Abstract
The genome of Chlamydophila caviae (formerly Chlamydia psittaci, GPIC isolate) (1 173 390 nt with a plasmid of 7966 nt) was determined, representing the fourth species with a complete genome sequence from the Chlamydiaceae family of obligate intracellular bacterial pathogens. Of 1009 annotated genes, 798 were conserved in all three other completed Chlamydiaceae genomes. The C.caviae genome contains 68 genes that lack orthologs in any other completed chlamydial genomes, including tryptophan and thiamine biosynthesis determinants and a ribose-phosphate pyrophosphokinase, the product of the prsA gene. Notable amongst these was a novel member of the virulence-associated invasin/intimin family (IIF) of Gram-negative bacteria. Intriguingly, two authentic frameshift mutations in the ORF indicate that this gene is not functional. Many of the unique genes are found in the replication termination region (RTR or plasticity zone), an area of frequent symmetrical inversion events around the replication terminus shown to be a hotspot for genome variation in previous genome sequencing studies. In C.caviae, the RTR includes several loci of particular interest including a large toxin gene and evidence of ancestral insertion(s) of a bacteriophage. This toxin gene, not present in Chlamydia pneumoniae, is a member of the YopT effector family of type III-secreted cysteine proteases. One gene cluster (guaBA-add) in the RTR is much more similar to orthologs in Chlamydia muridarum than those in the phylogenetically closest species C.pneumoniae, suggesting the possibility of horizontal transfer of genes between the rodent-associated Chlamydiae. With most genes observed in the other chlamydial genomes represented, C.caviae provides a good model for the Chlamydiaceae and a point of comparison against the human atherosclerosis-associated C.pneumoniae. This crucial addition to the set of completed Chlamydiaceae genome sequences is enabling dissection of the roles played by niche-specific genes in these important bacterial pathogens.
INTRODUCTION
Genome sequencing of the Chlamydiaceae, a distinctive Gram-negative family of uncertain evolutionary origin, has demonstrated that profound differences in host range and disease process can be produced by subtle variations in gene content. Recently divided into two genera, Chlamydia and Chlamydophila, by 16S rRNA phylogeny (1), the Chlamydiaceae cause ubiquitous infection and disease in humans and animals (2). Chlamydia trachomatis is the agent of chronic genital and ocular infections whose sequelae include pelvic inflammatory disease, ectopic pregnancy and infertility in women, and trachoma, the major cause of infectious blindness. The more recently characterized Chlamydia pneumoniae is a prevalent cause of community-acquired pneumonia, bronchitis and pharyngitis (3–5). Disseminated and chronic C.pneumoniae infections have been associated with high morbidity/high mortality diseases with significant public health impact, such as chronic obstructive pulmonary disease, lung cancer, stroke and coronary heart disease (6–9). In particular, serologic, ultrastructural and molecular data have revealed an association between C.pneumoniae infection and atherosclerosis, the number one cause of fatal disease in humans (10). Chlamydial infection is also widely distributed in many wild and domesticated animal species (11) with relatively rare, but potentially severe zoonotic disease (12).
In contrast to the observed host diversity, wide tissue tropism and varied disease pathology, the Chlamydiaceae are united by the relative uniformity of their infectious processes, whereby members survive and multiply within eukaryotic cells via a unique developmental cycle (13). A metabolically dormant form termed the elementary body (EB), specialized for infection and dissemination, defines the start and end of chlamydial development. The metabolically active form, termed the reticulate body (RB), is a ‘regular’ replicating bacterium, with the caveat that it resides strictly inside a membrane-bound vacuole within the cytosol of the infected eukaryotic cell. Transition from one form to the other involves major structural rearrangements involving developmentally regulated genes.
The recent availability of six Chlamydiaceae genome sequences (14–17), representing three species (C.trachomatis, Chlamydia muridarum and C.pneumoniae) has considerably refined and redefined our understanding of chlamydial biology (18,19) (Table 1). The genome sequence of C.trachomatis serovar D first highlighted the compact nature and plural origin of the Chlamydia genome, revealing important new features. Notably, a large polymorphic membrane protein multigene family (pmp), the full complement of genes required for a functional type III secretion system, and genes encoding a predicted unique peptidoglycan layer have all captured the imagination of the research community. Each new genome sequence has a high degree of conservation, both in gene order and in composition. This conservation underlines the constraints of the uniquely adapted evolutionary path and developmental life style of the Chlamydiaceae.
Table 1. General features of chlamydial genomes.
| C.caviae | C.muridarum | C.trachomatis (serovar D) | C.pneumoniae (AR39) | |
|---|---|---|---|---|
| Chromosome (nt) | 1 173 390 | 1 072 950 | 1 042 519 | 1 229 858 |
| Plasmid/*phage (nt) | 7966 | 7501 | 7493 | *4524 |
| GC (%) | 39.2 | 40.3 | 41.3 | 40.6 |
| Total ORFs | 1009 | 921 | 894 | 1130 |
| Conserved hypothetical (%) | 320 (31.7) | 281 (30.5) | nda | 285 (25.2) |
| Hypothetical (%) | 84 (8.3) | 77 (8.3) | nd | 263 (23.2)b |
| tRNAs | 38 | 37 | 37 | 38 |
| rRNA operons | 1 | 2 | 2 | 1 |
| Amino acid biosynthesis | 19 | 15 | nd | 14 |
| Purine, pyrimidine, nucleoside and nucleotide metabolism | 17 | 15 | nd | 16 |
| Fatty acid and phospholipid metabolism | 22 | 24 | nd | 24 |
| Biosynthesis of cofactors, prosthetic groups and carriers | 41 | 36 | nd | 41 |
| Central intermediary metabolism | 12 | 12 | nd | 13 |
| Energy metabolism | 60 | 64 | nd | 65 |
| Transport and binding proteins | 61 | 58 | nd | 61 |
| DNA metabolism | 56 | 53 | nd | 51 |
| Transcription | 22 | 23 | nd | 23 |
| Protein synthesis | 100 | 100 | nd | 101 |
| Protein fate | 49 | 58 | nd | 63 |
| Regulatory functions | 18 | 15 | nd | 15 |
| Cell envelope | 54 | 43 | nd | 61 |
| Cellular processes | 45 | 36 | nd | 35 |
| Other | 0 | 0 | nd | 2 |
aTIGR role categories are not assigned to genomes not sequenced at TIGR.
bHigher as C.pneumoniae was annotated before C.caviae.
In spite of systematic experimental difficulties, such as the lack of a genetic system, one of the strengths of chlamydial research lies in the wealth of animal model systems. Infections of the mouse lung or genital tract by C.muridarum are well established models for C.trachomatis infection of humans. However, although such models are particularly useful for the study of specific immune parameters of infection, there is little resemblance to either the disease process or associated pathology in humans. In contrast, C.caviae, the agent of guinea pig inclusion conjunctivitis (GPIC), despite being phylogenetically distant, provides an excellent model for naturally occurring C.trachomatis infection and disease in humans. This is highlighted by the similarity of the mechanisms of transmission (e.g. sexual), chronic immune-mediated disease progression (e.g. pannus formation and tubal salpingitis during ocular and female genital infections, respectively), and highly similar pathologic endpoints (e.g. corneal damage and tubal blockage) (20). Chlamydophila caviae is also extensively studied with regard to cellular and molecular pathogenesis. Given these similarities and the demonstrated utility of additional chlamydial genome sequences (15,16), we have undertaken to obtain the complete genome sequence of C.caviae. This new genome represents the fourth species sequenced within the family, establishing the Chlamydiaceae as a leading system for the study of the evolution of bacterial pathogens through comparative genomics.
MATERIALS AND METHODS
Library preparation and random sequencing of C.caviae
Chlamydophila caviae GPIC was provided by Dr Roger Rank. The organism was propagated in HeLa 229 cells. EBs were harvested and purified by step gradient density centrifugation. Purified EBs were lysed with 10% SDS and proteinase K. The DNA was extracted twice with buffered phenol and once with 25:24:1 phenol:chloroform:isoamyl alcohol and precipitated with alcohol.
Cloning, sequencing and assembly were as described previously for genomes sequenced by TIGR (21–24). One small-insert plasmid library (1.5–2.5 kb) was generated by random mechanical shearing of genomic DNA. One large-insert library was generated by partial Tsp5091 digestion and ligation to the λ-DASHII/EcoRI vector (Stratagene). In the initial random sequencing phase, ∼8-fold sequence coverage was achieved with 16 523 sequences (average read length 581 bases). The plasmid and λ sequences were jointly assembled using TIGR Assembler. Sequences from both ends of 423 λ clones served as a genome scaffold, verifying the orientation, order and integrity of the contigs. Sequence gaps were closed by editing the ends of sequence traces and/or primer walking on plasmid clones. Physical gaps were closed by direct sequencing of genomic DNA or combinatorial PCR followed by sequencing of the PCR product. The final genome sequence is based on 19 962 sequences.
ORF prediction and gene family identification
An initial set of ORFs likely to encode proteins was identified by GLIMMER 2.0 (25) and those shorter than 30 codons eliminated. ORFs that overlapped were visually inspected and, in some cases, removed. ORFs were searched against a non-redundant protein database as previously described (21–24). Frameshifts and point mutations were detected and corrected where appropriate as described previously. Remaining frameshifts and point mutations are considered authentic and corresponding regions were annotated as ‘authentic frameshift’ or ‘authentic point mutation’, respectively. Annotation was completed using the methodology described previously (22). Two sets of hidden Markov models (HMMs) were used to determine ORF membership in families and superfamilies. These included 721 HMMS from Pfam v2.0 and 631 HMMS from the TIGR ortholog resource. TopPred46 was used to identify membrane-spanning domains in proteins.
Comparative genomics
For each C.caviae protein, we obtained BLAST P-values for the alignment against itself and the most similar protein in each of C.pneumoniae and C.muridarum. These P-values were converted using a –ln function and the BLAST score ratio (BSR) obtained for the self-alignment versus each genome.
The Chlamydiaceae genomes were rotated based on the results of GC skew analysis (26) so that the first base was near the hemB genes. All genes and predicted proteins from each genome, as well as from all other completed genomes, were compared using BLAST. For determination of the presence and absence of particular genes in each Chlamydiaceae genome, protein comparisons were used to better detect distantly related homologs. A gene was considered absent from a genome if there was no match to that gene with a P-value <10–5. For comparisons of chromosome organization between two genomes, gene (i.e. DNA) comparisons were used. Each gene in species 1 was paired with its most similar gene (as measured by P-value) in species 2. Frameshifts and small unique ORFs (<30 amino acids) were excluded from the analysis. For the identification of recent gene duplications, all genes from all chlamydial genomes were compared to each other. A gene was considered recently duplicated if the most similar gene (as measured by P-value) was another gene within the same genome (relative to genes from the two other genomes).
Conservation of gene order was established using position effect analysis (http://www.tigr.org/software), where matches between chlamydial genes were calculated using BLASTP with a cutoff E-value of 10–15. The query and hit gene from each match were defined as anchor points in gene sets composed of adjacent genes. Up to 10 genes upstream and downstream from each anchor gene were used in creating the gene set. An optimal alignment was calculated between the ordered gene sets using BLASTP percent similarity scores and a linear gap penalty. Low-scoring alignments with a cumulative percent similarity <100 were not used. Each optimal alignment provided a list of matching genes with conserved order between chlamydiae.
Database submission
The nucleotide sequence of the whole genome of C.caviae was submitted to GenBank under accession numbers AE015925 (chromosome) and AE015926 (plasmid).
RESULTS
Niche-specific genes in C.caviae
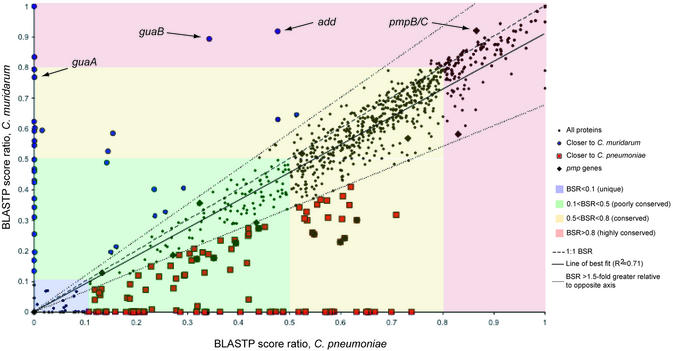
The C.caviae genome sequence (1 173 390 nt, with a plasmid of 7966 nt) was determined using a random shotgun approach (16,21). The distribution of conservation of C.caviae proteins was visualized by comparing BSR obtained from C.muridarum and C.pneumoniae orthologs (Fig. 1). Sub stituting C.trachomatis for C.muridarum on the y-axis yields a similar plot (data not shown), owing to the almost identical gene content of the two genomes. The line of best fit in Figure 1 (R2 = 0.71) indicates a bias for C.caviae proteins to be more similar to those in C.pneumoniae versus C.trachomatis and C.muridarum. This is consistent with the predicted phylogenetic relationship. The BSR plot also identifies proteins with an unusual relationship of similarity between genomes, as indicated by the guaBA and add genes. In this case, this may be because of recent transfer of genes into C.caviae from a Chlamydia sp. strain (discussed below).
Figure 1.
Plot of BSRs of C.caviae proteins against C.pneumoniae and C.muridarum. For each predicted C.caviae protein, BLASTP P-values against itself and for the most similar proteins in C.pneumoniae and C.muridarum are obtained. The P-values are converted using a –ln function and the BSR calculated using the C.caviae self-alignment value. Each ratio pair (C.pneumoniae:C.caviae and C.muridarum:C.caviae) is then plotted for each C.caviae gene. Certain classes of similarity are highlighted—regions of the plot with highly conserved proteins (BSR ≥ 0.8) in red, conserved (0.5 < BSR < 0.8) in yellow, poorly conserved in green (0.1 < BSR < 0.5) and non-conserved (unique) in blue (BSR <0.1). Where the ratio of the score for a gene in one genome was at least 1.5-fold greater than the other (as delineated by the dotted lines), proteins have been marked as red squares (closer to C.pneumoniae) or blue circles (closer to C.muridarum). The pmp proteins are also highlighted (black diamond), showing their wide distribution of similarity and particularly pmpB/C, unusual for its high conservation in other chlamydial genomes. The positions of members of the guaBA-add gene cluster, found in the C.caviae replication termination region, are also indicated.
The BSR plot (Fig. 1) combined with position effect analysis (see Materials and Methods) was further used to categorize the C.caviae proteins by their conservation in other chlamydial genomes (BSR > 0.1; summarized in Fig. 2 and Table S1A–D). Approximately three-quarters of C.caviae genes encoded a set of functions conserved across the four chlamydial species with complete genomes. The remaining ORFs appeared to encode ‘niche-specific’ functions. We define niche specificity as those functions that arise from unique genes (or gene combinations) that are presumably necessary for survival and virulence of chlamydiae in specific sites or hosts. A proportion of C.caviae genes had no orthologs in other Chlamydiaceae (Fig. 1, BSR < 0.1; Table S1A). Some were conserved in C.pneumoniae (Fig. 1, red squares closer to x-axis) or C.muridarum (Fig. 1, blue circles closer to y-axis).
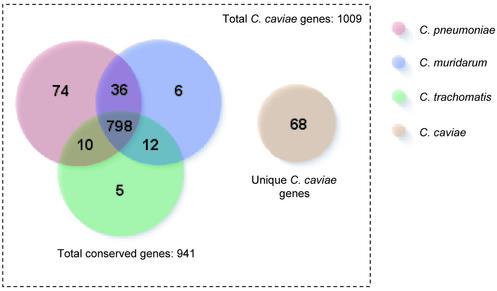
Figure 2.
Breakdown of orthologous C.caviae genes in other Chlamydiaceae genomes. Numbers represent a count of the most similar genes in particular genomes to C.caviae.
Sixty-eight predicted genes in C.caviae have no orthologs in other Chlamydiaceae genomes (Fig. 2 and Table S1A). Several genes within this unique group encode potential virulence determinants such as kynureninase and tryptophan biosynthesis proteins. A novel ABC transporter not present in the other sequenced chlamydial species (CCA00626-0628), distantly related to the toluene tolerance system from Pseudomonas putida (27), has also been identified. Nevertheless, for all the unique genes in C.caviae, there is no evidence typical of recent horizontal acquisition from non-Chlamydiaceae, such as altered gene content or trinucleotide composition, flanking IS elements, or inverted repeats.
Of 1009 annotated C.caviae genes, 798 had orthologs in all three other completed Chlamydiaceae species (Fig. 2); furthermore, 787 of these orthologs formed perfect clusters of orthologous groups (28). These most likely represent the ‘minimal gene content’ required for the developmental cycle and intracellular survival of the Chlamydiaceae. Genes for essential functions such as DNA replication, structural components of the type III secretion system necessary for host interaction as well as numerous genes of unknown function are found within this set. Only three of the 103 highly conserved genes (BSR > 0.8; Fig. 1 and Table S1B) are hypothetical, while 62 of 115 0.1 < BSR < 0.5 genes are in this group. Moreover, the C.caviae genes conserved in the other chlamydial genomes are functionally and phylogenetically distinct from non-chlamydial organisms. Comparing the 798 conserved gene products by BLASTP against the protein sets of 70 published complete genomes in the TIGR Comprehensive Microbial Resource (http://www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl) showed 183 had no match to any genome (below a threshold E-value of 10–5). In the remaining conserved proteins, similarity to other genomes was widely spread with no one genome outstanding, reflecting the evolutionary separation of the Chlamydiaceae (29).
The C.caviae niche-specific genes may encode functions not absolutely required for the primary cellular activities that define the Chlamydiaceae. The largest set of conserved genes shared exclusively with another chlamydial genome were those shared with C.pneumoniae (Fig. 2 and Table S1C), reflecting the closer phylogenetic relationship and potentially encapsulating any differences between Chlamydia and Chlamydophila. Orotate phosphoribosyltransferase (CCA00132; Table S1C), involved in pyrimidine biosynthesis, and DNA-3-methyladenine glycosylase (CCA00239; Table S1C) involved in DNA repair are identified within this set, however the majority of the remainder are hypothetical genes. A smaller number of C.caviae genes are shared with C.trachomatis and/or C.muridarum but not C.pneumoniae (Fig. 2 and Table S1D). In most cases, it is likely this distribution arose from loss of orthologous genes by deletion from the C.pneumoniae genome.
A number of genes found in the other Chlamydiaceae genomes have no significant match to C.caviae genes, although many of these are small hypothetical ORFs with low confidence of being real genes. Of 141 chlamydial gene products >100 amino acids in length with no apparent orthologs in C.caviae, only five have a known function (Table 2). One is a helicase found in C.muridarum, two are genes involved in pyrimidine nucleotide scavenging (udk and upp) and two are unique biotin biosynthesis genes in the replication termination region (RTR) of C.pneumoniae. Nonetheless, the finding that so many of the functionally assigned Chlamydiaceae genes are represented in the C.caviae genome underscores its value as a model organism for chlamydial infections.
Table 2. Genes encoding proteins of known function not found in C.caviae but found in the other three completely sequenced Chlamydiaceae species (P < 1 × 10–5).
| Name | Description |
|---|---|
| TC0602 | Helicase, putative |
| TC0833 | Uracil phosphoribosyltransferase (upp) |
| CP0011 | Uridine kinase (udk) |
| CP0613 | KDO-transferase 2, putative |
| CP0615 | KDO-transferase 2 |
| CP0808 | Biotin synthase (bioB) |
| CP0810 | Dethiobiotin synthase BioD, putative |
| CPA0002 | Capsid protein VP2-related protein |
| CT229 | Nonmuscle myosin heavy chain B |
| CT813 | MFP1 protein |
TC, C.muridarum; CP, C.pneumoniae; CT, C.trachomatis.
Comparative genomics
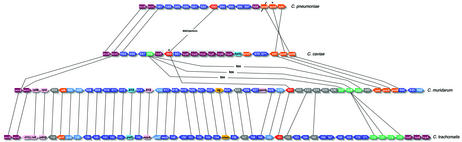
The availability of the complete genome sequences from multiple chlamydial species enables unprecedented in-depth comparative analyses. In addition to shedding light on the Chlamydiaceae, these sequences also provide information about the roles of genes in the evolution of bacterial genomes in general. Dot-plot comparison (30) of the genome sequences of C.pneumoniae, C.trachomatis and C.muridarum showed the presence of symmetrical inversions around the origins of replication and the putative termination region (Fig. 3). This region was previously termed the ‘plasticity zone’ (16), however this region surrounding the replication terminus is now more broadly recognized as a hot spot for genome rearrangement in prokaryotes (30,31). Thus, we use the term RTR in lieu of ‘plasticity zone’ in recognition of this broader phenomenon. However, in the context of the Chlamydiaceae, these terms should be considered interchangeable.
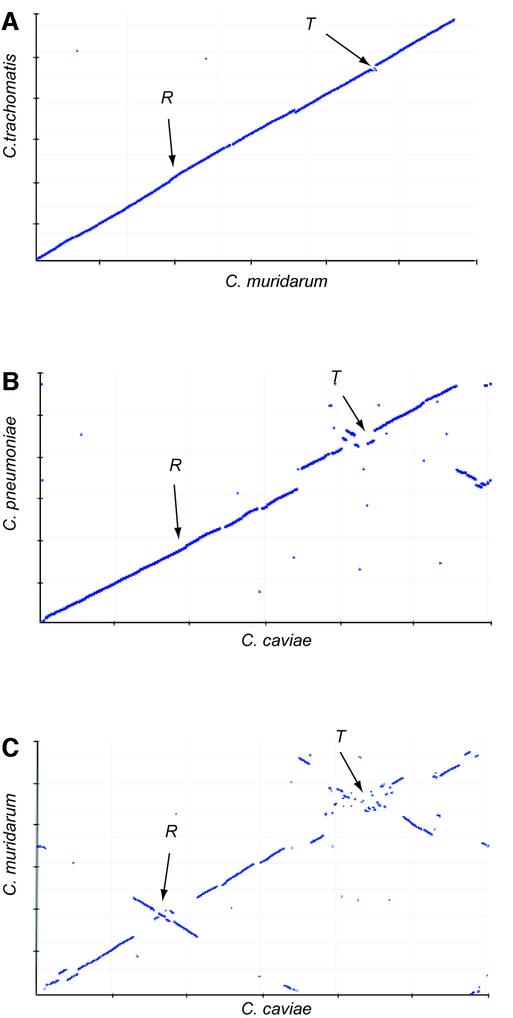
Figure 3.
Comparison of chromosomal synteny in the Chlamydiaceae. (A) Chlamydia trachomatis versus C.muridarum; (B) C.pneumoniae versus C.caviae; (C) C.muridarum versus C.caviae. Each panel is the plot of the most similar proteins (using BLASTP P-values), ordered by chromosomal position for each genome pair. Axes are marked with 200 kb graduations. R and T are the locations of the origin of replication and the termination region, respectively. Genomes have been rotated so that the replication origin is at ∼275 kb and the termination region at ∼850 kb.
The Chlamydiaceae provided good models for visualizing the roles of these inflexion points in bacterial evolution because of the low incidence of non-origin associated rearrangements, owing to the highly conserved gene content and the lack of IS elements. We plotted, by chromosome order, the position of highest similarity (judged by BLASTP E-value) between C.caviae predicted proteins and gene products of C.pneumoniae and C.muridarum (Fig. 3B and C, and Fig. 4). The comparison of C.caviae with C.muridarum produced the now familiar double-cross inversions around the inflexion points (16). However, in the C.caviae/C.pneumoniae plot, inversions were noticeable only at the terminus, not the replication origin. This plot also showed an unusual asymmetrical translocation of a gene block between approximate coordinates 600 kb in C.pneumoniae and 1100 kb in C.caviae. These dot-plot comparisons are snapshots of the progression of origin-associated recombination events between the Chlamydiaceae. The most closely related species, C.muridarum and C.trachomatis, give an almost linear plot (16) (Fig. 3A). As evolutionary time progresses, inversions accumulate more frequently around the terminus and then the replication origin.
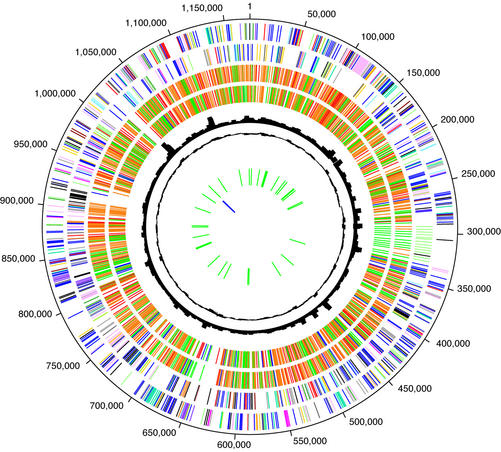
Figure 4.
Circular representation of the C.caviae chromosome. Data are from outermost circle to innermost. Circles 1 and 2: tick marks represent predicted coding sequences on the plus strand (circle 1) and minus strand (circle 2) colored by cellular role. Role categories and colors are as follows: amino acid biosynthesis, violet; biosynthesis of cofactors, prosthetic groups and carriers, light blue; cell envelope, light green; cellular processes, red; central intermediary metabolism, brown; DNA metabolism, gold; energy metabolism, light gray; fatty acid and phospholipid metabolism, magenta; protein synthesis and fate, pink; biosynthesis of purines, pyrimidines, nucleosides and nucleotides, orange; regulatory functions and signal transduction, olive; transcription, dark green; transport and binding proteins, blue–green; other categories, salmon; unknown function, gray; conserved hypothetical proteins, blue; hypothetical proteins, black. Circles 3 and 4: genes in C.caviae that are homologous to genes in C.pneumoniae (circle 3) and C.muridarum (circle 4) colored by normalized BLAST score: 0.8 < score, red; 0.5 < score < 0.8, yellow; 0.1 < score < 0.5, green; score < 0.1, blue. Circle 5: chi-square of trinucleotide skew. Circle 6: codon adaptation index (CAI) score for each gene. Circle 7: predicted tRNAs. Circle 8: predicted rRNAs.
Chlamydophila caviae niche-specific genes in the replication termination region
A region surrounding genes (tox) encoding proteins with striking similarity to EHEC adherence factor appeared particularly prone to rearrangement (Fig. 5). In C.caviae, an almost complete tryptophan biosynthesis operon (only the components of anthranilate synthase, trpE and trpG, are missing), and a guaBA-add cluster for purine nucleotide interconversion are both in this region. Together with the tryptophan operon and the tox gene, the guaB and add genes represent key differences between sequenced members of the Chlamydiaceae (Table 3). Chlamydia muridarum lacks the tryptophan operon downstream from multiple tox genes but the guaBA-add cluster is present (Fig. 5 and Table 3). The guaBA-add genes are also found in the RTRs of C.caviae and C.pneumoniae (Fig. 5) but are not in the C.trachomatis D genome (14,16). A third gene in the cluster, guaA, has an in-frame internal deletion of ∼270 bp in C.pneumoniae; however, the key C-terminal GMP synthase and the N-terminal glutamine amidotransferase domains are maintained. The nucleotide sequences of the C.caviae genes are 91–98% identical to C.muridarum but only 60–65% identical to C.pneumoniae. One possibility is that the guaBA-add cluster may have recently crossed between RTRs of the rodent-associated species C.muridarum and C.caviae. However, there is no evidence for typical flanking gene-transfer structures, such as long inverted repeats or integrases.
Figure 5.
Schematic comparing chlamydial replication termination regions with lymphostatin, tryptophan and purine biosynthesis determinants. View depicts gene order in the replication termination regions of C.pneumoniae AR39 (top; locus prefix CP0), C.caviae (middle; locus prefix CCA00) and C.muridarum (bottom; locus prefix TC0). Genes are colored according to role-category assignments (see legend to Fig. 4) and labeled either with the appropriate gene symbol or with the published locus numbers. Lines connect orthologs of predicted proteins between genomes. Elements of the C.caviae and C.trachomatis tryptophan operon are highlighted with gray shading. The guaA gene of C.pneumoniae and the MAC/perforin gene of C.caviae are marked with an asterisk (*) to denote the internal deletion (see text) and a frameshift, respectively. The truncation of the C.pneumoniae guaB gene is denoted with a diagonal line.
Table 3. Distribution of key ‘niche-specific’ genes with suspected virulence phenotypes in sequenced Chlamydiaceae genomes.
| Genome | Known cytotoxic effectors | Tryptophan metabolism | Nucleotide salvaging |
|---|---|---|---|
| C.pneumoniae AR39 | – | tph | add guaBA udk pyrE |
| C.muridarum MoPn | toxa | – | add guaBA upp |
| C.trachomatis D | toxb | trpABCR | – |
| C.caviae GPIC | toxc | trpABCDER kynU prsA tph | add guaBA pyrE |
tox, chlamydial toxin; trp, tryptophan biosynthesis; kynU, kynureninase; prsA, ribose-phosphate pyrophosphokinase; tph, tryptophan hydroxylase; add, adenosine deaminase; gua, purine biosynthesis; upp, uracil phosphororibosyl transferase; pyrE, UMP synthase; udk, uridine kinase.
aThree orthologs of C.caviae tox; each contains glucosyltransferase, UDP-glucose binding and YopT domains.
bFour ORF fragments; only CT166 contains the glucosyltransferase and UDP-glucose binding domains.
cSingle tox gene; contains glucosyltransferase, UDP-glucose binding and YopT domains.
The C.caviae genome has the most complete set of tryptophan biosynthesis determinants yet seen in the Chlamydiaceae, including several genes not previously seen in this group and, in addition, has genes involved in nucleotide scavenging and purine interconversion (Table 3). The trpABFCDR operon will not allow the first step of tryptophan biosynthesis (chorismate to anthranilate) but should allow for production of tryptophan given anthranilate (32). There is an additional copy of the trpB gene (trpB-2) adjacent to the trp operon in the C.caviae RTR—this duplicated gene has approximately the same amino acid identity to its C.caviae paralog (62%) as to the C.trachomatis trpB ortholog (65%). This suggests an ancient duplication event, or perhaps that trpB-2 is a remnant of a previously inserted trp operon.
Located immediately upstream of the trpABFCDR operon are the kynU and prsA genes, encoding kynureninase and ribose-phosphate pyrophosphokinase, respectively. These are unique to C.caviae among the Chlamydiaceae genomes sequenced to date. Phospho-ribosylpyrophosphate (PRPP), whose formation is catalyzed by ribose-phosphate pyrophosphokinase, is a key intermediate in both nucleoside and tryptophan biosynthesis. The absence of prsA in the other Chlamydiaceae genomes has been a puzzle for investigators (33). In the absence of prs, it has been suggested that indole, a readily transported tryptophan intermediate, could rescue IFN-γ-mediated tryptophan restriction of genital isolates of C.trachomatis (34). Kynureninase catalyses the conversion of kynurenin to anthranilate, an intermediate in tryptophan biosynthesis that combines with PRPP via the TrpD enzyme. Kynurenin is a product of the IFN-γ-mediated breakdown of tryptophan. The presence of kynU may offer the possibility of salvage of this essential amino acid in hostile environments and may allow the effect of the absence of the trpE/G gene product, which catalyzes the formation of anthranilate from chorismate, to be circumvented. There is no known dedicated kynurenin importer in C.caviae, however there is an aromatic amino acid transporter of unknown specificity (CCA00787), potentially responsible for uptake of this metabolite.
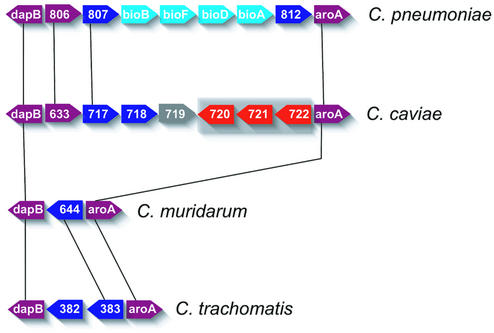
The RTR was also the site of phage DNA insertions in both C.caviae and C.pneumoniae. Free-living ssDNA microviruses have been reported infecting C.caviae, C.pneumoniae, C.psittaci and C.abortus (16,35–39)—these viruses are very similar at the nucleotide level (40). Scrambled segments of three chlamydiaphage genes were found upstream of the aroA gene (3-phosphoshikimate 1-carboxyvinyl transferase; Fig. 6) in C.caviae. The phage gene fragments in C.pneumoniae are in a different location than C.caviae; between tgt (encoding queuing tRNA ribosyl transferase) and dsbB (encoding a disulfide bond oxidoreductase) (15–17), providing strong evidence that the C.caviae and C.pneumoniae insertions were distinct events.
Figure 6.
Variable genes found between the conserved aspartate and aromatic amino acid biosynthesis operons. View depicts the region around these genes in the replication termination region of C.pneumoniae, C.caviae, C.muridarum and C.trachomatis (descending order). Lines connect orthologs between the genomes. Chlamydophila caviae genes with homology to truncated chlamydiaphage proteins are highlighted with gray shading. Genes are colored according to role-category assignments (see legend to Fig. 4) and labeled either with the appropriate gene symbol or with the published locus numbers [C.pneumoniae AR39 (locus prefix CP0), C.caviae (locus prefix CCA00), C.muridarum (locus prefix TC0) and C.trachomatis (locus prefix CT)]. The conserved genes flanking the variable segments are dihydropicolinic reductase (dapB; CCA00715) and 3-phosphoshikimate 1-carboxyvinyl transferase (aroA; CCA00723). CP0806 is a biopterin-dependent amino acid hydroxlase. CP0808-0811 form a portion of a biotin biosynthesis operon, encoding biotin synthase (bioB), 8-amino 7-oxononanoate synthase (bioF), dethiobiotin synthase (CP0810) and adenosylmethionine 8-amino 7-oxononanoate aminotransferase (bioA), respectively.
Interestingly, this region of the C.caviae RTR (with the C.caviae phage insertion sandwiched between two conserved amino acid biosynthesis operons) contained different niche-specific genes between chlamydial genomes (Fig. 6). Both C.caviae and C.pneumoniae have putative tryptophan hydroxylase genes at this location but C.pneumoniae additionally possesses a unique biotin biosynthesis operon (bioBFDA). A number of unrelated genes of unknown function are observed in this region across the sequenced chlamydial genomes. Chlamydophila caviae unique niche-specific genes involved in the biosynthesis and/or salvage of the cofactors biotin and thiamine (thiM, thiE and apbE) were found in other locations in the genome.
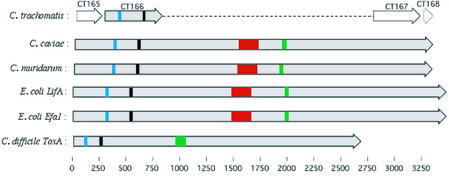
Chlamydial toxin genes
The 10038 nt CCA00558 encodes a 3346 amino acid residue product (tox) very similar to cytotoxic enterobacterial genes encoding EHEC adherence factor (Efa1), lymphostatin (LifA) and clostridial large cytotoxins (LCT). Three orthologs of tox are clustered within the RTR of C.muridarum. Truncated ORFs matching portions of the N- and C-terminal regions of tox are found within C.trachomatis serovar D (CT165–168). No tox orthologs are identifiable in C.pneumoniae.
Glycosyltransferase and UDP-glucose binding domains found in LCTs (41,42) are present in the C.caviae and C.muridarum tox orthologs and in one of the truncated C.trachomatis serovar D ORFs (CT166) (43). The glycosyltransferase activity within LCTs mono-glycosylates and thus functionally inactivate Rho GTPases leading to disassembly of the actin cytoskeleton with distinct associated cytopathic effects (42). A central hydrophobic region in LCTs, identified as a transmembrane segment likely involved in translocation into the cytosol (44), is also present in the C.caviae and C.muridarum tox genes. In contrast, the C-terminal repeats found in LCTs, involved with cell surface receptor binding (44), are not found in the chlamydial tox genes. Analysis of the cytotoxicity of C.muridarum and C.trachomatis has demonstrated cytopathic effects in epithelial cells indistinguishable from that of LCTs and thus possibly attributable to the tox genes (43). Moreover, it has recently been confirmed that interaction with an intact host actin cytoskeleton is a requirement for entry for chlamydial invasion, with substantial reorganization and remodeling of the cytoskeleton at the point of entry (45,46). The tox gene products, representing the first chlamydial proteins to be identified with probable activity against the host cell actin network, may play a role in invasion-related actin remodeling events.
The C.muridarum tox genes have also recently been identified as members of the YopT effector family of type III-secreted cysteine proteases (47). Members of this family derive from bacterial pathogens of plant or animal cells. YopT induces a cytotoxic effect in mammalian cells, characterized in vitro by disruption of the actin cytoskeleton and rounding up of the cells (48). Shao et al. (47) demonstrate that this effect arises from YopT-mediated proteolytic C-terminal cleavage of Rho GTPases. This cleavage leads to the release of the GTPases from the cell membrane with concomitant disruption of the actin cytoskeleton. The cysteine protease domain with the invariant catalytic triad (CHD) that defines this family (47) is absent in C.trachomatis but is present in the C.caviae tox gene (Fig. 7).
Figure 7.
Schematic depicting alignment of known and putative toxin genes with key domains highlighted. The relative locations of the four truncated C.trachomatis genes with homology to the N- and C-terminal regions of the C.caviae (CCA00558) and one of the three C.muridarum (TC0438) tox genes are shown. Escherichia coli LifA and Efa1, and Clostridium difficile ToxA are also shown. Blue: UDP-glucose binding domain; black: glycosyltransferase domain; red: YopT domain; green: transmembrane domain. Genes shaded gray indicate known or putative cytotoxic function.
Consequently, the proteins encoded by the tox genes of C.muridarum and C.caviae are likely to be secreted by the type III machinery and possess two distinct domains that inhibit actin polymerization by the inactivation of Rho GTPases by different strategies. It has been suggested that the presence of an actin-disrupting toxin is a key difference between C.trachomatis serovars, possibly accounting for the different sites of infection between serovars by mediating trafficking within early endosomes and thus affecting the degree of systemic dissemination (43). The presence of an additional, distinct actin-disrupting domain in the C.muridarum and C.caviae tox genes extends the hypothesis to these mucosal-restricted species, strengthening the concept that the presence (or absence) of chlamydial tox genes contributes to niche specificity (Table 3).
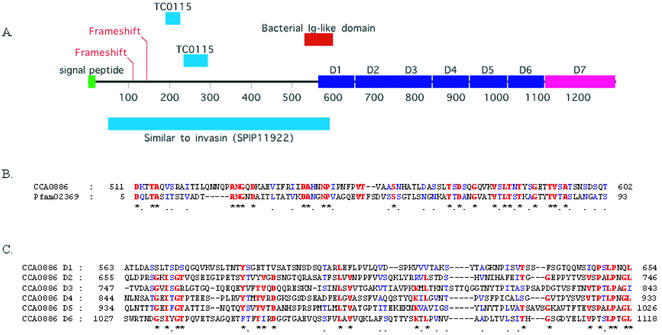
A novel member of the invasin/intimin family in C.caviae
The 3874 nt CCA00886 encodes a 1291 amino acid residue product, identified as a member of the virulence-associated invasin/intimin family (IIF) of outer membrane proteins of Gram-negative bacteria (Fig. 8). This family currently includes the prototypic yersinial invasins and intimins of enterobacterial pathogens [e.g. the Bvg phase protein BipA of Bordetella bronchoseptica (49)], and other genome sequence-revealed hypothetical genes of Salmonella enterica serotype Typhimurium (sinH), Salmonella typhi (sivH), Escherichia coli O157:H7 and K12) (50–52). However, CCA00886 is interrupted by two frameshifts early in the ORF. One of these lies within a string of 13 Gs, and may have occurred by slipped strand misrepair during replication. While authentic frameshifts are observed in other chlamydial genes (often hypothetical genes), the possible virulence function of this gene renders it particularly interesting. One interpretation is that this gene is not essential for infection of cultured HeLa cells and thus is in the process of being lost. Given the intense reductive pressures associated with the genomes of obligate intracellular pathogens (53), such inactivation of CCA00886 could be considered an evolutionarily recent event and may be evidence of long term in vitro culture altering the genomic composition of chlamydiae. This is potentially significant in light of the reliance of chlamydial research on in vitro culture.
Figure 8.
Detail of the novel C.caviae invasin-like gene. (A) Schematic indicating relative location if the key domains within the invasin predicted protein sequence. The N-terminal half is most similar to Yersinia pseudotuberculosis invasin (SP:P11922) (72). A bacterial immunoglobulin-like domain also found in invasins and intimins is indicated. The C-terminal half shows no similarity to other invasins or intimins, lacking particularly several described sequences motifs and a disulfide loop required for binding to eukaryotic β1 chain integrins (73). Nonetheless, the presence of six repeats of approximately 90 amino acids (D1–D6) in this half suggests a similar structure and function in projecting a functional domain (D7, putative) away from the chlamydial outer membrane and into the milieu. The sequence encompassing the tandem repeats D2–D6 and some of the putative C-terminal functional domain D7 display sequence homology to the tandem repeat region of Bhp of Staphylococcus epidermidis, a paralog of Bap (74), involved in biofilm formation and virulence. The positions of the frameshifts in the nucleotide sequence and the location of the subsequences with similarity to C.muridarum TC0115 (see text) are also indicated. (B) Alignment of the bacterial immunoglobulin-like domain to the Pfam consensus sequence (75). Identical and conserved residues are indicated by red and blue, respectively. (C) Alignment of the intragenic repeats D1–D6 to each other. Identical and conserved residues are indicated by red and blue, respectively.
Despite CCA00886 being absent from all other chlamydial genomes, two segments (114 and 141 bp) of the C.muridarum genome exhibit 77 and 60% identity respectively to intragenic regions of CCA00886. These two segments are located in TC0115, a small hypothetical C.muridarum ORF and represent 72% of TC0115. As with CCA00886, the C.muridarum segment lies downstream and adjacent to the NADH: ubiquinone oxidoreductase β subunit gene. Moreover, TC0115 occupies the same relative position and orientation in C.muridarum as CCA00886 in C.caviae, supporting the notion that it is a vestigial CCA00886 ortholog once present in C.muridarum. The presence of these remnants in C.muridarum supports the notion that CCA00886 is not required for in vitro cultivation of C.muridarum and C.caviae and is in the process of being lost. Further investigations of wild-type and less intensively passaged chlamydiae is required to examine this possibility, and to determine the extent and nature of gene loss or inactivation under long term in vitro culture conditions.
Variable multi-gene families
Certain chlamydial multi-gene families have orthologs conserved across the other chlamydial genomes but many exhibit short-term variability. The most numerous paralogous gene families in the Chlamydiaceae sequenced to date are the polymorphic outer membrane proteins (pmp genes), encoding an important group of surface proteins first discovered in C.caviae (54) and more recently shown to resemble autotransporters (55). In C.caviae, there are 17 pmp genes clustered in four regions, divided into six families by sequence homology. The pmpG and pmpE/F families are expanded in C.pneumoniae and C.caviae relative to the C.trachomatis and C.muridarum genomes.
Members of the pmp family stand out as major regions of variability in the otherwise very similar genomes of C.caviae and C.pneumoniae. The 17 pmp genes of C.caviae typically have a low BSR relative to their best match in other genomes compared to the norm for C.caviae (Fig. 1). This probably reflects selection for variability in response to host factors (e.g. the immune response). Highly variable poly-G tracts are frequently found upstream of the pmp genes in the genomes, suggesting a mechanism for transcriptional silencing via slip-strand repair. A significant number of pmp genes in the different Chlamydiaceae genomes also contain frameshifts. The complex relationship between pmp genes across chlamydial genomes, especially in the pmpG family could be due, in part, to frequent expansions by tandem duplications and contractions by deletion.
Many other diverged C.pneumoniae proteins fall into a broad group predicted to consist mostly of α-helices by structural prediction. In a manner analogous to the pmp gene family, these proteins appear to be evolving rapidly. Comparison of differences between the C.pneumoniae AR39, J138 and CWL029 genomes (which are >99.9% similar) found that ∼40% of ∼600 SNPs were located in two hypothetical genes encoding coiled proteins (56). The coiled nature of the proteins suggests that they may be secreted effectors of the type III secretion system (57). A recent FACS-based study found a number of these coiled hypothetical proteins to be on the surface of C.pneumoniae EBs (58).
DISCUSSION
Possibly the greatest challenge emerging from the chlamydial sequencing projects is to reconcile the observed variations in the biology of the chlamydiae with the relatively small differences in their genomes. As chlamydial genomes are so conserved in terms of content and gene order, we have focused on the few differences between genomes in order to speculate on the observed biological traits. These differences are likely to account for much of the phenotypic variation between chlamydial species, although subtle differences in expression of genes shared across the chlamydiae are also likely to play a role in this variation. The tracking and characterization of such niche-specific genes is also a credible pathway toward more efficient diagnostic tools and may offer targets for therapeutic drug development.
Chlamydophila caviae emerges from this study as a useful model for all the Chlamydiaceae pathogens examined to date. Relatively few genes found in other chlamydial species are missing from this genome. Additionally, C.caviae has good representation of pathways for biosynthesis and/or salvaging of key metabolic compounds such as nucleotides, tryptophan, thiamine and biotin. The C.caviae sequence also provides an interesting genomic comparison with the sequence from the human pathogen C.pneumoniae, however, the reasons why C.pneumoniae causes persistent and invasive infection in humans compared to C.caviae (which instead resembles the C.trachomatis mode of infection) are difficult to explain from the genome comparison alone. The presence of novel niche-specific genes within C.pneumoniae may contribute to the unique infection processes. There are 171 genes on the C.pneumoniae chromosome with no orthologs in any other Chlamydiaceae genome (BLAST score E-value ≤ 10–5). Of these, only three share similarity with functionally characterized proteins (uridine kinase and biotin biosynthesis determinants). Sequencing of C.pneumoniae strains that do not cause human infection [such as koala (59) or equine strains] may reveal that many of these 171 ‘unique’ genes in human and animal strains are shared. The C.pneumoniae sequence is unusual in that it lacks a number of apparently key niche-specific virulence determinants compared to other Chlamydiaceae, particularly in the nucleotide salvaging pathways, tryptophan biosynthesis and tox genes (Fig. 5 and Table 3). Subtle variations in the combinations of niche-specific genes, paralogous families and their expression may be the key determining features in these differences. The observation of hypothetical genes being in the fastest evolving group suggests a mechanism of adaptation by the Chlamydiaceae to meet particular requirements of their niche, for example, the host specificity of the organism. In this context, C.caviae, with a relatively large array of apparent chlamydial niche-specific genes, might have a particularly stringent natural host or may be able to occupy a range of different niches within that host.
Understanding the mechanisms of chlamydial genome variation will be vital for tracing the genetic causes of pathogenesis. The obligate intracellular lifestyle of members of the Chlamydiaceae leads to two major constraints on their evolutionary path. First, as highly adapted pathogens, there is strong conservative (purifying) selection on the maintenance of the determinants necessary for long-term survival in the host. Secondly, in the restricted evolutionary niche of the intracellular vacuole, there may be limited opportunities for uptake of extraneous genes. Given these constraints, what are the mechanisms for the variations in gene content amongst the Chlamydiaceae genomes sequenced to date?
Differences in niche-specific gene content may simply be due to lineage-specific deletion events. This parsimonious explanation of gene loss through deletional bias (i.e. ‘use it or lose it’) is widely observed in other bacteria, particularly those with pathogenic lifestyles (53,60,61). However, deletion in the absence of uptake of new genes from other Chlamydiaceae means each individual lineage would be an evolutionary dead-end if, as we predict, niche-specific genes are important in host-specific adaptations. The close nucleotide similarity of the guaBA-add cluster in C.caviae and C.muridarum found in this study provides evidence that the processes driving evolution of chlamydial genomes are more dynamic than simple lineage-specific gene loss. It is notable that some of the niche-specific genes that distinguish C.caviae from C.pneumoniae are also found in the murine C.muridarum, such as remnants of the invasin/intimin-like gene, and the guaBA-add cluster. Possibly these genes have increased importance for infection in the similar natural rodent hosts of the bacteria. The observations in this study of gene loss possibly attributable to long term in vitro culture warrants more extensive genomic investigations of wild-type chlamydiae.
Niche-specific genes are often found in chromosomal locations apparently ‘inserted’ in C.caviae (for example, Fig. 6). In the case of the biotin biosynthesis genes, different loci appear in the same relative position in the C.caviae and C.pneumoniae genomes. If chlamydiae do acquire genes horizontally from close relatives, a probable mechanism for integration into the chromosome would be recombination with DNA containing homologous flanking sequences. A potential barrier to niche-specific gene uptake by this mechanism is the mismatch repair system of the host strain (62) and the need to maintain the observed synteny—the donor DNA should have flanking homologous genes. For the Chlamydiaceae, which lack typical agents of chromosomal rearrangement such as insertion sequences, synteny appears to be a direct function of evolutionary distance (Fig. 3). The predominant force breaking synteny between chlamydiae appears to be symmetric inversion around the axes of the replication origin and the termination regions, a phenomenon that seems to be general in bacteria (31).
A further barrier to gene movement is the lack of a mechanism for entry of foreign DNA into the cell; the Chlamydiaceae have proven to be highly refractory to in vitro transformation (63). In this context, chlamydiaphage represent a possible natural conduit for gene flow. The isolation of phages from a range of Chlamydia species and strains raises the question of the possible impact of chlamydiaphage infection on chlamydial biology. Apart from a possible role of phage infection in attenuating and/or exacerbating chlamydial disease (35), chlamydiaphage may represent active genetic elements capable of exerting changes on the phenotype of their chlamydial hosts. Although not a currently recognized property of lytic phages such as φX174, phage genome integration/excision into the chlamydial genome may provide a mechanism for horizontal gene transfer between susceptible chlamydiae. This possibility is supported by evidence of insertion of phage sequences in the genomes of C.pneumoniae and C.caviae (Fig. 6).
An unexpected theme uncovered from the recent chlamydial genome sequencing projects has been the identification of a number of genes with similarity to virulence determinants in enterobacterial pathogens. These genes include YopT-like virulence factors that interfere with the host actin cytoskeleton and intimins/invasins (this study), and type III secretion systems (64,65). In the enterobacteria, these are classic virulence factors, found only in some strains and typically associated with pathogenicity islands or virulence plasmids. Chlamydial EB attachment is known to cause host cell actin reorganization; recently, the formation of pedestal-like structures and hypertrophic microvilli on the host cell surface have also been associated with EB attachment and actin reorganization (45). This mode of attachment is reminiscent of the ‘attaching and effacing’ (A/E) histopathology typified by enteropathogenic E.coli (EPEC) (66), a human pathogen that causes disease by subversion of the host cell cytoskeleton and signaling pathways through the injection of type III secreted virulence factors. The concept that chlamydiae may form A/E-like lesions upon attachment is further strengthened as the chlamydial genes identified above correspond to several of the key components critical to EPEC pathogenicity. A characteristic feature of EPEC pathogenesis—the insertion of the bacteria’s own attachment receptor into the host cell membrane [e.g. translocated intimin receptor (Tir) (67)]—has not yet been identified in chlamydia. Nevertheless, that chlamydial proteins can be exported to host cell membranes is well established with several proteins known to be secreted and subsequently associated with the inclusion membrane (68–70). Moreover, the identification of a novel chlamydial intimin/invasin family member in this study suggests a corresponding Tir-like activity may exist. Further examination of the identity and role of chlamydial proteins in host membrane structures is warranted.
This theme of virulence genes similar to enterobacterial pathogens raises the question: could the chlamydiae have been reservoirs for enterobacterial virulence genes? If this were the case, any gene transfer events would have been in the distant past, as there is little nucleotide sequence similarity between likely enterobacterial and chlamydial orthologs. The recent finding of a virus of Bdellovibrio, with strong similarity to chlamydiaphage in both gene organization and sequence (71) opens up the intriguing possibility of chlamydial genes transferred via phage to these parasitic bacteria, followed by horizontal gene transfer to the Bdellovibrio natural hosts: the Enterobacteriaceae.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENT
This work was funded by NIAID grant AI43359 to C.M.F.
REFERENCES
- 1.Everett K.D., Bush,R.M. and Andersen,A.A. (1999) Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species and standards for the identification of organisms. Int. J. Syst. Bacteriol., 49, 415–440. [DOI] [PubMed] [Google Scholar]
- 2.Schachter J. (1999) Infection and disease epidemiology. In Stephens,R.S. (ed.), Chlamydia: Intracellular Biology, Pathogenesis and Immunity. ASM Press, Washington, DC, pp. 139–169.
- 3.Grayston J.T. (1992) Chlamydia pneumoniae, strain TWAR pneumonia. Annu. Rev. Med., 43, 317–323. [DOI] [PubMed] [Google Scholar]
- 4.Kuo C.C., Jackson,L.A., Campbell,L.A. and Grayston,J.T. (1995) Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev., 8, 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahn D.L., Azenabor,A.A., Beatty,W.L. and Byrne,G.I. (2002). Chlamydia pneumoniae as a respiratory pathogen. Front Biosci., 7, e66–e76. [DOI] [PubMed] [Google Scholar]
- 6.Sethi S. and Murphy,T.F. (2001). Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev., 14, 336–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson L.A., Wang,S.P., Nazar-Stewart,V., Grayston,J.T. and Vaughan,T.L. (2000) Association of Chlamydia pneumoniae immunoglobulin A seropositivity and risk of lung cancer. Cancer Epidemiol. Biomarkers Prev., 9, 1263–1266. [PubMed] [Google Scholar]
- 8.Elkind M.S., Lin,I.F., Grayston,J.T. and Sacco,R.L. (2000) Chlamydia pneumoniae and the risk of first ischemic stroke: The Northern Manhattan Stroke Study. Stroke, 31, 1521–1525. [DOI] [PubMed] [Google Scholar]
- 9.Campbell L.A., Kuo,C.C. and Grayston,J.T. (1998) Chlamydia pneumoniae and cardiovascular disease. Emerg. Infect. Dis., 4, 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saikku P. (1999) Epidemiology of Chlamydia pneumoniae in atherosclerosis. Am. Heart J., 138, S500–S503. [DOI] [PubMed] [Google Scholar]
- 11.Fukushi H. and Hirai,K. (1993) Chlamydia pecorum—the fourth species of genus Chlamydia. Microbiol. Immunol., 37, 516–522. [PubMed] [Google Scholar]
- 12.Gregory D.W. and Schaffner,W. (1997) Psittacosis. Semin. Respir. Infect., 12, 7–11. [PubMed] [Google Scholar]
- 13.Rockey D.D. and Matsumoto,A. (1999) The chlamydial development cycle. In Brun,Y.V. and Shimkets,L.J. (eds), Prokaryotic Development. ASM Press, Washington, DC, pp. 403–425.
- 14.Stephens R.S., Kalman,S., Lammel,C., Fan,J., Marathe,R., Aravind,L., Mitchell,W., Olinger,L., Tatusov,R.L., Zhao,Q. et al. (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science, 282, 754–759. [DOI] [PubMed] [Google Scholar]
- 15.Kalman S., Mitchell,W., Marathe,R., Lammel,C., Fan,J., Hyman,R.W., Olinger,L., Grimwood,J., Davis,R.W. and Stephens,R.S. (1999) Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nature Genet., 21, 385–389. [DOI] [PubMed] [Google Scholar]
- 16.Read T.D., Brunham,R.C., Shen,C., Gill,S.R., Heidelberg,J.F., White,O., Hickey,E.K., Peterson,J., Utterback,T., Berry,K. et al. (2000) Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res., 28, 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirai M., Hirakawa,H., Kimoto,M., Tabuchi,M., Kishi,F., Ouchi,K., Shiba,T., Ishii,K., Hattori,M., Kuhara,S. et al. (2000) Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res., 28, 2311–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephens R.S. (1999) Genomic autobiographies of Chlamydiae. In Stephens,R.S. (ed.), Chlamydia: Intracellular Biology, Pathogenesis and Immunity. ASM Press, Washington, DC, pp. 9–27.
- 19.Hatch T. (1998) Chlamydia: old ideas crushed, new mysteries bared. Science, 282, 638–639. [DOI] [PubMed] [Google Scholar]
- 20.Rank R.G. and Sanders,M.M. (1992) Pathogenesis of endometritis and salpingitis in a guinea pig model of chlamydial genital infection. Am. J. Pathol., 140, 927–936. [PMC free article] [PubMed] [Google Scholar]
- 21.Fleischmann R.D., Adams,M.D., White,O., Clayton,R.A., Kirkness,E.F., Kerlavage,A.R., Bult,C.J., Tomb,J.-F., Dougherty,B.A., Merrick,J.M. et al. (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science, 269, 496–512. [DOI] [PubMed] [Google Scholar]
- 22.Nelson K.E., Clayton,R.A., Gill,S.R., Gwinn,M.L., Dodson,R.J., Haft,D.H., Hickey,E.K., Peterson,J.D., Nelson,W.C., Ketchum,K.A. et al. (1999) Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature, 399, 323–329. [DOI] [PubMed] [Google Scholar]
- 23.Fraser C.M., Gocayne,J.D., White,O., Adams,M.D., Clayton,R.A., Fleischmann,R.D., Bult,C.J., Kerlavage,A.R., Sutton,G., Kelley,J.M. et al. (1995) The minimal gene complement of Mycoplasma genitalium. Science, 270, 397–403. [DOI] [PubMed] [Google Scholar]
- 24.Fraser C.M., Casjens,S., Huang,W.M., Sutton,G.G., Clayton,R., Lathigra,R., White,O., Ketchum,K.A., Dodson,R., Hickey,E.K. et al. (1997) Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature, 390, 580–586. [DOI] [PubMed] [Google Scholar]
- 25.Salzberg S.L., Delcher,A.L., Kasif,S. and White,O. (1998) Microbial gene identification using interpolated Markov models. Nucleic Acids Res., 26, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobry J.R. (1996) Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol., 13, 660–665. [DOI] [PubMed] [Google Scholar]
- 27.Kim K., Lee,S., Lee,K. and Lim,D. (1998) Isolation and characterization of toluene-sensitive mutants from the toluene-resistant bacterium Pseudomonas putida GM73. J. Bacteriol., 180, 3692–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatusov R.L., Koonin,E.V. and Lipman,D.J. (1997) A genomic perspective on protein families. Science, 278, 631–637. [DOI] [PubMed] [Google Scholar]
- 29.Tanner M.A., Harris,J.K. and Pace,N.R. (1999) Molecular phylogeny of Chlamydia and relatives. In Stephens,R.S. (ed.), Chlamydia: Intracellular Biology, Pathogenesis and Immunity. American Society for Microbiology, Washington, DC, pp. 1–8.
- 30.Eisen J.A., Heidelberg,J.F., White,O. and Salzberg,S.L. (2000) Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol., 1, research0011.1–0011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suyama M. and Bork,P. (2001) Evolution of prokaryotic gene order: genome rearrangements in closely related species. Trends Genet., 17, 10–13. [DOI] [PubMed] [Google Scholar]
- 32.Pittard A.J. (1996) Biosynthesis of the aromatic amino acids. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella, 2nd Edn. ASM Press, Washington, DC, Vol. 1, pp. 458–484.
- 33.McClarty G. (1999) Chlamydial metabolism as inferred from the complete genome sequence. In Stephens,R.S. (ed.), Chlamydia: Intracellular Biology, Pathogenesis and Immunity. American Society for Microbiology, Washington, DC, pp. 69–100.
- 34.Fehlner-Gardiner C., Roshick,C., Carlson,J.H., Hughes,S., Belland,R.J., Caldwell,H.D. and McClarty,G. (2002) Molecular basis defining human Chlamydia trachomatis tissue tropism: a possible role for tryptophan synthase. J. Biol. Chem., 13, 13. [DOI] [PubMed] [Google Scholar]
- 35.Hsia R., Ohayon,H., Gounon,P., Dautry-Varsat,A. and Bavoil,P.M. (2000) Phage infection of the obligate intracellular bacterium, Chlamydia psittaci strain guinea pig inclusion conjunctivitis. Microbes Infect., 2, 761–772. [DOI] [PubMed] [Google Scholar]
- 36.Hsia R.C., Ting,L.M. and Bavoil,P.M. (2000) Microvirus of Chlamydia psittaci strain guinea pig inclusion conjunctivitis: isolation and molecular characterization. Microbiology, 146, 1651–1660. [DOI] [PubMed] [Google Scholar]
- 37.Storey C.C., Lusher,M. and Richmond,S.J. (1989) Analysis of the complete nucleotide sequence of Chp1, a phage which infects avian Chlamydia psittaci. J. Gen. Virol., 70, 3381–3390. [DOI] [PubMed] [Google Scholar]
- 38.Storey C.C., Lusher,M., Richmond,S.J. and Bacon,J. (1989) Further characterization of a bacteriophage recovered from an avian strain of Chlamydia psittaci. J. Gen. Virol., 70, 1321–1327. [DOI] [PubMed] [Google Scholar]
- 39.Liu B.L., Everson,J.S., Fane,B., Giannikopoulou,P., Vretou,E., Lambden,P.R. and Clarke,I.N. (2000) Molecular characterization of a bacteriophage (Chp2) from Chlamydia psittaci. J. Virol., 74, 3464–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Read T.D., Fraser,C.M., Hsia,R.C. and Bavoil,P.M. (2000) Comparative analysis of Chlamydia bacteriophages reveals variation localized to a putative receptor binding domain. Microb. Comp. Genomics, 5, 223–231. [DOI] [PubMed] [Google Scholar]
- 41.Busch C., Hofmann,F., Gerhard,R. and Aktories,K. (2000) Involvement of a conserved tryptophan residue in the UDP-glucose binding of large clostridial cytotoxin glycosyltransferases. J. Biol. Chem., 275, 13228–13234. [DOI] [PubMed] [Google Scholar]
- 42.Busch C., Hofmann,F., Selzer,J., Munro,S., Jeckel,D. and Aktories,K. (1998) A common motif of eukaryotic glycosyltransferases is essential for the enzyme activity of large clostridial cytotoxins. J. Biol. Chem., 273, 19566–19572. [DOI] [PubMed] [Google Scholar]
- 43.Belland R.J., Scidmore,M.A., Crane,D.D., Hogan,D.M., Whitmire,W., McClarty,G. and Caldwell,H.D. (2001) Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl Acad. Sci. USA, 98, 13984–13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richard J.F., Petit,L., Gibert,M., Marvaud,J.C., Bouchaud,C. and Popoff,M.R. (1999) Bacterial toxins modifying the actin cytoskeleton. Int. Microbiol., 2, 185–194. [PubMed] [Google Scholar]
- 45.Carabeo R.A., Grieshaber,S.S., Fischer,E. and Hackstadt,T. (2002) Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect. Immun., 70, 3793–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coombes B.K. and Mahony,J.B. (2002) Identification of MEK- and phosphoinositide 3-kinase-dependent signalling as essential events during Chlamydia pneumoniae invasion of HEp2 cells. Cell Microbiol., 4, 447–460. [DOI] [PubMed] [Google Scholar]
- 47.Shao F., Merritt,P.M., Bao,Z., Innes,R.W. and Dixon,J.E. (2002) A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell, 109, 575–588. [DOI] [PubMed] [Google Scholar]
- 48.Iriarte M. and Cornelis,G.R. (1998) YopT, a new Yersinia Yop effector protein, affects the cytoskeleton of host cells. Mol. Microbiol., 29, 915–929. [DOI] [PubMed] [Google Scholar]
- 49.Stockbauer K.E., Fuchslocher,B., Miller,J.F. and Cotter,P.A. (2001). Identification and characterization of BipA, a Bordetella Bvg-intermediate phase protein. Mol. Microbiol., 39, 65–78. [DOI] [PubMed] [Google Scholar]
- 50.McClelland M., Sanderson,K.E., Spieth,J., Clifton,S.W., Latreille,P., Courtney,L., Porwollik,S., Ali,J., Dante,M., Du,F. et al. (2001) Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature, 413, 852–856. [DOI] [PubMed] [Google Scholar]
- 51.Parkhill J., Dougan,G., James,K.D., Thomson,N.R., Pickard,D., Wain,J., Churcher,C., Mungall,K.L., Bentley,S.D., Holden,M.T. et al. (2001) Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature, 413, 848–852. [DOI] [PubMed] [Google Scholar]
- 52.Frankel G., Candy,D.C., Fabiani,E., Adu-Bobie,J., Gil,S., Novakova,M., Phillips,A.D. and Dougan,G. (1995) Molecular characterization of a carboxy-terminal eukaryotic-cell-binding domain of intimin from enteropathogenic Escherichia coli. Infect. Immun., 63, 4323–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moran N.A. (2002) Microbial minimalism: genome reduction in bacterial pathogens. Cell, 108, 583–586. [DOI] [PubMed] [Google Scholar]
- 54.Cevenini R., Donati,M., Brocchi,E., De Simone,F. and La Placa,M. (1991) Partial characterization of an 89-kDa highly immunoreactive protein from Chlamydia psittaci A/22 causing ovine abortion. FEMS Microbiol. Lett., 65, 111–115. [DOI] [PubMed] [Google Scholar]
- 55.Henderson I.R. and Lam,A.C. (2001) Polymorphic proteins of Chlamydia spp—autotransporters beyond the Proteobacteria. Trends Microbiol., 9, 573–578. [DOI] [PubMed] [Google Scholar]
- 56.Daugaard L., Christiansen,G. and Birkelund,S. (2001) Characterization of a hypervariable region in the genome of Chlamydophila pneumoniae. FEMS Microbiol. Lett., 203, 241–248. [DOI] [PubMed] [Google Scholar]
- 57.Pallen M.J., Dougan,G. and Frankel,G. (1997) Coiled-coil domains in proteins secreted by type III secretion systems. Mol. Microbiol., 25, 423–425. [DOI] [PubMed] [Google Scholar]
- 58.Montigiani S., Falugi,F., Scarselli,M., Finco,O., Petracca,R., Galli,G., Mariani,M., Manetti,R., Agnusdei,M., Cevenini,R. et al. (2002) Genomic approach for analysis of surface proteins in Chlamydia pneumoniae. Infect. Immun., 70, 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Girjes A.A., Hugall,A.F., Timms,P. and Lavin,M.F. (1988) Two distinct forms of Chlamydia psittaci associated with disease and infertility in Phascolarctos cinereus (koala). Infect. Immun., 56, 1897–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andersson J.O. and Andersson,S.G. (2001) Pseudogenes, junk DNA and the dynamics of Rickettsia genomes. Mol. Biol. Evol., 18, 829–839. [DOI] [PubMed] [Google Scholar]
- 61.Mira A., Ochman,H. and Moran,N.A. (2001) Deletional bias and the evolution of bacterial genomes. Trends Genet., 17, 589–596. [DOI] [PubMed] [Google Scholar]
- 62.Matic I., Rayssiguier,C. and Radman,M. (1995) Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell, 80, 507–515. [DOI] [PubMed] [Google Scholar]
- 63.Tam J.E., Davis,C.H. and Wyrick,P.B. (1994) Expression of recombinant DNA introduced into Chlamydia trachomatis by electroporation. Can. J. Microbiol., 40, 583–591. [DOI] [PubMed] [Google Scholar]
- 64.Bavoil P.M. and Hsia,R.C. (1998) Type III secretion in Chlamydia: a case of deja vu? Mol. Microbiol., 28, 860–862. [DOI] [PubMed] [Google Scholar]
- 65.Hsia R.C., Pannekoek,Y., Ingerowski,E. and Bavoil,P.M. (1997) Type III secretion genes identify a putative virulence locus of Chlamydia. Mol. Microbiol., 25, 351–359. [DOI] [PubMed] [Google Scholar]
- 66.Vallance B.A. and Finlay,B.B. (2000) Exploitation of host cells by enteropathogenic Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 8799–8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kenny B., DeVinney,R., Stein,M., Reinscheid,D.J., Frey,E.A. and Finlay,B.B. (1997) Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell, 91, 511–520. [DOI] [PubMed] [Google Scholar]
- 68.Fields K.A. and Hackstadt,T. (2000) Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol., 38, 1048–1060. [DOI] [PubMed] [Google Scholar]
- 69.Fling S.P., Sutherland,R.A., Steele,L.N., Hess,B., D’Orazio,S.E., Maisonneuve,J., Lampe,M.F., Probst,P. and Starnbach,M.N. (2001) CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. Proc. Natl Acad. Sci. USA, 98, 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rockey D.D., Lenart,J. and Stephens,R.S. (2000) Genome sequencing and our understanding of chlamydiae. Infect. Immun., 68, 5473–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brentlinger K.L., Hafenstein,S., Novak,C.R., Fane,B.A., Borgon,R., McKenna,R. and Agbandje-McKenna,M. (2002) Microviridae, a family divided: isolation, characterization and genome sequence of phiMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovirus. J. Bacteriol., 184, 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Isberg R.R., Voorhis,D.L. and Falkow,S. (1987) Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell, 50, 769–778. [DOI] [PubMed] [Google Scholar]
- 73.Isberg R.R., Hamburger,Z. and Dersch,P. (2000) Signaling and invasin-promoted uptake via integrin receptors. Microb. Infect., 2, 793–801. [DOI] [PubMed] [Google Scholar]
- 74.Cucarella C., Solano,C., Valle,J., Amorena,B., Lasa,I. and Penades,J.R. (2001) Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol., 183, 2888–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.