Abstract
The E1 and E2 proteins are both required for papillomavirus DNA replication, and replication efficiency is controlled by the abundance of these factors. In human papillomaviruses (HPVs), the regulation of E1 and E2 expression and its effect on viral replication are not well understood. In particular, it is not known if E1 and E2 modulate their own expression and how posttranscriptional mechanisms may affect the levels of the replication proteins. Previous studies have implicated splicing within the E6 open reading frame (ORF) as being important for modulating replication of HPV type 31 (HPV31) through altered expression of E1 and E2. To analyze the function of the E6 intron in viral replication more specifically, we examined the effects of E6 splicing mutations in the context of entire viral genomes in transient assays. HPV31 genomes which had mutations in the splice donor site (E6SD) or the splice acceptor site (E6SA), a deletion of the intron (E6ID), or substituted heterologous intron sequences (E6IS) were constructed. Compared to wild-type (wt) HPV31, pHPV31-E6SD, -E6SA, and -E6IS replicated inefficiently while pHPV31-E6ID replicated at an intermediate level. Cotransfection of the E6 mutant genomes with an E1 expression vector strongly activated their replication levels, indicating that efficient expression of E1 requires E6 internal splicing. In contrast, replication was activated only moderately with an E2 expression vector. Replacing the wt E6 intron in HPV31 with a heterologous intron from simian virus 40 (E6SR2) resulted in replication levels similar to that of the wt in the absence of expression vectors, suggesting that mRNA splicing upstream of the E1 ORF is important for high-level replication. To examine the effects of E6 intron splicing on E1 and E2 expression directly, we constructed reporter DNAs in which the luciferase coding sequences were fused in frame to the E1 (E1Luc) or E2 (E2Luc) gene. Reporter activities were then analyzed in transient assays with cotransfected E1 or E2 expression vectors. Both reporters were moderately activated by E1 in a dose-dependent manner. In addition, E1Luc was activated by low doses of E2 but was repressed at high doses. In contrast, E2 had little effect on E2Luc activity. These data indicate that E1 expression and that of E2 are interdependent and regulated differentially. When the E6 splicing mutations were analyzed in both reporter backgrounds, only E1Luc activities correlated with splicing competence in the E6 ORF. These findings support the hypothesis that the E6 intron primarily regulates expression of E1. Finally, in long-term replication assays, none of the E6 mutant genomes could be stably maintained. However, cotransfection of the E6 splicing mutant genomes with pHPV31-E7NS, which contains a nonsense mutation in the E7 coding sequence, restored stable replication of some mutants. Our observations indicate that E1 expression and that of E2 are differentially regulated at multiple levels and that efficient expression of E1 is required for transient and stable viral replication. These regulatory mechanisms likely act to control HPV copy number during the various phases of the viral life cycle.
Papillomaviruses are small DNA viruses that infect the epithelia of their vertebrate hosts. To date, more than 85 types of human papillomaviruses (HPVs) have been identified, of which a small number, high-risk types HPV16, -18, -31, and -33, are causally associated with neoplasia. HPVs infect basal keratinocytes and maintain their genomes as low-copy-number nuclear plasmids. The production of viral progeny occurs only in suprabasal cells following terminal differentiation (16). Under physiological conditions, viral DNA replication requires the action of both E1 and E2 (4, 55, 58), which form a complex at the replication origin (ori) near the 3" terminus of the upstream regulatory region (URR; Fig. 1A) (8, 27, 34, 35, 43, 45). While it has been determined that viral replication efficiency depends on the abundance of the E1 and E2 proteins (3, 18, 27, 30, 33, 38, 60), it is not clear how the expression of these viral replication factors is regulated during the life cycle of HPVs. In particular, it has not been established how the abundance of the E1 and E2 proteins is affected by transcriptional or posttranscriptional mechanisms of regulation. Recently, experimental systems have been developed which permit the analysis of the replication properties of HPV31 in cell culture models throughout the three phases of the viral life cycle: establishment, maintenance, and DNA amplification (9, 26, 52). Short-term replication assays closely mimic the establishment phase of replication, while long-term replication assays mimic viral maintenance in undifferentiated basal cells. Finally, DNA amplification and virion assembly, which are hallmarks of productive viral infection, can be analyzed following keratinocyte differentiation either in organotypic raft cultures or by suspension in semisolid medium.
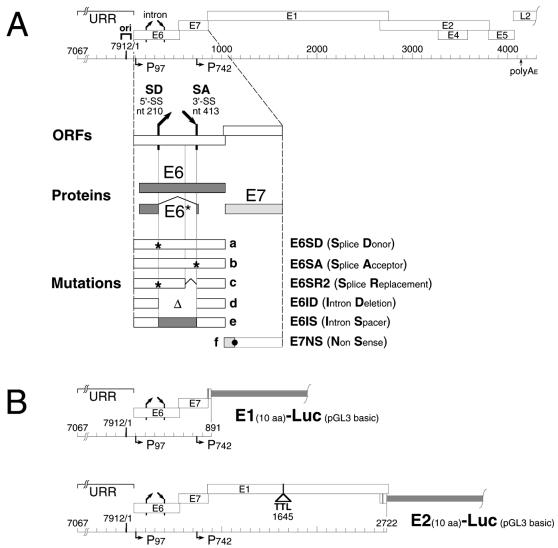
FIG. 1.
Diagrams for plasmids and mutations used in this study. (A) Early region of the HPV31 genome (top) with the URR, replication origin (ori), ORFs (open boxes), promoters (horizontal arrows), and selected mRNA splice sites (slanted arrows). The enlargement (bottom) shows ORFs E6 and E7 (open boxes), their proteins (stippled boxes), and the location of the mutations. Nucleotide positions refer to HPV31 unless stated otherwise. a, inactivated splice donor (nt 212T→C); b, inactivated splice acceptor (nt 411A→C); c, inserted splice replacement (HPV31 nt 344→415 replaced with SV40 small t antigen intron from pGL2 control [Promega; pGL2 nt 2157→2228], nt 212T→C, and nt 342T→C); d, E6 intron deletion including nt 211→412; e, inserted E6 intron spacer replacing HPV31 nt 344→415 with amino phosphotransferase coding sequences from pREP10 (Stratagene; pREP10, nt 248→449); f, E7 nonsense mutation (nt 610T→C and nt 611G→T, E7 Glu18→amber). (B) Schematic maps of HPV31 E1- and E2-specific luciferase reporters E1Luc and E2Luc, respectively, in the pGL3 basic parental plasmid (Promega). In each reporter, the firefly luciferase gene is fused in frame to the first 10 codons of the specified HPV gene. E1Luc contains nt 7067 to 892 from HPV31. E2Luc contains nt 7067 to 2723 from HPV31. The E1 gene in E2Luc is inactivated with a six-frame translational termination linker at nt 1645. aa, amino acids.
Analysis of viral gene expression in undifferentiated cells has identified a major transcriptional promoter in HPV31, designated P97, which is activated by cellular proteins that bind to the keratinocyte enhancer in the URR (28). mRNAs expressed from P97 are polycistronic and abundant in undifferentiated as well as differentiated cells (19, 20). Among the transcripts expressed from P97 are those encoding E1 and E2. By binding to an E2 binding site located proximal to the TATA box of P97, the E2 protein has been shown to negatively regulate this promoter as well as viral replication (54). This modulation of P97 activity has been hypothesized to contribute to copy number control in basal cells. It is not known, however, if P97 is the only promoter which directs the expression of E1 and E2 or if other promoters are involved (36). Furthermore, it has not been established with HPVs how the levels of E1 and E2 affect their own expression and if posttranscriptional mechanisms contribute to regulation.
Many DNA tumor viruses, such as simian virus 40 (SV40) (7, 24, 59) and human adenoviruses (types 2, 11, and 15), use mRNA splicing to modulate viral gene expression (5, 23, 32). In HPV31, the splicing of an intron located at nucleotides (nt) 211 to 412 in the E6 open reading frame (ORF) could potentially modulate E1 and E2 gene expression since these mRNA splice sites lie upstream of the coding sequences for both replication factors. This hypothesis is supported by the findings of recent replication studies which have demonstrated that mutations in the E6 ORF splicing signals decrease the transient replication levels of HPV31 genomes (57). Modulating the splicing efficiency of the E6 intron could thus provide an additional mechanism for regulating the abundance of the E1 and E2 proteins.
In this study, we examined the role of the splicing of the E6 intron in the regulation of HPV31 DNA replication during viral establishment and maintenance. In addition, the transcriptional regulation of E1 and E2 expression was analyzed in transient reporter assays. We determined that splicing of the intron located at nt 211 to 412 is essential for efficient viral replication during the early phase of viral infection and that it primarily affects E1 expression. We also found that E1 expression and that of E2 are regulated differentially by the replication factors themselves. These transcriptional and posttranscriptional control mechanisms could contribute to the regulation of the HPV31 plasmid copy number in all phases of the viral life cycle by differentially modulating the levels of the replication factors.
MATERIALS AND METHODS
Plasmids.
Wild-type (wt) pBRmin-HPV31 DNA was used as the parental viral genome. It contains a minimal pBR322 vector at the unique EcoRI site in the E2 coding sequence (18). All HPV31 nucleotide references are based on the published wt sequence (11). The locations and types of E6 mutations generated for this study are shown schematically in Fig. 1A. The viral splice mutants pHPV31-E6SD (splice donor; nt 212T→C) and pHPV31-E6SA (splice acceptor; nt 411A→C) contain inactivated 5" and 3" splice sites in the E6 coding sequence at nt 210 and 413, respectively, and have been described previously (57). A set of HPV31 genomes containing new E6 intron mutations were generated by overlap extension PCR (2): pHPV31-E6SR2, splice replacement; pHPV31-E6ID, intron deletion; pHPV31-E6IS, intron spacer. Mutated DNA fragments were generated and joined with oligonucleotide primers which contain DNA sequences from two heterologous PCR templates. The mutated PCR products were cloned into pBRmin-HPV31 (18) by SpeI-to-BanII fragment exchange. All PCR-generated fragments in the HPV31 genomes were sequenced. In the pHPV31-E6SR2 genome, the intron from the small t antigen of SV40 (pGL2 control nt 2157 to 2228; Promega) was combined with pHPV31-E6SD sequences (nt 7557 to 343) at the 5" end and wt HPV31 sequences (nt 416 to 811) at the 3" end. In addition to the small t antigen intron, pHVP31-E6SR2 also contains a point mutation (nt 342T→C) to inhibit potential mRNA splicing from a cryptic HPV31 site. The splice replacement fragment is of the same size as the wt one. The E6SR2 mutation inhibits expression of full-length E6 as well as E6*. pHPV31-E6ID was generated with a primer that spanned the E6* splice junction. The deleted HPV31 sequences include nt 211 through 412. The E6ID mutation inhibits full-length E6 expression but not E6* expression. pHPV31-E6IS contains wt HPV31 DNA (nt 7557 to 210) fused in frame to sequences from the hygromycin resistance gene (pREP10 nt 248 to 449; Invitrogen-Life Technologies) and again wt HPV31 sequences (nt 413 to 811). The intron spacer fragment is of the same size as the wt one. The E6IS mutation inhibits expression of functional full-length E6 and E6*. In the pHPV31-E7NS (nonsense) genome, point mutations were generated in the E7 coding sequence by PCR (nt 610T→C and nt 611G→T). These mutations change a wt Bsu36I site to a unique AvrII site and introduce an amber stop codon in place of the codon for amino acid position 18 of E7. An HpaI-to-BanII fragment with the nonsense mutation was replaced in pBRmin-HPV31. pHPV31-E7NS is competent for the expression of full-length E6 and E6* but not E7.
The HPV31-based luciferase reporter DNAs are both based on pPro983 (22), a pGL3 basic (Promega) derivative, and are shown schematically in Fig. 1B. The E1-specific luciferase reporter (E1Luc) contains wt HPV31 sequences from nt 7045 to 892, and the first 10 codons of the E1 coding sequence are fused in frame to the firefly luciferase gene. The E2-specific luciferase reporter (E2Luc) contains wt HPV31 sequences from nt 7045 to 2723, and the first 10 codons of the E2 coding sequence are fused to the luciferase gene. To inhibit potential E1 expression in E2Luc, a 14-bp translational termination linker (39) with stop codons in all six reading frames was inserted in the E1 coding sequence at the unique SwaI site (nt 1641). All PCR-generated fragments in the luciferase reporters were sequenced. The E6 coding sequence mutations described above (E6SD, E6SA, E6SR2, E6ID, and E6IS) were transferred from the HPV31 genomes into the E1Luc and E2Luc reporter DNAs as SpeI-to-BanII fragments, and the resulting reporters were designated accordingly.
Transient DNA replication analysis.
The short-term replication assay was performed essentially as described previously (18). Briefly, 3 μg (based on HPV31; 7,912 bp) of unimolecularly ligated HPV31 DNAs was transfected into HPV-negative human squamous cell carcinoma 13 (SCC13) cells (42) (5 × 106 cells) by electroporation. SCC13 cells were cocultured with mitomycin-treated fibroblast feeders throughout the experiment. As indicated in the figure legends, viral genomes were also cotransfected with pSG-E1 and pSG-E2 vectors for heterologous expression of the viral replication factors (8). Low-molecular-weight DNA was isolated 5 days posttransfection by a modified Hirt (15) extraction method and analyzed by Southern blotting and hybridization as previously described (18). The relative transient replication levels in Fig. 2A were determined by quantitative analysis of a phosphorimage (Molecular Dynamics). The integrated areas from the replicated DNA bands of the E6 intron mutants (Fig. 2A, bars 2 through 6) were each divided by the area of the wt band. For example, the value for bar 2 is equal to the intensity of lane 2 divided by the intensity of lane 1. The relative activation of transient replication, as determined by complementing the HPV31 genomes with either the pSG-E1 (Fig. 2B) or pSG-E2 (Fig. 2C and D) expression vector in trans, was calculated for the wt and each mutant separately. For any given HPV31 DNA, the replicated DNA amount with the expression vector was divided by the respective DNA amount replicated without the vector (autonomous level). For example, in Fig. 2C, the value for bar 8 is equal to the amount of replicated DNA from Fig. 2C, lane 8, divided by the amount of replicated DNA from Fig. 2B, lane 2.
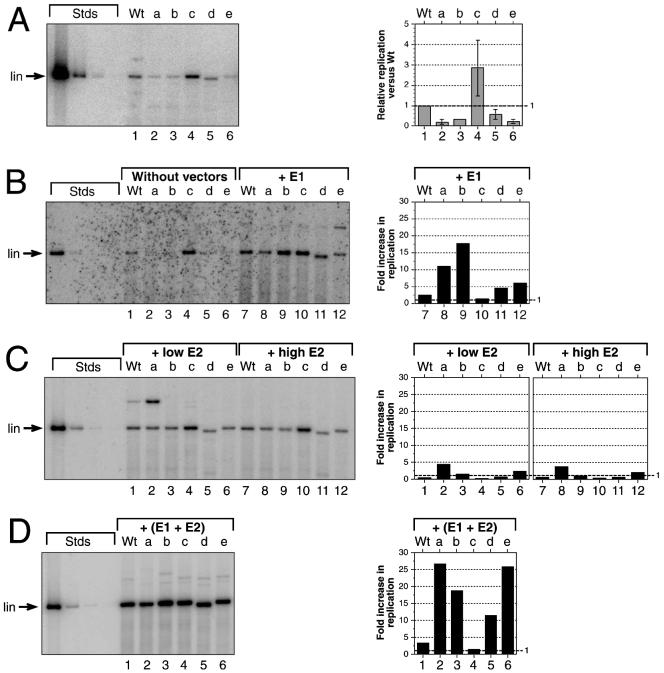
FIG. 2.
The effects of splicing mutations in the E6 ORF on transient replication of HPV31 genomes. SCC13 cells were either transfected with ligated viral DNAs by themselves or cotransfected with HPV31 DNA and constant amounts of expression vectors for E1 or E2 or both. The amounts of replicated DNAs were quantified and graphed for each panel. Wt, wt HPV31. Mutations in the HPV31 genome background: a, pHPV31-E6SD; b, pHPV31-E6SA; c, pHPV31-E6SR2; d, pHPV31-E6ID; e, pHPV31-E6IS. Each autoradiogram contains HPV31 standards (Stds): 500, 25, 2.5, and 0.5 pg of linearized DNA (7,912 bp) (lin, migratory position in 0.8% agarose gels). The DNA bands migrating more slowly than the linearized samples in panels B through D are due to multimeric forms generated during the initial ligation and transfection. (A) Autoradiogram of replicating of viral genomes without expression vectors. Graphs indicate the relative replication levels of mutants versus wt and represent the averages ± standard deviations of samples quantified from panels A and B, lanes 1 through 6. (B) Autoradiogram of replicating viral genomes without expression vectors (lanes 1 through 6) or in the presence of equimolar amounts (versus the HPV genome) of the E1 expression vector (lanes 7 through 12). Bars 7 through 12 indicate how the replication level of each DNA is increased by the cotransfected E1 vector. (C) Autoradiogram of replicating viral genomes in the presence of small (0.1 M) or large (equimolar) amounts of the E2 expression vector. The graph indicates how the replication level of each DNA is increased by the small (bars 1 through 6) and large (bars 7 through 12) amounts of cotransfected E2 vector. (D) Autoradiogram of replicating viral genomes with equimolar amounts of E1 and E2 expression vectors. The graph indicates how the replication level of each DNA is increased by the presence of both vectors.
Transient reporter analysis.
SCC13 cells (5 × 105 cells) were transfected with 0.5 μg (based on wt E1Luc; 6,513 bp) of wt and E6 mutant E1Luc or E2Luc DNA and Lipofectamine essentially as prescribed by the manufacturer (Invitrogen-Life Technologies). pSG-E1 and pSG-E2 expression vectors (8) were cotransfected at molar ratios indicated in Fig. 3 and 4. To rule out nonspecific effects potentially induced by the presence of various amounts of heterologous promoter sequences, the molar amounts of vector were kept constant by addition of the parental pSG5 expression vector (Stratagene). Lysates were prepared from transfected cells 40 h posttransfection, and luciferase assays were performed according to the manufacturer's instructions (Promega). Protein concentrations of lysates were determined by the method of Bradford (Bio-Rad). Relative luciferase activities in Fig. 3 were determined separately for each wt reporter (E1Luc and E2Luc) and expression vector (pSG-E1 and pSG-E2) combination. Luciferase activities at a given molar ratio of expression vector to reporter were divided by the basal activity of the reporter DNA (no expression vector, molar ratio of 0.001 in Fig. 3). The relative luciferase activities of the reporter DNAs which contain E6 intron mutations in Fig. 5A were calculated separately for E1Luc and E2Luc. The mutant reporter activities were divided by their respective wt reporter activities.
FIG. 3.
The response of E1Luc and E2Luc reporters to increasing amounts of E1 or E2. Shown are activities from transient reporter assays. SCC13 cells were cotransfected with constant amounts of reporter DNA and increasing amounts of either an E1 or E2 expression vector. Mutations in both reporter backgrounds: a, E6SD; b, E6SA; c, E6SR2; d, E6ID; e, E6IS. Each datum represents the average specific luciferase activity of two independent transfections (relative light units[RLU] per microgram of lysate protein) versus the molar ratio of expression vector to reporter DNA. Error bars, standard deviations (at some data points, bars are obscured by the symbol). The value labels show the relative activities versus that for transfection without the expression vector (basal activity). To facilitate graphing on a log scale, the datum of the zero expression vector was assigned a molar ratio of 0.001. (A) Dose response of E1Luc versus E1 (0 to 10 molar ratio). (B) Dose response of E1Luc versus E2 (0 to 5 molar ratio). (C) Dose response of E2Luc versus E1 (0 to 10 molar ratio). (D) Dose response of E2Luc versus E2 (0 to 5 molar ratio).
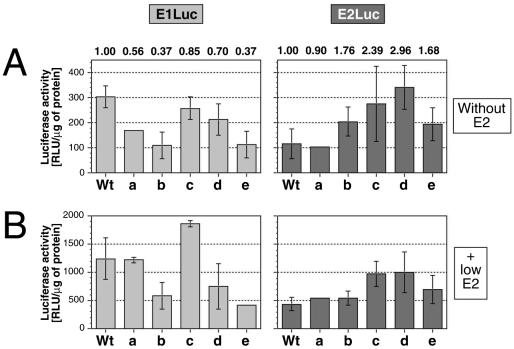
FIG. 5.
The response of E1Luc and E2Luc reporters containing mutations in the E6 splice sites to increasing amounts of E1 or E2. The graphs show the specific E1Luc and E2Luc reporter activities from transient transfection assays with SCC13 cells. Transfection mixtures contained a reporter DNA, and transfections were performed without the E2 vector (basal) or with a 0.1 molar ratio of E2 expression vector as indicated. Mutations in both reporter backgrounds: a, E6SD; b, E6SA; c, E6SR2; d, E6ID; e, E6IS. Each datum represents the average specific luciferase activity of two independent transfections (relative light units [RLU] per microgram of lysate protein). Error bars, standard deviations. (A) Absolute basal activities of the E1Luc and E2Luc reporters. The value labels show the relative activities of mutants versus wt in each reporter background. (B) Absolute E2-induced activities of the E1Luc and E2Luc reporters in the presence of 0.1 M amounts of the E2 expression vector.
Stable DNA replication analysis.
The long-term replication assay was performed essentially as described previously (18). Briefly, 3 μg (based on HPV31; 7,912 bp) of unimolecularly ligated HPV31 DNAs and equimolar amounts of pSV2-neo (46) were cotransfected into subconfluent cultures of primary human foreskin keratinocytes (HFK) using Lipofectamine (Invitrogen-Life Technologies). As indicated in the legend of Fig. 6, viral genomes were also cotransfected with a threefold molar excess of unimolecularly ligated pHPV31-E7NS DNA. Transfected cells were selected briefly with G-418 (Invitrogen-Life Technologies), and drug-resistant colonies were expanded as mass culture cell lines. Total cellular DNA and low-molecular-weight DNA were prepared from parallel cultures at 4 to 6 weeks posttransfection and analyzed by Southern blotting and hybridization as previously described (18). The relative stable replication levels of the E6 intron mutant genomes in Fig. 6B were determined by quantitative analysis of a phosphorimage (Molecular Dynamics). The integrated areas from the replicated low-molecular-weight DNA bands of the mutants (bars 8 through 12) were each divided by the area of the wt band. For example, the value for bar 8 is equal to the intensity of lane 8 divided by the intensity of lane 7. The relative stable replication level of mutant f in Fig. 6D (bar 14) was obtained similarly by dividing the amount of replicated DNA from bar 14 of Fig. 6D by the amount of replicated DNA from bar 7 of Fig. 6B. The relative activation of stable replication (Fig. 6D), as determined by complementing the HPV31 wt and E6 intron mutant genomes with pHPV31-E7NS in trans was calculated for the wt and each mutant separately. For any given HPV31 DNA, the integrated area of the replicated DNA band with pHPV31-E7NS was divided by the respective area of the DNA band without pHPV31-E7NS (autonomous level). For example, for Fig. 6B, the value for bar 7 is equal to the amount of replicated DNA from the lower band (mutant) of lane 7 in Fig. 6D divided by the amount of replicated DNA from lane 8 in Fig. 6B.
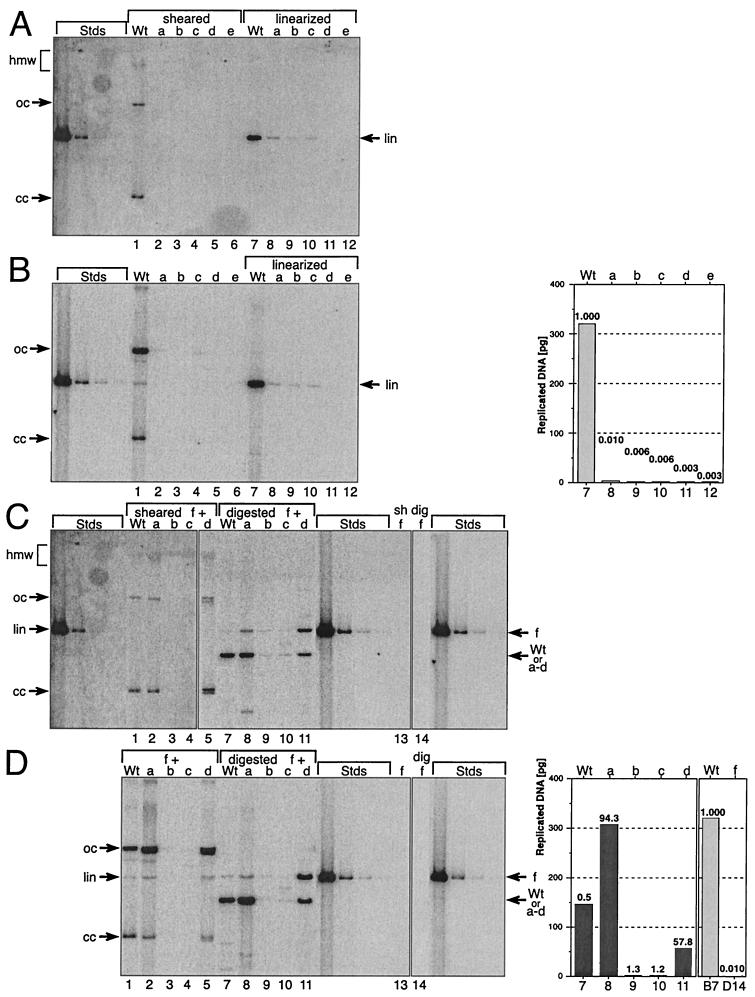
FIG. 6.
Stable replication of HPV31 genomes containing mutant E6 splice sites. Shown is the stable replication of wt and mutant HPV31 genomes: Wt, wt HPV31; a, pHPV31-E6SD; b, pHPV31-E6SA; c, pHPV31-E6SR2; d, pHPV31-E6ID; e, pHPV31-E6IS. Primary HFK were stably transfected with individual viral DNAs (A and B) or paired DNAs (C and D), where the wt and mutants a through e were each cotransfected with mutant f (pHPV31-E7NS). Each autoradiogram contains HPV31 standards (Stds): 500, 25, 2.5, and 0.5 pg of linearized DNA (7,912 bp). Aliquots (5 μg) of total cellular DNA were analyzed in panels A and C; therefore, the DNA standards represent about 100, 5, 0.25, and 0.1 viral genomes per cell, respectively. Arrows, migration of the different topological forms of the viral DNA. A high-molecular-weight (hmw) smear indicates that the HPV DNA is either integrated into the cellular genome or replicates as a plasmid multimer. The covalently closed (cc) and open (oc) formsof HPV DNA result from autonomously replicating plasmid monomers. Linearized (lin) HPV DNA bands were used to quantify the replicated viral DNA. (A) Autoradiogram from transfections with individual HPV31 genomes (wt and mutants a through e). DpnI-resistant, sheared-total-DNA samples are shown on the left (lanes 1 to 6), and the linearized ones are shown on the right (lanes 7 to 12). (B) Autoradiogram showing low-molecular-weight (Hirt) DNA isolated and analyzed from the same cell lines as in panel A. DpnI-resistant samples are shown on the left (lanes 1 to 6), and linearized ones are shown on the right (lanes 7 to 12). The linearized sample bands were quantified and graphed. The value labels indicate the relative replication levels of mutants versus wt. (C) Autoradiogram from transfections with paired HPV31 genomes (wt and mutants a through d, each paired with f). Lanes 1 to 5, DpnI-resistant-, sheared-total-DNA samples; lanes 7 to 12, digested-DNA samples; lanes 13 (sheared) and 14 (digested), stable replication of pHPV31-E7NS (f) by itself. Arrows, positions of pHPV31-E7NS (f) and the cotransfected DNA (wt and a to d). (D) Autoradiogram showing low-molecular-weight (Hirt) DNA isolated and analyzed from the same cell lines as in panel C. DpnI-resistant samples (left; lanes 1 to 5) and digested ones (middle; lanes 7 to 11) are shown. Lanes 13 and 14, stable replication of pHPV31-E7NS (f) by itself (digested). Arrows, positions of pHPV31-E7NS (f) and cotransfected DNA (wt and a to d). The digested lower sample bands (wt, a to d) were quantified and graphed (right). The value labels show the relative increases of stable replication levels of the sample DNAs with pHPV31-E7NS (f) compared to the autonomous levels (panel B). The replication of wt (panel B, lane 7) and pHPV31-E7NS (panel D, lane 14) in single DNA transfections are compared (far right). The value labels in graphs 7 and 14 indicate the relative replication levels of pHPV31-E7NS versus wt.
RESULTS
mRNA splicing within the E6 ORF is required for high-level transient HPV31 replication.
To investigate how E1 and E2 expression is regulated during the productive life cycle of HPV31, we examined how splicing in the E6 ORF affects the efficiency of viral DNA replication. Previous studies indicated that the splicing of the E6 intron was required for efficient transient replication of viral genomes. To analyze the specific role of E6 intron splicing in viral regulation, a series of HPV31 genomes which contain altered mRNA splice sites were constructed. A mutant which contains an inactivated 5" splice site at nt 210 (pHPV31-E6SD) and one which contains an inactivated 3" splice site at nt 413 (pHPV31-E6SA; Fig. 1) (57) have been described previously. Additional mutant genomes which contain a deletion of the entire E6 intron (pHPV31-E6ID) or a replacement of the entire intron with heterologous spacer sequences (pHPV31-E6IS) were also generated. These constructs were used in experiments to address whether the process of splicing by itself is important for efficient replication or if the sequences within the intron exert a negative effect on expression of the replication proteins. The replication efficiency of the HPV31 mutants was first tested under conditions where both replication factors are expressed from the viral genome. For the transient replication assay, the bacterial vector sequences were removed from the HPV31 genomes by restriction enzyme digestion, and the viral DNAs were transfected into HPV-negative SCC13 cells. Five days posttransfection, low-molecular-weight DNA was isolated, and the replicated (DpnI-resistant) viral DNA was analyzed by Southern blotting and hybridization as described in Materials and Methods (18).
Consistent with previous observations (57), the replication efficiency of splicing-deficient pHPV31-E6SD and pHPV31-E6SA was found to be significantly reduced compared to that of wt HPV31 (Fig. 2A and B). Splicing-deficient mutant pHPV31-E6IS also replicated at a low level (20% of the wt level; Fig. 2A), while intron deletion mutant pHPV31-E6ID replicated at approximately 50% of the wt HPV31 level. The results with the pHPV31-E6SD, -E6SA, and -E6IS genomes indicate that the splicing of the E6 intron is required for high-level replication of the viral genome. The inefficient replication phenotype of these E6 splicing mutants does not correlate with E6 expression since only pHPV31-E6IS and -E6ID are E6 deficient. Furthermore, it has been shown previously that E6 is not required for transient viral replication (57). To test if the introduction of a heterologous intron could restore the replication efficiency of E6 splicing-defective HPV31 mutants, the intron from the small t antigen of SV40 was inserted into the pHPV31-E6SD genome to generate pHPV31-E6SR2. In the transient replication assays, virus carrying splicing-competent pHPV31-E6SR2 was found to replicate with a higher efficiency than wt HPV31 (Fig. 2A). pHPV31-E6SR2 is deficient for the expression of full-length E6 because of an ochre stop codon within the heterologous intron. It is also deficient for E6* expression because of the E6SD mutation. Therefore, the high replication efficiency of the E6SR2 genome may, in part, be the result of increased mRNA stability or more-efficient translation of downstream ORFs.
The most plausible explanation for the low-replication phenotype of the E6 splice mutants is that efficient expression of the viral replication factors requires mRNA splicing in the E6 ORF. To analyze if pHPV31-E6SD, -E6SA, and -E6IS were unable to express sufficient levels of either E1 or E2, we cotransfected all E6 splicing mutants with E1 or E2 expression vectors as described in Materials and Methods (18). In the first set of experiments, equimolar amounts of the E1 vector were cotransfected with the HPV31 genomes. The addition of exogenous E1 activated replication of the splicing-deficient pHPV31-E6SD, -E6SA, and -E6IS genomes by 5- to 17-fold (Fig. 2B) compared to their autonomous replication levels. These results indicate that addition of E1 alone can restore replication efficiency of the splicing-defective mutant genomes. Exogenous E1 also elevated the replication efficiencies of the splicing-competent wt HPV31 and pHPV31-E6SR2 but to a much lesser degree, 2.0- and 1.2-fold, respectively (Fig. 2B). We then tested if the replication ability of the splicing mutants could be restored by cotransfecting an E2 expression vector with the viral genomes. In the presence of either large or small amounts of E2 vector (0.1 or 0.5 molar ratio of vector to viral genome), replication of the splicing-deficient genomes was only moderately activated compared to their autonomous levels (Fig. 2C). While exogenous E2 increased the efficiency of pHPV31-E6SD fourfold, little activation was seen with the other genomes (Fig. 2C). Interestingly, the splicing-competent wt HPV31 and pHPV31-E6SR2 actually replicated at a lower level in the presence of exogenous E2 (Fig. 2C). Since addition of E1 expression vectors could restore the replication efficiencies of splicing-defective mutants pHPV31-E6SD, -E6SA, and -E6ID, we conclude that the splicing of the E6 intron has a pronounced effect on E1 expression but not on E2 expression.
To exclude the possibility that the mutations introduced into E6 might have a cis-acting effect which diminishes the efficiency of the viral replication origin, we also analyzed replication of the HPV31 mutants under conditions of excess E1 and E2. Consistent with previous observations (18, 57), cotransfection of equimolar amounts of E1 and E2 vectors led to overall high replication levels (Fig. 2D). The presence of both expression vectors increased replication of the splicing-deficient mutants 10- to 25-fold. The replication levels of the tested HPV31 genomes were comparable, indicating that the E6 splicing mutations had little effect on the viral origin.
E1 gene expression and that of E2 are autoregulated, interdependent, and differentially modulated by DNA replication.
The preceding analysis determined the effects of E1 and E2 expression indirectly by assessing the ability of mutant genomes to replicate in short-term assays. We next sought to examine the regulatory circuits that govern the expression of either E1 or E2 more directly by means of reporter assays. For these studies, E1- and E2-specific luciferase gene fusion reporters were constructed. The E1Luc and E2Luc reporters contain all upstream DNA sequences from the 5" terminus of the URR to the start of each protein coding sequence. Specifically, the luciferase gene is fused in frame to the first 10 codons of either the E1 or E2 coding sequence. To prevent potential expression of E1 from the E2Luc reporter, the E1 gene was inactivated by inclusion of a translational termination linker (see Materials and Methods) (39). Therefore, since neither the E1Luc nor the E2Luc reporter can produce any replication factors, their activities are only regulated by exogenous sources of E1 and E2.
In the first set of experiments, we examined the dose response of the reporters in the presence of only one replication factor. SCC13 cells were cotransfected with reporter DNAs and increasing amounts of expression vectors for either E1 or E2 (Fig. 3). Expression of heterologous E1 moderately activated both reporter activities in a dose-dependent manner (Fig. 3A and C). In contrast, expression of E2 increased E1Luc activity only at low vector doses (molar ratio of vector versus reporter below 0.25), while at high vector dose (up to 5.0 molar excess), the E1Luc activity declined to nearly basal levels (Fig. 3B). Interestingly, E2 did not significantly alter E2Luc activity at any tested vector dose (Fig. 3D). These findings indicate that E1 and E2 differentially regulate their expression. Furthermore, E2 modulates E1 expression but not on its own expression.
To test if replication modulates reporter activity, E1Luc and E2Luc were cotransfected with both expression vectors. Transfection mixtures contained constant amounts of a reporter as well as one replication factor and increasing levels of the other replication factor. In the presence of constant E2, E1Luc activity was increased moderately by E1 in a dose-dependent manner (Fig. 4A and C, respectively). Similarly, the activity of E2Luc in the presence of constant E2 increased moderately with added E1. The response profiles of the E1- and E2Luc reporters in the presence of increasing E1 and constant E2 were similar to those with E1 alone, although the reporter activities were increased under replicating conditions. In response to high doses of E2 (Fig. 4B and D) at constant E1 levels, the activity of the E1Luc reporter and that of the E2Luc reporter decreased in a similar manner. Interestingly, the activity of E1Luc appeared to be increased more strongly under replicating conditions (10-fold) than that of E2Luc (3-fold) (compare Fig. 4A and B with C and D). We conclude from these data that E1 expression and that of E2 in HPV31 are interdependent and generally activated by replication.
FIG. 4.
Response of E1Luc and E2Luc reporters to the combination of E1 and E2. SCC13 cells were transfected in transient assays, and the resulting reporter activities were graphed. All cotransfection mixtures contained a constant amount of reporter DNA (E1Luc or E2Luc) and increasing amounts of one expression vector (E1 or E2 vector). In addition, transfections were performed either without or with a constant amount of the second expression vector as indicated (molar ratio versus reporter). Mutations in both reporter backgrounds: a, E6SD; b, E6SA; c, E6SR2; d, E6ID; e, E6IS. In each panel, the graph shows the specific luciferase activities resulting from transfections with three DNAs (reporter plus E1 and E2 DNAs [solid lines and symbols]) or with two DNAs (reporter plus one vector [dashed lines and open symbols]) to assess the contribution of replication to reporter activity. Each datum represents the average specific luciferase activity of two independent transfections (relative light units [RLU] per microgram of lysate protein) versus the molar ratio of expression vector versus reporter DNA. Error bars, standard deviations (at some data points, bars are obscured by symbols). To facilitate graphing on a log scale, the datum of the zero expression vector was assigned a molar ratio of 0.001. (A) Dose response of E1Luc versus E1 (0 to 10 molar ratio) in the absence or presence of constant 0.1 M amounts of E2 vector. (B) Dose response of E1Luc versus E2 (0 to 5 molar ratio) in the absence or presence of constant 0.2 M amounts of E1 vector. (C) Dose response of E2Luc versus E1 (0 to 10 molar ratio) in the absence or presence of constant 0.1 M amounts of E2 vector. (D) Dose response of E2Luc versus E2 (0 to 5 molar ratio) in the absence or presence of constant 0.2 M amounts of E1 vector.
E6 intron splicing differentially regulates E1 and E2 expression.
We next investigated the role of E6 internal mRNA splicing in the regulation of E1 and E2 expression by using luciferase assays as described above. For these studies, the splicing mutations were transferred from the HPV31 genomes into both E1Luc and E2Luc reporter backgrounds. SCC13 cells were then transfected with wt and mutant reporters and assayed for activity. The splicing-deficient E1Luc reporters with the splice donor (E6SD), splice acceptor (E6SA), and intron spacer (E6IS) mutations showed a significant decrease in activity compared to wt E1Luc (Fig. 5A, left). The activity of the splicing-competent E6SR2-E1Luc reporter was similar to that of wt E1Luc, while that of E6ID-E1Luc was reduced to an intermediate level. The lower activities of the splicing-deficient E1Luc reporters (Fig. 5A, left) thus closely mirror the impaired replication capacities of the viral genomes containing the same mutations (Fig. 2A). Cotransfection of the splicing mutant E1Luc reporters with low doses of E2 expression vector (0.1 molar ratio of vector versus reporter) increased the overall reporter activities three- to sevenfold (Fig. 5B, left) but did not change the relative responses. Furthermore, addition of large amounts of E2 vector (0.5 molar ratio) did not further increase the reporter activities (W. G. Hubert and L. A. Laimins, unpublished data). Since the activities of the splicing-defective E1Luc reporters closely correlated with the replication efficiencies of the corresponding mutant viral genomes, we conclude that the splicing of the E6 intron is important for efficient expression of E1, which in turn affects replication ability. Our data also indicate that addition of E2 does not alter the effect of the splicing mutations on E1 expression.
In contrast to the results from the E1Luc analysis, introduction of the E6SD, E6SA, and E6IS mutations into the E2Luc reporter background only moderately increased the respective reporter activity compared to that of wt E2Luc in the absence or presence of the E2 vector (Fig. 5, right). The expression levels of the mutant E2Luc reporters did not correlate with the replication efficiencies of the correspondingly mutated genomes. We conclude, therefore, that the splicing of the E6 intron is primarily required for the efficient expression of E1. These results also show that E1 expression and that of E2 are differentially regulated by E6 internal mRNA splicing.
Stable replication requires efficient E1 and E2 expression as well as functional E6 and E7.
Stable replication of HPV31 under physiological conditions requires expression of viral replication factors E1 and E2 (18, 54). Furthermore, expression of fully functional viral oncoproteins E6 and E7 is also necessary for stable replication of viral plasmid DNA (57), presumably to create a cellular environment conducive to HPV31 maintenance. To test the capacity of the E6 splicing mutants to replicate stably, long-term replication assays were performed with primary HFK. HPV31 genomes were first unimolecularly ligated after removing the bacterial vector sequences and then cotransfected into HFK with a selectable marker plasmid. After selection for 5 days, the drug-resistant colonies were pooled and then grown as mass cultures. Total cellular and low-molecular-weight DNAs were isolated 4 to 6 weeks posttransfection and were analyzed for the presence of viral DNA by Southern blotting and hybridization as described previously (18) (see Materials and Methods).
Consistent with previous observations, any alterations in E6 which affect mRNA splicing invariably reduced stable-replication capacity. No hybridization signals indicative of normal stable plasmid replication could be observed in the sheared-total-DNA samples from transfections with mutant pHPV31-E6SD, -E6SA, -E6SR2, -E6ID, and -E6IS genomes (Fig. 6A, center). When unsheared sample DNAs were digested with a unique restriction enzyme, faint bands could be detected at the position of linearized genome-length viral DNA (Fig. 6A, right). In contrast, wt HPV31 replicated extrachromosomally at about 20 copies per cell. To increase the sensitivity for detection of extrachromosomal viral genomes, low-molecular-weight DNAs from parallel cell cultures were also analyzed. As with the total-DNA analysis, only small amounts of closed-circle and open-circle viral DNA could be detected in the samples from transfections with E6 splice mutants (Fig. 6B). These results demonstrate that genomes containing mutated E6 splice sites are unable to replicate stably.
We next wanted to test if the E6 splicing mutants could be complemented in trans by another HPV31 DNA and, therefore, constructed another genome that contained a mutated E7 ORF, pHPV31-E7NS. This mutant is competent for the expression of E1, E2, and full-length E6 but cannot produce a functional E7 protein owing to the presence of an amber stop codon near the 5" terminus of the E7 coding sequences. Consistent with earlier findings that most alterations in E7 also reduce the capacity for stable DNA replication (57), pHPV31-E7NS by itself was unable to replicate stably, as indicated by the lack of monomeric HPV bands in the total-cellular and low-molecular-weight DNA samples (Fig. 6C and D, right). When each of the E6 splicing mutants was cotransfected with pHPV31-E7NS and replication was analyzed in long-term assays, some of the splicing mutants could be complemented successfully. pHPV31-E6SD and -E6ID were able to replicate stably in the presence of pHPV31-E7NS (Fig. 6C and D, left). Southern analysis following digestion with restriction enzymes that generate different-size bands for each genome identified both types of viral DNA in the same cell population. pHPV31 wt- and splice mutant-specific bands migrated below the pHPV31- E7NS-derived ones (Fig. 6C and D, center). Overall, these data demonstrate that trans-complementation of viral genomes in long-term replication assays can occur and that the stable replication defects of the E6 splicing mutant genomes are caused by insufficient expression of trans-acting factors.
DISCUSSION
In this study, we have examined how the expression of replication factors E1 and E2 is regulated during the life cycle of HPV31. We determined that, during the establishment and maintenance phases, the replication factors regulate their own expression and thus modulate the efficiency of viral DNA replication. The levels of E1 and E2 proteins are so low that their direct measurement is not possible. For these reasons we performed replication assays with genetically altered HPV31 genomes and examined viral gene expression by means of E1- and E2-specific reporter assays. These functional studies demonstrated that expression of the viral replication factors is regulated transcriptionally by E1 and E2 and posttranscriptionally by mRNA splicing in the E6 ORF. Regulation by E1, E2, and E6 intron splicing primarily modulates the expression of E1 during the various modes of HPV replication.
Transcriptional autoregulation by E1 and E2.
Our studies examined specifically how the expression of the HPV31 replication factors is regulated by either E1 or E2. We observed that E2 slightly activates E1 expression at low levels but represses it at high concentrations. These findings are consistent with the results of earlier studies in which the role of E2 in the regulation of HPV early gene expression was analyzed (3, 6, 51, 56). Moderate activation of E1 expression by E2 has also been observed with bovine papillomavirus type 1 (BPV1) (17). Surprisingly, we found that HPV31 E2 did not strongly modulate its own expression, which is in contrast to the E2-mediated regulation of BPV1 transcription. The BPV1 E2 (E2TA) protein activates viral early gene expression (13, 49), including its own expression (14, 47, 48). E2TA-mediated trans-activation is antagonized by multiple E2 repressor proteins of BPV1 (29, 31). E2 repressor E8/E2C, also recently identified in HPV31, contains several amino acids from E8, a short alternative reading frame overlapping the E1 ORF, fused to the C-terminal DNA-binding domain of E2. Much like its BPV1 counterpart, HPV31 E8/E2C antagonizes full-length E2 function in viral transcription and DNA replication (53). It is, therefore, likely that E8/E2C plays a role in augmenting the repressor function of full-length E2 in viral regulation.
When we analyzed how E1 regulates the expression of the replication factors in HPV31, we found that E1 moderately activates both its own expression as well as that of E2. While E1 of BPV1 has been shown to be a transcriptional repressor for early viral transcription (44), its specific effect on E1 or E2 expression is not known. The elevated E1- and E2-specific reporter activities in the presence of E1 in our studies, particularly at high doses of the expression vector, could potentially be induced by E2-independent replication of these reporters. E2-independent replication has been observed with HPV1 when a large molar excess of the E1 expression vector was cotransfected with an origin plasmid (12). Our studies did not formally address this possibility because replication of small amounts of reporter DNAs could not be detected reliably.
Posttranscriptional regulation of E1 and E2 expression.
In addition to the transcriptional regulation by E1 and E2, the expression of these proteins is regulated differentially by mRNA splicing. The results of our studies suggest that most E1-encoding transcripts of HPV31 are spliced in the E6 ORF and that viral replication is down-regulated by reduced levels of E1 in the absence of splicing. Similar to the genome of HPV31-E6SA, which has a low-transient-replication phenotype, a BPV1 genome with a comparable mutation in the E6 ORF has a lower stable copy number than wt BPV1 (21), which is primarily caused by reduced expression of E1 (17). It is unlikely that the intron sequences themselves exert a negative cis-acting effect on E1 expression, since the lack of all E6 intron sequences in pHPV31-E6ID does not lead to a high replication efficiency for this mutant. Furthermore, pHPV31-E6SR2, which contains most of the 5" sequences of the E6 intron, in addition to the heterologous intron, replicates as efficiently as a wt HPV31 genome. These results indicate that the process of mRNA splicing in the E6 ORF is necessary for efficient E1 expression, possibly by elevating the steady-state levels of E1-encoding mRNAs through an increase in RNA transport or stability, as has been suggested for E7 expression (1, 50). Alternatively, the E6-spliced transcripts may form a highly efficient template for E1 translation, as has been suggested by a recent biochemical study. Cell-free translation of E1 from HPV18 was found to be more efficient when the E6 intron was absent from the mRNA template (41). It is possible that a certain level of E1 protein is required for viral DNA replication to take place and that reduction of E1 expression below such a threshold could result in loss of replication. Our studies indicate that inhibition of E6 intron splicing significantly reduces E1 expression as determined by reporter assay. The apparent inability of our splicing mutant genomes to replicate could, therefore, be caused by their insufficient expression of the E1 protein.
In contrast to what was found for E1 expression, our reporter studies with E6 splicing mutants determined that expression of E2 is not dependent on splicing within E6. Our results indicate that the splicing of the E6 intron may not be necessary for E2 expression or that other mechanisms regulate the abundance of the E2 protein. E2 expression may involve the utilization of other minor promoters rather than P97, as has been suggested in recent studies (36, 37). Alternatively, E2 expression may primarily be regulated by RNA stability or by translational mechanisms. Our experiments demonstrate that some E6 splicing mutants can be complemented in trans with mutants defective in E7. Such stable trans-complementations of HPV mutant genomes have not previously been described. Since E6 intron splicing appears to modulate the expression of the E1 and E7 genes and perhaps that of other downstream genes, the regulation of viral gene expression under trans-complementing conditions is rather complex. It is therefore possible that some E6 intron mutants can be more efficiently complemented than others in these assays due to the nature of the mutation and the downstream targets it affects.
Potential mechanisms for regulating HPV plasmid copy number and switching replication modes.
During the viral life cycle, papillomaviruses switch between different modes of replication. Upon viral entry into the undifferentiated host cells, HPV genomes replicate more frequently than the cellular genome to establish an optimal plasmid copy number. Then, during the maintenance phase, the viral genomes replicate only once per cell cycle, on average, to keep the viral copy number constant (10, 40). Finally, during the productive phase, high levels of viral replication occur in differentiated cells, which leads to thousands of copies per cell. The mechanisms which regulate these switches in viral replication mode are not fully understood but likely involve changes in the levels or activities of the E1 and E2 proteins.
Our studies have identified transcriptional and posttranscriptional mechanisms which differentially regulate E1 and E2 expression. Based on our observations and the findings of others, the following model of HPV copy number control can be proposed. During the establishment phase, rapid viral replication is required to quickly reach an optimal copy number. This could be achieved by increased E1 expression through transcriptional activation at low E2 levels and E1 mRNA splicing in E6. In the maintenance phase, viral replication proceeds at a moderate level and is synchronized to cellular proliferation. Such stable replication could be facilitated by reduced E1 expression through transcriptional repression by higher levels of E2 and unspliced E1 mRNAs. Finally, high levels of DNA replication are required during amplification to allow for the synthesis of several thousand copies of viral genomes for virion assembly. The process of amplification likely requires high levels of both E1 and E2 proteins. However, since high levels of E2 repress E1 expression from major promoter P97, additional E1-specific transcripts could initiate from differentiation-specific promoter P742 of HPV31, as has been suggested (25). Furthermore, this model for HPV copy number control is compatible with the functions of the E8/E2C repressor protein in modulating viral replication.
In summary, our studies demonstrate that E2 can up- or down-regulate E1 expression depending on its abundance. E1, on the other hand, up-regulates its own expression as well as that of E2. We also show that the splicing of the E6 intron is essential for efficient expression of E1 and that viral replication during establishment correlates with the abundance of E1. The combined actions of these transcriptional and posttranscriptional mechanisms contribute to the regulation of the HPV copy number throughout the viral life cycle.
Acknowledgments
This research project was supported by grants from the National Cancer Institute (NCI) to L.A.L. (CA59665 and CA74202). W.G.H. was supported by grant F32-CA73087 from the NCI.
We thank the former and current members of the Laimins laboratory for their generous help.
REFERENCES
- 1.Belaguli, N. S., M. M. Pater, and A. Pater. 1995. Splice sites of human papillomavirus type 16 E6 gene or heterologous gene required for transformation by E7 and accumulation of E7 RNA. J. Med. Virol. 47:445-453. [DOI] [PubMed] [Google Scholar]
- 2.Butz, K., and F. Hoppe-Seyler. 1993. Transcriptional control of human papillomavirus (HPV) oncogene expression: composition of the HPV type 18 upstream regulatory region. J. Virol. 67:6476-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiang, C. M., G. Dong, T. R. Broker, and L. T. Chow. 1992. Control of human papillomavirus type 11 origin of replication by the E2 family of transcription regulatory proteins. J. Virol. 66:5224-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang, C. M., M. Ustav, A. Stenlund, T. F. Ho, T. R. Broker, and L. T. Chow. 1992. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl. Acad. Sci. USA 89:5799-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow, L. T., T. R. Broker, and J. B. Lewis. 1979. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J. Mol. Biol. 134:265-303. [DOI] [PubMed] [Google Scholar]
- 6.Cripe, T. P., T. H. Haugen, J. P. Turk, F. Tabatabai, P. G. d. Schmid, M. Durst, L. Gissmann, A. Roman, and L. P. Turek. 1987. Transcriptional regulation of the human papillomavirus-16 E6-E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J. 6:3745-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fanning, E. 1998. Introduction to simian virus 40: getting by with more than a little help from its host cell. Dev. Biol. Stand. 94:3-8. [PubMed] [Google Scholar]
- 8.Frattini, M. G., and L. A. Laimins. 1994. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology 204:799-804. [DOI] [PubMed] [Google Scholar]
- 9.Frattini, M. G., H. B. Lim, and L. A. Laimins. 1996. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. USA 93:3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert, D. M., and S. N. Cohen. 1987. Bovine papilloma virus plasmids replicate randomly in mouse fibroblasts throughout S phase of the cell cycle. Cell 50:59-68. [DOI] [PubMed] [Google Scholar]
- 11.Goldsborough, M. D., D. DiSilvestre, G. F. Temple, and A. T. Lorincz. 1989. Nucleotide sequence of human papillomavirus type 31: a cervical neoplasia-associated virus. Virology 171:306-311. [DOI] [PubMed] [Google Scholar]
- 12.Gopalakrishnan, V., and S. A. Khan. 1994. E1 protein of human papillomavirus type 1a is sufficient for initiation of viral DNA replication. Proc. Natl. Acad. Sci. USA 91:9597-9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haugen, T. H., T. P. Cripe, G. D. Ginder, M. Karin, and L. P. Turek. 1987. Trans-activation of an upstream early gene promoter of bovine papilloma virus-1 by a product of the viral E2 gene. EMBO J. 6:145-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermonat, P. L., B. A. Spalholz, and P. M. Howley. 1988. The bovine papillomavirus P2443 promoter is E2 trans-responsive: evidence for E2 autoregulation. EMBO J. 7:2815-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 16.Howley, P. M. 1996. Papillomavirinae: the viruses and their replication, p. 947-978. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 17.Hubert, W. G. 1995. Ph.D. dissertation. University of Wisconsin, Madison.
- 18.Hubert, W. G., T. Kanaya, and L. A. Laimins. 1999. DNA replication of human papillomavirus type 31 is modulated by elements of the upstream regulatory region that lie 5" of the minimal origin. J. Virol. 73:1835-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hummel, M., J. B. Hudson, and L. A. Laimins. 1992. Differentiation-induced and constitutive transcription of human papillomavirus type 31.b in cell lines containing viral episomes. J. Virol. 66:6070-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hummel, M., H. B. Lim, and L. A. Laimins. 1995. Human papillomavirus type 31b late gene expression is regulated through protein kinase C-mediated changes in RNA processing. J. Virol. 69:3381-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jareborg, N., A. Alderborn, and S. Burnett. 1992. Identification and genetic definition of a bovine papillomavirus type 1 E7 protein and absence of a low-copy-number phenotype exhibited by E5, E6, or E7 viral mutants. J. Virol. 66:4957-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanaya, T., S. Kyo, and L. A. Laimins. 1997. The 5" region of the human papillomavirus type 31 upstream regulatory region acts as an enhancer which augments viral early expression through the action of YY1. Virology 237:159-169. [DOI] [PubMed] [Google Scholar]
- 23.Kanopka, A., O. Muhlemann, S. Petersen-Mahrt, C. Estmer, C. Ohrmalm, and G. Akusjarvi. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393:185-187. [DOI] [PubMed] [Google Scholar]
- 24.Khoury, G., J. C. Alwine, R. Dhar, P. Gruss, C. J. Lai, S. Segal, and I. Seif. 1980. Regulation of SV40 gene expression through RNA splicing. Cold Spring Harbor Symp. Quant. Biol. 44:41-54. [DOI] [PubMed] [Google Scholar]
- 25.Klumpp, D. J., and L. A. Laimins. 1999. Differentiation-induced changes in promoter usage for transcripts encoding the human papillomavirus type 31 replication protein E1. Virology 257:239-246. [DOI] [PubMed] [Google Scholar]
- 26.Klumpp, D. J., F. Stubenrauch, and L. A. Laimins. 1997. Differential effects of the splice acceptor at nucleotide 3295 of human papillomavirus type 31 on stable and transient viral replication. J. Virol. 71:8186-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo, S. R., J. S. Liu, T. R. Broker, and L. T. Chow. 1994. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J. Biol. Chem. 269:24058-24065. [PubMed] [Google Scholar]
- 28.Kyo, S., D. J. Klumpp, M. Inoue, T. Kanaya, and L. A. Laimins. 1997. Expression of AP1 during cellular differentiation determines human papillomavirus E6/E7 expression in stratified epithelial cells. J. Gen. Virol. 78:401-411. [DOI] [PubMed] [Google Scholar]
- 29.Lambert, P. F., N. L. Hubbert, P. M. Howley, and J. T. Schiller. 1989. Genetic assignment of multiple E2 gene products in bovine papillomavirus-transformed cells. J. Virol. 63:3151-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambert, P. F., B. C. Monk, and P. M. Howley. 1990. Phenotypic analysis of bovine papillomavirus type 1 E2 repressor mutants. J. Virol. 64:950-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambert, P. F., B. A. Spalholz, and P. M. Howley. 1987. A transcriptional repressor encoded by BPV-1 shares a common carboxy-terminal domain with the E2 transactivator. Cell 50:69-78. [DOI] [PubMed] [Google Scholar]
- 32.Larsson, S., J. P. Kreivi, and G. Akusjarvi. 1991. Control of adenovirus alternative RNA splicing: effect of viral DNA replication on RNA splice site choice. Gene 107:219-227. [DOI] [PubMed] [Google Scholar]
- 33.Liu, J. S., S. R. Kuo, T. R. Broker, and L. T. Chow. 1995. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell-free DNA replication. J. Biol. Chem. 270:27283-27291. [DOI] [PubMed] [Google Scholar]
- 34.Lusky, M., J. Hurwitz, and Y. S. Seo. 1993. Cooperative assembly of the bovine papilloma virus E1 and E2 proteins on the replication origin requires an intact E2 binding site. J. Biol. Chem. 268:15795-15803. [PubMed] [Google Scholar]
- 35.Mohr, I. J., R. Clark, S. Sun, E. J. Androphy, P. MacPherson, and M. R. Botchan. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699. [DOI] [PubMed] [Google Scholar]
- 36.Ozbun, M. A., and C. Meyers. 1998. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31.b. J. Virol. 72:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozbun, M. A., and C. Meyers. 1999. Two novel promoters in the upstream regulatory region of human papillomavirus type 31b are negatively regulated by epithelial differentiation. J. Virol. 73:3505-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plumpton, M., N. A. Sharp, L. H. Liddicoat, M. Remm, D. O. Tucker, F. J. Hughes, S. M. Russell, and M. A. Romanos. 1995. A high capacity assay for inhibitors of human papillomavirus DNA replication. Bio/Technology 13:1210-1214. [DOI] [PubMed] [Google Scholar]
- 39.Rabson, M. S., C. Yee, Y. C. Yang, and P. M. Howley. 1986. Bovine papillomavirus type 1 3" early region transformation and plasmid maintenance functions. J. Virol. 60:626-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravnan, J. B., D. M. Gilbert, K. G. Ten Hagen, and S. N. Cohen. 1992. Random-choice replication of extrachromosomal bovine papillomavirus (BPV) molecules in heterogeneous, clonally derived BPV-infected cell lines. J. Virol. 66:6946-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remm, M., A. Remm, and M. Ustav. 1999. Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J. Virol. 73:3062-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rheinwald, J. G., and M. A. Beckett. 1981. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 41:1657-1663. [PubMed] [Google Scholar]
- 43.Sanders, C. M., and A. Stenlund. 2000. Transcription factor-dependent loading of the E1 initiator reveals modular assembly of the papillomavirus origin melting complex. J. Biol. Chem. 275:3522-3534. [DOI] [PubMed] [Google Scholar]
- 44.Sandler, A. B., C. C. Baker, and B. A. Spalholz. 1996. Sp1 is critical for basal and E2-transactivated transcription from the bovine papillomavirus type 1 P89 promoter. J. Gen. Virol. 77:189-198. [DOI] [PubMed] [Google Scholar]
- 45.Sedman, T., J. Sedman, and A. Stenlund. 1997. Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J. Virol. 71:2887-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Southern, P. J., and P. Berg. 1982. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J. Mol. Appl. Genet. 1:327-341. [PubMed] [Google Scholar]
- 47.Spalholz, B. A. 1993. Importance of the bovine papillomavirus P2443 promoter in the regulation of E2 and E5 expression. J. Virol. 67:6278-6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spalholz, B. A., S. B. Vande Pol, and P. M. Howley. 1991. Characterization of the cis elements involved in basal and E2-transactivated expression of the bovine papillomavirus P2443 promoter. J. Virol. 65:743-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spalholz, B. A., Y. C. Yang, and P. M. Howley. 1985. Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell 42:183-191. [DOI] [PubMed] [Google Scholar]
- 50.Stacey, S. N., D. Jordan, P. J. Snijders, M. Mackett, J. M. Walboomers, and J. R. Arrand. 1995. Translation of the human papillomavirus type 16 E7 oncoprotein from bicistronic mRNA is independent of splicing events within the E6 open reading frame. J. Virol. 69:7023-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steger, G., and S. Corbach. 1997. Dose-dependent regulation of the early promoter of human papillomavirus type 18 by the viral E2 protein. J. Virol. 71:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stubenrauch, F., A. M. Colbert, and L. A. Laimins. 1998. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J. Virol. 72:8115-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stubenrauch, F., M. Hummel, T. Iftner, and L. A. Laimins. 2000. The E8E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J. Virol. 74:1178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stubenrauch, F., H. B. Lim, and L. A. Laimins. 1998. Differential requirements for conserved E2 binding sites in the life cycle of oncogenic human papillomavirus type 31. J. Virol. 72:1071-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sverdrup, F., and S. A. Khan. 1994. Replication of human papillomavirus (HPV) DNAs supported by the HPV type 18 E1 and E2 proteins. J. Virol. 68:505-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thierry, F., and M. Yaniv. 1987. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J. 6:3391-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas, J. T., W. G. Hubert, M. N. Ruesch, and L. A. Laimins. 1999. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. USA 96:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1. requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whalen, B., J. Laffin, T. D. Friedrich, and J. M. Lehman. 1999. SV40 small T antigen enhances progression to G2 during lytic infection. Exp. Cell Res. 251:121-127. [DOI] [PubMed] [Google Scholar]
- 60.Yang, L., R. Li, I. J. Mohr, R. Clark, and M. R. Botchan. 1991. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature 353:628-632. [DOI] [PubMed] [Google Scholar]