Abstract
Werner syndrome is a premature aging disease caused by the mutation in the WRN gene. The cloning and characterization of the WRN gene and its product allows investigators to study the disease and the human aging process at molecular level. This review summarizes the recent progresses on various aspects of the WRN research including functional analysis of the protein, interactive cloning, complexes formation, mouse models, and SNPs (single nucleotide polymorphisms). These in depth investigations have greatly advanced our understanding of the disease and elucidated future research direction for Werner syndrome and the human aging process.
INTRODUCTION
Until recently, Werner syndrome (WS) had been considered a model of accelerated aging. The WRN gene was identified in 1996 [1], and was subsequently shown to act both as a DNA helicase and as an exonuclease [2, 3]. Since then, cancer researchers and those that study DNA metabolism have collaborated to further characterize WS and the function of the WRN gene. The more we understand about the WRN gene, the more we realize that WS is not merely involved in accelerated aging. WS certainly does not represent premature aging, in a sense that the characteristic aging phenotypes seen in WS are considerably different from those observed in normal elders. WS is now being more correctly recognized as a condition in which the lack of WRN protein (WRNp) results in an overall decline in the normal physiological functions of various organs, including those most frequently used to estimate the chronological age, such as skin and hair.
Since the identification of the WRN gene five years ago, various in vitro biochemical studies of WRNp have answered many of our initial questions regarding the helicase and exonuclease functions of this enzyme. Considerably more time will be required to answer the more difficult questions concerning the in vivo functions of WRN at the organism and cellular levels. The lack of mouse models of WS that mimic the human disorder currently limits our ability to carry out such studies. In this review, the known in vitro functions of WRNp and the cellular characteristics of WS cells will be summarized. From this, the putative in vivo functions of WRN will be extrapolated.
CLINICAL FEATURE OF WERNER SYNDROME
The clinical phenotypes of WS can be best summarized as onset of an aged-appearance and age-related common disorders [4, 5, 6, 7]. Unlike people with Hutchinson-Gilford progeria, WS patients usually develop normally until they reach the second decade of life. Some patients may present with flat feet. Generally, the first sign is a lack of the pubertal growth spurt during the teen years. Patients frequently recall that they were of average height when they entered grade school, but were the shortest ones in their class by the time they graduated from high school. In their 20s and 30s, patients begin to manifest skin atrophy, loss of hair, and graying hair. Subcutaneous fat tends to deposit on the trunk, and combined with osteoporosis of the limbs, patients exhibit a stocky appearance. The other most common age-related disorders seen in WS patients are bilateral cataracts and type II diabetes mellitus. Penetrances are 98% for cataracts and 90% for diabetes mellitus [7]. These numbers obviously depend on the age of the patient when clinical reports are made, and how rigorously patients were examined. Demographically, WS cases are most frequently reported in Japan. This can be partly attributed to an awareness of the syndrome among Japanese physicians, and the higher consanguinity among the Japanese population as compared to the US population. The chronological order of the onset of these complications is similar among Caucasian and Japanese WS patients [6, 7]. The International Registry of Werner Syndrome (Seattle, Wash, USA) has documented more than 100 cases of WS. New cases with a diagnosis confirmed by genetic analysis are mostly in patients over the age of 30, probably due to the lack of prominent symptoms prior to the age of 20.
Two common causes of death among WS patients are malignancy and myocardial infarction [6, 8]. Although these are also two common causes of death in the general population, unique characteristics are observed in WS patients. An extensive review of malignancies has been carried out in WS cases in Japan, where the prevalence of WRN mutations is relatively high [9]. Strikingly, the ratio between cancers of epithelial origin and sarcomas of mesenchymal origins is 1:1 in WS patients, whereas this ratio is approximately 10:1 in the general population. A possible explanation for the high representation of mesenchymal tumors in WS patients may be related to the mechanism by which telomere length is maintained [10]. Review of the pathological studies of these malignancies revealed unusual primary sites for cancers in WS patients. For example, melanomas in WS patients are of the acral lentigenous type in the mucosas, and are unrelated to sun exposure [9]. The primary sites of osteosarcoma in WS patients are more likely to be in the lower extremities, whereas these are more common in the upper extremities in the general population [11]. This variation may be related to the threshold of WRNp required to maintain DNA stability in each cell type within a given organ. Among WS patients, the specific cell type in which cancer develops may differ, depending on the type of mutation in the WRN gene. Papillary carcinoma has been associated with an N-terminal mutation, whereas follicular carcinoma is more frequently observed with a C-terminal mutation [5]. This finding clearly contradicts our original assumption that all identified mutations within WRN result in truncation of the nuclear localization signal of WRNp, and thereby act as null mutations (see Section “WRN mutations”). Further studies may reveal additional correlation between specific genotypes and phenotypes.
Several other important differences have been noted between WS patients and normal elders. Atherosclerosis exhibits unique characteristics in WS patients. Atherosclerotic lesions are more extensive in arterioles as compared to non-WS patients, who exhibit lesions primarily in major arteries. Calcification of cardiac valves is also sometimes observed in WS, possibly reflecting excessive cell death due to the constant pressure of the blood flow. Skin ulcers around the ankles and elbows that are more severe than those typically seen in the progression of diabetes mellitus are not uncommon in WS. Dementia of the Alzheimer type is relatively rare [12] despite the fact that WRNp is expressed in the brain [1]. Whereas in the general population, osteoporosis has a more pronounced effect on vertebrae, long bones, particularly those of the lower limbs, tend to be more affected by osteoporosis in WS patients [13].
Werner syndrome was first described by Dr Otto Werner at Kiel University in his doctoral thesis at the turn of the 20th century [14]. Two symptoms that drew his attention were bilateral cataracts and scleoderma-like skin. The latter turned out to provide insight into the interaction between WRNp and the Ku complex (see Section “Telomere maintenance”), the autoantigen that triggers scleroderma, an autoimmune disorder.
WRN GENE PRODUCT
Functional domains of WRN gene product
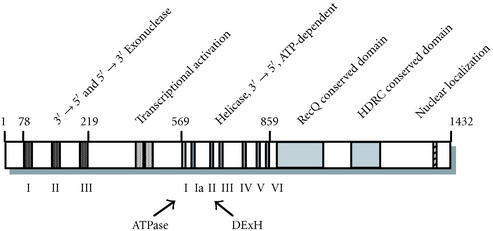
The WRN gene is comprised of 35 exons on the short arm of chromosome 8, and encodes protein of 1432 amino acids. It was identified in 1996 using a conventional positional cloning approach [1], and the methods that were available at that time [15]. Simple alignment in database searches and more complicated structural studies showed four defined regions of WRNp. These include exonuclease domains I, II, and III in the N-terminal region [16, 17]; RecQ-type helicase domains I, Ia, II, III, IV, V, and VI in the central region [18]; a RecQ conserved motif immediately following the helicase motifs [19]; and a helicase ribonuclease D C-terminal (HRDC) conserved motif in the C-terminal regions [19].
Biochemical studies confirmed the helicase and exonuclease activity of WRNp in vitro [2, 3, 20]. The function of the RecQ conserved motif has not been defined. The HRDC motif is thought to form a scaffold which interacts with substrate DNA with relatively low affinity, based on the three-dimensional structure, which resembles the auxiliary DNA-binding domains of bacterial DNA helicases [21]. Cell biological studies identified two other functional domains. One of these functions as a transcriptional activator within a highly acidic region between the exonuclease and helicase domains [22, 23]. The other functions as a nuclear localization signal in the extreme C-terminus of WRNp [24, 25]. The relative locations of these structural domains are shown in Figure 1.
Figure 1.
Functional domains of the WRN protein. Arabic numbers indicate amino acids of the WRN protein. The designated functions of the exonuclease domains (I–III), the acidic repeat transcriptional activator regions, helicase domains (I–VI), and nuclear localization signal have been demonstrated in various biological assays. The roles of the RecQ C-terminal conserved region and the helicase RNaseD C-terminal (HDRC) conserved regions are based on the structural studies.
WRN mutations
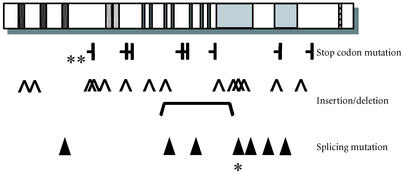
To date, at least 35 different WRN mutations have been reported from all over the world [1, 8, 26, 27, 28, 29] (see Figure 2). The known mutations correspond to either stop codons, insertions, or deletions that result in a frame shift, or splicing donor or acceptor site mutations, which cause an exon to be skipped, resulting in a frame shift. All of these mutations result in truncation of the nuclear localization signal. Unlike the BLM gene [30], no missense mutation has been identified in WS. As we expand WS screening, this type of mutation may be identified in future WS cases.
Figure 2.
WRN mutations in WS patients. The rectangular boxes indicate the functional domains of the WRN protein (see Figure 1). Known WRN mutations are grouped based upon the type of mutation, shown above and underneath the WRN protein diagram along with Registry codes. Parentheses indicate the heterozygous mutations. R367Stp (∗∗) is the most common mutation seen in Caucasian WS patients (approximately 25%) and the second most common mutation in Japanese WS patients (approximately 18%) [33]. Deletion of exon 26 (∗), which is due to a splice junction mutation, is the most common mutation in Japanese WS patients (approximately 52%) [33].
In addition to the loss of the nuclear localization signal in WRN mutations, the mutant mRNAs, and the resulting mutant proteins exhibit shorter half-lives than do the wild-type mRNA, and WRNp [31, 32]. Mutant products truncated N-terminal to the helicase regions were more labile than a mutant protein truncated C-terminal to the helicase region. The biological significance of low levels of mutant WRNp in the cytosol is unknown. Depending on the specific mutation, certain WRNp mutations can retain enzyme activity or other functions. These proteins can modify DNA during the mitotic phase or modify RNA during all phases of the cell cycles. It is possible that various mutant forms of WRNp, which may be retained in the cytosol, could contribute to the slight differences in phenotype observed with various WRN mutations. For example, thyroid cancer associated with the two major WRN mutations differs [5]. Papillary carcinoma is associated with the N-terminal mutation, while follicular carcinoma is seen more often with the C-terminal mutation.
WRN helicase activity—effect of single-strand binding proteins and substrate specificities
The helicase activity of WRNp is a RecQ-type helicase, named after the prototypic Escherichia coli RecQ. A number of laboratories have demonstrated that WRNp exhibits ATP-dependent 3′ → 5′ helicase activity in an oligo-displacement assay [2, 20, 34, 35, 36, 37]. One study showed that this activity was present in an immunoprecipitated sample of WRNp [38]. WRNp appears to first bind to the single-stranded portion of the longer strand of the DNA duplex and proceeds in a 3′ → 5′ direction with respect to the longer strand [35]. This 3′ → 5′ movement corresponds to the direction of proofreading, as opposed to the direction of DNA synthesis, which proceeds from 5′ → 3′. Thus, WRN has been proposed to play a role in some aspects of DNA strand repair, although the specific mechanisms involved are not known. WS cells are hypersensitive to 4-nitroquinoline-1-oxide (4NQO) [39], although WRNp does not have a higher binding affinity for DNA that has been damaged by 4NQO or by UV light [35]. The WRN helicase can also function to unwind DNA-RNA duplexes [20].
The helicase activity of WRNp can be enhanced by the presence of various single-strand binding proteins, such as Escherichia coli SSB, the T4 gene 32 product or, more efficiently, human replication protein A (hRPA) [2, 20, 35, 36, 37, 40]. These single-strand binding proteins are believed to facilitate WRN helicase activity by stabilizing single strand structures and by preventing their re-annealing. Several investigators have demonstrated that WRNp and hRPA are colocalized in Xenopus replication initiation complexes, and in Hela cells arrested in S-phase with hydroxyurea [41, 42, 43]. Moreover, recombinant WRNp and purified hRPA have been shown to co-immuno-precipitate [36]. hRPA might play an additional role, besides stabilizing the single-stranded DNA structure, in the WRN-catalyzed unwinding reaction, which requires direct interaction with WRNp.
Several unusual substrates have been tested as potential physiological targets for WRN helicase activity. WRN was able to efficiently unwind a G4 quartet made by two hairpin loops (G'2 biomolecular tetraplex) of d(CGG)n [44]. In this study, the G4 structure was generated under a high salt concentration in vitro. Although its presence has not been demonstrated in vivo, a G4 quartet can potentially be formed from two GC-rich regions of unwound single-stranded DNA (or RNA) during replication, repair, recombination, or transcription. Another interesting structure is a recombination intermediate, or α structure. WRNp was able to promote branch migration of a Holliday junction [42]. In fact, the WRN helicase appeared to dissociate the α structure better than a simple DNA duplex, as assessed by the length of the migration. This structure can be formed both during recombination and replication, such as break-induced DNA replication or repair of the stalled replication. The complex secondary structures described above were also substrates for the BLM helicase in vitro. Thus, in vivo, WRNp may participate in the dissociation of these structures as well.
Characteristics and significance of WRN exonuclease activity
The 3′ → 5′ exonuclease activity of WRN has been demonstrated by at least four independent laboratories [3, 40, 45, 46, 47, 48, 49]. One group measured 5′ → 3′ exonuclease activity [34], although no laboratory has demonstrated both 3′ → 5′ and 5′ → 3′ exonuclease activities in WRNp.
While exonuclease activity in the 3′ → 5′ direction does not require adenine triphosphate (ATP), the enzyme activity is enhanced in the presence of ATP and is specific for the 3′ recessed end of the duplex [3, 45, 46, 48]. The WRN exonuclease utilizes the 3′ end of the blunt end as well as the 3′ overhang end in presence of Ku70/80, the regulatory subunit of DNA protein kinase (DNA-PK) [47, 49]. Like its WRN helicase activity, the exonuclease activity of WRN also digests RNA-DNA heteroduplexes [48]. The exonuclease region of the mouse WRNp showed an activity [48].
Double-stranded DNA substrates with multiple base-pair mismatches within the duplex (termed bubbles) were more susceptible than those without mismatches to digestion by the WRN exonuclease, but the presence of more than two mismatches at the end of the substrate made them relatively more resistant to digestion [46, 48]. Structures resembling Holliday junction are also known to be more susceptible to WRN exonuclease digestion than are simple DNA duplexes [46] similar to human WRNp.
Relative substrate specificity suggests that, like the WRN helicase, WRN exonuclease activity may be involved in repairing DNA damage. The exonuclease and helicase activities of WRN are physically and functionally separable [3]. How exonuclease and helicase functions are specifically coordinated during the DNA repair process remains unknown.
Regulation of transcriptional activation by WRN
Transcriptional activation of the WRN protein was originally suggested by findings in the yeast one-hybrid system [1]. Subsequently, transactivation of the adenovirus major late promoter was used as a reported plasmid to demonstrate that WRN can mediate transcription activation [23]. This deficit was reversed by the addition of recombinant WRN protein. Interestingly, a mutant WRN protein defective in helicase activity (K577M) was unable to complement the reduced transactivation in WS nuclear lysates, indicating that the transactivation function of WRN is coupled with its helicase/ATPase activities [23].
WRN PROTEIN COMPLEXES AND CELLULAR FUNCTION OF WRN
Identification of proteins that interact with WRN has helped to shed light on the in vivo functions of WRNp. Some laboratories have characterized the association of WRN with specific candidate interacting proteins, while others have screened cDNA libraries by the yeast two-hybrid system, isolated the WRN complex from the cell extracts, or captured the interacting proteins by binding to a WRNp affinity column. Interestingly, each method identified different sets of WRN interacting proteins. Thus, WRN may associate with different protein complexes, depending on the status of DNA metabolism during the time that the WRN protein was recruited.
Cell biological studies also suggested that WRN has multiple functions. Two characteristics of WS cells that had been well known, even before the WRN gene was identified, were that these cells exhibit a shortened replicative lifespan and genomic instability. More recently, studies of drug sensitivities and telomere metabolism led us to a better understanding of the specific functions of WRNp. Interacting proteins combined with cellular and subnuclear studies suggests that WRN may be involved in a wide variety of DNA metabolic processes.
DNA replication
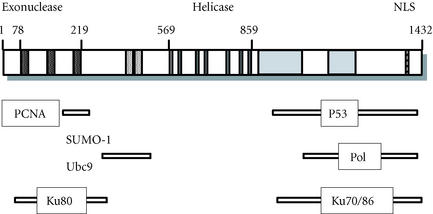
FFA-1 is a Xenopus ortholog of WRN, and was originally identified in the replication initiation complex of Xenopus oocytes as being required for the formation of replication foci during DNA replication [41]. FFA-1 and the p70 subunit of RPA have also been co-immunoprecipitated from lysates of Xenopus oocytes [43]. WRNp interacts with several components of the DNA replication complex, including proliferating cell nuclear antigen (PCNA) and topoisomerase I [50]. The N-terminal region within the exonuclease domain of the WRNp, particularly the region including amino acids 168–246, strongly interacted with the C-terminal portion of PCNA [50], as indicated in Figure 3. The N-terminal region of recombinant WRNp appears to form mostly trimers, as does PCNA, suggesting that the interaction between WRN and PCNA involves quaternary structure [48].
Figure 3.
Proteins that interact with the WRN protein. Proteins known to interact with WRNp at specific sites are shown schematically. The approximate regions of protein-protein interaction are indicated.
DNA polymerase δ
DNA polymerase δ (pol δ) participates in DNA replication and repair of DNA damage. This enzyme is also found in the telomere complex. Evidence of physical interaction between WRN and pol δ initially came from a yeast two-hybrid screen, in which the C-terminal region of WRNp was used as the “bait” to capture the p50 subunit of pol δ, as shown in Figure 3 [51]. Immunoprecipitation with an anti-p50 antibody resulted in a complex including p120, the catalytic subunit of pol δ. In yeast, physical, and functional interaction between WRN, and pol δ requires the third subunit pol32p [52].
Functional interaction of WRNp, and pol δ appears to be unilateral. In a simple primer extension assay, the addition of WRNp enhanced pol δ activity, whereas the addition of pol δ did not stimulate exonulcease or helicase activities [52]. In contrast, WRNp did not stimulate either pol α- or pol ɛ-mediated DNA synthesis [52, 53]. The presence of WRNp also enabled pol δ to transverse hairpin and G'2 bimolecular tetraplex structures to complete DNA synthesis [53]. One of the functions of WRNp may be to recruit pol δ to the complex secondary DNA structure and to alleviate stalled DNA synthesis [51, 53].
Homologous recombination
One of the important biological functions of E coli RecQ is to suppress homologous recombination by disrupting its intermediate structure [54]. The yeast homolog of RecQ, Sgs1, is involved in both homologous recombination and in illegitimate recombination or nonhomologous end joining (NHEJ). Both the WRN and BLM helicases are able to complement increased homologous recombination and illegitimate recombination of an Sgs1 deletion mutant [55]. However, the cytogenetic characterization of cells from Bloom patients shows increased exchanges of sister chromatids, whereas cells from WS patients exhibit translocation mosaicism [56]. These findings suggest that the BLM helicase may play a more prominent role in the suppression of homologous recombination in vivo.
Repair of breaks in double-stranded DNA
DNA-PK is a protein complex including a protein kinase catalytic subunit, DNA-PKcs, and a regulatory subunit, Ku70/80 [57]. This protein complex is involved in the initial stages of NHEJ. WRNp has been shown to directly interact with DNA-PKcs and Ku80 [47, 49, 57]. Association of WRN with the Ku complex enhances its exonuclease activity, but has no effect on its helicase activity [49]. Upon binding to Ku, WRN is able to degrade DNA from both the 5′ recessed end (5′ → 3′ exonuclese activity), and from the blunt end. This may explain why both 3′ → 5′ [3, 45, 58], and 5′ → 3′ [34] exonuclease activities were observed in recombinant WRNp. The presence of exonuclease activity has been speculated to be a necessary step for DNA-PK mediated NHEJ prior to the polymerization, and ligation step mediated by XRCC 4 and ligase IV [57]. In addition, the WRN helicase may unwind broken ends of dsDNA in the search for microhomology.
Telomere maintenance
In a high quality immunofluorescence study, Shiratori et al [59] identified nuclear dots suggesting that WRNp is localized on telomeres. Johnson et al [10] demonstrated WRNp co-localizes with various telomere components, including TRFI and TRFII in six ALT cell lines. ALT cells lack telomerase activity, and the telomeres in these cells are presumably maintained by recombination. Recombinant WRNp did not unwind a G'2 tetraplex containing a telomere repeat sequence [44]. However, WRNp did appear to unwind up to 23 kb of a PCR-generated telomere repeat sequence to ssDNA, and this process was stabilized by hRPA [60]. These findings collectively suggest that WRNp plays an important role in telomere maintenance.
During the serial passage of primary WS fibroblasts, telomeres became shortened more quickly, and telomeres in cells that had stopped dividing were longer than those in control fibroblasts [61]. Telomeres of WS Lymphoblastoid cell lines (LCLs) were unstable, and telomere length varied more widely in LCLs from WS patients as compared to those from normal subjects [62]. The telomere length at which LCLs from WS patients went into crisis also varied widely. It has been suggested that one way in which WRNp may be involved in the ALT pathway is by regulating the number of extra-chromosomal telomere repeats [63]. This may explain why the catalytic subunit of human telomerase (hTERT) is able to extend the replicative lifespan of WS cells indefinitely [64, 65, 66]. hTERT may be able to circumvent the early halt of cell division in WS cells because the WRNp complex functions in pathways that are distinct from hTERT-dependent telomere maintenance [63]. These findings provided significant insight into the possible mechanism of mesenchymal tumorigenesis in WS patients, as ALT cell lines are frequently mensenchymal in origin [10].
However, other important factors may also be involved in regulation of telomere function. Hisama et al [67] showed that both WRN and hTERT could complement 4NQO sensitivity in SV40 transformed WS fibroblasts. Introduction of hTERT appeared to reprogram gene expression [66]. WRNp may play an additional role in telomere maintenance, aside from repair of damaged telomeres and immortalizing cells that lack telomerase activity.
p53
As indicated in Figure 3, the C-terminal region of WRNp interacts with the p53 tumor suppressor, as these proteins can be co-immunoprecipitated [68, 69]. In the absence of WRN, p53-mediated apoptosis is attenuated [68]. Overexpression of WRN enhances p53-dependent transcriptional activation of p21Waf1 [69], and potentiates p53-mediated apoptosis [69]. Synergistic actions of p53 and WRN have been observed in mouse models of WS [70, 71].
SUMO-1
A yeast two-hybrid screen using mouse WRN as the bait identified Ubc9 and SUMO-1 as WRN interacting proteins [72]. The N-terminal domain (amino acids 272–514) of WRN interacts with both Ubc9 and SUMO-1.
MOUSE MODELS OF WERNER SYNDROME
There are currently three mouse models of Werner syndrome:
• WRN helicase domain deletion mice (WRNΔhel/Δhel) [73],
• WRN truncation mutant mice (presumably null, WRN−/−) [70], and
• transgenic mice expressing dominant negative WRN (K577M-WRN) [74].
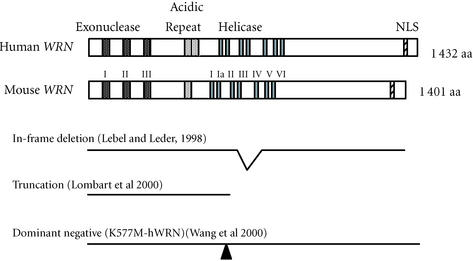
These various genetic mutations are depicted schematically in Figure 4.
Figure 4.
Mouse models of Werner syndrome. Structures of human and mouse WRN proteins and the alterations in the WRN gene product in the three mouse models of WS are shown.
WRN deficient mice carry a WRN gene with a frame deletion of the helicase domains III and IV, as described by Lebel and Leder [73]. These mice express a mutant WRN, WRNΔhel/Δhel, which retains the exonuclease domains and nuclear localization signal. Mouse ES cells showed hypersensitivity to campthothecin, and mitomycin C and the growth rate of embryonic fibroblasts was progressively reduced as the cells were passaged. The mice exhibited a normal phenotype at least until the age of 12 months [71].
Another mouse line expresses a mutant WRN protein that is truncated in the middle of the helicase region, resulting in mice that are functionally null (WRN−/−) [70]. This mutation resembles many of the mutations in WS patients. Embryonic fibroblasts showed accelerated replicative senescence, and this was enhanced in cells from mice on a BLM+/− background. They did not show hypersensitivity to campthothecin or 4NQO. Histopathological studies failed to show any unusual lesions in these mice up to the age of 17 months [70].
Mice with either a deletion or a truncation in the WRN helicase domain showed gross abnormalities when they were crossed to mice on a p53−/− background. WRN+/− mice showed increased rates of mortality, as assessed by both median survival and maximum survival of the cohort of 48 WRN−/−; p53−/− and 59 WRN+/−; p53−/− mice [70]. Median lifespan of WRN−/−; p53−/− mice was approximately 20% shorter than that of WRN+/−; p53−/− mice. Interestingly, WRNΔhel/Δhel; p53−/− mice exhibited increased numbers of tumors, and a larger variety of tumor types at an earlier age, as compared to WRN+/+; p53−/− mice [71]. Both studies suggest that WRN and p53 play a synergistic role in the maintenance of genomic stability.
The dominant negative WRN transgenic mouse line, K577M-WRN, was developed in my laboratory [74]. These mice express both a full length mutant (K577M) human WRN and endogenous mouse WRN. The K577M mutation abolishes the helicase and ATP-ase activity of WRN in vitro, but not the exonuclease activity [2, 3]. Tail fibroblasts from K577M-WRN mice showed hypersensitivity to 4NQO and a reduced replicative lifespan, as determined by clone size distribution [74]. These mice did not show any histopathological abnormalities at least up to the age of 18 months.
To date, there is no animal model of WS that accurately mimics the human disease. This is also true for some, if not all, other mouse models of RecQ helicase-deficient disorders. This lack of success in reproducing human disease states in rodents may be due to the difference between humans and mice in maintaining genomic instability. Nevertheless, these mouse models have greatly facilitated the characterization of the functional interaction of WRNp with other proteins and of the pathogenesis of WS phenotypes.
WRN POLYMERASE AND ITS ROLE OF WRN IN “NORMAL” AGING
A systematic search of the WRN polymorphism across a variety of ethnic groups identified 58 SNPs, single nucleotide polymorphisms, in 35 exons, and flanking introns. Of these, 15 are localized within the coding region and 11 of these result in alterations in amino acid sequence [75, 76]. A limited association study of the two known WRN SNPs with relatively high heterogeneity suggested that WRN may be involved in determining longevity, possibly by modulating the risk of a variety of common age-related disorders, including atherosclerosis. Statistically significant associations with longevity and atherosclerosis were observed for the 1074Leu/Phe mutation [77]. In addition, the 1367Cys/Arg mutation was associated with myocardial infarction [78], atherosclerosis [77, 79], and long-term hemodialysis [80]. These two polymorphic sites are in linkage. The functional manifestations of these two SNPs, with regard to WRN function, are unknown. However, it is of interest to note that 1074Leu/Phe resides within the RecQ consensus domain and 1367Cys/Arg is four amino acids away from the nuclear localization signal. Interestingly, in 1367Cys/Arg, the beneficial allele, Arg, is a minor allele. This might be explained by the presence of linkage disequilibrium between 1074 Leu/Phe and 1367 Cys/Arg in some populations [76, 77]. The relative contribution of WRN polymorphisms to the risk of age-related disorders or longevity may not be nearly as high as other major known genetic risk factors, for example, polymorphisms in ApoE [75]. However, these studies are beginning to provide hints that WRN may indeed be involved in the “normal” aging process in the general population.
SUMMARY
Following the initial biochemical characterization of the helicase and exonuclease activities of WRNp, the identification of WRN-interacting proteins has led to a variety of studies exploring various ways in which WRNp may be involved in DNA metabolism. These include DNA repair at the site of stalled replication and NHEJ. WRN may also have a special role in transcription by RNA polymerases I and II, as well as in homologous recombination. Cell biological studies raise an interesting question regarding the role of WRN in telomere maintenance. If WRN is capable of all these functions, how do cells regulate the functions of WRN at any given time? How is the switching of these roles mediated? How are the relationships with other RecQ helicases regulated? Since symptoms of WS are relatively mild (not lethal), WRN may have evolved for the “fine tuning” of these various DNA metabolisms. If this hypothesis is true, what are the driving forces for the origin of WRN? At the animal level, how is WRN involved in the progression of “normal” aging phenotypes? I look forward to learning the answers to these and other important questions during the coming decades.
Acknowledgments
ACKNOWLEDGMENT
The author thanks Mr Kevin Fung and Mr Nhut Vuong for the editorial assistance.
References
- Yu C E, Oshima J, Fu Y H, et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272(5259):558–562. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- Gray M D, Shen J C, Kamath-Loeb A S, et al. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997;17(1):100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- Huang S, Li B, Gray M D, Oshima J, Mian I S, Campisi J. The premature ageing syndrome protein, WRN, is a 3′ → 5′ exonuclease. Nat Genet. 1998;20(2):114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefsbol T O, Cohen H J. Werner's syndrome: an underdiagnosed disorder resembling premature aging. Age. 1984;7:75–88. [Google Scholar]
- Ishikawa Y, Sugano H, Matsumoto T, Furuichi Y, Miller R W, Goto M. Unusual features of thyroid carcinomas in Japanese patients with Werner syndrome and possible genotype-phenotype relations to cell type and race. Cancer. 1999;85(6):1345–1352. [PubMed] [Google Scholar]
- Epstein C J, Martin G M, Schultz A L, Motulsky A G. Werner's syndromea review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine(Baltimore) 1966;45(3):177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- Goto M, Imamura O, Kuromitsu J, et al. Analysis of helicase gene mutations in Japanese Werner's syndrome patients. Hum Genet. 1997;99(2):191–193. doi: 10.1007/s004390050336. [DOI] [PubMed] [Google Scholar]
- Goto M. Hierarchical deterioration of body systems in Werner's syndrome: implications for normal ageing. Mech Ageing Dev. 1997;98(3):239–254. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- Goto M, Miller R W, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5(4):239–246. [PubMed] [Google Scholar]
- Johnson F B, Marciniak R A, McVey M, Stewart S A, Hahn W C, Guarente L. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001;20(4):905–913. doi: 10.1093/emboj/20.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y, Miller R W, Machinami R, Sugano H, Goto M. A typical osteosarcomas in Werner Syndrome (adult progeria) Jpn J Cancer Res. 2000;91(12):1345–1349. doi: 10.1111/j.1349-7006.2000.tb00924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postiglione A, Soricelli A, Covelli E M, et al. Premature aging in Werner's syndrome spares the central nervous system. Neurobiol Aging. 1996;17(3):325–330. doi: 10.1016/0197-4580(96)00033-4. [DOI] [PubMed] [Google Scholar]
- Rubin C D, Zerwekh J E, Reed-Gitomer B Y, Pak C Y. Characterization of osteoporosis in a patient with Werner's syndrome. J Am Geriatr Soc. 1992;40(11):1161–1163. doi: 10.1111/j.1532-5415.1992.tb01808.x. [DOI] [PubMed] [Google Scholar]
- Werner O Kiel University. Schmidt and Klauning; Kiel: 1904. On Cataract Associated in Conjunction with Scleroderma, [doctoral dissertation] [Google Scholar]
- Hisama F M, Oshima J, Yu C E, et al. Comparison of methods for identifying transcription units and transcription map of the Werner syndrome gene regio. Genomics. 1998;52(3):352–357. doi: 10.1006/geno.1998.5475. [DOI] [PubMed] [Google Scholar]
- Mushegian A R, Bassett D E. Jr, Boguski M S, Bork P, Koonin E V. Positionally cloned human disease genes: patterns of evolutionary conservation and functional motifs. Proc Natl Acad Sci USA. 1997;94(11):5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M J, Holley W R, Chatterjee A, Mian I S. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25(24):5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov V, Mushegian A R, Koonin E V, Bork P. A putative nucleic acid-binding domain in Bloom's and Werner's syndrome helicases. Trends Biochem Sci. 1997;22(11):417–418. doi: 10.1016/s0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Shimamoto A, Imamura O, et al. DNA helicase activity in Werner's syndrome gene product synthesized in a baculovirus system. Nucleic Acids Res. 1997;25(15):2973–2978. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Macias M J, Bottomley M J, et al. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure Fold Des. 1999;7(12):1557–1566. doi: 10.1016/s0969-2126(00)88346-x. [DOI] [PubMed] [Google Scholar]
- Ye L, Nakura J, Morishima A, Miki T. Transcriptional activation by the Werner syndrome gene product in yeast. Exp Gerontol. 1998;33(7-8):805–812. doi: 10.1016/s0531-5565(98)00044-8. [DOI] [PubMed] [Google Scholar]
- Balajee A S, Machwe A, May A, et al. The Werner syndrome protein is involved in RNA polymerase II transcription. Mol Biol Cell. 1999;10(8):2655–2668. doi: 10.1091/mbc.10.8.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Shimamoto A, Goto M, Furuichi Y. Impaired nuclear localization of defective DNA helicases in Werner's syndrome. Nat Genet. 1997;16(4):335–336. doi: 10.1038/ng0897-335. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Imamura O, Goto M, Furuichi Y. Characterization of the nuclear localization signal in the DNA helicase involved in Werner's syndrome. Int J Mol Med. 1998;1(1):71–76. doi: 10.3892/ijmm.1.1.71. [DOI] [PubMed] [Google Scholar]
- Oshima J, Yu C E, Piussan C, et al. Homozygous and compound heterozygous mutations at the Werner syndrome locus. Hum Mol Genet. 1996;5(12):1909–1913. doi: 10.1093/hmg/5.12.1909. [DOI] [PubMed] [Google Scholar]
- Yu C E, Oshima J, Wijsman E M, et al. Mutations in the consensus helicase domains of the Werner syndrome gene. Werner's syndrome collaborative group. Am J Hum Genet. 1997;60(2):330–341. [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Imamura O, Yamabe Y, et al. Mutation and haplotype analyses of the Werner's syndrome gene based on its genomic structure: genetic epidemiology in the Japanese population. Hum Genet. 1997;100(1):123–130. doi: 10.1007/s004390050477. [DOI] [PubMed] [Google Scholar]
- Moser M J, Oshima J, Monnat R J. Jr. WRN mutations in Werner syndrome. Hum Mutat. 1999;13(4):271–279. doi: 10.1002/(SICI)1098-1004(1999)13:4<271::AID-HUMU2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Ellis N A, Groden J, Ye T Z, et al. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83(4):655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Yamabe Y, Sugimoto M, Satoh M, et al. Down-regulation of the defective transcripts of the Werner's syndrome gene in the cells of patients. Biochem Biophys Res Commun. 1997;236(1):151–154. doi: 10.1006/bbrc.1997.6919. [DOI] [PubMed] [Google Scholar]
- Goto M, Yamabe Y, Shiratori M, et al. Immunological diagnosis of Werner syndrome by down-regulated and truncated gene products. Hum Genet. 1999;105(4):301–307. doi: 10.1007/s004399900151. [DOI] [PubMed] [Google Scholar]
- Satoh M, Imai M, Sugimoto M, Goto M, Furuichi Y. Prevalence of Werner's syndrome heterozygotes in Japan. Lancet. 1999;353(9166):1766. doi: 10.1016/S0140-6736(98)05869-3. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Shiratori M, Goto M, Furuichi Y. Werner syndrome helicase contains a 5′ → 3′ exonuclease activity that digests DNA and RNA strands in DNA/DNA and RNA/DNA duplexes dependent on unwinding. Nucleic Acids Res. 1999;27(11):2361–2368. doi: 10.1093/nar/27.11.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren D K, Brosh R M. Jr, Nehlin J O, Machwe A, Gray M D, Bohr V A. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27(17):3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R M. Jr, Orren D K, Nehlin J O, et al. Functional and physical interaction between WRN helicase and human replication protein A. J Biol Chem. 1999;274(26):18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- Shen J C, Gray M D, Oshima J, Loeb L A. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res. 1998;26(12):2879–2885. doi: 10.1093/nar/26.12.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M J, Kamath-Loeb A S, Jacob J E, Bennett S E, Oshima J, Monnat R J. Jr. WRN helicase expression in Werner syndrome cell lines. Nucleic Acids Res. 2000;28(2):648–654. doi: 10.1093/nar/28.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogburn C E, Oshima J, Poot M, et al. An apoptosis-inducing genotoxin differentiates heterozygotic carriers for Werner helicase mutations from wild-type and homozygous mutants. Hum Genet. 1997;101(2):121–125. doi: 10.1007/s004390050599. [DOI] [PubMed] [Google Scholar]
- Machwe A, Ganunis R, Bohr V A, Orren D K. Selective blockage of the 3′ → 5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA. Nucleic Acids Res. 2000;28(14):2762–2770. doi: 10.1093/nar/28.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Chen C Y, Kobayashi R, Newport J. Replication focus-forming activity 1 and the Werner syndrome gene product. Nat Genet. 1998;19(4):375–378. doi: 10.1038/1263. [DOI] [PubMed] [Google Scholar]
- Constantinou A, Tarsounas M, Karow J K, et al. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1(1):80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C Y, Graham J, Yan H. Evidence for a replication function of ffa-1, the xenopus orthologue of Werner syndrome protein. J Cell Biol. 2001;152(5):985–996. doi: 10.1083/jcb.152.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry M, Loeb L A. Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274(18):12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- Shen J C, Gray M D, Oshima J, Kamath-Loeb A S, Fry M, Loeb L A. Werner syndrome protein. I. DNA helicase and DNA exonuclease reside on the same polypeptide. J Biol Chem. 1998;273(51):34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- Shen J C, Loeb L A. Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 2000;28(17):3260–3268. doi: 10.1093/nar/28.17.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Comai L. Functional interaction between Ku and the Werner syndrome protein in DNA end processing. J Biol Chem. 2000;275(37):28349–28352. doi: 10.1074/jbc.C000289200. [DOI] [PubMed] [Google Scholar]
- Huang S, Beresten S, Li B, Oshima J, Ellis N A, Campisi J. Characterization of the human and mouse WRN 3′ → 5′ exonuclease. Nucleic Acids Res. 2000;28(12):2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Comai L. Requirements for the nucleolytic processing of DNA ends by the Werner syndrome protein-Ku70/80 complex. J Biol Chem. 2001;276(13):9896–9902. doi: 10.1074/jbc.M008575200. [DOI] [PubMed] [Google Scholar]
- Lebel M, Spillare E A, Harris C C, Leder P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J Biol Chem. 1999;274(53):37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- Szekely A M, Chen Y H, Zhang C, Oshima J, Weissman S M. Werner protein recruits DNA polymerase delta to the nucleolus. Proc Natl Acad Sci USA. 2000;97(21):11365–11370. doi: 10.1073/pnas.97.21.11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb A S, Johansson E, Burgers P M, Loeb L A. Functional interaction between the Werner syndrome protein and DNA polymerase delta. Proc Natl Acad Sci USA. 2000;97(9):4603–4608. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb A S, Loeb L A, Johansson E, Burgers P M, Fry M. Interactions between the Werner syndrome helicase and DNA polymerase δ specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J Biol Chem. 2001;276(19):16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- Harmon F G, Kowalczykowski S C. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12(8):1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc Natl Acad Sci USA. 1998;95(15):8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehn H, Bryant E M, Au K, Norwood T H, Boman H, Martin G M. Variegated translocation mosaicism in human skin fibroblast cultures. Cytogenet Cell Genet. 1975;15(5):282–298. doi: 10.1159/000130526. [DOI] [PubMed] [Google Scholar]
- Karran P. DNA double strand break repair in mammalian cells. Curr Opin Genet Dev. 2000;10(2):144–150. doi: 10.1016/s0959-437x(00)00069-1. [DOI] [PubMed] [Google Scholar]
- Kamath-Loeb A S, Shen J C, Loeb L A, Fry M. Werner syndrome protein. II. Characterization of the integral 3′ → 5′ DNA exonuclease. J Biol Chem. 1998;273(51):34145–34150. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- Shiratori M, Sakamoto S, Suzuki N, et al. Detection by epitope-defined monoclonal antibodies of Werner DNA helicases in the nucleoplasm and their upregulation by cell transformation and immortalization. J Cell Biol. 1999;144(1):1–9. doi: 10.1083/jcb.144.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsugi I, Tokutake Y, Suzuki N, Ide T, Sugimoto M, Furuichi Y. Telomere repeat DNA forms a large non-covalent complex with unique cohesive properties which is dissociated by Werner syndrome DNA helicase in the presence of replication protein A. Nucleic Acids Res. 2000;28(18):3642–3648. doi: 10.1093/nar/28.18.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz V P, Zakian V A, Ogburn C E, et al. Accelerated loss of telomeric repeats may not explain accelerated replicative decline of Werner syndrome cells. Hum Genet. 1996;97(6):750–754. doi: 10.1007/BF02346184. [DOI] [PubMed] [Google Scholar]
- Tahara H, Tokutake Y, Maeda S, et al. Abnormal telomere dynamics of B-lymphoblastoid cell strains from Werner's syndrome patients transformed by Epstein-Barr virus. Oncogene. 1997;15(16):1911–1920. doi: 10.1038/sj.onc.1201377. [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Ide T, Goto M, Furuichi Y. Reconsideration of senescence, immortalization and telomere maintenance of Epstein-Barr virus-transformed human B-lymphoblastoid cell lines. Mech Ageing Dev. 1999;107(1):51–60. doi: 10.1016/s0047-6374(98)00131-6. [DOI] [PubMed] [Google Scholar]
- Wyllie F S, Jones C J, Skinner J W, et al. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nat Genet. 2000;24(1):16–17. doi: 10.1038/71630. [DOI] [PubMed] [Google Scholar]
- Ouellette M M, McDaniel L D, Wright W E, Shay J W, Schultz R A. The establishment of telomerase-immortalized cell lines representing human chromosome instability syndromes. Hum Mol Genet. 2000;9(3):403–411. doi: 10.1093/hmg/9.3.403. [DOI] [PubMed] [Google Scholar]
- Choi D, Whittier P S, Oshima J, Funk W D. Telomerase expression prevents replicative senescence but does not fully reset mRNA expression patterns in Werner syndrome cell strains. FASEB J. 2001;15(6):1014–1020. doi: 10.1096/fj.00-0104com. [DOI] [PubMed] [Google Scholar]
- Hisama F M, Chen Y H, Meyn M S, Oshima J, Weissman S M. WRN or telomerase constructs reverse 4-nitroquinoline 1-oxide sensitivity in transformed Werner syndrome fibroblasts. Cancer Res. 2000;60(9):2372–2376. [PubMed] [Google Scholar]
- Spillare E A, Robles A I, Wang X W, et al. p53-mediated apoptosis is attenuated in Werner syndrome cells. Genes Dev. 1999;13(11):1355–1360. doi: 10.1101/gad.13.11.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Kipnis J, Leal J F, Yu C E, Schellenberg G D, Oren M. Physical and functional interaction between p53 and the Werner's syndrome protein. J Biol Chem. 1999;274(41):29463–29469. doi: 10.1074/jbc.274.41.29463. [DOI] [PubMed] [Google Scholar]
- Lombard D B, Beard C, Johnson B, et al. Mutations in the WRN gene in mice accelerate mortality in a p53-null background. Mol Cell Biol. 2000;20(9):3286–3291. doi: 10.1128/mcb.20.9.3286-3291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel M, Cardiff R D, Leder P. Tumorigenic effect of nonfunctional p53 or p21 in mice mutant in the Werner syndrome helicase. Cancer Res. 2001;61(5):1816–1819. [PubMed] [Google Scholar]
- Kawabe Y, Seki M, Seki T, Wang W S, et al. Covalent modification of the Werner's syndrome gene product with the ubiquitin-related protein, SUMO-1. J Biol Chem. 2000;275(28):20963–20966. doi: 10.1074/jbc.C000273200. [DOI] [PubMed] [Google Scholar]
- Lebel M, Leder P. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc Natl Acad Sci USA. 1998;95(22):13097–13102. doi: 10.1073/pnas.95.22.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ogburn C E, Ware C B, et al. Cellular Werner phenotypes in mice expressing a putative dominant-negative human WRN gene. Genetics. 2000;154(1):357–362. doi: 10.1093/genetics/154.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro E, Ogburn C E, Hunt K E, et al. Polymorphisms at the Werner locus: I. Newly identified polymorphisms, ethnic variability of 1367Cys/Arg, and its stability in a population of Finnish centenarians. Am J Med Genet. 1999;82(5):399–403. [PubMed] [Google Scholar]
- Passarino G, Shen P, Van Kirk J B, et al. The Werner syndrome gene and global sequence variation. Genomics. 2001;71(1):118–122. doi: 10.1006/geno.2000.6405. [DOI] [PubMed] [Google Scholar]
- Castro E, Edland S D, Lee L, et al. Polymorphisms at the Werner locus: II. 1074Leu/Phe, 1367Cys/Arg, longevity, and atherosclerosis. Am J Med Genet. 2000;95(4):374–380. [PubMed] [Google Scholar]
- Ye L, Miki T, Nakura J, et al. Association of a polymorphic variant of the Werner helicase gene with myocardial infarction in a Japanese population. Am J Med Genet. 1997;68(4):494–498. doi: 10.1002/(sici)1096-8628(19970211)68:4<494::aid-ajmg30>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Morita H, Kurihara H, Sugiyama T, Hamada C, Yazaki Y. A polymorphic variant C1367R of the Werner helicase gene and atherosclerotic diseases in the Japanese population. Thromb Haemost. 1999;82(1):160–161. [PubMed] [Google Scholar]
- Yamada H, Yamada Y, Fukatsu A, et al. Polymorphism of Werner helicase-associated gene in long-term hemodialysis patients. Nephron. 2000;86(4):43. doi: 10.1159/000045863. [DOI] [PubMed] [Google Scholar]