Hypoxia
Hypoxia refers to below-normal levels of oxygen in air, blood, or tissue. Tissue hypoxia leads to cellular dysfunction and ultimately can lead to cell death. Causes of tissue hypoxia include (a) decreased blood oxygenation (such as occurs in certain pulmonary disorders), (b) altered oxygen release from hemoglobin (associated with some hemoglobinopathies), and (c) impaired blood delivery leading to localized anemia (i.e., ischemia) as a result of low cardiac output or vascular obstruction. In order to adapt to hypoxia, mammals use a number of physiological responses. These include (a) increased production of erythropoietin (EPO), which augments the production of red blood cells; (b) induction of tyrosine hydroxylase, which facilitates the control of ventilation through the carotid body; and (c) the stimulation of new blood vessels by upregulation of VEGF (1). At the cellular level, hypoxia induces a number of metabolic changes that allow for continued energy generation despite decreased oxygen availability.
Hypoxia-inducible factor
One of the most important factors in the cellular response to hypoxia is hypoxia-inducible factor (HIF), which transcriptionally activates genes encoding proteins that mediate adaptive responses to reduced oxygen availability. HIF is a heterodimer consisting of one of three α subunits (HIF1-α, HIF2-α, or HIF3-α) bound to the aryl hydrocarbon receptor nuclear translocator (ARNT), which is also known as HIF1-β. HIF-α is a member of the basic helix-loop-helix (bHLH) superfamily, in which the HLH domain mediates subunit dimerization while the basic domain binds to DNA. HIF-α, like some other bHLH family members, contains a Per/Arnt/Sim (PAS) domain that facilitates the heterodimerization of HIF-α with ARNT (2–4).
HIF target genes play critical roles in metabolism, angiogenesis, cell proliferation, and cell survival. Examples of HIF target genes include VEGF, glucose transporter 1 (GLUT1), and EPO. HIF binds to the hypoxia-responsive element, which contains the core recognition sequence 5′-TACGTG-3′ (5), in the cis-regulatory regions of hypoxia-inducible genes. Transcriptional activation by HIF is linked to its ability to recruit coactivator proteins such as CREB-binding protein (CBP), p300, steroid receptor coactivator-1, and translation initiation factor 2 (6–8).
Whereas changes in oxygen levels do not affect ARNT protein levels, hypoxia markedly increases the abundance of the HIF-α subunits (9–11). This increase is due primarily to protein stabilization. In some settings, hypoxia also leads to increased HIF1-α mRNA accumulation (12, 13).
Von Hippel–Lindau Disease
von Hippel–Lindau (VHL) disease is a hereditary cancer syndrome that affects approximately 1 in 35,000 individuals. The syndrome was first described in 1894 by Collins, who observed two siblings with bilateral retinal angiomas (now also called hemangioblastomas) (14). Later, the disease was mapped to chromosome 3p25 (15), and the VHL gene itself was isolated in 1993 (16).
VHL behaves as a classical tumor suppressor gene. Almost all individuals with a clinical diagnosis of VHL disease can be shown to harbor a germline mutation of the VHL gene, with tumor development linked to somatic inactivation or loss of the remaining wild-type allele (17). The cardinal features of VHL disease are blood vessel tumors of the retina and central nervous system. Other tumors associated with VHL disease include clear cell carcinomas of the kidney, pheochromocytoma, endolymphatic sac tumors, pancreatic islet cell tumors, and papillary cystadenomas of the epididymis (in males) or broad ligament (in females) (18). In addition, patients with VHL often develop multiple visceral cysts involving organs such as the pancreas and kidneys (18). Biallelic VHL inactivation also occurs commonly in patients with sporadic hemangioblastoma or renal cell carcinoma, in keeping with Knudson’s “2-hit” model for tumor suppressor gene inactivation and cancer (19–26).
The VHL gene is ubiquitously expressed. It contains three exons and encodes an approximately 4.5-kb mRNA (16, 27). Alternative splicing gives rise to a shorter mRNA isoform that lacks exon 2 (26). The longer VHL mRNA isoform encodes two proteins because of alternative translational initiation. The larger protein contains 213 amino acid residues and migrates with an apparent molecular weight of 24–30 kDa (28). The second and smaller protein arises from translational initiation from the second in-frame methionine (codon 54) and migrates with an apparent molecular weight of 18 kDa (29–31). Both protein isoforms behave similarly in the majority of assays described to date and hence are often collectively referred to as pVHL. Less is known about the shorter VHL mRNA isoform, although it is presumed to be at least partially defective with respect to tumor suppression, since some renal carcinomas exclusively produce this mRNA isoform (26).
Control of HIF by pVHL
In 1996, Iliopoulos and colleagues implicated pVHL in hypoxia-inducible gene regulation when they observed that renal carcinoma cells lacking wild-type pVHL overproduced the hypoxia-inducible mRNAs encoding VEGF, GLUT1, and PDGF B under both normoxic and hypoxic conditions. This abnormality was corrected after restoration of pVHL function by stable transfection (32). In parallel, and in keeping with these findings, several groups showed that pVHL-defective tumor cells overproduce VEGF (33–35).
pVHL is ubiquitous and forms a cellular complex that contains, at a minimum, Elongin B, Elongin C, Cul2, and Rbx1 (RING box protein 1; also called regulator of cullins 1 [ROC1] or high level expression reduces TYtransposition 1 [Hrt1]) (36–41). The architecture of this complex resembles SCF (Skp1/Cdc53/F-box) complexes in yeast, which serve as E3 ubiquitin ligases. In such complexes, the F-box protein (so-named because of a short motif first identified in cyclin F) binds to the target destined to be destroyed. This similarity provided an earlier suggestion that pVHL played a role in protein polyubiquitination. Elucidation of the crystal structure of pVHL bound to Elongin B and Elongin C strengthened this view insofar as (a) the structure of Elongin C was found, as predicted, to resemble that of Skp1, and (b) the Elongin C binding domain within pVHL, called the α domain, resembled an F-box motif (42). The crystal structure of pVHL also revealed the presence of a frequently mutated subdomain (the β domain) with features indicating a possible substrate docking site. Further support for a role of pVHL in polyubiquitination was provided by the finding that anti-pVHL immunoprecipitates could support the polyubiquitination of anonymous coimmunoprecipitated proteins (43, 44).
In 1999, Maxwell and colleagues demonstrated that cells lacking pVHL were unable to degrade HIF-α subunits and, furthermore, that pVHL and HIF-α subunits can physically associate (45). Subsequently, it was shown that pVHL binds directly, through the β domain, to a region of HIF called the oxygen-dependent degradation domain (ODD), which had previously been shown to render HIF unstable in the presence of oxygen (9–11). The binding of pVHL to HIF1-α directs the polyubiquitination and degradation of the latter (46–50).
Prolyl hydroxylation earmarks HIF for destruction
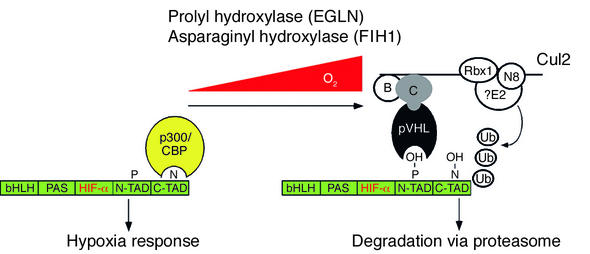
Why does pVHL only recognize HIF-α in the presence of oxygen? Recently, three groups reported that pVHL only binds to HIF1-α after the latter is enzymatically hydroxylated on conserved prolyl residues within the ODD (51–53). This modification is inherently oxygen dependent, because the oxygen atom of the hydroxyl group is derived from molecular oxygen. Moreover, this reaction requires the cofactors 2-oxoglutarate, vitamin C, and iron. The requirement of this last cofactor explains the hypoxia-mimetic effects of iron chelators (such as deferoxamine mesylate) and iron antagonists (such as cobalt chloride). Prolyl hydroxylation of HIF is carried out by the product of the egg laying abnormal-9 (Egl-9) gene in Caenorhabditis elegans and Drosophila melanogaster (54–56). The human genome contains Egl-9 homologues that are named egg laying nine 1 (EGLN1), EGLN2, and EGLN3 (also called prolyl hydroxylase domain–containing proteins PHD2, PHD1, and PHD3, respectively) (57, 58). All three proteins can hydroxylate HIF1-α at one of two proline sites within the ODD (Pro-402 and Pro-564), with some evidence for site specificity when the three enzymes are compared (54–56, 59). Analogous prolyl residues are present in HIF2-α and HIF3-α. In the presence of oxygen, the EGLN proteins are active and hydroxylate the ODD domain of HIF1-α, which allows pVHL to bind and polyubiquitinate HIF. This in turn leads to proteasomal degradation of HIF. Under hypoxic conditions, the PHD enzyme cannot hydroxylate HIF, and therefore HIF is not recognized by pVHL. As a result, HIF accumulates in the cell and is available to activate transcription (Figure 1).
Figure 1.
Hydroxylation of HIF-α. HIF-α contains bHLH and PAS DNA-binding and heterodimerization domains as well as two modular transcriptional activation domains (N-TAD and C-TAD). In the presence of oxygen, HIF-α is hydroxylated on conserved prolyl residues within the ODD (note that only one of two potential hydroxylation sites is shown for simplicity) by members of the EGLN family and on a conserved asparaginyl residue within the C-TAD by FIH1. The former leads to the recruitment of the pVHL ubiquitin ligase complex and the latter to displacement of p300 and CBP coactivator proteins. Polyubiquitination by the pVHL complex, which contains Elongin B, Elongin C, Rbx1, Cul2, and an unidentified E2 ubiquitin-conjugating enzyme, leads to proteasomal degradation of HIF. Recruitment of the E2 is linked to neddylation of the Cul2 scaffolding protein. Note that the HIF-α residues that are ubiquitinated are not known. B, Elongin B; C, Elongin C; ?E2, unidentified E2 ubiquitin-conjugating enzyme; N8, Nedd8; OH, hydroxy group; P, prolyl residue; N, aspariginyl residue; Ub, ubiguitin; ODD, oxygen-dependent degradation domain.
The recently solved crystal structure of a hydroxylated HIF1-α–derived peptide bound to the pVHL–Elongin B–Elongin C complex reveals that HIF1-α interacts, as predicted, with the β domain of pVHL. The β domain is largely hydrophobic but contains two hydrophilic residues, His-115 and Ser-111, which can interact with water in the absence of HIF. This water is displaced upon binding to the HIF peptide, which would be energetically unfavorable were it not for the fact that the hydroxyl group of the hydroxylated prolyl ring can form — in place of the water — two hydrogen bonds with His-115 and Ser-111. The absence of these interactions is presumed to account for the lack of binding to the nonhydroxylated HIF peptide (60, 61) (Figure 1).
Coactivator recruitment by HIF is regulated by asparagine hydroxylation
Modulation of protein stability is just one means by which HIF activity is induced by hypoxia. HIF1-α contains two modular transcriptional activation domains: (a) the N-terminal transactivation domain (N-TAD, which overlaps with the ODD), and (b) the C-terminal transactivation domain (C-TAD) (62). Earlier studies showed that transcriptional activation by C-TAD, when fused to a heterologous DNA-binding domain, was hypoxia inducible without an accompanying change in protein abundance. Lando and colleagues reported that C-TAD is enzymatically hydroxylated on a conserved asparagine residue (Asn-803) in the presence of oxygen, and this modification prevents the recruitment of coactivators such as p300 and CBP (63). The enzyme responsible for this modification is factor-inhibiting HIF1 (FIH1), which was previously shown to bind to HIF and to repress HIF-dependent transcription (64–66). FIH1, like the members of the EGLN family, is a member of the 2-oxoglutarate and iron-dependent dioxygenase superfamily. The recently solved crystal structures of the HIF C-TAD bound to p300 or CBP reveals how asparagine hydroxylation causes a steric clash that prevents coactivator recruitment, despite the large surface areas involved in these interactions (67, 68) (Figure 1).
Therapeutic implications
Dysregulation of HIF appears to play a central role in VHL-associated neoplasms (69, 70). Accordingly, small molecules that inhibit HIF or the actions of its downstream targets, including VEGF, PDGF, and TGF-α, are being investigated as potential therapeutic agents for such tumors. For example, preliminary data suggest a possible role for inhibitors of VEGF, or the VEGF receptor KDR (kinase insert domain receptor), in the treatment of renal carcinoma and hemangioblastoma (71, 72).
Conversely, drugs that activate HIF might theoretically be beneficial in diseases characterized by ischemia, such as myocardial infarction, stroke, and peripheral vascular disease. In this regard, there is currently no evidence that dysregulation of HIF is sufficient to cause tumorigenesis (73, 74). Moreover, the risk of inducing tumors (if such a risk exists) could presumably be minimized by limiting the duration of therapy (for example, by short-term administration immediately after a myocardial infarction). 2-Oxoglutarate and iron-dependent dioxygenases can clearly be inhibited by small molecules with drug-like properties (51, 55, 56). In one study, administration of a compound that, in retrospect, inhibits EGLN1 led to preservation of myocardial function when administered 48 hours after myocardial infarction in rats (56, 75). One could also envision, in time, the development of small molecules that specifically inhibit pVHL-associated ubiquitin ligase activity or that directly or indirectly (for example, allosterically) alter the binding properties of the pVHL β domain.
Future questions
The study of the interaction of pVHL with HIF has led to new insights into the mammalian oxygen-sensing pathway and has demonstrated a heretofore unappreciated role for enzymatic protein hydroxylation in intracellular signaling. The discovery of this modification, as well as the enzymes responsible (the EGLN family and FIH1), raises the question of whether this modification occurs on other intracellular proteins and, if so, to what end. In this regard, the work of Aravind and Koonin suggests that the human genome contains additional members of the 2-oxoglutarate and iron-dependent dioxygenase superfamily and hence additional potential protein hydroxylases (58). This implies that protein hydroxylation might be more common than previously thought.
This prompts one to question why the human genome contains three EGLN family members. Although all three can hydroxylate HIF in vitro, it will be important to establish which regulate HIF in vivo and whether they can be selectively regulated. Likewise, it will be important to establish whether any of the three EGLN family members have substrates in addition to, or perhaps instead of, HIF and to relate these substrates to the biological effects of hypoxia. The answers to such questions should provide new insights into cellular adaptation to hypoxia as well as provide a platform for new pharmacologic approaches to diverse diseases characterized by tissue ischemia.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: erythropoietin (EPO); hypoxia-inducible factor (HIF); aryl hydrocarbon receptor nuclear translocator (ARNT); basic helix-loop-helix (bHLH); Per/Arnt/Sim (PAS); glucose transporter 1 (GLUT1); CREB-binding protein (CBP); von Hippel–Lindau (VHL); RING box protein 1 (Rbx1); Skp1/Cdc53/F-box (SCF); oxygen-dependent degradation domain (ODD); egg laying nine (EGLN); prolyl hydroxylase domain (PHD); N-terminal transactivation domain (N-TAD); C-terminal transactivation domain (C-TAD); factor-inhibiting HIF1 (FIH1).
References
- 1.Bunn HF, Poyton RO. Oxygen sensing and molecular adaptation to hypoxia. Physiol. Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 2.Crews ST, Fan CM. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr. Opin. Genet. Dev. 1999;9:580–587. doi: 10.1016/s0959-437x(99)00003-9. [DOI] [PubMed] [Google Scholar]
- 3.Hogenesch JB, et al. The basic helix-loop-helix-PAS protein MOP9 is a brain-specific heterodimeric partner of circadian and hypoxia factors. J. Neurosci. 2000;20:RC83. doi: 10.1523/JNEUROSCI.20-13-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza GL. HIF1: mediator of physiological and pathophysiological responses to hypoxia. J. Appl. Physiol. 2000;88:1474–1480. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- 5.Semenza GL, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 6.Carrero P, et al. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1α. Mol. Cell Biol. 2000;20:402–415. doi: 10.1128/mcb.20.1.402-415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ema M, et al. Molecular mechanisms of transcription activation by HLF and HIF1α in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arany Z, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its α subunit. J. Biol. Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 11.Kallio PJ, Pongratz I, Gradin K, McGuire J, Poellinger L. Activation of hypoxia-inducible factor 1α: posttranscriptional regulation and conformational change by recruitment of the Arnt transcription factor. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5667–5672. doi: 10.1073/pnas.94.11.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu AY, et al. Temporal, spatial, and oxygen-regulated expression of hypoxia-inducible factor-1 in the lung. Am. J. Physiol. 1998;275:L818–L826. doi: 10.1152/ajplung.1998.275.4.L818. [DOI] [PubMed] [Google Scholar]
- 13.Wiener CM, Booth G, Semenza GL. In vivo expression of mRNAs encoding hypoxia-inducible factor 1. Biochem. Biophys. Res. Commun. 1996;225:485–488. doi: 10.1006/bbrc.1996.1199. [DOI] [PubMed] [Google Scholar]
- 14.Collins ET. Intra-ocular growth (two cases, brother and sister, with peculiar new growth, probably retinal, affecting both eyes) Trans. Ophthalmol. Soc. U. K. 1894;14:141–149. [Google Scholar]
- 15.Seizinger BR, et al. Von Hippel-Lindau disease maps to the region of chromosome 3 associated with renal cell carcinoma. Nature. 1988;332:268–269. doi: 10.1038/332268a0. [DOI] [PubMed] [Google Scholar]
- 16.Latif F, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 17.Stolle C, et al. Improved detection of germline mutations in the von Hippel-Lindau disease tumor suppressor gene. Hum. Mutat. 1998;12:417–423. doi: 10.1002/(SICI)1098-1004(1998)12:6<417::AID-HUMU8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 18.Maher ER, Kaelin WG., Jr Von Hippel-Lindau disease. Medicine. 1997;76:381–391. doi: 10.1097/00005792-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 19.Shuin T, et al. Germline and somatic mutations in von Hippel-Lindau disease gene and its significance in the development of kidney cancer. Contrib. Nephrol. 1999;128:1–10. doi: 10.1159/000059976. [DOI] [PubMed] [Google Scholar]
- 20.Shuin T, et al. Frequent somatic mutations and loss of heterozygosity of the von Hippel-Lindau tumor suppressor gene in primary human renal cell carcinomas. Cancer Res. 1994;54:2852–2855. [PubMed] [Google Scholar]
- 21.Foster K, et al. Somatic mutations of the von Hippel-Lindau disease tumour suppressor gene in non-familial clear cell renal carcinoma. Hum. Mol. Genet. 1994;3:2169–2173. doi: 10.1093/hmg/3.12.2169. [DOI] [PubMed] [Google Scholar]
- 22.Whaley JM, et al. Germ-line mutations in the von Hippel-Lindau tumor-suppressor gene are similar to somatic von Hippel-Lindau aberrations in sporadic renal cell carcinoma. Am. J. Hum. Genet. 1994;55:1092–1102. [PMC free article] [PubMed] [Google Scholar]
- 23.Clifford SC, Prowse AH, Affara NA, Buys CH, Maher ER. Inactivation of the von Hippel-Lindau (VHL) tumour suppressor gene and allelic losses at chromosome arm 3p in primary renal cell carcinoma: evidence for a VHL-independent pathway in clear cell renal tumourigenesis. Genes Chromosomes Cancer. 1998;22:200–209. doi: 10.1002/(sici)1098-2264(199807)22:3<200::aid-gcc5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 24.Kanno H, et al. Somatic mutations of the von Hippel-Lindau tumor suppressor gene in sporadic central nervous system hemangioblastomas. Cancer Res. 1994;54:4845–4847. [PubMed] [Google Scholar]
- 25.Tse JY, et al. Molecular genetic analysis of the von Hippel-Lindau disease tumor suppressor gene in familial and sporadic cerebellar hemangioblastomas. Am. J. Clin. Pathol. 1997;107:459–466. doi: 10.1093/ajcp/107.4.459. [DOI] [PubMed] [Google Scholar]
- 26.Gnarra JR, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 27.Renbaum P, et al. Isolation and characterization of the full-length 3′ untranslated region of the human von Hippel-Lindau tumor suppressor gene. Hum. Genet. 1996;98:666–671. doi: 10.1007/s004390050281. [DOI] [PubMed] [Google Scholar]
- 28.Iliopoulos O, Kibel A, Gray S, Kaelin WG., Jr Tumour suppression by the human von Hippel-Lindau gene product. Nat. Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 29.Iliopoulos O, Ohh M, Kaelin WG., Jr pVHL19 is a biologically active product of the von Hippel-Lindau gene arising from internal translation initiation. Proc. Natl. Acad. Sci. U. S. A. 1998;95:11661–11666. doi: 10.1073/pnas.95.20.11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoenfeld A, Davidowitz EJ, Burk RD. A second major native von Hippel-Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8817–8822. doi: 10.1073/pnas.95.15.8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blankenship C, Naglich JG, Whaley JM, Seizinger B, Kley N. Alternate choice of initiation codon produces a biologically active product of the von Hippel Lindau gene with tumor suppressor activity. Oncogene. 1999;18:1529–1535. doi: 10.1038/sj.onc.1202473. [DOI] [PubMed] [Google Scholar]
- 32.Iliopoulos O, Levy AP, Jiang C, Kaelin WG, Jr, Goldberg MA. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gnarra JR, et al. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siemeister G, et al. Reversion of deregulated expression of vascular endothelial growth factor in human renal carcinoma cells by von Hippel-Lindau tumor suppressor protein. Cancer Res. 1996;56:2299–2301. [PubMed] [Google Scholar]
- 35.Krieg M, Marti HH, Plate KH. Coexpression of erythropoietin and vascular endothelial growth factor in nervous system tumors associated with von Hippel-Lindau tumor suppressor gene loss of function. Blood. 1998;92:3388–3393. [PubMed] [Google Scholar]
- 36.Pause A, et al. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl. Acad. Sci. U. S. A. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pause A, Peterson B, Schaffar G, Stearman R, Klausner RD. Studying interactions of four proteins in the yeast two-hybrid system: structural resemblance of the pVHL/elongin BC/hCUL-2 complex with the ubiquitin ligase complex SKP1/cullin/F-box protein. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9533–9538. doi: 10.1073/pnas.96.17.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kibel A, Iliopoulos O, DeCaprio JA, Kaelin WG., Jr Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 39.Lonergan KM, et al. Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol. Cell. Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamura T, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 41.Kishida T, Stackhouse TM, Chen F, Lerman MI, Zbar B. Cellular proteins that bind the von Hippel-Lindau disease gene product: mapping of binding domains and the effect of missense mutations. Cancer Res. 1995;55:4544–4548. [PubMed] [Google Scholar]
- 42.Stebbins CE, Kaelin WG, Jr, Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 43.Iwai K, et al. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. U. S. A. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 46.Ohh M, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the β-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 47.Kamura T, et al. Activation of HIF1α ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10430–10435. doi: 10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cockman ME, et al. Hypoxia inducible factor-α binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 2000;275:25733–25741. doi: 10.1074/jbc.M002740200. [DOI] [PubMed] [Google Scholar]
- 49.Tanimoto K, Makino Y, Pereira T, Poellinger L. Mechanism of regulation of the hypoxia-inducible factor-1α by the von Hippel-Lindau tumor suppressor protein. EMBO J. 2000;19:4298–4309. doi: 10.1093/emboj/19.16.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu F, White SB, Zhao Q, Lee FS. Dynamic, site-specific interaction of hypoxia-inducible factor-1α with the von Hippel-Lindau tumor suppressor protein. Cancer Res. 2001;61:4136–4142. [PubMed] [Google Scholar]
- 51.Jaakkola P, et al. Targeting of HIFα to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 52.Ivan M, et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 53.Yu F, White SB, Zhao Q, Lee FS. HIF1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 55.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 56.Ivan M, et al. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor MS. Characterization and comparative analysis of the EGLN gene family. Gene. 2001;275:125–132. doi: 10.1016/s0378-1119(01)00633-3. [DOI] [PubMed] [Google Scholar]
- 58.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2:RESEARCH0007.1–0007. doi: 10.1186/gb-2001-2-3-research0007. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Min JH, et al. Structure of an HIF1α-pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 61.Hon WC, et al. Structural basis for the recognition of hydroxyproline in HIF1α by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 62.Bruick RK, McKnight SL. Transcription. Oxygen sensing gets a second wind. Science. 2002;295:807–808. doi: 10.1126/science.1069825. [DOI] [PubMed] [Google Scholar]
- 63.Lando DP, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 64.Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF1α and VHL to mediate repression of HIF1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lando D, et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hewitson KS, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 67.Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for HIF1α/CBP recognition in the cellular hypoxic response. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Freedman SJ, et al. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1α. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 70.Maranchie JK, et al. The contribution of VHL substrate binding and HIF1-α to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1:247–255. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 71.Drevs J, et al. Effects of PTK787/ZK 222584, a specific inhibitor of vascular endothelial growth factor receptor tyrosine kinases, on primary tumor, metastasis, vessel density, and blood flow in a murine renal cell carcinoma model. Cancer Res. 2000;60:4819–4824. [PubMed] [Google Scholar]
- 72.Yang J, Haworth S, Steinberg S, Rosenberg S, Novotny W. A randomized double-blind placebo-controlled trail of bevacizumab (anti-VEGF antibody) demonstrating a prolongation in time to progression in patients with metastatic renal cancer. Proc. Am. Soc. Clin. Oncol. 2002;21:15 . (Abstr.) [Google Scholar]
- 73.Vincent KA, et al. Angiogenesis is induced in a rabbit model of hindlimb ischemia by naked DNA encoding an HIF1α/VP16 hybrid transcription factor. Circulation. 2000;102:2255–2261. doi: 10.1161/01.cir.102.18.2255. [DOI] [PubMed] [Google Scholar]
- 74.Elson DA, et al. Induction of hypervascularity without leakage or inflammation in transgenic mice overexpressing hypoxia-inducible factor-1α. Genes Dev. 2001;15:2520–2532. doi: 10.1101/gad.914801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nwogu JI, et al. Inhibition of collagen synthesis with prolyl 4-hydroxylase inhibitor improves left ventricular function and alters the pattern of left ventricular dilatation after myocardial infarction. Circulation. 2001;104:2216–2221. doi: 10.1161/hc4301.097193. [DOI] [PubMed] [Google Scholar]