombinant MjUDG protein. Each step was analyzed by staining the 15% SDS–polyacrylamide gel with Coomassie brilliant blue. Lane 1, control (crude extract of cells harboring vector pET28a); lane 2, soluble extract of cells harboring pET28a-MjUDG after 6 h induction with IPTG; lane 3, protein fraction eluted from the Ni-NTA affinity column; lane 4, MjUDG protein eluted from Superdex-75 gel filtration chromatography after thrombin treatment; M, protein molecular weight markers.
UDG activity of MJ1434
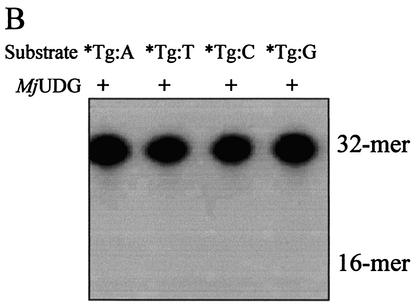
We investigated whether purified MJ1434 protein had EndoIII activity. MJ1434 protein was tested for its ability to excise Tg using a Tg-containing oligonucleotide duplex that is a typical substrate of EndoIII (28), and no activity was detected for this substrate (Fig. 3A). To confirm that MJ1434 does not have EndoIII activity, other duplex substrates containing four common bases at the site opposite to Tg (Tg:A, Tg:T, Tg:C or Tg:G) were also used as substrates (Fig. 3B). Again no detectable cleavage of the oligonucleotide was observed with each of the substrates even with excess amounts of the purified protein. When the activity assay of MJ1434 was also performed at different temperatures (37–90°C), EndoIII activity was not detected (data not shown). From these results, we could prove that Tg was not a substrate for MJ1434.
Figure 3.
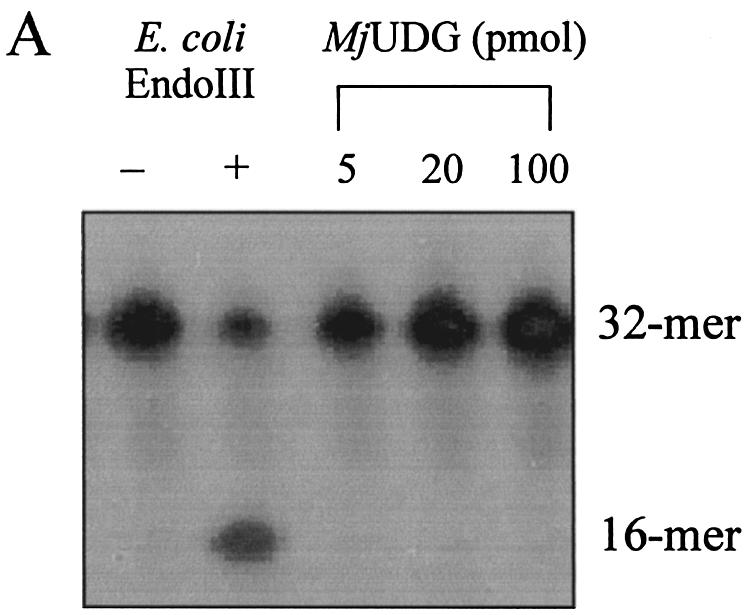
Activity assay of MjUDG protein on thymine glycol (Tg)-containing DNA substrate. Each reaction mixture was analyzed on a 15% denaturing polyacrylamide gel and the bands were visualized on a BAS 2500 image analyzer. The intact 32mer and 16mer are indicated as the substrates and products, respectively. (A) Different amounts of MjUDG (5 pmol, lane 3; 20 pmol, lane 4; 100 pmol, lane 5) were incubated with 1 pmol of 5′-labeled 32mer oligonucleotide duplex containing Tg at 55°C for 20 min. The E.coli EndoIII reaction, as a control, was performed at 37°C for 20 min. (B) Activity assay of MjUDG on four different substrates containing a Tg site. The reaction was carried out with duplex substrates containing four common bases opposite a Tg site at 55°C for 20 min. The asterisk indicates the 5′-end-labeled strand.
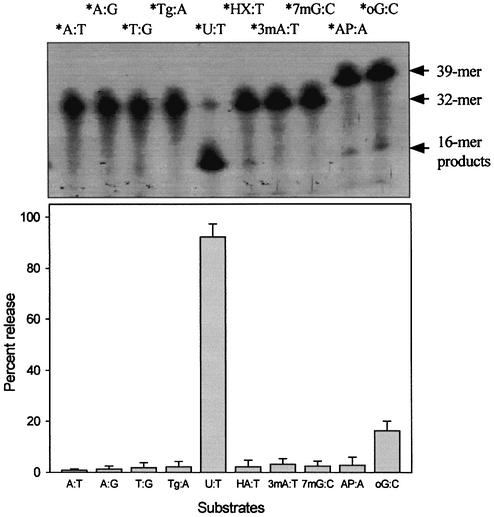
Several oligonucleotide duplexes were used for determination of the enzymatic activity of MJ1434 protein. First, the DNA glycosylase activity assay was carried out using defined double-stranded oligonucleotides with all possible matches and mismatches of A, T, C, G, U, Tg, HX, 3-mA, 7-mG, 8-oxoG and an AP site with the four common bases. Each substrate was incubated with MJ1434 protein and then its product was analyzed by PAGE. The reactions were performed at 55°C with MJ1434 to prevent thermal denaturation of the oligonucleotide duplex substrates. After the reaction of these substrates with MJ1434, samples were treated with NaOH for cleavage at the internal AP site following removal of the mismatched base. Among various substrates, to our surprise, MJ1434 protein efficiently processed uracil-containing DNA and also had a weak activity on the 8-oxoG-containing oligonucleotide duplex (Fig. 4). The activity of the MJ1434 protein was also measured in a single-stranded substrate containing uracil. Like other thermophilic UDGs (16,17), MJ1434 protein was also capable of removing uracil from a single-stranded oligonucleotide (Fig. 5A). In order to eliminate the possibility that the observed uracil-excising activity was due to contamination by the endogenous E.coli UDG, purified sample was heated at 80°C for 30 min. Since MJ1434 protein is a thermostable protein, it was expected that high temperature would not alter the enzyme activity. Even after the heating, no significant loss of uracil-releasing activity of MJ1434 was observed (data not shown). Maximum enzyme activity was observed at between 55 and 65°C. MJ1434 was more active at 65°C than at 37°C, while E.coli UDG activity was greatly decreased at high temperature (Fig. 5B). This observation provides additional evidence that the activity of MJ1434 was not a contaminant from E.coli UDG. However, at temperatures higher than 70°C, MJ1434 activity was decreased, probably due to partial denaturation of the DNA substrate (data not shown). The optimal pH for the activity of MJ1434 protein was near pH 6.0, and no significant difference in activity was observed between pH 5.0 and 6.5. Most UDGs are known to be monofunctional and hydrolyze uracil without AP lyase activity (8,29). After the reaction of MJ1434 protein with the uracil-containing oligonucleotide duplex, reaction mixtures were treated in the presence or absence of NaOH. MJ1434 also displayed a monofunctional activity. The DNA backbone was cleaved at the AP site only when treated with NaOH at high temperature (Fig. 5C). This backbone cleavage was not observed without NaOH treatment. The same result was observed in experiments using 8-oxoG-containing substrate (Fig. 5D). These results convinced us that MJ1434 protein was the enzyme denoted as uracil-DNA glycosylase (UDG), and we therefore termed it MjUDG.
Figure 4.
DNA glycosylase activity of MjUDG protein. Purified MjUDG protein (5 pmol) was incubated with 1 pmol of 5′-end-labeled oligonucleotide duplex at 55°C for 20 min. The reaction mixture was treated with 100 mM NaOH and incubated at 95°C for 10 min. The cleavage products were analyzed by 15% denaturing PAGE and using a BAS 2500 image analyzer. The asterisk indicates the 5′-end-labeled strand. Oligonucleotide strands containing the apurinic/apyrimidinic (AP) site or 8-oxoguanine (oG) were 39mer, and strands containing adenine (A), thymine (T), thymine glycol (Tg), uracil (U), hypoxanthine (HX), 3-methyladenine (3mA) or 7-methylguanine (7mG) were 32mer. Only a reaction mixture of AP- containing substrate was not treated with NaOH and heating after reaction with MjUDG. Each damaged base in the oligonucleotide sequences was at position 16. In the bottom graph, the quantitative data were obtained from at least three independent experiments.
Figure 5.
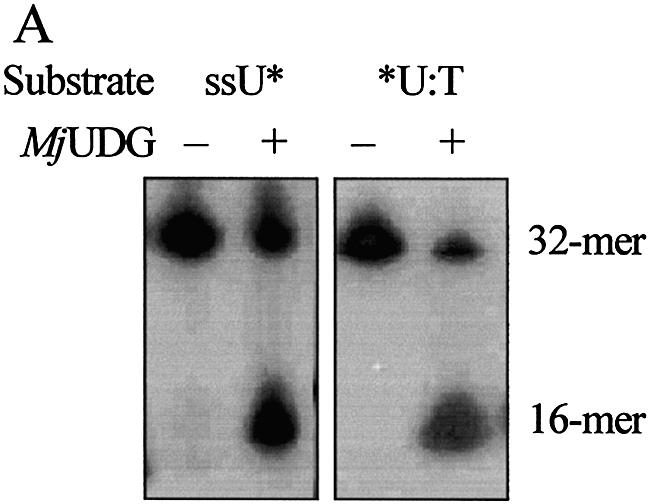
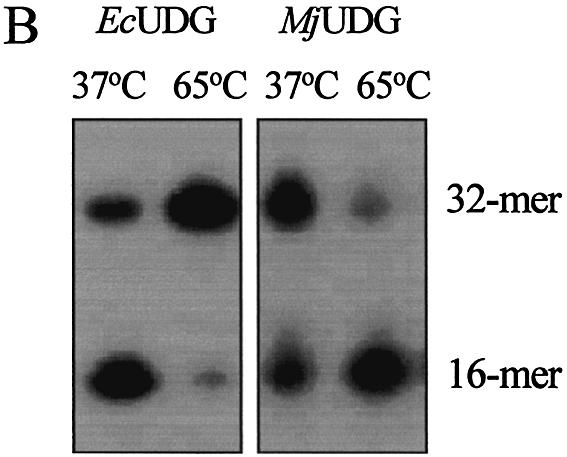
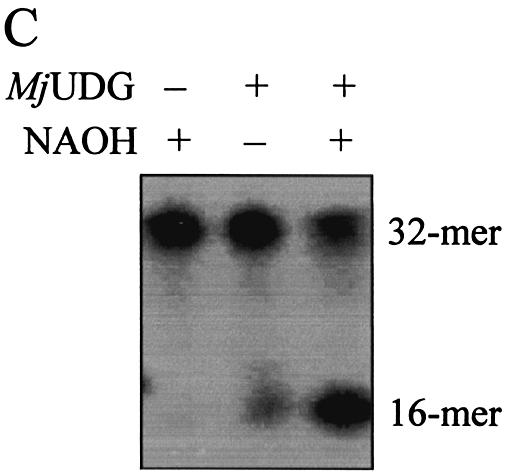
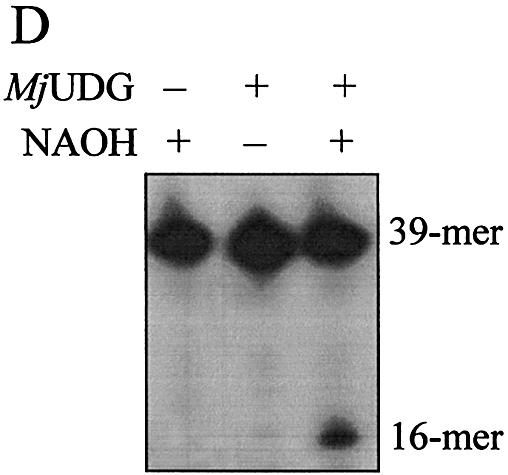
Uracil-DNA glycosylase activity of MjUDG. (A) Uracil-processing activity of MjUDG on the single- or double-stranded DNA substrates. The 32mer substrates (*U-containing single strand or *U:T mismatch-containing duplex) were incubated with 5 pmol of MjUDG at 55°C for 20 min. The reaction mixture was treated with 100 mM NaOH at 95°C for 10 min. The products were separated on a 15% denaturing polyacrylamide gel, and the bands were visualized using a BAS 2500 image analyzer. The asterisk indicates the 5′-end-labeled strand. (B) Comparison of uracil-DNA glycosylase activity at different temperature of MjUDG and E.coli UDG (EcUDG). MjUDG was incubated with 5′-end-labeled *U:T mismatch-containing oligonucleotide duplex at 37 or 65°C, respectively. The reaction mixture was treated with 100 mM NaOH at 95°C for 10 min. Activity assay of EcUDG was also performed with the instruction manual. (C) Monofunctional activity of MjUDG. MjUDG protein (5 pmol) was incubated with 1 pmol of 5′-end-labeled oligonucleotide duplex (*U:T mismatch) at 55°C for 20 min. The reaction mixture was then incubated at 95°C for 10 min in the absence (lane 2) or presence (lane 3) of NaOH. (D) Monofunctional activity of MjUDG on 8-oxoG-containing substrate was also examined under the same conditions. After reacting 1 pmol of 8-oxoG-containing 39mer duplex (*oG:C) with MjUDG for 20 min at 55°C, the reaction mixture was treated at 95°C for 10 min without (lane 2) or with (lane 3) NaOH.
Substrate specificity of MjUDG
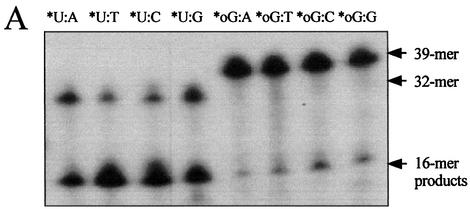
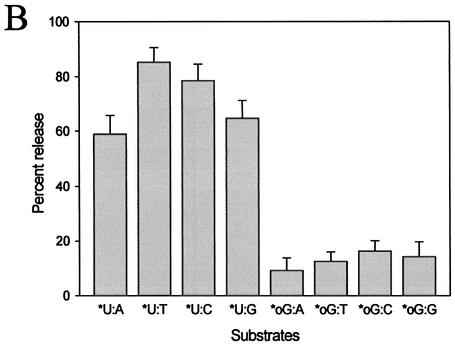
To evaluate the specificity of uracil excision from a U:N (N = A, T, C or G) mismatch, MjUDG activity was compared using DNA duplexes containing a uracil residue opposite to each of the four common bases (A, T, G and C). These mismatched duplex substrates (U:A, U:T, U:C and U:G) were incubated with MjUDG at 55°C for 20 min. The uracil-releasing activity of MjUDG was observed in the following order U:T>U:C>U:G>U:A, showing insignificant differences in activity level on each of the substrates (Fig. 6A and B). The opposite base-dependent efficacy of uracil excision by MjUDG is similar to that of previously reported A.fulgidus UDG (AfUDG) (30). Although MjUDG also showed some activities with oligonucleotide duplexes containing 8-oxoG, the activities were 5-fold lower than that of oligonucleotide duplex containing U:T (Fig. 6A and B). Originally, the 8-oxoG residues are excised from oxidatively damaged DNA by the DNA repair enzymes known as MutM (31) and 8-oxoG DNA glycosylase (Ogg) (32). This activity of MjUDG protein is unique, and is not found in other UDGs.
Figure 6.
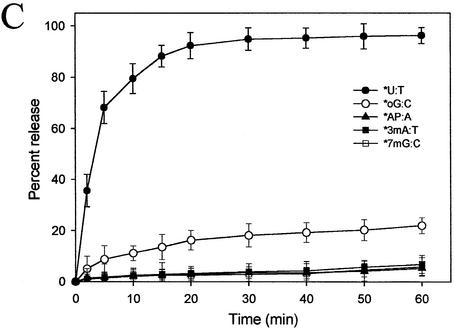
Substrate specificity of MjUDG. (A) Substrate specificity of MjUDG on the 5′-end-labeled 32mer oligonucleotide substrates containing uracil (*U:A, *U:T, *U:C and *U:G) and 8-oxoG-containing 39mer substrates (*oG:A, *oG:T, *oG:C and *oG:G). After treating 1 pmol of substrate with MjUDG for 20 min at 55°C, the reaction mixture was treated with NaOH and electrophoresed on a 15% denaturing polyacrylamide gel. The bands were visualized on a BAS 2500 image analyzer. (B) The average values were obtained from at least three independent experiments. (C) The MjUDG reaction was carried out at 55°C with various oligonucleotide duplexes (containing uracil, *U:T; 8-oxoG, *oG:C; apurinic/apyrimidinic site, *AP:A; 3-methyladenine, *3mA:T; 7-methylguanine, *7mG:C, respectively, where the asterisk indicates the 5′-end-labeled strand). At the indicated time points, the reaction mixture was removed and treated with 100 mM NaOH at 95°C for 10 min. The cleavage products were analyzed by 15% denaturing polyacrylamide gel electrophoresis and using a BAS 2500 image analyzer. All experiments were carried out independently three times.
In order to confirm that MjUDG could specifically process the uracil-containing DNA substrate, we also performed reactions with different time courses. The results are summarized in Fig. 6C. MjUDG processed the uracil-containing DNA substrate with high efficacy, whereas it was less effective on 8-oxoG-containing DNA substrate. Besides the duplex substrates containing uracil or 8-oxoG, oligonucleotide substrates containing 3-mA, 7-mG and an AP site also were examined as substrates for MjUDG, and they were not hydrolyzed by MjUDG (Fig. 6C). In addition, oligonucleotide duplex substrates containing HX, Tg and base mismatches (T:G and A:G) were also tested. Again, no activities were detected under identical experimental conditions (data not shown). Reactions were repeated under the various buffer conditions with a broad pH range, and with various complementary base pairs with the residues mentioned above. In no case was MjUDG activity observed.
UDG activity of homologous protein of MjUDG
Through the analysis of multiple sequence alignments, genes homologous to MjUDG were found in several organisms (Fig. 1). Although their functions are unknown to date, interestingly, the amino acid sequences, especially those of the HhH motif and a [4Fe–4S] site, are highly conserved. This suggests that the biochemical function of these homologous members is probably identical to that of MjUDG. Among them, a hypothetical protein, (AAC06526) annotated as EndoIII from A.aeolicus (termed AaeUDG) (33), that is homologous to MjUDG, has been purified (Fig. 7A). All the procedures to express and purify AaeUDG protein were the same as those for MjUDG, as described in Materials and Methods. The UDG activity of AaeUDG protein was also tested using several oligonucleotide substrates containing a uracil residue. As expected, AaeUDG actually had UDG activity and its specificity was similar to that of MjUDG (Fig. 7B). AaeUDG protein also showed monofuctional activity like MjUDG, and displayed maximum activity under weak acidic conditions (data not shown). This result suggests that other MjUDG homologs described in Figure 1 could potentially have UDG activity.
Figure 7.
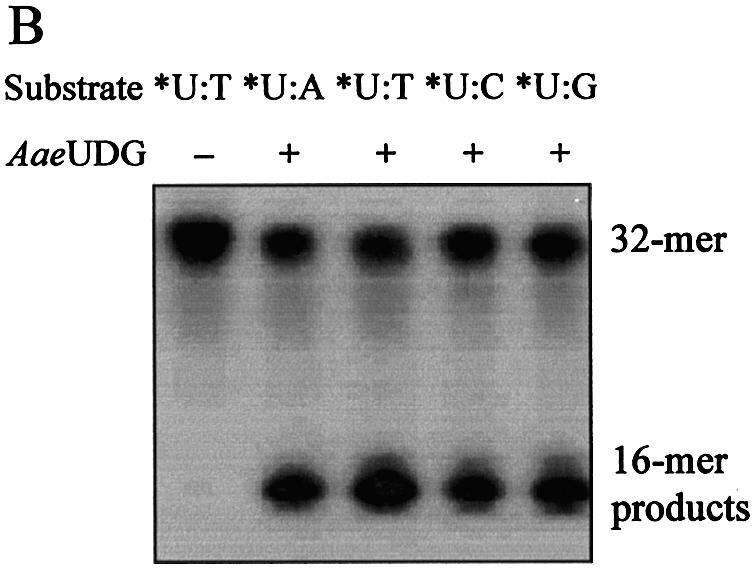
Purification and uracil-DNA glycosylase activity of the putative UDG (AaeUDG) from A.aeolicus. (A) Expression and purification of the recombinant AaeUDG protein. Lane M, protein molecular weight markers; lane 1, soluble extract of cells harboring pET28a-AaeUDG after 6 h induction with IPTG; lane 2, protein fraction eluted from an Ni-NTA affinity column; lane 3, AaeUDG protein eluted by Superdex-75 gel filtration chromatography after thrombin treatment. (B) Activity assay of AaeUDG on four different oligonucleotide substrates containing a uracil base (*U:A, *U:T, *U:C and *U:G). The asterisk indicates the 5′-end-labeled strand. The reaction was carried out with 32mer duplex substrates containing four common bases opposite a uracil at 55°C for 20 min. Each reaction mixture was treated with NaOH at 95°C for 10 min, and then samples were analyzed by 15% denaturing polyacrylamide gel electrophoresis and using a BAS 2500 image analyzer.
Sequence comparison and phylogenetic analysis
Bacterial and archaeal homologs of MjUDG have been identified in several organisms through the BLASTP search (Fig. 1). The MjUDG amino acid sequence was aligned with its homologous sequences using the program Clustal W. The amino acid sequences of members of other UDG families have been also obtained from the GenBank database. From the analysis of partial sequence alignment, several regions of MjUDG and homologs are highly conserved, and two motifs (the HhH motif and the [4Fe–4S] cluster site) are shared with members of the EndoIII and MIG family as shown in Fig. 8A. MjUDG does not appear to be closely related to other known UDGs at the level of the amino acid sequence. MjUDG family proteins are similar to the MIG and EndoIII family but differ from them in having a glutamic acid (Glu132 in the case of MjUDG) at the position corresponding to lysine in EndoIII or tyrosine in MIG (Fig. 8A).
Figure 8.
(Opposite) Partial amino acid sequence alignment and phylogenetic analysis. (A) Partial amino acid sequence alignment of five UDG families: the MjUDG family (this work), MIG family and endonuclease III (Endo III) family. The active site motifs A and B of five UDG families are shown in the upper panel (families I, II, III, IV and V, respectively). The conserved amino acid residues within each family are shaded. Highly conserved amino acid residues among all five UDG families are shaded in squares. Unique conserved amino acids within each family are shown in bold. The highly conserved HhH motif and [4Fe–4S]-binding cluster of the MjUDG, MIG and EndoIII family are represented in the bottom panel. The conserved amino acid residues within the MjUDG, MIG and Endo III families are shaded. Strictly conserved amino acids (-G-V/I-G-) among the three families are shaded in squares, and unique amino acids within each family are also shown in bold. The cysteine residues involved in binding of the [4Fe–4S] cluster are shown in black boxes. The conserved lysine residue within the EndoIII family is marked with a dot. Putative aspartic acid residues required for the catalytic activity in the HhH motif are also conserved in the three families, and they are marked with an asterisk. (B) Phylogenetic analysis of members of the UDG, MjUDG, MIG and EndoIII families. The proteins shown are M.jannaschii UDG (this work, NP_248438), A.aeolicus (AAC06526), T.volcanium (BAB60438), S.solfataricus (AAK42620), H.pylori (AAD07668), E.coli EndoIII (AAC74705), A.fulgidus EndoIII (NP_070520), T.maritima (AAD35453), M.thermautotrophicus MIG (P29588), P.aerophilum MIG (AAF37270), Aeropyrum pernix (BAA79857), M.thermautotrophicus (AAB85267), human UDG (P13051), E.coli UDG (P12295), Saccharomyces cerevisiae UDG (P12887), human TDG (AAC50540), E.coli MUG (P43342), Schizosaccharomyces pombe Thp1 (CAB93678), human sMUG1 (AAD17301), Xenopus laevis UDG (AAD17300), Mus musculus UDG (NP_082161), A.fulgidus UDG (NP_071102), T.maritima UDG (AAD35596), T.thermophilus UDG (CAD29336), P.aerophilum UDG (NP_558739), T.thermophilus UDG-B (CAD29337), P.aerophilum UDG-B (NP_559226) and Mycobacterium tuberculosis (Q11059). Proteins whose biochemical functions were not determined are designated by only the name of the organism.
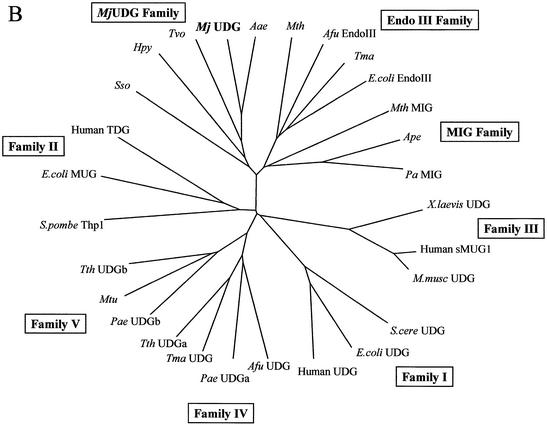
In the phylogenetic analysis, there are roughly five branches that contain one or more DNA glycosylase families (Fig. 8B). MjUDG is closer to Endo III proteins than other UDGs. A branch containing closely related proteins (MjUDG, MIG and EndoIII) is classified into three distinct groups, and MjUDG falls into the category of a novel family. This result suggests that these proteins may have had a common ancestor during evolution.
DISCUSSION
Almost universally, when the function of a new gene is determined, its DNA (or amino acid) sequence is compared with that of known proteins using basic alignment search tools such as BLAST (34). However, in some cases, the inference of a gene function by sequence homology can result in misinterpretation. To date, even with the completed genome sequences from M.jannaschii, functions were assigned to about half of all gene products based on sequence homologies, leaving a significant fraction of genes coding for hypothetical proteins with no known functions (21). Furthermore, several genes essential to the DNA repair mechanism have not yet been identified. Also, uracil-removing repair proteins have not been found in M.jannaschii to date. However, a UDG (AfUDG) of A.fulgidus, known to be closely related to M.jannaschii in evolutionary tree, has been identified and characterized. (17,30). Until recently, no homologs of all known UDG families have been detected in the genome sequences of M.jannaschii using BLAST searches (35). In this work, we identified and characterized a novel UDG from M.jannaschii belonging to a new UDG family.
We have expressed and purified the soluble recombinant protein of ORF MJ1434 of a hyperthermophilic archaeon, M.jannaschii, which has been annotated as a putative EndoIII. In most cases, EndoIII family proteins possess two highly conserved regions; an HhH motif corresponding to the putative active site and a C-X6-C-X2-C-X4-5-C segment containing a [4Fe–4S]-binding center near the C-terminus (36). MJ1434 possesses both a HhH motif and a [4Fe-4S]-binding site and shows 21% amino acid sequence identity with E.coli EndoIII. First, we had to determine whether MJ1434 functioned as an EndoIII. EndoIII activity of MJ1434 was examined using an oligonucleotide duplex containing Tg (Tg:A), and no endonuclease activity was detected for this substrate (Fig. 3). To characterize the biochemical activity of this protein, we examined its ability to excise bases from oligonucleotide duplexes containing base pairs comprised of several modified bases and four common normal bases (A, T, G or C) (Fig. 4). The result shows that MJ1434 possesses substrate specificity. In other words, it removed uracil most efficiently when uracil was paired with thymine. Although the activity was 5-fold lower compared with uracil-containing substrates, it also had an activity on oligonucleotide duplexes containing 8-oxoG, which is not observed in other UDGs (Fig. 4). Like most UDGs, MJ1434 protein is active on single-stranded DNA containing uracil and a monofunctional DNA glycosylase (Fig. 5A and C). In order to eliminate the possibility that the observed uracil-excising activity was due to contamination with the endogenous E.coli UDG, purified MJ1434 protein was heated to 80°C for 30 min. Since MJ1434 is a thermostable protein, it was expected that high temperature would not alter the enzyme activity. Even after heating, no significant loss of uracil release activity of MJ1434 was observed. Thus, we concluded that MJ1434 was a uracil-DNA glycosylase (MjUDG). Additional evidence that the activity of MjUDG was not from E.coli UDG contamination came from the observation that the maximum activity of MjUDG was achieved at a temperature of 55–65°C while E.coli UDG activity was greatly reduced as the temperature increased (Fig. 5B).
Until comparatively recently, six different families of DNA glycosylases (five UDG families and one MIG family) that remove uracil from damaged DNA have been classified (8–10). The DNA glycosylases belonging to the five UDG families (family I–V) share two commonly conserved motifs, motif A (-GX2PX7-8GX2F-) and motif B (-HPS/AXn-), thought to be responsible for the enzyme activity (Fig. 8A). However, the amino acid sequences of the motifs of five UDG families are somewhat different, and thus their biochemical properties might be classified into distinct families (8,9).
Although the three-dimensional structure of MjUDG protein has not yet been determined, its sequence analysis shows that it has very little sequence similarity to other UDGs and lacks the conserved active site motifs of a typical UDG; this suggests that MjUDG differs from the other UDG families (Fig. 8A). In addition, in terms of the amino acid sequence, MjUDG and its homologs are closer to the EndoIII family than to the typical UDG family. Thus, these proteins, including MjUDG, branch off as a novel cluster of UDGs in phylogenetic analysis (Fig. 8B). MjUDG also has a HhH motif and [4Fe–4S]-binding cluster found in a diverse range of structurally related DNA repair proteins such as EndoIII and MutY DNA glycosylase (36,37). A conserved aspartic acid residue in the HhH motif that is likely to be implicated in the catalytic activity of the EndoIII enzymes is also present in MjUDG, suggesting that the catalytic mechanism of MjUDG is similar to that of other EndoIII proteins. However, the MjUDG family has a glutamic acid residue in the HhH motif whereas EndoIII has lysine at the corresponding position (Fig. 8A). This different amino acid sequence of MjUDG might be inefficient for the AP lyase activity observed in EndoIII. Indeed, like other UDG families, no AP lyase activity was observed in MjUDG (Fig. 5C).
The MIG family is also a member of the HhH superfamily of DNA glycosylases. MIG family DNA glycosylases are T:G mismatch-specific enzymes without AP lyase activity, and their HhH motifs contain a conserved tyrosine residue instead of the lysine that is found in HhH motifs of EndoIII (10,38). Several unique conserved amino acid residues such as tyrosine, valine and asparagine are also found in the MIG family HhH motif (Fig. 8A). Recently, by structural and mutagenesis analysis of Methanobacterium thermoautotrophicum MIG (MthMIG), the molecular mechanism of substrate specificity and catalysis for target nucleotides has been reported (39). It was demonstrated that the tyrosine residue (Tyr126) of the MthMIG HhH motif was a key to the specificity for thymine bases. Asn146 was also an important residue for the catalytic activity of MthMIG. These two amino acid residues are conserved in MIG family enzymes (Fig. 8A).
MjUDG shares common features such as the HhH motif and [4Fe–4S] cluster with the MIG family but differs functionally due to distinctive substrate specificity and activity on single-stranded DNA (10,38). The HhH motif of MjUDG differs from that of MIG in that both tyrosine and asparagine, which play an important role in MIG substrate recognition, are replaced by glutamic acid and tyrosine, respectively. The highly conserved region (-TADSILLYALD-) also exists downstream of the -GV/IG- sequence within the HhH motif of the MjUDG family (Fig. 8A). Interestingly, MjUDG family enzymes have unique phenylalanines (Phe147 and Phe201 in the case of MjUDG) that can interact through π–π interactions in the binding pocket of the enzyme with the flipped-out base. These uniquely conserved amino acids in the motifs of the MjUDG family might be what distinguish the MjUDG family from the MIG family as well as the EndoIII family. Again, the substrate specificity of the members of the MjUDG family probably differentiates them from MIG family proteins.
Unlike any other UDGs, MjUDG also shows activity for 8-oxoG. Recently, Matsumoto et al. showed that E.coli and human EndoIII removed 8-oxoG from DNA duplexes (40). However, it is unclear whether MjUDG recognizes 8-oxoG mismatches in vivo and facilitates BER under specific conditions. As illustrated in Figure 8A, unique amino acid residues within the HhH motif are probably important factors determining substrate recognition and specificity of MjUDG family proteins.
So far, in addition to MjUDG, its homologs have been identified in several organisms such as A.aeolicus, T.volcanium, S.solfataricus, Desulfovibrio desulfuricans and Helicobacter pylori (Figs 1 and 8A). Among those, a putative UDG from A.aeolicus (AaeUDG) was expressed and purified. Indeed, as in the case of MjUDG, AaeUDG also showed uracil excision activity from DNA substrates (Fig. 7B). With all these data collected, we could confirm the biochemical function of MjUDG. Furthermore, MjUDG and its homologs including AaeUDG are probably an indispensable part of the uracil-removing DNA repair systems in various organisms. Further experiments to solve the three-dimensional structure using the MjUDG protein–DNA substrate complex and mutant proteins are now under way to clarify the recognition mechanism and biochemical means of this novel UDG. In this study, we have established that MjUDG and its homologous proteins could be categorized under the new family of UDGs, the MjUDG family.
Acknowledgments
ACKNOWLEDGEMENTS
This study was supported by a grant of the Korea Health Research Foundation 21 R&D Project, Ministry of Health and Welfare, Republic of Korea (00-PJ3-PG6-GN01-0001).
REFERENCES
- 1.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham R.P. (1997) DNA glycosylases. Mutat. Res., 383, 189–196. [DOI] [PubMed] [Google Scholar]
- 3.Krokan H.E., Standal,R. and Slupphaug,G. (1997) DNA glycosylases in the base excision repair of DNA. Biochem. J., 325, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindahl T. (1974) An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl Acad. Sci. USA, 71, 3649–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savva R., McAuley-Hecht,K., Brown,T. and Pearl,L.H. (1995) The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature, 373, 487–493. [DOI] [PubMed] [Google Scholar]
- 6.Percival K.J., Klein,M.B. and Burgers,P.M. (1989) Molecular cloning and primary structure of the uracil-DNA glycosylase gene from Saccharomyces cerevisiae. J. Biol. Chem., 264, 2593–2598. [PubMed] [Google Scholar]
- 7.Slupphaug G., Markussen,F.H., Olsen,L.C., Aasland,R., Aarsaether,N., Bakke,O., Krokan,H.E. and Helland,D.E. (1993) Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the same gene. Nucleic Acids Res., 21, 2579–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearl L.H. (2000) Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res., 460, 165–181. [DOI] [PubMed] [Google Scholar]
- 9.Sartori A.A., Fitz-Gibbon,S., Yang,H., Miller,J.H. and Jiricny,J. (2002) A novel uracil-DNA glycosylase with broad substrate specificity and an unusual active site. EMBO J., 21, 3182–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang H., Fitz-Gibbon,S., Marcotte,E.M., Tai,J.H., Hyman,E.C. and Miller,J.H. (2000) Characterization of a thermostable DNA glycosylase specific for U/G and T/G mismatches from the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Bacteriol., 182, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Putnam C.D., Shroyer,M.J., Lundquist,A.J., Mol,C.D., Arvai,A.S., Mosbaugh,D.W. and Tainer,J.A. (1999) Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol., 287, 331–346. [DOI] [PubMed] [Google Scholar]
- 12.Mol C.D., Arvai,A.S., Slupphaug,G., Kavli,B., Alseth,I., Krokan,H.E. and Tainer,J.A. (1995) Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell, 80, 869–878. [DOI] [PubMed] [Google Scholar]
- 13.Gallinari P. and Jiricny,J. (1996) A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature, 383, 735–738. [DOI] [PubMed] [Google Scholar]
- 14.Nedderman P. and Jiricny,J. (1993) The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem., 268, 21218–21224. [PubMed] [Google Scholar]
- 15.Haushalter K.A., Stukenberg,P.T., Kirschner,M.W. and Verdine,G.L. (1999) Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol., 9, 174–185. [DOI] [PubMed] [Google Scholar]
- 16.Sandigursky M. and Franklin,W.A. (1999) Thermostable uracil-DNA glycosylase from Thermotoga maritima, a member of a novel class of DNA repair enzymes. Curr. Biol., 9, 531–534. [DOI] [PubMed] [Google Scholar]
- 17.Sandigursky M. and Franklin,W.A. (2000) Uracil-DNA glycosylase in the extreme thermophile Archaeoglobus fulgidus. J. Biol. Chem., 275, 19146–19149. [DOI] [PubMed] [Google Scholar]
- 18.Hinks J.A., Evans,M.C., De Miguel,Y., Sartori,A.A., Jiricny,J. and Pearl,L.H. (2002) An iron–sulfur cluster in the family 4 uracil-DNA glycosylases. J. Biol. Chem., 277, 16936–16940. [DOI] [PubMed] [Google Scholar]
- 19.Starkuviene V. and Fritz,H.J. (2002) A novel type of uracil-DNA glycosylase mediating repair of hydrolytic DNA damage in the extremely thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res., 30, 2097–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koulis A., Cowan,D.A., Pearl,L.H. and Savva,R. (1996) Uracil-DNA glycosylase activities in hyperthermophilic micro-organisms. FEMS Microbiol. Lett., 143, 267–271. [DOI] [PubMed] [Google Scholar]
- 21.Bult C.J., White,O., Olsen,G.J., Zhou,L., Fleischmann,R.D., Sutton,G.G., Blake,J.A., FitzGerald,L.M., Clayton,R.A., Gocayne,J.D. et al. (1996) Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science, 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- 22.Lindahl T. and Nyberg,B. (1974) Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry, 13, 3405–3410. [DOI] [PubMed] [Google Scholar]
- 23.Jones W.J., Leigh,J.A., Mayer,F., Woese,C.R. and Wolfe,R.S. (1993) Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch. Microbiol., 136, 254–261. [Google Scholar]
- 24.Friedmann T. and Brown,D.M. (1978) Base-specific reactions useful for DNA sequencing: methylene blue-sensitized photooxidation of guanine and osmium tetraoxide modification of thymine. Nucleic Acids Res., 5, 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page R.D.M. (1996) TreeView: an application to display phylogenetic trees on personal computers. CABIOS, 12, 357–358. [DOI] [PubMed] [Google Scholar]
- 28.Ide H. (2001) DNA substrates containing defined oxidative base lesions and their application to study substrate specificities of base excision repair enzymes. Prog. Nucleic Acid Res. Mol. Biol., 68, 207–221. [DOI] [PubMed] [Google Scholar]
- 29.Parikh S.S., Putnam,C.D. and Tainer,J.A. (2000) Lessons learned from structural results on uracil-DNA glycosylase. Mutat. Res., 460, 183–199. [DOI] [PubMed] [Google Scholar]
- 30.Knaevelsrud I., Ruoff,P., Anensen,H., Klungland,A., Bjelland,S. and Birkeland,N.K. (2001) Excision of uracil from DNA by the hyperthermophilic Afung protein is dependent on the opposite base and stimulated by heat-induced transition to a more open structure. Mutat. Res., 487, 173–190. [DOI] [PubMed] [Google Scholar]
- 31.Michaels M.L., Pham,L., Cruz,C. and Miller,J.H. (1991) MutM, a protein that prevents G·C→T·A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res., 19, 3629–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tchou J., Kasai,H., Shibutani,S., Chung,M.H., Laval,J., Grollman,A.P. and Nishimura,S. (1991) 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl Acad. Sci. USA, 88, 4690–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deckert G., Warren,P.V., Gaasterland,T., Young,W.G., Lenox,A.L., Graham,D.E., Overbeek,R., Snead,M.A., Keller,M., Aujay,M. et al. (1998) The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature, 392, 353–358. [DOI] [PubMed] [Google Scholar]
- 34.Altschul S.F., Gish,W., Miller,W., Meyers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 35.Aravind L. and Koonin,E.V. (2000) The α/β fold uracil-DNA glycosylases: a common origin with diverse fates. Genome Biol., 1, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thayer M.M., Ahern,H., Xing,D., Cunningham,R.P. and Tainer,J.A. (1995) Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J., 14, 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu A.L., Li,X., Gu,Y., Wright,P.M. and Chang,D.Y. (2001) Repair of oxidative DNA damage: mechanisms and functions. Cell Biochem. Biophys., 35, 141–170. [DOI] [PubMed] [Google Scholar]
- 38.Horst J.P. and Fritz,H.J. (1996) Counteracting the mutagenic effect of hydrolytic deamination of DNA 5-methylcytosine residues at high temperature: DNA mismatch N-glycosylase Mig.Mth of the thermophilic archaeon Methanobacterium thermoautotrophicum THF. EMBO J., 15, 5459–5469. [PMC free article] [PubMed] [Google Scholar]
- 39.Mol C.D., Arvai,A.S., Begley,T.J., Cunningham,R.P. and Tainer,J.A. (2002) Structure and activity of a thermostable thymine-DNA glycosylase: evidence for base twisting to remove mismatched normal DNA bases. J. Mol. Biol., 315, 373–384. [DOI] [PubMed] [Google Scholar]
- 40.Matsumoto Y., Zhang,Q.M., Takao,M., Yasui,A. and Yonei,S. (2001) Escherichia coli Nth and human hNTH1 DNA glycosylases are involved in removal of 8-oxoguanine from 8-oxoguanine/guanine mispairs in DNA. Nucleic Acids Res., 29, 1975–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]