Abstract
The baculovirus replication factors LEF-1 and LEF-2 of the Autographa californica multinucleocapsid nucleopolyhedrovirus were overexpressed as fusions containing a hemagglutinin (HA) epitope and a HIS6 tag using recombinant baculoviruses. LEF-1 was purified to near homogeneity and found to have primase activity in an indirect assay employing Escherichia coli DNA polymerase I (Klenow enzyme) and poly(dT) template. The LEF-1 primase products were also directly characterized by electrophoresis in 20% polyacrylamide-8 M urea gels and agarose gels. Primer synthesis was time dependent, and products of several hundred nucleotides or more were observed from the M13 single-stranded DNA (ssDNA) template. The LEF-1 primase was absolutely dependent on divalent cations (Mg2+), and optimal activity was supported by 10 mM MgCl2. An alkaline pH (8.8 to 9.4) was optimal, whereas monovalent salt (KCl) was inhibitory. Mutation of an invariant aspartic acid in a putative primase domain caused LEF-1 activity to be abolished. Upon ultracentrifugation in glycerol gradients, LEF-1 was found to have a sedimentation coefficient of 3S that is consistent with its being present as a monomer. Elution profiles of LEF-1 and LEF-2 from ssDNA-cellulose and DEAE resin suggested that LEF-2 may bind to both DNA and LEF-1.
The Baculoviridae are a large and diverse family of rod-shaped, enveloped, occluded viruses that are pathogenic for invertebrates, particularly members of the Insecta. They have been reported from over 600 species, most of which are members of the Lepidoptera, Diptera, and Hymenoptera (34). Two genera of baculoviruses have been characterized, and they include the nucleopolyhedroviruses (NPVs) (47), which have numerous virions within large polyhedron-shaped occlusion bodies, and the granuloviruses (52), which commonly have a single virion within small granular occlusion bodies. Baculovirus genomes consist of double-stranded, circular, supercoiled DNA of 100 to 180 kb, depending on the strain of virus (20). Although evidence suggests that baculovirus genomes may replicate via a rolling-circle intermediate (31, 42), the mechanisms of initiation, elongation, processing, and maturation have not been determined.
Baculovirus DNA replication has been shown to be associated with discrete replication factories in the nuclei of infected cells (41). In addition, a conserved set of genes that are essential or highly stimulatory for transient DNA replication have been identified for Autographa californica multinucleocapsid NPV (AcMNPV) (26, 33), Orgyia pseudotsugata MNPV(OpMNPV) (reviewed in reference 1), and Lymantria dispar MNPV (44). These include genes encoding a DNA polymerase homolog, a DNA helicase homolog, ie-1, a transactivator of early gene transcription, and late expression factors (LEFs) encoded by lef-1, -2, and -3. The DNA polymerase and DNA helicase homologs were subsequently shown to have activities associated with these enzymes (35, 36). In addition, lef-3 encodes a product with the properties of a single-stranded DNA (SSB) binding protein (16, 37), interacts with helicase (14), and is required for transfer of helicase to the nucleus (53).
In addition to its ability to transactivate early gene transcription, the product of ie-1 has been demonstrated to bind to homologous regions which act as replication origins in transient assays (9, 46) and colocalizes with LEF-3 in nuclear replication factories (41). AcMNPV genes lef-1 and lef-2 potentially encode basic proteins with isoelectric points (pI) of 8.84 and 9.38, respectively, and calculated molecular masses of 30,780 and 23,926 Da, respectively (4). Possible roles for LEF-1 and LEF-2 have not been elucidated, although LEF-1 contains several motifs related to eukaryotic primases. When the putative primase domain in LEF-1, WVVDAD, was altered to WVVQAD, transient DNA replication was abolished (13). In addition, it was found in yeast two-hydrid analyses that LEF-1 and LEF-2 interact (13). These data suggested that LEF-1 may function as a primase and that LEF-2 may be associated with this activity.
In this report we describe the overexpression and purification of LEF-1 and demonstrate that it has DNA primase activity. The parameters of the LEF-1 primase activity were optimized, and the primase products were characterized by electrophoresis in polyacrylamide and agarose gels. We also discuss a possible role of LEF-2 in the baculovirus replication complex.
MATERIALS AND METHODS
Chemicals and enzymes.
Nucleoside triphosphates (NTPs) and deoxy-NTPs (dNTPs) were purchased from Roche Molecular Biochemicals. 7-Deaza-2"-deoxyadenosine 5"-triphosphate and poly(dT) were from Amersham Pharmacia Biotech. Radiolabeled nucleotides [α-32P]ATP, [γ-32P]ATP, and [α-32P]dATP were from Perkin-Elmer. Escherichia coli DNA polymerase I (Klenow Exo−) and other modifying enzymes were from New England Biolabs.
Cells.
Spodoptera frugiperda 9 (Sf9) cells were cultured in Sf900II serum-free medium (Gibco-BRL) supplemented with 10% fetal bovine serum, penicillin G (50 U/ml), streptomycin (50 μg/ml; Whittaker Bioproducts), and Fungizone (amphotericin B; 375 ng/ml; Flow Laboratories) as previously described (17).
Transfer plasmids and virus construction.
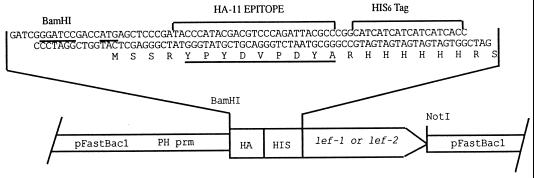
The transfer plasmids were constructed as follows. Plasmids pHSEpiHisLef-1 and pHSEpiHisLef-2 (45) were digested with BglII, treated with calf intestinal phosphatase, and modified using two oligonucleotides that were phosphorylated by treatment with polynucleotide kinase and ATP, gel purified, annealed (48), and inserted into the BglII site. The oligonucleotide names and sequences are 5bamhahisc (GATCGGGATC CGACCATGAG CTCCCGATAC CCATACGACG TCCCAGATTA CGCCCGGCAT CATCATCATC ATCATCACC) and 3bamhahisc (GATCGGTGAT GATGATGATG ATGCCGGGCG TAATCTGGGA CGTCGTATGG GTATCGGGAG CTCATGGTCG GATCCC). This resulted in the creation of an ATG translation start site followed by the HA epitope and HIS tag upstream of each open reading frame (ORF) but downstream of a new BamHI site (Fig. 1).
FIG. 1.
Schematic diagram of plasmids used to construct recombinant baculoviruses expressing LEF-1 and LEF-2. The two oligomers used to engineer the plasmids pHSEpiHisLef-1 and pHSEpiHisLef-2 (45) are shown. They were inserted into a BglII site and resulted in the elimination of this site and the creation of an upstream BamHI site. The inserts were then removed with BamHI and NotI and inserted into pFastBac1 as shown. The orientation of a generic lef gene is indicated by the open arrow.
Clones were screened using BamHI digestion (the correct orientation is about 70 nucleotides [nt] smaller than the reverse orientation), and presumptive correct plasmids were sequenced to confirm the correct orientation of the insert. The primers used for sequencing were AcMNPV nt 10971 to 10990 (lef1-330; CGGTATACATGACTCTTGAC) and nt 3480 to 3462 (lef2-373, AACCTCTTCCTGTACATAC) (4). The ORFs were then removed by digestion with BamHI and NotI, gel purified, and inserted into a pFastbac1 vector (Gibco-BRL) digested with the same enzymes and gel purified. Recombinant baculoviruses were produced using the Bac-to-Bac baculovirus expression system (Gibco-BRL) following the manufacturer's instructions.
A virus, vfbHAHISLef-1D76Q, in which the conserved primase motif WVVDAD was altered to WVVQAD was constructed by removing an MluI-NdeI fragment from a mutant plasmid described previously (13) and inserting it into pfbHAHISLef-1 digested with the same enzymes. The insertion of the mutant sequence was confirmed by DNA sequence analysis. The recombinant virus expressing the mutant lef-1 gene was then generated as described above.
Purification of *LEF-1.
Sf9 cells at a density of 1 × 106 to 1.5 × 106/ml in shaker flasks were coinfected with the recombinant baculoviruses vfbHAHISLef-1 and vfbHAHISLef-2 at a multiplicity of infection (MOI) of about 5 and incubated with shaking for 46 h at 28°C. LEF-1 was purified routinely from a 100-ml or 50-ml culture of infected cells by liquid chromatography sequentially on Ni-nitrilotriacetic acid (NTA)-agarose (Qiagen), DEAE-Toyopearl 650 (TosoHaas), and heparin-Sepharose CL-6B (Amersham Pharmacia Biotech) columns. The infected cells (100 ml) were pelleted by centrifugation for 5 min at 500 × g and resuspended in 8 ml of lysis buffer containing 50 mM Tris-HCl (pH 8.5), 200 mM KCl, 1% Nonidet P-40, 5 mM 2-mercaptoethanol, and a set of protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin, 5 μg of leupeptin per ml, 5 μg of aprotinin per ml, and 2 μg of E64 per ml). After extraction for 15 min at 4°C on a rotating shaker, the preparation was clarified by centrifugation at 150,000 × g for 30 min. The supernatant was loaded onto an Ni-NTA-agarose column (0.8 ml) equilibrated with buffer A (20 mM Tris-HCl [pH 8.5], 0.5 M KCl, 10% [vol/vol] glycerol, 5 mM 2-mercaptoethanol, 20 mM imidazole). The column was washed successively with 10 ml of buffer A, 4 ml of the same buffer containing 1 M KCl, 3 ml of buffer A, and finally with 3 ml of buffer B (20 mM Tris-HCl [pH 8.5], 100 mM KCl, 10% [vol/vol] glycerol, 5 mM 2-mercaptoethanol, 50 mM imidazole).
Proteins were eluted from the column with 4 ml of buffer B containing 150 mM imidazole. The sample was immediately mixed with an equal volume of dilution buffer containing 20 mM Tris-HCl (pH 7.5), 30% (vol/vol) glycerol, 2 mM dithiothreitol (DTT), 2 mM EDTA, and the set of protease inhibitors to the final concentrations listed above, and then passed at a rate of 4 ml per h through a DEAE-Toyopearl column (0.5 by 2.5 cm) equilibrated with a 1:1 mixture of buffer B and dilution buffer. The flowthrough was loaded on a 0.5-ml column of heparin-Sepharose. The column was washed with several volumes of buffer C (0.1 M KCl, 10 mM Tris-HCl [pH 7.5], 20% [vol/vol] glycerol, 1 mM DTT, 1 mM EDTA, and the set of protease inhibitors), and HAHISLEF-1 (*LEF-1) was then eluted with several volumes of the same buffer containing 0.2 M KCl. In some experiments, the heparin-Sepharose column was successively processed with 1-ml portions of buffer C containing KCl in final concentrations of 0.1, 0.15, 0.20, 0.25, 0.30, 0.40, and 0.5 M. Proteins from each fraction were analyzed by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE), followed by staining with silver or Coomassie brilliant blue. The presence of *LEF-1 was monitored by Western blotting with monoclonal antibody HA.11 (see below). Gelatin (0.5 mg per ml) was added to the fractions containing *LEF-1. The fractions were combined or dialyzed separately against buffer D (50 mM KCl, 10 mM Tris-HCl [pH 7.5], 50% [vol/vol] glycerol, 1 mM DTT, 0.1 mM EDTA), and stored at −20°C for periods of 2 to 3 weeks or at −80°C for longer terms.
PAGE and Western blotting.
SDS-12% PAGE was performed as described by Laemmli (29). Gels were either fixed and stained or electrophoretically transferred to PVDF-Plus transfer membrane (Micron Separations Inc) using a Trans-blot SD semidry transfer cell (Bio-Rad) according to the manufacturer's guidelines. Western blots were probed with a 1:1,000 dilution of monoclonal antibody HA.11 (BAbCO), washed, incubated with a 1:3,000 dilution of goat anti-mouse immunoglobulin G (IgG) conjugated to horseradish peroxidase (Bio-Rad Laboratories), and developed by using BM chemiluminescence blotting substrate (POD) (Roche Molecular Biochemicals) according to the manufacturer's instructions.
Assays for endonuclease and topoisomerase activity.
Reactions were carried out in three mixtures of different composition. Reaction mixture 1 contained 25 mM Tris-HCl (pH 7.5), mixture 2 contained 25 mM Tris-HCl (pH 7.5) and 50 mM KCl, and mixture 3 contained 25 mM Tris-HCl (pH 8.8). All reactions contained 0.3 μg of DNA (replicative form I [RFI]) of plasmid pHSEHLEF-1 (45), 10 mM MgCl2, 50 μg of bovine serum albumin (BSA) per ml, 1 mM DTT, and 1 mM ATP. *LEF-1 (0.1 μg in 3 μl of buffer D) was added to a final volume of 15 μl. After incubation for 1 h at 37°C, reactions were terminated by the addition of EDTA to 15 mM and SDS to 0.5%, and the samples were treated with proteinase K (100 μg/ml) for 30 min at 37°C. The samples were then analyzed by electrophoresis in a 0.7% agarose gel, followed by staining with ethidium bromide, and examined for the conversion of RFI into RFII and RFIII or topoisomers.
Primase assays. (i) Indirect assay.
Reaction mixtures (25 μl) contained 25 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 20 μg of poly(dT) per ml, 100 μg of BSA per ml, 1 mM DTT, 1 mM ATP, 2.5 μM 7-deaza-dATP, 2.5 μM [α-32P]dATP (10 μCi per ml), 5 U of E. coli DNA polymerase I (Klenow Exo−) per ml, and various amounts of *LEF-1 added with 5 μl of buffer D. Reactions were carried out at 30°C for 60 min and were terminated by chilling on ice and adding 5 μl of 0.5 M EDTA saturated with sodium pyrophosphate. The samples were transferred onto pieces of Whatman 3MM paper, which were washed in four changes of cold 5% trichloroacetic acid-1% sodium pyrophosphate. The acid-insoluble radioactivity was measured by Cerenkov counting.
(ii) Direct assays.
Reaction mixtures (10 μl) contained 25 mM Tris-HCl (pH 9.1), 10 mM MgCl2, 25 μg of poly(dT) per ml, 100 μg of BSA per ml, 1 mM DTT, 5 μM [α-32P]ATP (100 μCi per ml), and various amounts of *LEF-1 added with 3 μl of buffer D. In experiments with M13mp9 DNA (25 μg per ml) instead of poly(dT), the reaction mixtures were supplemented with 50 μM each GTP, CTP, and UTP. Reactions were carried out at 30°C for 2 h and were terminated by chilling on ice and adding 7 μl of stop solution (95% formamide, 20 mM EDTA, 0.05% each bromophenol blue and xylene cyanol). In initial experiments, the reactions were inactivated by heating at 75°C for 5 min and treated with proteinase K (100 μg/ml) at 37°C for 30 min prior to the addition of the stop solution. We routinely omitted the proteinase K treatment because it did not affect electrophoretic pattern under subsequent analysis of the reaction products in a polyacrylamide gel.
After heat denaturation for 5 min at 75°C, a portion of each reaction was loaded onto a 20% polyacrylamide-8 M urea slab gel (17 by 14.7 by 0.08 cm). Electrophoresis was performed in TBE (Tris-borate-EDTA) buffer (48) at 600 V for 2 to 2.5 h until the bromophenol blue had migrated 3 to 4 cm above the bottom of the gel. Size standards, generated by partial alkaline hydrolysis of 5"-end 32P-labeled A10 (12), were electrophoresed in a parallel lane on the same gel. The gel was transferred onto a polymer support and exposed to X-ray film at −80°C with an intensifying screen.
For analysis of RNA synthesized by *LEF-1 on single-stranded M13 DNA, the mixture containing 25 μg of M13mp9 ssDNA per ml, 25 mM Tris-HCl (pH 9.1), 10 mM MgCl2, 5 μM (100 μCi/ml) [α-32P]ATP, 50 μM each CTP, GTP, and UTP, 100 μg of BSA per ml, and 1 mM DTT was assembled on ice. *LEF-1 (1.3 μg in 39 μl of buffer D) was added to a final volume of 130 μl, and 30-μl portions were taken from the mixture immediately (time zero) or after incubation for 30, 60, and 120 min at 30°C. Reactions were terminated by the addition of EDTA to 15 mM and SDS to 0.5%, and the samples were treated with proteinase K (100 μg/ml) for 30 min at 37°C. An excess of unincorporated label was removed by spinning the samples in multispin tubes (AxyGen, Inc.) equipped with Bio-Gel P-4 medium (Bio-Rad Laboratories) according to the manufacturer's instructions. Polynucleotides were precipitated in cold 75% ethanol, then dissolved in TE (Tris-EDTA) buffer, and analyzed by electrophoresis in a 1.2% neutral agarose gel in Tris-acetate buffer. The gel was transferred onto Whatman 3MM paper, dried under vacuum, and exposed to X-ray film as described above. Size standards (0.3 μg of M13mp9 DNA and 1.5 μg of a 0.24- to 9.5-kb RNA ladder [Gibco-BRL]) were electrophoresed in parallel lanes on the same gel and visualized by ethidium bromide staining.
Sedimentation in glycerol gradients.
Linear 15 to 30% glycerol gradients were prepared in buffer E (0.4 M KCl, 10 mM Tris-HCl [pH 7.5], 1 mM DTT, 1 mM EDTA). Protein sample (0.1 ml), after dialysis against buffer E containing 5% glycerol, was layered over 4.9 ml of a glycerol gradient prepared in nitrocellulose tubes for an SW 50.1 rotor (Beckman). BSA (66 kDa, 4.3S), aldolase (158 kDa, 8.3S), and catalase (220 kDa, 11.3S) (0.2 mg of each) were centrifuged in individual tubes as sedimentation standards. After centrifugation in the SW 50.1 rotor at 48,000 rpm and 4°C for 22 h, the gradients were fractionated from the bottom with a peristaltic pump. The presence of *LEF-1 was monitored by SDS-12% PAGE followed by Western blotting with monoclonal antibody HA.11 (see above).
RESULTS
Purification of *LEF-1.
In order to facilitate the characterization of LEF-1, the LEF-1 ORF was fused at its N terminus with the HA epitope and a HIS6 tag and overexpressed in a recombinant baculovirus under the control of the polyhedrin promoter. The recombinant virus is called vfbHAHISLef-1 (Fig. 1), and the recombinant protein HAHISLEF-1 is designated *LEF-1 in this report. The HA epitope allows monitoring of LEF-1 by Western blot analysis, whereas the HIS6 tag allows purification of LEF-1 with Ni-NTA resin. Evidence suggests that LEF-1 may form a stable complex with another essential replication protein (LEF-2) in infected cells (13). Association with LEF-2 may be required for LEF-1 function as well as the proper processing of LEF-1 during the infection cycle. Therefore, we also overexpressed LEF-2 with a recombinant virus, vfbHAHISLef-2, that was constructed in the same way as vfbHAHISLef-1 (Fig. 1). For our studies, Sf9 cells were coinfected with both baculoviruses (vfbHAHISLef-1 and vfbHAHISLef-2), thereby ensuring expression of both *LEF-1 and *LEF-2.
Although infection with the recombinant viruses vfbHAHISLef-1 and vfbHAHISLef-2 induced accumulation of both proteins in infected cells, only a minor portion of the cellular pool of *LEF-1 and *LEF-2 appeared soluble upon extraction under nondenaturing conditions. An increase in KCl concentration in the lysis buffer up to 1.0 M elevated the yield of both proteins in the clarified extract (stage 1) but caused solubilization of chromatin, which markedly hampered further purification. We found that lysis buffer containing 0.2 M KCl was optimal for purification of *LEF-1.
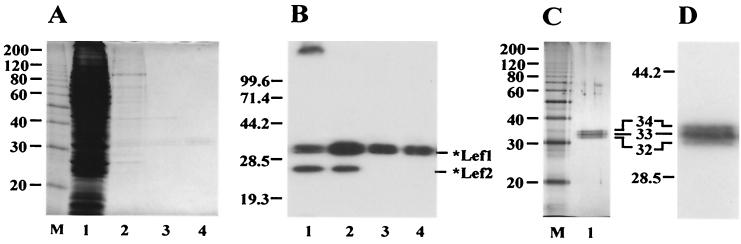
The purification procedure was monitored by SDS-PAGE followed by Coomassie staining (Fig. 2A) and Western blotting with antibody HA.11 (Fig. 2B). Most contaminating proteins were removed from the extract by chromatography on an Ni-NTA column (stage 2) (Fig. 2A, lane 2). However, this fraction contained contaminating proteins and a prominent soluble *LEF-2 component. Further chromatography on a DEAE column caused the separation of the *LEF-1 sample into two fractions: *LEF-1 associated with *LEF-2 was retained on the column, whereas the *LEF-1 fraction free of *LEF-2 appeared in the flowthrough (stage 3) (Fig. 2B, lane 3).
FIG. 2.
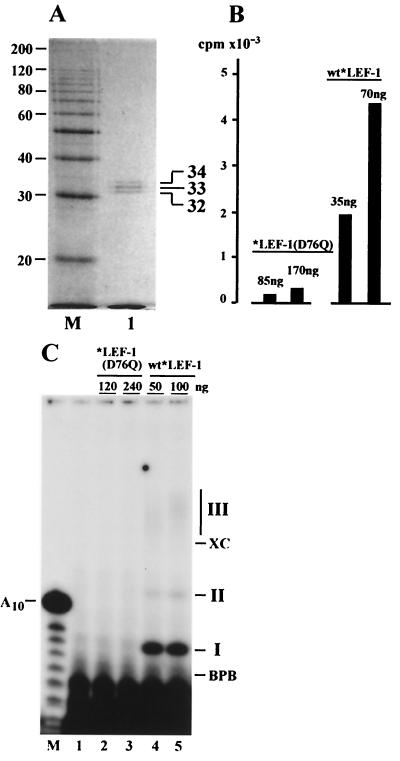
Purification of *LEF-1 from infected Sf9 cells. The samples obtained at different stages of the purification procedure were analyzed by SDS-12% PAGE, followed by staining with Coomassie brilliant blue (A) or by Western blotting with monoclonal antibody HA.11 (B). Lane 1, the high-speed supernatant (stage 1), 5 μl (A) and 2.5 μl (B); lane 2, the sample collected from Ni-NTA (stage 2), 10 μl (A) and 5 μl (B); lane 3, the flowthrough from DEAE-Toyopearl (stage 3), 10 μl (A) and 5 μl (B); lane 4, the final preparation from heparin-Sepharose (stage 4), 10 μl (A) and 5 μl (B); lane M, 10-kDa protein ladder. (C and D) SDS-12% PAGE of purified *LEF-1 followed by silver staining (C) or by Western blotting with monoclonal antibody HA.11 (D). (C) Lane 1, *LEF-1, 60 ng; lane M, 10-kDa protein ladder. (D) *LEF-1, 15 ng. A magnified section of the blot with *LEF-1 is shown. The molecular masses of protein markers (in kilodaltons) are shown on the left sides of panels A to D.
Besides contaminating proteins, the DEAE column removed nucleic acids from the sample. After final chromatography on a heparin-Sepharose column (stage 4), *LEF-1 was essentially free of contamination (Fig. 2A, lane 4). SDS-PAGE followed by silver staining (Fig. 2C) confirmed that *LEF-1 was purified to near homogeneity. Western blot analysis did not reveal contamination of the *LEF-1 samples with traces of *LEF-2 (Fig. 2B, lanes 3 and 4). The *LEF-1 purification protocol requires 1 day and yields about 30 μg of pure protein from a 100-ml culture of infected cells.
At all stages of purification, *LEF-1 was present as a set of three polypeptides with apparent molecular masses of 32, 33, and 34 kDa (Fig. 2B, C, and D). These values are close to the calculated molecular mass of *LEF-1 (33.4 kDa). The reason for the observed microheterogeneity of *LEF-1 remains unclear. All three polypeptides were immunoreactive with antibody HA.11 (Fig. 2D). Therefore, they retained intact N termini with the HA epitope. Two smaller polypeptides were unlikely to have been produced by limited proteolysis of the largest one during purification because the extracts analyzed immediately after extraction of infected cells showed the same tripartite pattern of *LEF-1 as the purified samples. Treatment of samples with calf intestinal phosphatase had no effect on the electrophoretic pattern of *LEF-1 when it was analyzed by SDS-PAGE, suggesting that the heterogeneity was not due to phosphorylation (data not shown). However, this result does not completely exclude modification by phosphorylation, e.g., if the phosphates are inaccessible to the enzyme.
Primase activity of *LEF-1.
Before purified *LEF-1 was assayed for primase activity, the *LEF-1 samples were tested for the presence of endonucleases and topoisomerases (see Materials and Methods). No conversion of RFI DNA into RFII and RFIII and no appearance of DNA topoisomers was observed after incubation of plasmid DNA with purified *LEF-1 (data not shown). This indicates that the *LEF-1 samples are essentially free of endonuclease and topoisomerase activity.
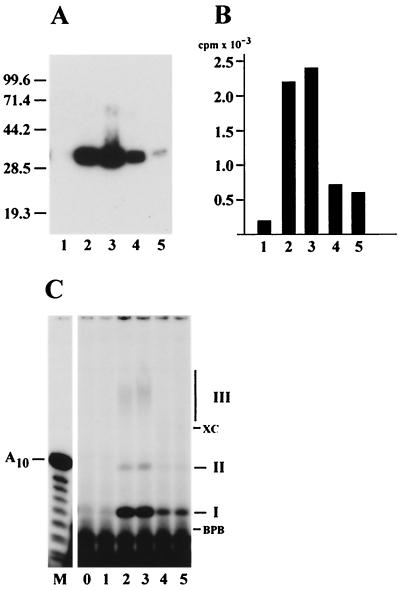
Purified *LEF-1 showed a primase activity in both the indirect and direct assays (Fig. 3). The indirect assay is based on the ability of primase to initiate DNA synthesis on an ssDNA template in the absence of exogenous primers. A typical reaction mixture contains Klenow enzyme, [α-32P]dATP, poly(dT) template, and ATP. There is no DNA synthesis and label incorporation in this system in the absence of primase. If RNA transcripts are synthesized by primase from ATP, they serve as primers for DNA polymerization by Klenow enzyme, thus resulting in the incorporation of [32P]dAMP into nascent DNA, which is monitored by trichloroacetic acid (TCA) precipitation and counting in a scintillation counter. The *LEF-1 fractions collected from a heparin-Sepharose column at the final stage in purification (Fig. 3A) were able to support DNA synthesis by Klenow enzyme on poly(dT) (Fig. 3B). The primase activity correlated well with the amount of *LEF-1 protein in the column fractions.
FIG. 3.
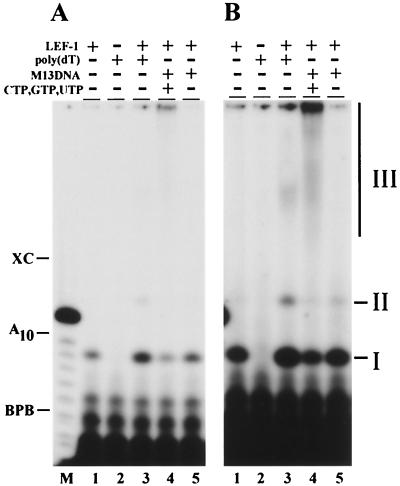
Correlation of primase activity with *LEF-1. The fractions eluted from heparin-Sepharose at KCl concentrations of 100 mM (1), 150 mM (2), 200 mM (3), 250 mM (4), and 300 mM (5) were analyzed by the following methods: (A) SDS-12% PAGE, followed by Western blotting with monoclonal antibody HA.11; (B) indirect primase assay with Klenow enzyme, followed by determination of the acid-insoluble radioactivity; (C) direct primase assay on the poly(dT) template, followed by electrophoresis in a 20% polyacrylamide-8 M urea gel. The assays were performed as described in Materials and Methods. Portions of 2.5 μl (A), 5 μl (B), and 3 μl (C) were taken from the fractions for analysis. Lane 0 in panel C represents a control reaction lacking a column fraction. The molecular masses of protein markers (in kilodaltons) are shown on the left side of panel A. The migration of xylene cyanol (XC), bromophenol blue (BPB), and the primase products (I, II, and III) is indicated on the right side of panel C. Lane M in panel C represents oligonucleotides generated by partial alkaline hydrolysis of 5"-end 32P-labeled A10.
In the direct primase assay, the radioactive products synthesized by *LEF-1 in the presence of [α-32P]ATP and poly(dT) were analyzed by electrophoresis in a 20% polyacrylamide-8 M urea gel (Fig. 3C). *LEF-1 produced three radioactive products of different electrophoretic mobilities, called products I, II, and III (Fig. 3C). The amount of all three products correlated with the amount of *LEF-1 (Fig. 3A) and the primase activity (Fig. 3B) in the column fractions. Treatment with 0.1 N NaOH caused degradation of product III, confirming that it is RNA (data not shown). In contrast, products I and II were insensitive to alkali, and their structure is currently under investigation.
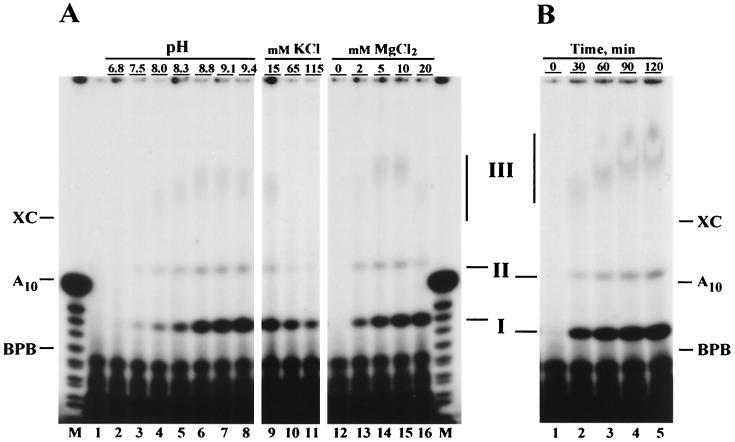
To find an optimum for the primase activity of LEF-1, we varied the pH and concentration of KCl and MgCl2 in the reaction mixture (Fig. 4A). *LEF-1 primase activity was highly dependent on pH and reached a maximum at alkaline conditions (pH 8.8 to 9.4) (Fig. 4A, lanes 2 to 8). The effect of KCl and MgCl2 was assayed at pH 7.5, 8.3, and 9.1. The results obtained at pH 9.1 are shown in Fig. 4A (lanes 9 to 11 and 12 to 16). At all three pHs, KCl was inhibitory. The primase activity of *LEF-1 was absolutely dependent on the presence of divalent cations in the reaction mixture and reached a maximum at 10 mM MgCl2. The magnesium dependence shown in Fig. 4A (lanes 12 to 16) clearly indicated that the optima for synthesis of products I and III are different. The amount of high-molecular-weight product III reached a maximum at 5 to 10 mM MgCl2, whereas the amount of product I was maximal at 10 to 20 mM. At the optimum conditions, *LEF-1 remained enzymatically active during a 2-h incubation at 30°C (Fig. 4B).
FIG. 4.
Optimization of primase activity of *LEF-1 on poly(dT). (A) Three parameters of the reaction mixture used in the direct primase assay were varied: the pH of Tris-HCl buffer (lanes 2 to 8), the KCl concentration (lanes 9 to 11), and the MgCl2 concentration (lanes 12 to 16). The reaction shown in lane 1 contained 50 mM Tris-HCl (pH 9.1), 15 mM KCl, and 10 mM MgCl2, but did not contain *LEF-1. Other reactions contained 200 ng of *LEF-1. The reactions contained the following: lanes 2 to 8, 15 mM KCl, 10 mM MgCl2, and 50 mM Tris-HCl buffer at the indicated pH; lanes 9 to 11, 30 mM Tris-HCl (pH 9.1), 10 mM MgCl2, and KCl at the indicated concentration; lanes 12 to 16, 30 mM Tris-HCl (pH 9.1), 15 mM KCl, and MgCl2 at the indicated concentration. Other ingredients in the reaction mixture [25 μg/ml poly(dT), 5 μM (100 μCi/ml) [α-32P]ATP, 100 μg of BSA per ml, and 1 mM DTT] were not varied. After incubation for 2 h at 30°C, the reactions were terminated and the samples were processed for electrophoresis in a 20% polyacrylimide-8 M urea gel as described in Materials and Methods. (B) Time course of reaction catalyzed by *LEF-1 on poly(dT) at the optimal conditions. The reaction mixture containing 25 mM Tris-HCl (pH 9.1), 10 mM MgCl2, 25 μg of poly(dT) per ml, 5 μM (100 μCi/ml) [α-32P]ATP, 100 μg of BSA per ml, and 1 mM DTT was assembled on ice. *LEF-1 (600 ng in 18 μl of buffer D) was added to a final volume of 60 μl, and 10-μl portions were taken from the mixture immediately (time zero) and after incubation for 30, 60, 90, and 120 min at 30°C. The samples were processed for further analysis in a 20% polyacrylamide-8 M urea gel as described in Materials and Methods. The migration of xylene cyanol (XC), bromophenol blue (BPB), and the primase products (I, II, and III) is indicated on the sides of panels A and B. Lanes M in panel A represent oligonucleotides generated by partial alkaline hydrolysis of 5"-end 32P-labeled A10.
The template dependence of *LEF-1 activity was analyzed in experiments with poly(dT) and ssDNA of phage M13mp9 (Fig. 5). The synthesis of products II and III by *LEF-1 was absolutely dependent on the presence of DNA template in the reaction mixture. In contrast, the synthesis of product I was highly stimulated in the presence of poly(dT), but was not absolutely dependent on the presence of DNA template (Fig. 5A and B, lane 1). The lack of strict template dependence for product I suggests that it might be synthesized by a mechanism other than the regular template-dependent polymerization. The synthesis of long RNA transcripts (product III) on poly(dT) and M13 DNA templates indicated that LEF-1 is capable of extensive nucleotide polymerization (Fig. 5A and B, lanes 3 and 4). However, the synthesis of nascent poly(A) chains on the poly(dT) template appeared to be less efficient than synthesis from M13 DNA templates. This may be because elongation of the poly(A) chain on the poly(dT) template can be blocked by the poly(dT) template's folding back to form Hoogsteen base pairs with the nascent poly(A) chain in the transient triplexes (38). The trapping of nascent poly(A) in such triplex structures prevents efficient elongation of poly(A) chains by the enzyme. Natural single-stranded DNAs do not form regular triplexes with nascent RNA, although stable secondary structures in DNA may serve as physical obstacles for polymerization. In the presence of M13 DNA and only one precursor, [α-32P]ATP, *LEF-1 synthesized the radioactive products I and II, but not product III (Fig. 5, lane 5). Addition of three other precursors, CTP, GTP, and UTP, allowed synthesis of long RNA polymers, some of which did not enter the 20% polyacrylamide gels that were employed in this assay (Fig. 5, lane 4). The long transcripts appeared to be a major product of the enzymatic activity of *LEF-1 in the complete system containing M13 DNA and four NTPs.
FIG. 5.
Template dependence of synthesis catalyzed by *LEF-1. The direct primase assay was carried out in 10-μl reaction mixtures containing 25 μg of poly(dT) (lanes 2 and 3) or 25 μg of ssDNA of phage M13mp9 (lanes 4 and 5) per ml. Lanes 1 and 2 represent control reactions lacking DNA template and *LEF-1, respectively. Other reactions contained 200 ng of *LEF-1. CTP, GTP, and UTP, each at a concentration of 50 μM, were added to the reaction shown in lane 4. Other ingredients in the reaction mixtures were 25 mM Tris-HCl (pH 9.1), 10 mM MgCl2, 5 μM (100 μCi/ml) [α-32P]ATP, 100 μg of BSA per ml, and 1 mM DTT. After incubation for 2 h at 30°C, the reactions were terminated and the samples were processed for electrophoresis in a 20% polyacrylamide-8 M urea gel as described in Materials and Methods. Panel B represents lanes 1 to 5 of panel A after a longer exposure to X-ray film. The migration of xylene cyanol (XC), bromophenol blue (BPB), and the primase products (I, II, and III) is indicated on the sides of panels A and B. Lane M in panel A represents oligonucleotides generated by partial alkaline hydrolysis of 5"-end 32P-labeled A10.
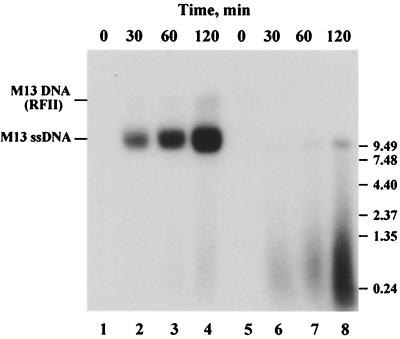
To estimate the size of the RNA transcripts made on M13 DNA, the reaction products were subjected to electrophoresis in a 1.2% agarose gel (Fig. 6). Prior to thermal denaturation, the radioactive products synthesized by *LEF-1 were associated with M13 DNA (lanes 2 to 4). Heat treatment caused liberation of heterogeneous radioactive polymers mostly in a range of several hundred bases in size (lanes 6 to 8). Thus, *LEF-1 is capable of synthesis in vitro of long RNA transcripts that greatly exceed the size of primers synthesized by DNA primases inside the cell.
FIG. 6.
RNA synthesis catalyzed by *LEF-1 on single-stranded DNA of phage M13mp9. Reactions were conducted as described in Materials and Methods. Aliquots were removed after 30, 60, and 120 min. Prior to electrophoresis in a 1.2% agarose gel, each sample was divided in two portions. The first portion was loaded directly onto the gel (lanes 1 to 4), while the second portion was loaded after heating for 3 min at 100°C (lanes 5 to 8). After electrophoresis, the gel was transferred onto Whatman 3MM paper, dried under vacuum, and exposed to X-ray film. M13 DNA and an RNA ladder were loaded onto separate lanes and visualized by staining with ethidium bromide. The migration of M13 DNA and RNA markers is indicated on the left and right sides of the gel, respectively.
Primase domain mutation in *LEF-1.
The ability to synthesize RNA transcripts on poly(dT) and natural ssDNA is in agreement with the primase function of LEF-1 predicted earlier (13) that was based on the presence of conserved primase motifs in baculovirus LEF-1 sequences. The conserved motif of Pri-type primases comprising two aspartate residues separated by a single hydrophobic amino acid and preceded by three hydrophobic amino acids (W[I/V][I/L/V]D[A/I/V]D) is found in the LEF-1 sequence of AcMNPV and several other baculoviruses (Fig. 7).
FIG. 7.
Sequence alignment of LEF-1 proteins of baculoviruses AcMNPV (ac) (4), OpMNPV (op) (2), Choristoneura fumiferana multinucleocapsid NPV (cf) (5), Lymantria dispar multinucleocapsid NPV (ld) (28), Spodoptera exigna multinucleocapsid NPV (se) (21), Heliothus armigera single-nucleocapsid NPV (8), Plutella xylostella granulovirus (px) (18), Xestia c-nigrum granulovirus (xc) (19), and Culex nigripalpus granulovirus (cuni) (40). Putative primase domains are boxed. Invariant amino acids in the putative primase domains are in white letters against black, and invariant aspartates in these domains are indicated by asterisks. Identical amino acids are shown at the bottom of each alignment, and dots indicate conservative changes. The last line within the boxed domains shows invariant amino acids from selected archaeal and eukaryotic primases (3). Dashes indicate gaps in the alignment. The numbers on the left and right indicate the amino acid sequence coordinates. The alignment was produced using MacVector DNA analysis software.
To confirm that the enzymatic activity observed in *LEF-1 samples was actually dependent on the predicted primase domain, we made a conserved change from aspartate to glutamine at amino acid 76 (REWVV DAD to REWVV QAD) in LEF-1 and expressed this construct in a recombinant baculovirus. In the previous study, we found that this mutation did not prevent interaction with LEF-2, but it eliminated the ability of LEF-1 to function in the transient-replication assay (13). The mutant *LEF-1(D76Q), fused at the N terminus with the HA epitope and HIS6 tag, was overexpressed in Sf9 cells and purified by the same method as the wild-type (wt) *LEF-1. When purified to near homogeneity, *LEF-1(D76Q) demonstrated three polypeptides with the same mobility as those from our wt *LEF-1 construct (Fig. 8A). However, the activity of the mutant *LEF-1(D76Q) in the indirect assay with Klenow enzyme was very low (Fig. 8B). In the direct assay, the mutant *LEF-1(D76Q) did not synthesize any of the three products, I, II, or III, typical of wt *LEF-1 (Fig. 8C). Thus, the enzymatic activity of LEF-1 requires the intact primase domain in the protein. These data confirm the dependence of DNA synthesis by Klenow enzyme in the indirect assay on RNA primers provided by *LEF-1. The minor stimulation of DNA synthesis caused by *LEF-1(D76Q) in the indirect assay may be due to a nonspecific stabilization effect of the added protein on Klenow enzyme.
FIG. 8.
Elimination of primase activity by a mutation in a putative primase domain of LEF-1. The mutated *LEF-1(D76Q) with the aspartate-to-glutamine change at amino acid 76 was purified from Sf9 cells as described in Materials and Methods. (A) SDS-12% PAGE of the purified *LEF-1(D76Q) followed by staining with Coomassie brilliant blue. Lane 1, *LEF-1(D76Q), 400 ng; lane M, 10-kDa protein ladder. (B) The indirect primase assay of *LEF-1(D76Q) and wild-type (wt) *LEF-1. The amount of proteins in each reaction is shown above the bars. After reaction for 1 h at 30°C, the samples were processed for determination of the acid-insoluble radioactivity as described in Materials and Methods. (C) The direct primase assay on the poly(dT) template of *LEF-1(D76Q) (lanes 2 and 3) and wt *LEF-1 (lanes 4 and 5). The amount of protein in each reaction is shown above the lanes. Lane 1 represents a control reaction lacking any protein. Reactions were carried out for 2 h at 30°C, and the samples were processed for electrophoresis in a 20% polyacrylamide-8 M urea gel as described in Materials and Methods. The migration of xylene cyanol (XC), bromophenol blue (BPB), and the primase products (I, II, and III) is indicated on the right side of panel C. Lane M in panel C represents oligonucleotides generated by partial alkaline hydrolysis of 5"-end 32P-labeled A10.
Sedimentation analysis of *LEF-1 in glycerol gradients.
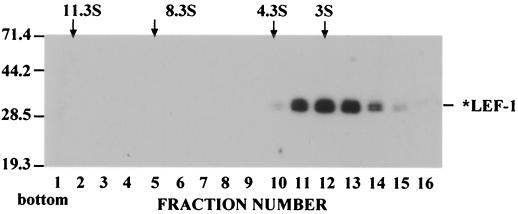
To elucidate the molecular structure of LEF-1, we analyzed the sedimentation of the purified protein in a 15 to 30% glycerol gradient under conditions that prevent nonspecific aggregation (Fig. 9). *LEF-1 sedimented in the gradient slower than BSA (4.3S, 66 kDa) and had a sedimentation coefficient of about 3S. This value suggests that the *LEF-1 samples consist of a mixture of protein monomers. The sedimentation rate of *LEF-1 in the glycerol gradients was much lower than that of AcMNPV RNA polymerase, another viral enzyme capable of synthesizing RNA products. In our experiments, the sedimentation pattern of purified AcMNPV RNA polymerase was consistent with a molecular mass of 300 to 400 kDa (data no shown), which is slightly less than has been reported previously (15).
FIG. 9.
Sedimentation analysis of *LEF-1. *LEF-1 (7 μg in 100 μl) was layered over 4.9 ml of a 15 to 30% glycerol gradient in buffer containing 0.4 M KCl, 10 mM Tris-HCl (pH 7.5), 1 mM DTT, and 1 mM EDTA and centrifuged in an SW 50.1 rotor at 48,000 rpm and 4°C for 22 h. The gradient was fractionated from the bottom into 16 fractions, and 5-μl portions from each fraction were analyzed by SDS-12% PAGE, followed by Western blotting with monoclonal antibody HA.11. The positions of the standards centrifuged in separate tubes are shown by arrows: catalase (220 kDa, 11.3S), aldolase (158 kDa, 8.3S), and BSA (66 kDa, 4.3S).
Chromatographic behavior of LEF-2.
LEF-2 presumably forms a functional complex with LEF-1 in infected cells (13), and it has been shown to be essential for plasmid replication in the transient-replication assay (26, 33). However, the function of LEF-2 in baculovirus replication is not known. The primase activity found in the purified *LEF-1 samples which were essentially free of contaminating *LEF-2 clearly indicated that LEF-2 is not required for the primase activity of LEF-1 in vitro. Some of our indirect data suggest that LEF-2 may serve as an accessory factor needed for incorporation of LEF-1 into replication complexes assembled on viral DNA.
We routinely overexpressed HIS-tagged proteins LEF-1 and LEF-2 together, and both proteins were expressed in Sf9 cells at almost equivalent levels. However, it was more difficult to solubilize *LEF-2 than *LEF-1, and thus the samples after passage through an Ni-NTA column appeared to enriched for *LEF-1 (Fig. 2B, lane 2). At the next chromatographic step, *LEF-2 bound tightly to a DEAE resin, whereas *LEF-1 appeared mostly in the flowthrough (Fig. 2B, lane 3). It was possible to elute *LEF-2 and a portion of *LEF-1 from DEAE-Toyopearl only by using buffers with relatively high concentrations of monovalent salt (0.3 to 0.5 M KCl) (data not shown). Optical densitometry revealed the presence of nucleic acids in this high-salt fraction. Because all free proteins were removed from the DEAE resin at lower salt concentration, this result suggested that *LEF-2 and *LEF-1 might be associated with traces of viral DNA.
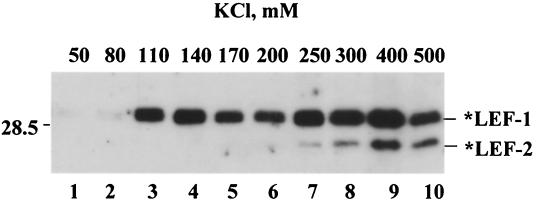
In another experiment, the sample of *LEF-1 and *LEF-2 collected from the Ni-NTA column was subjected to chromatography on an ssDNA-cellulose column (Fig. 10). Stepwise elution resulted in the *LEF-1 pool's being divided in two fractions. One fraction of *LEF-1 was free of *LEF-2 and eluted at a relatively low salt concentration (110 to 170 mM KCl), whereas the other fraction contained both proteins and eluted at much higher salt concentrations (300 to 500 mM KCl). This result suggests that LEF-1 might associate with LEF-2 to form a stable complex with single-stranded DNA and probably with other replicative proteins associated with DNA.
FIG. 10.
Chromatography of *LEF-1 and *LEF-2 on ssDNA-cellulose. A sample of *LEF-1 and *LEF-2 was obtained from a 50-ml culture of Sf9 cells infected with the recombinant viruses vfbHAHISLef-1 and vfbHAHISLef-2 by purification on an Ni-NTA column. After dialysis against buffer containing 25 mM KCl, 10 mM Tris-HCl (pH 7.5), 20% (vol/vol) glycerol, 1 mM DTT, and 1 mM EDTA, the sample was loaded onto an 0.5-ml column of ssDNA-cellulose (Sigma), which was processed with 1-ml portions of the same buffer containing KCl in the concentrations indicated above the lanes. Aliquots (5 μl) from each fraction were subjected to SDS-12% PAGE, followed by Western blotting with antibody HA.11. A portion of the blot with *LEF-1 and *LEF-2 is shown. The position of the 28.5-kDa protein marker is indicated on the left side.
DISCUSSION
The essential baculovirus replication protein LEF-1 was originally predicted to serve as a DNA primase, because a mutation in a conserved primase-like domain failed to support transient DNA replication (13). Therefore, we initiated a biochemical study of this protein to determine if it possesses primase activity. Upon testing a number of fractions obtained from AcMNPV-infected Sf9 cells in an indirect assay employing Klenow enzyme and poly(dT) template (see Materials and Methods), a strong primase activity was found associated with host cell DNA polymerase α (data not shown). However, we were unable to detect a virus-induced DNA primase in extracts from infected cells.
To increase the LEF-1 level in infected cells, we overexpressed it in recombinant baculovirus under the control of the strong polyhedrin promoter. In this construct, the HA epitope and a HIS6 tag were fused to LEF-1 at the N terminus. A similar construct was found to retain its activity in transient replication and transcription assays (45). This construct allowed the rapid and efficient purification of the recombinant protein, *LEF-1, to homogeneity, as judged by SDS-PAGE (Fig. 2). Biochemical analyses revealed the presence of primase activity by *LEF-1, providing evidence for its role as a primase in baculovirus replication that had been predicted earlier. The list of virally encoded enzymes involved in baculovirus DNA replication now includes a DNA primase in addition to the DNA polymerase (35, 39, 50) and DNA helicase (32, 36) characterized previously. A set of similar viral enzymes are required for genome replication by the Herpesviridae, another family of large DNA viruses (7).
The identification of LEF-1 as a DNA primase is based on its ability to synthesize primers from a poly(dT) template, which then allows initiation of DNA synthesis by exogenous DNA polymerase (Klenow enzyme) (Fig. 3B). The synthesis of primers was confirmed in a direct assay involving examination of the products by gel electrophoresis (Fig. 3C). Furthermore, analysis of a mutant LEF-1 confirmed the dependence of its activity on an intact domain conserved in archaeal and eukaryotic primases (Fig. 7 and 8).
The *LEF-1 primase activity is absolutely dependent on divalent cations (Fig. 4A), which may be directly involved in catalysis (see below). The most striking property of *LEF-1 is its ability to synthesize in vitro polynucleotides of a thousand nucleotides or more on ssDNA of phage M13 (Fig. 6). In this respect *LEF-1 resembles DNA primase from the archaeon Pyrococcus furiosus (6). The RNA transcripts synthesized by *LEF-1 greatly exceed the size of primers synthesized by known DNA primases during DNA replication inside eukaryotic cells. If baculovirus replication is similar to that of other systems, some other viral or host cell factors may limit the size of RNA transcripts synthesized by LEF-1 to produce physiologically relevant RNA primers.
Two distinct types of primases that lack sequence or structural relatedness have been characterized. The Pri-type primases are found in eukaryotes and archaea, whereas the DnaG type are found in eubacteria (23). Baculovirus LEF-1 contains three motifs (for AcMNPV, residues 69 to 78, 114 to 139, and 189 to 214) that are conserved in eukaryotic and archaeal primases (boxed in Fig. 7), suggesting that it is a Pri-type primase. These motifs presumably contribute to the structure of the major domain, which contains the primase active site (3). The lef-1 gene of AcMNPV encodes a protein of 266 amino acids with a calculated molecular mass of 30,780 Da. Other known lef-1 genes of baculoviruses encode proteins as small as 216 amino acids (Fig. 7). These represent the smallest RNA-synthesizing enzymes characterized so far. Due to its small size, baculovirus LEF-1 may be a valuable tool for structural research of Pri-type primases as well as for biochemical and mechanistic studies. The first conserved LEF-1 motif (REW[I/V][I/L/V]D[A/I/V]D) contains two aspartate residues separated by a single hydrophobic amino acid (DXD) and preceded by three hydrophobic amino acids. This motif is also found in primases from a number of herpesviruses, phage T7, Saccharomyces cerevisiae, archaea, and mammals (3, 24). Experimental data suggest that the two proximal aspartates in this motif are important for enzymatic activity. In herpes simplex virus type 1 primase (UL52), a conserved aspartate-to-glutamine (IILDLD to IILQLD) change at amino acid 628 completely eliminated the primase activity associated with the UL5-UL8-UL52 complex (25). In the catalytic subunit p49 of mouse primase, two aspartates (D109 and D111) of the respective domain and one distal aspartate (D306) are essential for primase activity and are thought to form the metal-binding core of the active site (10).
Aspartates D76 and D78 in the first conserved motif of AcMNPV LEF-1 and the distal invariant aspartate D193 in the third motif presumably play the same role. In agreement with this prediction, a conserved change from aspartate to glutamine at amino acid 76 (REWVVDAD to REWVVQAD) in AcMNPV LEF-1 completely abolished the LEF-1 primase activity (Fig. 8). Because this mutation eliminated the ability of LEF-1 to function in transient-replication assays (13), the essential function of LEF-1 in baculovirus replication is directly connected with the primase activity. Interestingly, the three aspartates in mouse primase (D109, D111, and D306) align precisely with similar residues in the 31-kDa domain of DNA polymerase β, suggesting that Pri-type primases are probably members of the Pol X polymerase family (24). The three aspartates (D95, D97, and D280) in the archaeon Pyrococcus furiosus (Pfu) primase could be superimposed with the active-site residues of four different DNA polymerases, including human DNA polymerase β, indicating similar three-dimensional arrangements in all five structures (3). These data suggest that Pri-type primases including baculovirus LEF-1, use the common two-metal ion mechanism of DNA polymerases (49).
Another conserved motif in DNA primases is the Zn2+ binding domain located near the active site. In bacterial DnaG-type primases, this domain is thought to play a role in template sequence recognition (43). The Pfu primase structure confirms the presence of a Zn2+ ion at a location quite close to the putative active site, suggesting that this ion may also play a role in the activity of Pri-type primases (3). However, the zinc binding site (C/HX2-5C/HX11-13C/HX2-5C/H) and its derivatives are absent in some archaeal primases (3) and in baculovirus LEF-1. This raises doubts about whether the zinc ion is essential for the function of Pri-type primases.
Although *LEF-1 itself has relatively low affinity for ssDNA and was eluted from ssDNA-cellulose at 110 to 170 mM KCl, a portion of the cellular pool of *LEF-1 copurified with *LEF-2 as a fraction possessing a high affinity for ssDNA (Fig. 10). Since it has been demonstrated that LEF-1 interacts with LEF-2 (13), this observation suggests that LEF-2 may stabilize binding of LEF-1 to DNA template and probably to other baculovirus replication proteins. In this respect, LEF-2 resembles subunit p58 of eukaryotic DNA primase. Although p58 is not required for enzymatic activity of the catalytic subunit p49, it binds to ssDNA, tethers p49 to the 180-kDa subunit of DNA polymerase α, and stabilizes p49 (11).
All known DNA primases act in close association with either replicative DNA polymerases or DNA helicases. Both subunits of simian primase interact with host cell replication protein A and simian virus 40 large T antigen, which serves as the helicase in virus replication (51). E. coli primase (DnaG) interacts with the replicative DnaB helicase, SSB protein, and DNA polymerase III holoenzyme (27). In T7 phage, whose helicase and primase are homologous to the equivalent eubacterial proteins (22), the COOH-terminal region of the primase is directly linked to the NH2-terminal region of the helicase, forming a single polypeptide. Replication of baculoviruses may proceed by a mechanism similar to that of herpesviruses. In herpesviruses, the 114-kDa primase subunit UL52 is tightly associated with UL5 and UL8, forming a heterotrimeric primosome possessing DNA-dependent ATPase, DNA helicase, and DNA primase activities (for a review, see reference 30). The functional partner of the LEF-1 and LEF-2 complex in the replisome of baculoviruses is not known. Further analysis of the complexes of LEF-1 and LEF-2 with ssDNA observed in this study may provide a more detailed picture of the structure and organization of the replication machinery of baculoviruses.
ADDENDUM IN PROOF
After this paper was submitted, Eugene Koonin informed us that his group had detected a low level of homology between baculovirus LEF-2 and the large subunit (p58) of eukaryotic DNA primases (E. V. Koonin, Y. I. Wolf, A. S. Kondrashov, and L. Aravind, J. Mol. Microbiol. Biotechnol. 2:509-512, 2000). We appreciate this information, which is in agreement with our suggestion that LEF-2 plays an accessory role in baculovirus DNA primase activity, analogous to that of p58 in eukaryotic primases.
Acknowledgments
The suggestions of Sam Bennett, Rebecca Russell, and Margot Pearson related to this project and the gift of plasmids from Lois Miller, Joyce Wilson, and Lorena Passarelli are gratefully acknowledged.
This research was supported by grants from the NSF (9982536) and NIH (GM60404) to G.F.R., a supplement from the NSF Central and Eastern Europe and Program, and a grant from the Russian Foundation for Basic Research (00-04-49237) to V.S.M.
Footnotes
Technical report no. 11820 from the Oregon State University Agricultural Experiment Station.
REFERENCES
- 1.Ahrens, C. A., D. J. Leisy, and G. F. Rohrmann. 1996. Baculovirus DNA replication, p. 855-872. In M. De Pamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Ahrens, C. H., R. Russell, C. J. Funk, J. T. Evans, S. H. Harwood, and G. F. Rohrmann. 1997. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology 229:381-399. [DOI] [PubMed] [Google Scholar]
- 3.Augustin, M., R. Huber, and J. Kaiser. 2001. Crystal structure of a DNA-dependent RNA polymerase (DNA primase). Nat. Struct. Biol. 8:57-61. [DOI] [PubMed] [Google Scholar]
- 4.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 5.Barrett, J., H. Lauzon, P. Mercuri, P. Krell, S. Sohi, and B. Arif. 1996. The putative LEF-1 proteins from two distinct Choristoneura fumiferana multiple nucleopolyhedroviruses share domain homology to eukaryotic primases. Virus Genes 13:229-237. [DOI] [PubMed] [Google Scholar]
- 6.Bocquier, A. A., L. Liu, I. K. Cann, K. Komori, D. Kohda, and Y. Ishino. 2001. Archaeal primase: bridging the gap between RNA and DNA polymerases. Curr. Biol. 11:452-456. [DOI] [PubMed] [Google Scholar]
- 7.Boehmer, P. E., and I. R. Lehman. 1997. Herpes simplex virus DNA replication. Annu. Rev. Biochem. 66:347-384. [DOI] [PubMed] [Google Scholar]
- 8.Chen, X., W. IJkel, R. Tarchini, X. Sun, H. Sandbrink, H. Wang, S. Peters, D. Zuidema, R. Lankhorst, J. Vlak, and Z. Hu. 2001. The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. J. Gen Virol. 82:241-257. [DOI] [PubMed] [Google Scholar]
- 9.Choi, J., and L. A. Guarino. 1995. The baculovirus transactivator IE1 binds to viral enhancer elements in the absence of insect cell factors. J. Virol. 69:4548-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copeland, W., and X. Tan. 1995. Active site mapping of the catalytic mouse primase subunit by alanine scanning mutagenesis. J. Biol. Chem. 270:3905-3913. [DOI] [PubMed] [Google Scholar]
- 11.Copeland, W., and T. Wang. 1993. Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J. Biol. Chem. 268:26179-26189. [PubMed] [Google Scholar]
- 12.Donis-Keller, H., A. Maxam, and W. Gilbert. 1977. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 4:2527-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, J. T., D. J. Leisy, and G. F. Rohrmann. 1997. Characterization of the interaction between the baculovirus replication factors, LEF-1 and LEF-2. J. Virol. 71:3114-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, J. T., G. S. Rosenblatt, D. J. Leisy, and G. F. Rohrmann. 1999. Characterization of the interaction between the baculovirus ssDNA-binding protein (LEF-3) and putative helicase (P143). J. Gen. Virol. 80:493-500. [DOI] [PubMed] [Google Scholar]
- 15.Guarino, L. A., B. Xu, J. Jin, and W. Dong. 1998. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol. 72:7985-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hang, X., W. Dong, and L. A. Guarino. 1995. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J. Virol. 69:3924-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harwood, S. H., L. Li, P. S. Ho, A. K. Preston, and G. F. Rohrmann. 1998. Ac MNPV late expression factor-5 interacts with itself and contains a zinc ribbon domain that is required for maximal late transcription activity and is homologous to elongation factor TFIIS. Virology 250:118-134. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto, Y., T. Hayakawa, Y. Ueno, T. Fugita, Y. Sano, and T. Matsumoto. 2000. Sequence analysis of the Plutella xylostella granulovirus genome. Virology 275:358-372. [DOI] [PubMed] [Google Scholar]
- 19.Hayakawa, T., R. Ko, K. Okano, S. Seong, C. Goto, and S. Maeda. 1999. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology 262:277-297. [DOI] [PubMed] [Google Scholar]
- 20.Hayakawa, T., G. F. Rohrmann, and Y. Hashimoto. 2000. Patterns of genome organization and content in lepidopteran baculoviruses. Virology 278:1-12. [DOI] [PubMed] [Google Scholar]
- 21.Ijkel, W. F. J., E. A. van Strien, J. G. M. Jeldens, R. Broer, D. Zuidema, R. W. Goldbach, and J. M. Vlak. 1999. Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus genome. J. Gen. Virol. 80:3289-3304. [DOI] [PubMed] [Google Scholar]
- 22.Ilyina, T., A. Gorbalenya, and E. Koonin. 1992. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J. Mol. E vol. 34:351-357. [DOI] [PubMed] [Google Scholar]
- 23.Keck, J., and J. Berger. 2001. Primus inter pares (first among equals). Nat. Struct. Biol. 8:2-4. [DOI] [PubMed] [Google Scholar]
- 24.Kirk, B., and R. Kuchta. 1999. Arg304 of human DNA primase is a key contributor to catalysis and NTP binding: primase and the family X polymerases share significant sequence homology. Biochemistry 38:7727-7736. [DOI] [PubMed] [Google Scholar]
- 25.Klinedinst, D. K., and M. D. Challberg. 1994. Helicase-primase complex of herpes simplex virus type 1: a mutation in the UL52 subunit abolishes primase activity. J. Virol. 68:3693-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kool, M., C. Ahrens, R. W. Goldbach, G. F. Rohrmann, and J. M. Vlak. 1994. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc. Natl. Acad. Sci. USA 91:11212-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Koonin, E. V., Y. I. Wolf, A. S. Kondrashov, and L. Aravind. 2000. Bacterial homologs of the small subunit of eukaryotic DNA primase. J. Mol. Microbiol. Biotechnol. 2:509-512. [PubMed] [Google Scholar]
- 27.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2nd ed. W. H. Freeman and Company, New York, N.Y.
- 28.Kuzio, J., M. N. Pearson, S. H. Harwood, C. J. Funk, J. T. Evans, J. Slavicek, and G. F. Rohrmann. 1999. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology 253:17-34. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lehman, I., and P. Boehmer. 1999. Replication of herpes simplex virus DNA. J. Biol. Chem. 274:28059-28062. [DOI] [PubMed] [Google Scholar]
- 31.Leisy, D. J., and G. F. Rohrmann. 1993. Characterization of the replication of plasmids containing hr sequences in baculovirus-infected Spodoptera frugiperda cells. Virology 196:722-730. [DOI] [PubMed] [Google Scholar]
- 32.Lu, A., and E. B. Carstens. 1991. Nucleotide sequence of a gene essential for viral DNA replication in the baculovirus Autographa californica nuclear polyhedrosis virus. Virology 181:336-347. [DOI] [PubMed] [Google Scholar]
- 33.Lu, A., and L. K. Miller. 1995. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J. Virol. 69:975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martignoni, M. E., and P. J. Iwai. 1986. A catalog of viral diseases of insects, mites, and ticks, 4th ed. USDA Forest Service publication PNW-195. U.S. Department of Agriculture, Portland, Ore.
- 35.McDougal, V. V., and L. A. Guarino. 1999. Autographa californica nuclear polyhedrosis virus DNA polymerase: measurements of processivity and strand displacement. J. Virol. 73:4908-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDougal, V. V., and L. A. Guarino. 2000. The Autographa californica nuclear polyhedrosis virus p143 gene encodes a DNA helicase. J. Virol. 74:5273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikhailov, V. 2000. Helix-destabilizing properties of the baculovirus single-stranded DNA-binding protein (LEF-3). Virology 270:180-189. [DOI] [PubMed] [Google Scholar]
- 38.Mikhailov, V., and D. Bogenhagen. 1996. Termination within oligonucleotide (dT) tracts in template DNA by DNA polymerase gamma occurs with formation of a DNA triplex structure and is relieved by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 271:30774-30780. [DOI] [PubMed] [Google Scholar]
- 39.Mikhailov, V., K. Marlyev, J. Ataeva, P. Kullyev, and A. Atrzhev. 1986. Characterization of 3"→5" exonuclease associated with DNA polymerase of silkworm nuclear polyhedrosis virus. Nucleic Acids Res. 14:3841-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moser, B., J. Becnel, S. White, C. Afonso, G. Kutish, S. Shanker, and E. Almira. 2001. Morphological and molecular evidence that Culex nigripalpus baculovirus is an unusual member of the family Baculoviridae. J. Gen Virol. 82:283-297. [DOI] [PubMed] [Google Scholar]
- 41.Okano, K., V. S. Mikhailov, and S. Maeda. 1999. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J. Virol. 73:110-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oppenheimer, D. I., and L. E. Volkman. 1997. Evidence for rolling circle replication of Autographa californica M nucleopolyhedrovirus genomic DNA. Arch. Virol. 142:2107-2113. [DOI] [PubMed] [Google Scholar]
- 43.Pan, H., and D. Wigley. 2000. Structure of the zinc-binding domain of Bacillus stearothermophilus DNA primase. Struct. Fold. Des. 8:231-239. [DOI] [PubMed]
- 44.Pearson, M. N., and G. F. Rohrmann. 1998. Characterization of a baculovirus-encoded ATP-dependent DNA ligase. J. Virol. 72:9142-9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rapp, J. C., J. A. Wilson, and L. K. Miller. 1998. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J. Virol. 72:10197-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodems, S. M., and P. D. Friesen. 1995. Transcriptional enhancer activity of hr5 requires dual-palindrome half sites that mediate binding of a dimeric form of the baculovirus transregulator IE1. J. Virol. 69:5368-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohrmann, G. F. 1999. Nuclear polyhedrosis viruses, p. 146-152. In R. G. Webster and A. Granoff (ed.), Encyclopedia of virology, 2nd ed. Academic Press, London, England.
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 49.Steitz, T., S. Smerdon, J. Jager, and C. Joyce. 1994. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science 266:2022-2025. [DOI] [PubMed] [Google Scholar]
- 50.Tomalski, M. D., J. Wu, and L. K. Miller. 1988. The location, sequence, transcription, and regulation of a baculovirus DNA polymerase gene. Virology 167:591-600. [PubMed] [Google Scholar]
- 51.Weisshart, K., H. Forster, E. Kremmer, B. Schlott, F. Grosse, and H. Nasheuer. 2000. Protein-protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system. J. Biol. Chem. 275:17328-17337. [DOI] [PubMed] [Google Scholar]
- 52.Winstanley, D., and D. O'Reilly. 1999. Granuloviruses, 2nd ed. Academic Press, London, England.
- 53.Wu, Y., and E. B. Carstens. 1998. A baculovirus single-stranded DNA binding protein, LEF-3, mediates the nuclear localization of the putative helicase P143. Virology 247:32-40. [DOI] [PubMed] [Google Scholar]