Abstract
The use of Cre and FLP recombinases to analyze embryogenesis and organogenesis in Xenopus has not been applied so far. We report on the generation of transgenic Xenopus animals containing a Cre-activated reporter gene cassette expressing blue fluorescent protein that can be switched over to yellow fluorescent protein expression upon Cre-mediated recombination. By injecting Cre mRNA into the two-cell stage embryo we show that Cre-mediated activation of the yellow fluorescent protein gene occurs. In addition, we observe upon injection an extinction of blue fluorescence in animals expressing the transgene and the induction of blue fluorescence in larvae containing a silent reporter gene. By crossing the reporter strains with animals expressing a muscle-specific Cre transgene we obtained an efficient and specific recombination of the reporter gene that leads to yellow fluorescence in myotomes and myofibrils of the developing larvae. Removal of the tail tips of these larvae allows the continuous recording of muscle cell differentiation in the regenerating tail. We detect a dramatic increase in transgene expression at the site of tissue removal in the tail stump. In the regenerated tail, yellow fluorescence is restricted to the myotomes thus excluding transdifferentiation of muscle cells.
INTRODUCTION
The recent sequencing of the human and mouse genome has given a wealth of new information on genes and their structure in mammals. Now the most challenging work is to identify the regulation and function of genes and this requires efficient model organisms as functional analysis in mammals is time consuming and expensive. Clearly, the amphibian species Xenopus is an attractive model to analyze gene function in development and organogenesis of vertebrates, as embryonic development occurs outside the female and the developing larvae are highly transparent allowing an easy observation and manipulation of the organism during development (1). Furthermore, embryos are available in large amounts and need a relatively low infrastructure for breeding. The attractiveness of Xenopus has been further increased by the method to generate transgenic frogs (2,3) and several groups succeeded in establishing stable transgenic Xenopus lines transmitting transgenes to the next generations (4–7).
In mice the DNA recombinases Cre and FLP have been used most successfully for conditional manipulation of gene expression allowing the dissection of regulatory networks in an entire organism and to label cell lineages (reviewed in 8,9). The application of these techniques to Xenopus is of great importance and we started to establish the use of DNA recombination in Xenopus (10). We succeeded in showing that Cre as well as FLP recombinases recombine coinjected reporters in developing Xenopus efficiently. More importantly, we observed no sign of interference with normal Xenopus development. We now report on the use of Cre recombinase to label cell lineages in stable transgenic Xenopus.
MATERIALS AND METHODS
Plasmids
The reporter plasmid LCMV:ECFP(loxP)(FRT)EYFP is similar to the reporter construct pCMV:ECFP(FRT)EYFP (10), but contains in addition loxP sites flanking the ECFP cassette. Furthermore, we introduced by linker insertion a NotI and a SalI site at the 5′ end of the CMV promoter allowing the excision of the bacterial sequences by NotI and the replacement of the CMV promoter by SalI and HindIII digestion. The construct LCMV:ECFP(loxP)EYFP lacks the FRT sites. The full sequence data of these reporter plasmids can be obtained on request. The Cre expression vector CAR-Cre was made by exchanging the GFP gene in CAR-GFP (kindly provided by Kristen Kroll) as a HindIII NotI fragment by the corresponding Cre gene from pCSCre2. pCSCre2 was constructed by replacing the BamHI XbaI GFP sequence in the pCSGFP2 vector by the open reading frame of Cre derived as a BglII XbaI PCR fragment from pCSCre (10) using the forward primer 5′-CGGAAGATCTCCGATATGTCCAATTTACTG-3′ and the reverse primer 5′-CTGCTGGAAGATGGCG-ATGGTTCTCTAGAGCA-3′. pCSRed2 was derived by replacing the BamHI XbaI GFP fragment in pCSGFP2 with the corresponding DsRed2 fragment of pDsRed2 (Clontech).
Generating transgenic Xenopus
The protocol used previously (11) was modified by using frozen sperm nuclei (12) and omitting the egg extract to get more normal developing larvae (13). For the reporter lines C and Y the plasmids LCMV:ECFP(loxP)(FRT)EYFP and LCMV:ECFP(loxP)EYFP were used, respectively. The reporter DNA was digested with Asp700 and NotI and the NotI fragment lacking the bacterial plasmid sequence was purified. The CAR-Cre vector was linearized by NotI digestion without removing bacterial sequences. Tadpoles were grown up by feeding with sera micron (www.sera.de) and after metamorphosis the froglets were fed with a rich diet including fresh tubifex, frozen red larvae and dried gammarus (obtained from a local pet shop) as well as with living worms (Dendrobena, www.superwurm.de) and chopped pig liver.
Cre mRNA injection
For mRNA injections pCSCre2 and pCSDsRed2 were linearized with NotI and NarI, respectively, and RNA was synthesized in vitro using SP6 RNA polymerase. One nanogram of RNA was injected into one blastomere of the two-cell stage embryo as described (14). Fluorescence microscopy was done with a Leica MZ/FLIII stereomicroscope with the appropriate filters as described (10).
RESULTS
Function of Cre on stable reporter genes in double transgenic Xenopus
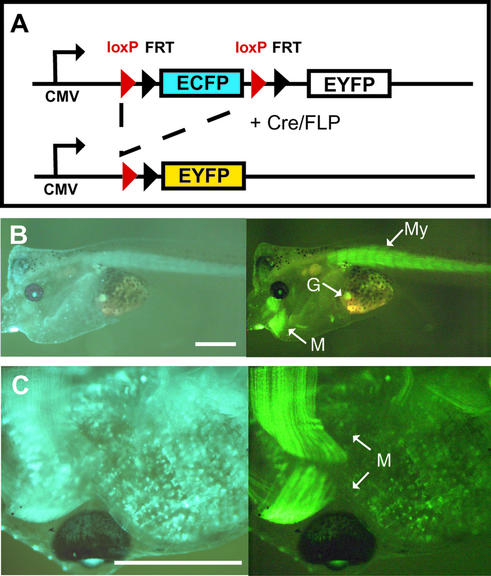
To explore whether DNA recombinases act on stable integrated transgenes in Xenopus, we generated in a single step transgenic frogs containing both a ubiquitously expressed reporter and a tissue-specifically expressed Cre recombinase. As a ubiquitously expressed reporter gene we used the construct LCMV:ECFP(loxP)(FRT)EYFP that contains the CMV promoter expressing the blue fluorescent protein ECFP. Upon Cre or FLP recombinase action the ECFP cassette is replaced by the EYFP gene encoding the yellow fluorescent protein (Fig. 1A). The yellow fluorescent protein yields a light green color that is similar to the green fluorescence of GFP, although both proteins are characterized by distinct excitation and emission spectra (10). In previous experiments we had shown that the combination of ECFP and EYFP is suitable for simultaneous observation of blue and yellow fluorescence using appropriate filters of the stereomicroscope (10). However, using the filter set to detect yellow fluorescence, bleeding through of the blue fluorescence of ECFP occurs. Therefore, we always compared the pictures obtained with the filter set for yellow and blue fluorescence throughout this analysis, thus allowing a meaningful interpretation. As Cre expression vector we used CAR-Cre that contains the coding sequence for Cre recombinase driven by the muscle-specific actin promoter (CAR). Integrating both genes into the Xenopus genome we obtained blue fluorescent Xenopus embryos that showed at later stages yellow fluorescence restricted to the muscle of the tail and the jaw (Fig. 1B). At higher magnification the myofibrils of the jaw muscle could specifically be recognized due to their strong yellow fluorescence (Fig. 1C). These results establish that Cre recombinase acts in a muscle-specific way on stable integrated reporter gene copies in these double transgenic Xenopus. However, using this approach only a limited number of healthy animals can be generated and each individual transgenic animal is distinct, as the copy number and integration sites differ. To circumvent these problems we generated separate Xenopus strains containing either the Cre-activated reporter DNA or the CAR-Cre vector as transgene.
Figure 1.
Muscle-specific activation of the EYFP gene. (A) Schematic drawing of the reporter gene LCMV:ECFP(loxP)(FRT)EYFP containing the CMV promoter driving the expression of the ECFP and EYFP genes. The ECFP gene with its polyadenylation signal is flanked by loxP and FRT sites allowing Cre and FLP recombinase-mediated deletion of the ECFP gene and thus leading to the expression of the EYFP gene. (B and C) Transgenic Xenopus generated by simultaneously introducing the reporter gene LCMV:ECFP(loxP)(FRT)EYFP and the Cre expression vector CAR-Cre that drives Cre activity by the muscle-specific actin promoter (11). (B) Lateral view of a living larvae using filters to detect blue (ECFP, left) or yellow (EYFP, right) fluorescence. (C) Detailed ventral view of another larvae using the same filters. M, myofibrils of the jaws; My, myotomes of the tail; G, gall bladder with autofluorescence. The scale bars are 1 mm. The two double transgenic animals shown were derived from an experiment using 2900 injected eggs. We obtained 118 larvae of stage 35 (20) and 51 larvae were transgenic as assayed by ECFP fluorescence. Fifteen transgenic animals reached the swimming larval stage and two showed EYFP expression in the muscle. The larvae in (B) has abnormal morphology. The use of egg extract in this experiment explains the low frequency of healthy tadpoles.
Establishment of Cre-activated reporter strains
We established two different types of reporter animals each expressing the blue fluorescent protein ECFP as a selectable marker and the yellow fluorescent protein (EYFP) as a recombinase-activated marker: the Y-animals received the transgene LCMV:ECFP(loxP)EYFP that contains the ECFP gene flanked by loxP sites and are thus targets for Cre recombinase. In contrast, the C-animals contain the construct LCMV:ECFP(loxP)(FRT)EYFP where loxP and FRT sites are flanking the ECFP gene and are thus potentially useful with Cre as well as FLP recombinases.
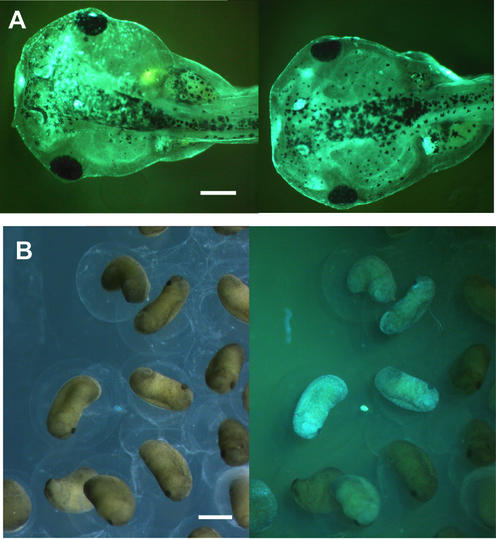
The expression of the transgene in the founder animals was established by fluorescence microscopy of the larvae and only larvae with a homogenous fluorescence as exemplified in Figure 2A were selected for growth to sexual maturity. In all cases the fluorescence disappeared more or less completely after metamorphosis, possibly reflecting the fact that the frogs are not very transparent. Five transgenic females (Y1, Y2, C3, C4 and C5) were tested for their potential to transmit the transgene to their offspring by in vitro fertilization with sperm from wild-type males. The presence of the ECFP transgene in F1 animals was analyzed by the presence of the corresponding fluorescence protein and/or the expected ECFP-encoding DNA by PCR. Whereas three females (Y2, C4 and C5) transmit an active transgene to their progeny as seen by fluorescent microscopy (see Fig. 2B for an example), two females (Y1 and C3) transmit the transgene in an inactive form that could only be detected by PCR (data not shown). In all cases no expression of the EYFP gene was observed indicating no leaky transcription of the EYFP gene in the non-induced state. This tight control is in agreement with our previous data analyzing similar constructs in transient assays in developing Xenopus embryos (10).
Figure 2.
Founder animals at larval stage and F1 embryos. (A) Blue fluorescence of two Xenopus larvae derived from a transgenic reaction with LCMV:ECFP(loxP) (FRT)EYFP. (B) Early tail bud stage embryos derived from the founder female C5 under normal light or blue fluorescence show that ∼30% of the embryos are blue. The intensity of the blue fluorescence in F1 animals differs. The scale bars are 1 mm.
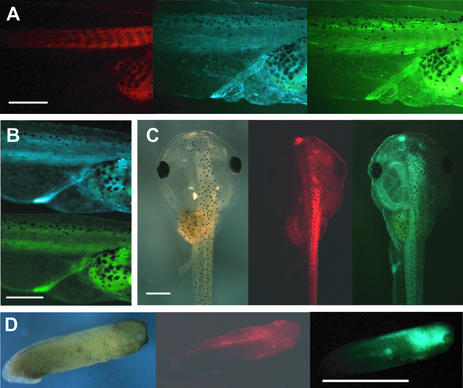
To analyze whether the transgenes transmitted through the germ line can be targeted by the recombinase, we injected Cre recombinase mRNA into two-cell stage embryos that were obtained from crossing the reporter female C5 with wild-type males. To monitor the translation of the injected Cre mRNA we coinjected DsRed2 mRNA encoding red fluorescent protein as a marker. Figure 3A illustrates in the red fluorescent part of the larvae some myotomes that are much stronger in the yellow fluorescent light than in the blue fluorescent light. However, the high background fluorescence of ECFP, resulting in bleeding through when looking for yellow fluorescence, precludes a detailed analysis. As yellow fluorescent myotomes were restricted to the region targeted by the injected RNA and were never seen in uninjected larvae (Fig. 3B), it represents a recombination of the transgene specifically mediated by Cre. In other larvae from the same batch we observed an extinction of the blue fluorescence in the injected side that was identified by the red fluorescence of the coinjected DsRed2 (Fig. 3C). A third phenotype was observed in embryos derived from C5 that lacked blue fluorescence: in these embryos a strong blue fluorescence was induced that colocalized with the DsRed2 marker (Fig. 3D). In none of these animals was yellow fluorescence present. This ‘blue induction’ phenotype was also found in injected eggs of females C3 and Y1, both of which lack any blue fluorescent F1 embryos, but transmit a silent ECFP transgene as assayed by PCR.
Figure 3.
Cre action in F1 larvae of C5 containing EYFP as an activated marker gene. Eggs of female C5 were fertilized with wild-type sperm and injected with a mixture of Cre (one part) and DsRed2 (three parts) mRNA at the two-cell stage. To get a high level of recombination we injected a high amount of Cre mRNA known to interfere with development in the tail bud stage. Injecting 364 two-cell stage embryos we obtained 42 swimming tadpoles (stage 45) (20) expressing the marker DsRed2. Pictures of living larvae are given. (A) Red, blue and yellow fluorescence of the lateral view of the middle part of an injected larvae. Twelve tadpoles showed such yellow myotomes. (B) Blue and yellow fluorescence of the lateral view of the middle part of an uninjected larvae. (C) Dorsal view of a swimming larvae in normal light as well as in red and blue fluorescence. Eight tadpoles showed this phenotype of extinction of blue fluorescence in the injected part. (D) Lateral view of a tail bud embryo in normal light as well as in red and blue fluorescence. This ‘blue-induction’ phenotype was seen in 21 animals from a sample of 30 larvae derived from injected non-fluorescent two-cell stage embryos. All these animals died before reaching the swimming tadpole stage. The scale bars are 2 mm.
These data establish that in our reporter strains the recombinase acts on the transgene transmitted through the germ line.
Crossing of reporter strains with recombinase strains proves recombinase-mediated gene activation
To explore whether a recombinase introduced as a transgene would be more efficient in recombining the transgenic reporter we established Xenopus strains expressing transgenic recombinases. To express Cre in a tissue-specific way we used the construct with the muscle actin promoter (CAR) driving the expression of Cre. The presence of the transgene was verified at late larval stages by PCR of tail DNA (data not shown). Positive larvae were grown up to obtain sexually mature CAR-Cre males.
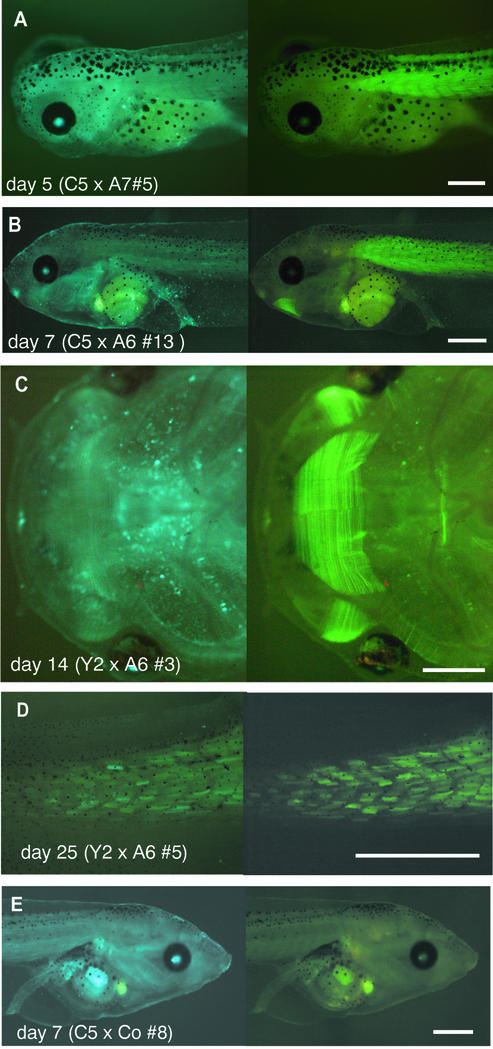
Two sexually mature males (A6 and A7) were used to prepare sperm suspensions for in vitro fertilization of the eggs from the five females known to transmit the transgene to the F1 progeny. Female C4 gave eggs of bad quality that did not develop normally thus precluding any conclusion. In crossings with females Y1 and C3, both containing a silent transgene, we failed to see any induction of fluorescent protein expression. However, we observed a Cre-mediated effect in the offspring of females Y2 and C5: analyzing blue larvae we detected yellow tail muscle with distinct myotomes in the blue general fluorescence. This yellow fluorescence of the tail muscle gradually develops with time starting at ∼5 days of development (Fig. 4A) and reaches a high intensity at day 7 (Fig. 4B) of development. At later stages, yellow fluorescence could also be seen in the jaw muscle constituting fine myofibrils that cannot be seen in normal light (Fig. 4C). As the tail of Xenopus larvae grows up to metamorphosis, there is a progressive differentiation of myotomes in an anterior to posterior direction. In the tail tip, the myotomes initially show blue fluorescence and this turns into yellow fluorescence only gradually (Fig. 4D). This allows visualization of the gradient of muscle differentiation along the tadpole tail.
Figure 4.
Muscle-specific recombination in crossings of Xenopus strains. (A–C) Eggs from reporter females containing the LCMV:ECFP (loxP)(FRT)EYFP transgene were fertilized with sperm of transgenic Cre founder males expressing Cre under the control of the muscle actin promoter. Lateral (A and B) and ventral (C) views of larvae are given. The age of the larvae and the name of the founder animals are indicated for each panel. (D) Tail tip of a 28 day larvae in the region of gradual muscle differentiation. (E) Lateral view of a larvae obtained by the crossing of the reporter founder female C5 containing the LCMV:ECFP(loxP)(FRT)EYFP transgene with a control male lacking Cre recombinase. The left and right panel show the blue and yellow fluorescence, respectively. The scale bars are 1 mm.
A quantification of the larvae showing muscle-restricted Cre action is given in Table 1. In the F1 generation of Y2 ∼50% of the larvae are blue and in the crossing with male A6 ∼20% of the blue larvae (12 from 52) showed yellow muscle in the blue general fluorescence (Table 1). In contrast, the F1 generation of C5 represents ∼30% of blue larvae and about a third of these blue larvae (8 from 28) show muscle-specific recombination (Table 1). Based on this finding we conclude that at least some of the sperm of male A6 transmit a Cre recombinase transgene that is expressed in a muscle cell-specific way. Using the sperm of male A7 only, an inefficient fertilization was observed with eggs derived from Y2 and C5. However, two of the five blue larvae obtained from C5 developed yellow fluorescence in the muscle indicating the transmission of an active Cre transgene in A7, too.
Table 1. Crossing between the transgenic males A6 or A7 containing CAR-Cre as a transgene and the reporter females Y2 or C5 containing the Cre inducible EYFP marker.
| Female | Male | ||||
|---|---|---|---|---|---|
| A6 (CAR-Cre) | A7 (CAR-Cre) | Control | |||
| Y2 | Eggs | 270 | 180 | 280 | |
| Blastulae | 119 | 5 | 155 | ||
| Larvae (day 7) | Non-blue | 49 | 3 | 67 | |
| Blue | 52 | 2 | 67 | ||
| Yellow muscle in blue | 12 | 0 | 0 | ||
| C5 | Eggs | 220 | 190 | 240 | |
| Blastulae | 132 | 22 | 112 | ||
| Larvae (day 7) | Non-blue | 59 | 14 | 46 | |
| Blue | 28 | 5 | 20 | ||
| Yellow muscle in blue | 8 | 2 | 0 | ||
The females Y2 and C5 contain the LCMV:ECFP(loxP)EYFP and LCMV:ECFP(loxP)(FRT)EYFP reporter genes as transgenes, respectively. Number and phenotypes of animals at various stages are given. At day 7, non-blue and blue larvae are given. The blue larvae include the blue larvae with yellow muscle that are given in bold numbers. The control refers to fertilizations with sperm of a male lacking the Cre transgene.
Comparing the time of appearance and the extent of yellow fluorescence in the muscle we observed a mostly homogenous effect in the various larvae. Apparently in all three successful crossings saturating levels of Cre were obtained. This contrasts with the Cre mRNA injections where only partial recombination with extensive variability between different animals was observed (Fig. 3).
As some of the blue larvae obtained by fertilization with sperm of males A6 and A7 failed to develop yellow fluorescence in the muscle, we analyzed such animals by PCR for the presence of the Cre-encoding transgene. Four non-activated blue larvae lacked the Cre-encoding DNA, whereas 11 blue larvae with yellow muscle gave a PCR fragment specific for Cre (data not shown). Therefore, we deduce that the transmission of the CAR-Cre transgene leads strictly to the recombination of the reporter gene as seen by muscle-restricted yellow fluorescence.
The effect seen in the offspring using the sperm from the CAR-Cre males A6 and A7 was specific, as no yellow muscle was seen in larvae derived from eggs fertilized with sperm of a control male lacking a Cre transgene (Fig. 4E and Table 1). In conclusion, these data establish that successful muscle-specific Cre gene activation has occurred by crossing corresponding Xenopus strains.
Muscle cell lineage labeling in the regenerating Xenopus tail
It is known that the tail tip of Xenopus larvae is regenerated upon removal (15,16). This regeneration process involves the formation of an undifferentiated blastema that gradually differentiates into the various cell types of the tail tip. It has been postulated that the cells of the blastema are recruited from undifferentiated cells as well as from differentiated cells close to the amputated region. We wondered whether in this process muscle cells are recruited to form other cell types and thus would undergo transdifferentiation.
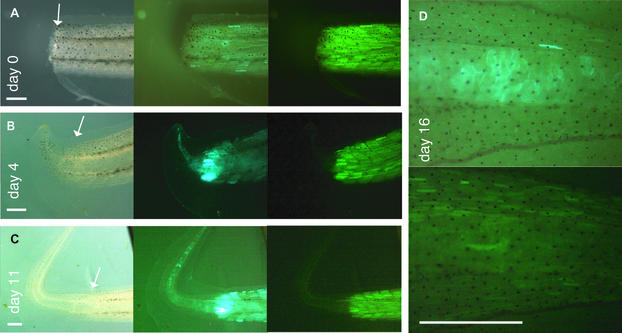
Therefore, we cut-off the tail tips of larvae with myotomes that were labeled by yellow fluorescence. The tail tip (Fig. 4D) as well as the stump of the tail (Fig. 5A) contained a mosaic pattern of blue fluorescent cells including myotomes and a more homogenous yellow fluorescence restricted to the myotomes, indicating the progressive muscle differentiation in this region. Four days after removing the tail tip we observed an intensive blue fluorescence in the zone close to tail removal indicating activation of the transgene (Fig. 5B). As this blue fluorescence was most evident in the myotomes and the yellow fluorescence of the myotomes remained essentially unchanged, we deduce that primarily nuclei with unrecombined transgenes were activated. The newly formed blastema showed blue fluorescence lacking any yellow fluorescent protein expression. This pattern of fluorescent protein expression remained stable during the following days when the regenerate continued to grow (Fig. 5C). Notably, all cells in the regenerate showed blue fluorescence at this stage of regeneration. At day 16 we observed some yellow fluorescent myotomes in the regenerate indicating muscle cell-restricted Cre activity in the differentiating myotomes, whereas the vacuolar cells of the chorda were blue (Fig. 5D). During the whole process of regeneration we could not detect any yellow fluorescent non-muscle cells in the regenerated tail. Therefore, we exclude a major transdifferention of differentiated myocytes recruited from the tail stump.
Figure 5.
Muscle cell labeling in the regenerating Xenopus tail. Larvae derived from crossings between C5 and A6 that contain yellow muscle were grown up to stage 52 (day 28) and the tail tip was removed (see Fig. 4D). (A–C) Pictures in normal light, blue and yellow fluorescence are given at various times after tail removal. The border between the tail stump and the regenerate is marked by an arrow. (D) Close-up of a regenerated tail tip in blue (top) and yellow fluorescence (bottom) at day 16. The scale bars are 1 mm.
DISCUSSION
As the generation of sexually mature Xenopus laevis frogs takes about a year, we initially introduced the reporter gene and the Cre expression vector as stable transgenes simultaneously (Fig. 1). Using this approach immediate evaluation of Cre recombinase action is possible in the founder generation. However, the number of healthy transgenic animals is limited and each individual will be unique as it has its own copy number and integration site of the transgene. Nevertheless, such double transgenic founder animals are potentially most valuable in experiments aimed at interfering with gene function. We envisage to select double transgenic founder animals with Cre-mediated activation of the cotransgenic fluorescence reporter in the most optimal fashion. These animals could then be crossed with animals containing a Cre-activated gene to interfere with gene function in the developing organism. This would lead to a more predictable outcome, as the Cre activity would already be defined and the cotransgenic reporter would be a most valuable marker to look for the functional consequence of the Cre-induced expression of the interfering gene. Based on the observation that different transgenes introduced simultaneously are frequently integrated at the same locus and show a coordinate expression (5), we exclude a segregation of the Cre transgene and the reporter in the F1 generation. However, we cannot exclude that the Cre transgene is expressed in the germ line leading to the transmission of a recombined reporter.
Clearly, strains with separate reporter DNA and Cre-expressing transgenes have several advantages, too. It would allow the crossing in many combinations, an undertaking most promising in the long run, especially if the efforts of different laboratories are combined. This approach is also very attractive as it generates many animals with most similar phenotypes as the transgenes involved are integrated in identical copy number and at the same chromosomal locus between the various animals.
Analyzing five reporter females selected in the larval stage for its transgene expression, we obtained two females that transmitted a silent transgene. As the injection of Cre mRNA into the two-cell stage embryo results in an efficient activation of the reporter gene (Fig. 3D), we exclude the possibility that the transgene has been inactivated in a permanent way as, for example, by deletion of some crucial regulatory DNA element. In the three other transgenic reporter lines we obtained expression of the transgene in the offspring. Assuming one integration site, we would expect 50% of the F1 animals to be transgenic. Indeed, this was found in the larvae derived from Y2 (Table 1) and all the non-expressing larvae lacked any transgene as assayed by PCR. But curiously, we observed only 30% blue fluorescent larvae in the F1 generation of female C5 (Table 1). This unexpectedly low number may be due to a mosaic germ line of C5 or some uneven selection occurring during oogenesis that depends on differential transgene integration. Obviously, a detailed analysis of the F1 and F2 generation of the C5 founder female is required to sort out these various possibilities.
To rapidly score whether the reporter transgene can be targeted by Cre recombinase we injected Cre mRNA into two-cell stage embryos of the F1 generation. We observed in blue fluorescent larvae a mosaic pattern of yellow fluorescent myotomes. This partial effect possibly reflects the fact that the recombinase introduced as a mRNA into the two-cell stage embryo has a limited stability. We exclude the possibility that only a fraction of the cell nuclei can be targeted by the recombinase, as in the same strains we observed extensive muscle-specific recombination in crossing experiments (Table 1). Injecting mRNA encoding Cre we also observed a silencing of the blue fluorescent protein expression without the appearance of yellow fluorescence. As frequently multiple copies are integrated into one locus in transgenic Xenopus (2), we assume that the copy number of the reporter has been reduced by recombination without generating an active EYFP expression cassette. Possibly this transgene silencing is not observed in crossing experiments, as the continuous production of transgenic Cre will ultimately recombine all loxP sites and thus lead to an active EYFP gene. Most surprisingly, we detected upon Cre mRNA injection an induction of ECFP expression in non-fluorescent larvae. We speculate that in these non-fluorescent transgenic larvae a high copy number of the transgene leads to gene silencing and that Cre-mediated reduction of the copy number activates the transgene. Such a transgene activation upon Cre-mediated copy number reduction has been reported in transgenic mice (17).
Our observation that the silent reporter in the females Y1 and C3 can be activated by Cre mRNA injection into the two-cell stage but not in crossing experiments using muscle-specific expressed Cre may indicate that the silent transgene can only be activated early in embryogenesis. Thus, this perplexing effect does not interfere in crossing experiments.
Tail regeneration in amphibians is an easy model to address the question how blastema formation occurs and how the fully differentiated regenerate arises (15,16). In the axolotl (Ambystoma mexicanum) it has been established that muscle fibers injured by amputation are dedifferentiated to form mononucleated cells that populate the newly formed blastema (18). By fluorescence labeling of single muscle fibers it has been estimated that up to 30% of the cells in the blastema are derived from mature muscle cells. As in this analysis the fluorescent label just marked the cell content, it was not possible to analyze whether the dedifferentiated muscle cell nuclei would redifferentiate into muscle cells or also transdifferentiate into non-muscle cells. In our approach muscle cells are irreversibly marked due to the Cre-mediated activation of the EYFP gene. Therefore, we conclude from the absence of any non-muscle cells with yellow fluorescence that transdifferentiation of muscle cells is not a major event in tail regeneration.
In general, at early stages of tail regeneration we observed an extensive reactivation of transgene expression at the site of amputation. We assume that this reflects the proliferative stimulus acting on the transgenic CMV promoter whose activity is known to be sensitive to proliferative stimuli in transgenic mice (19). In transgenic larvae containing Cre-induced yellow muscle, transgene activation at the site of tail amputation involves predominantly the expression of blue fluorescent protein in the multinucleate myotome. This indicates a preferential transcriptional activation of nuclei with unrecombined transgene. It is not clear whether this represents myotomes that are not yet fully differentiated or whether nuclei with unrecombined transgene are preferentially activated within a myotome. Consistent with the finding that blue fluorescence predominates at the site of amputation we detect blue fluorescence in the newly formed blastema and this is maintained throughout the outgrowth of the regenerating tail. Only at later stages of development can yellow fluorescence be detected in the myotomes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Heike Thomas and Kallal Pramanik for critically reviewing our manuscript and Gero Hilken for taking care of the Xenopus facility. This work was supported by the Volkswagen Foundation.
REFERENCES
- 1.Sive H., Grainger,R.M. and Harland,R.M. (2000) Early Development of Xenopus laevis, A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 2.Kroll K.L. and Amaya,E. (1996) Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development, 122, 3173–3183. [DOI] [PubMed] [Google Scholar]
- 3.Amaya E. and Kroll,K.L. (1999) A method for generating transgenic frog embryos. Methods Mol. Biol., 97, 393–414. [DOI] [PubMed] [Google Scholar]
- 4.Bronchain O.J., Hartley,K.O. and Amaya,E. (1999) A gene trap approach in Xenopus. Curr. Biol., 9, 1195–1198. [DOI] [PubMed] [Google Scholar]
- 5.Marsh-Armstrong N., Huang,H., Berry,D.L. and Brown,D.D. (1999) Germ-line transmission of transgenes in Xenopus laevis. Proc. Natl Acad. Sci. USA, 96, 14389–14393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Offield M.F., Hirsch,N. and Grainger,R.M. (2000) The development of Xenopus tropicalis transgenic lines and their use in studying lens developmental timing in living embryos. Development, 127, 1789–1797. [DOI] [PubMed] [Google Scholar]
- 7.Hartley K.O., Nutt,S.L. and Amaya,E. (2002) Targeted gene expression in transgenic Xenopus using the binary Gal4-UAS system. Proc. Natl Acad. Sci. USA, 99, 1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewandoski M. (2001) Conditional control of gene expression in the mouse. Nature Rev. Genet., 2, 743–755. [DOI] [PubMed] [Google Scholar]
- 9.Kwan K.M. (2002) Conditional alleles in mice: practical considerations for tissue-specific knockouts. Genesis, 32, 49–62. [DOI] [PubMed] [Google Scholar]
- 10.Werdien D., Peiler,G. and Ryffel,G.U. (2001) FLP and Cre recombinase function in Xenopus embryos. Nucleic Acids Res., 29, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryffel G.U. and Lingott,A. (2000) Distinct promoter elements mediate endodermal and mesodermal expression of the HNF1alpha promoter in transgenic Xenopus. Mech. Dev., 90, 65–75. [DOI] [PubMed] [Google Scholar]
- 12.Huang H., Marsh-Armstrong,N. and Brown,D.D. (1999) Metamorphosis is inhibited in transgenic Xenopus laevis tadpoles that overexpress type III deiodinase. Proc. Natl Acad. Sci. USA, 96, 962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sparrow D.B., Latinkic,B. and Mohun,T.J. (2000) A simplified method of generating transgenic Xenopus. Nucleic Acids Res., 28, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nastos A., Pogge v.Strandmann,E., Weber,H. and Ryffel,G.U. (1998) The embryonic expression of the tissue-specific transcription factor HNF1alpha in Xenopus: rapid activation by HNF4 and delayed induction by mesoderm inducers. Nucleic Acids Res., 26, 5602–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsonis P.A. (2000) Regeneration in vertebrates. Dev. Biol., 221, 273–284. [DOI] [PubMed] [Google Scholar]
- 16.Brockes J.P. and Kumar,A. (2002) Plasticity and reprogramming of differentiated cells in amphibian regeneration. Nature Rev. Mol. Cell Biol., 3, 566–574. [DOI] [PubMed] [Google Scholar]
- 17.Garrick D., Fiering,S., Martin,D.I. and Whitelaw,E. (1998) Repeat-induced gene silencing in mammals. Nature Genet., 18, 56–59. [DOI] [PubMed] [Google Scholar]
- 18.Echeverri K., Clarke,J.D. and Tanaka,E.M. (2001) In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Dev. Biol., 236, 151–164. [DOI] [PubMed] [Google Scholar]
- 19.Loser P., Jennings,G.S., Strauss,M. and Sandig,V. (1998) Reactivation of the previously silenced cytomegalovirus major immediate-early promoter in the mouse liver: involvement of NFkappaB. J. Virol., 72, 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nieuwkoop P.D. and Faber,J. (1975) Normal Table of Xenopus laevis (Daudin). Elsevier/North-Holland Publishing Co., Amsterdam, The Netherlands.