Abstract
Cellular BCL-2 family proteins can inhibit or induce programmed cell death in part by counteracting the activity of other BCL-2 family members. All sequenced gammaherpesviruses encode a BCL-2 homologue that potently inhibits apoptosis and apparently escapes some of the regulatory mechanisms that govern the functions of their cellular counterparts. Examples of these protective proteins include BHRF1 of Epstein-Barr virus (EBV) and KSBcl-2 of Kaposi's sarcoma-associated herpesvirus, also known as human herpesvirus 8. The gamma-1 subgroup of these viruses, such as EBV, encodes a second BCL-2 homologue. We have now found that this second BCL-2 homologue encoded by EBV, BALF1, inhibits the antiapoptotic activity of EBV BHRF1 and of KSBcl-2 in several transfected cell lines. However, BALF1 failed to inhibit the cellular BCL-2 family member, BCL-xL. Thus, BALF1 acts as a negative regulator of the survival function of BHRF1, similar to the counterbalance observed between cellular BCL-2 family members. Unlike the cellular BCL-2 family antagonists, BALF1 lacked proapoptotic activity and could not be converted into a proapoptotic factor in a manner similar to cellular BCL-2 proteins by caspase cleavage or truncation of the N terminus. Coimmunoprecipitation experiments and immunofluorescence assays suggest that a minimal amount, if any, of the BHRF1 and BALF1 proteins colocalizes inside cells, suggesting that mechanisms other than direct interaction explain the suppressive function of BALF1.
Apoptosis is a highly regulated, energy-dependent form of cell suicide. The apoptotic machinery is conserved among metazoans from worms to humans (33), and choreographed cell death is required for proper development (53) and tissue homeostasis (60). In addition, apoptosis serves to protect organisms by eliminating damaged or superfluous cells. Dysregulation of the death program has been implicated in a variety of diseases, ranging from Parkinson's disease in the case of inappropriate death of dopaminergic neurons, to cancer where genetically unstable cells resist the signal to die (72). Proteolysis drives the apoptotic death program. The executioners are a family of cysteine proteases known as caspases for their unique predilection for cleavage after aspartate residues (20). Caspases are ordered into a loose hierarchy of initiators and executioners based on their structure and substrate specificity. Caspase activation leads to the stereotypical morphological changes associated with apoptotic cell death (66).
The apoptotic program can be initiated by a large number of stimuli, but it appears that higher eukaryotes rely on two main pathways for activating caspases (9, 66). The extrinsic pathway is receptor mediated and directly activates an initiator caspase bound to the receptor complex. The intrinsic pathway monitors stress inside the cell and inflicts damage to mitochondria, resulting in the release of proapoptotic factors that in turn accelerate the caspase cascade. This mitochondrial step is regulated by the BCL-2 protein family (7, 38). BCL-2 family members may either induce or repress apoptosis and are defined by the presence of one or more of the four BCL-2 homology (BH) domains as well as a C-terminal hydrophobic domain. It has been proposed that BCL-2 family members can bind one another in a complex web of dimers and that dimerization serves as a regulatory rheostat controlling the pro- and antiapoptotic BCL-2 functions through titration (1, 15, 64, 77, 79). However, this model has been challenged by the findings that many of these dimers may not occur inside cells (36) and that dimerization defective mutants retain their functions (12). An unstructured loop region between BH4 and BH3 near the N terminus of many BCL-2 family proteins is an important regulatory domain, containing sites for phosphorylation and proteolytic cleavage (23). Caspase cleavage of antiapoptotic BCL-2 releases a proapoptotic C-terminal fragment that may act as a trigger for the death pathway or as a feed-forward loop to ensure the death of committed cells (11, 16, 22, 28). The proapoptotic C-terminal fragment induces cytochrome c release from mitochondria and from synthetic lipid vesicles similar to the proapoptotic BCL-2 family member BAX (3, 4, 41).
The host employs apoptosis as a defense against virus infections. The extrinsic pathway can be engaged by virus-induced expression of death receptors or as part of the cytotoxic T-cell response (58, 70, 73), while the intrinsic pathway can be used as an early-warning system to induce suicide of the infected cell before the virus has a chance to replicate and escape to other sites (17). In response, viruses have acquired from their host cell a number of antiapoptotic genes, often stripped down to their essential elements, for use against the cell (21, 29). Large DNA viruses such as poxviruses or herpesviruses are especially adept at arming themselves with an arsenal of antiapoptotic proteins, presumably because of the amount of time required to replicate their genomes.
Epstein-Barr virus (EBV) is a human gammaherpesvirus that is widely disseminated and has two distinct phases to its life cycle. EBV infects oral epithelium, where it undertakes lytic replication, and resting B cells, where it establishes a largely latent infection for the life of the host. EBV is closely associated with a variety of malignancies, such as Burkitt's lymphoma, nasopharyngeal carcinoma, and Hodgkin's disease, and a number of lymphoproliferative diseases in immunocompromised hosts (18). In addition, the gammaherpesviruses all encode a functional BCL-2 homologue (5, 32, 54, 62). BHRF1 of EBV is an early lytic cycle gene product that inhibits apoptosis induced by a variety of stimuli and may prevent cell death during viral lytic replication, although other possibilities remain (25, 27, 32, 37, 40, 71, 75). Interestingly, B-cell-associated mutant viruses lacking BHRF1 are still able to replicate in cell culture (43, 51), raising the possibility of redundant lytic cycle antideath genes in the EBV genome. Alternatively, this system may not accurately reflect the situation in vivo. Marshall et al. (52) identified a second BCL-2 homologue encoded by EBV, BALF1. We show here that BALF1 is conserved in other gamma-1 viruses and that the BALF1 map position in these genomes is analogous to that of the antiapoptotic BCL-2 homologues present in gamma-2 herpesviruses like herpesvirus saimiri and Kaposi's sarcoma-associated herpesvirus (also known as human herpesvirus 8) genomes. However, BALF1 proteins are more distantly related to the antiapoptotic herpesvirus BCL-2 proteins than to BALF1 proteins from other viruses, forming a subgroup of BALF1-like proteins (75).
In contrast to the BHRF1-like herpesvirus BCL-2 homologues, we found that EBV BALF1 lacks antiapoptotic activity. Furthermore, BALF1 impairs the ability of BHRF1 to inhibit programmed cell death. Therefore, like their cellular counterparts where antiapoptotic cellular BCL-2 proteins modulate the function of proapoptotic cellular BCL-2 family members, EBV also encodes an antagonist of its own BCL-2 protein.
MATERIALS AND METHODS
Cloning BALF1 from primate viruses.
Genomic libraries were generated from DNA extracted from baboon, chimpanzee, and gorilla lymphoblastoid B-cell lines latently infected with cercopithecine herpesvirus 12, pongine herpesvirus 1, and pongine herpesvirus 3, respectively, inserted into the Lambda FIX II vector (Stratagene) and amplified in Escherichia coli XL-1 Blue MRA (P2). Plaque lifts on Nitran+ membranes were screened using EBV BALF1 at low stringency. Three clones from each virus were further subcloned and sequenced.
Plasmids, cells, viruses, and transfections.
All indicated open reading frames were generated by PCR, confirmed by DNA sequence analysis, and expressed from the pSG5 plasmid (Stratagene) or plasmid derivatives containing an N-terminal hemagglutinin (HA) tag or FLAG tag. BALF1 and BHRF1 were cloned from cloned fragments of the M-ABA strain of EBV (provided by Nancy Raab-Traub) and are identical in sequence to B958. Cell lines were maintained in Dulbecco's modified Eagle's medium with glutamine (BHK [baby hamster kidney], CHO [Chinese hamster ovary], COS-1 [African green monkey kidney], Rat-1 [fibroblasts], and HeLa [human cervical carcinoma] cells) or RPMI 1640 (DG75 [EBV-negative Burkitt's lymphoma]) supplemented with 100 IU of penicillin/ml, 100 μg of streptomycin/ml, and 10% heat-inactivated fetal bovine serum (Life Technologies). The murine epidermal cell line 308 (67) was maintained in mKer medium (61). CHO and COS-1 cells were transfected with FuGENE 6 (Roche) according to the manufacturer's instructions. BHK cells were transfected with Lipofectamine Plus (Life Technologies). DG75 cells were transfected by electroporation with a Gene Pulser (Bio-Rad). Briefly, 5 × 106 log-phase cells were resuspended in 0.5 ml of growth medium with 5 μg of plasmid DNA and were given a single pulse (270 V, 960 μF). Recombinant Sindbis viruses were generated by transfection of BHK cells with infectious RNA prepared from the double subgenomic dsTE12Q plasmid vector encoding the indicated open reading frames as previously described (30). Cells were infected with a multiplicity of infection of 5 PFU per cell, except for coinfections that were infected at an multiplicity of infection of 10 for each recombinant virus.
Viability assays.
At the indicated times after infection, cells were trypsinized and resuspended in phosphate-buffered saline. Immediately prior to analysis, propidium iodide (Sigma) was added to a final concentration of 100 ng/ml. Cells were analyzed for viability with a FACSCaliber (Becton Dickinson) flow cytometer. Viability assays with β-galactosidase or trypan blue were performed as described previously (5).
Immunofluorescence, immunoprecipitation, and immunoblot assays.
For fluorescence microscopy, cells were seeded at a density of 2 × 104 (COS-1) or 5 × 104 (CHO) per well in a two-well chamber slide (Falcon) and transfected with plasmids containing BALF1, BALF0, and BHRF1 as indicated. At 24 h posttransfection, the cells were treated with 100 nM chloromethyl-X-rosamine (CMX-Ros) for 1 h, fixed with 4% paraformaldehyde, permeabilized with a 1:1 mixture of methanol and acetone on ice for 5 min as previously described (10, 50), and incubated with monoclonal antibodies to HA (12CA5; Sigma) or FLAG (M2; Sigma) or polyclonal antibodies to HA (Y-11; Santa Cruz Biotechnology). After incubation with fluorescein-conjugated anti-mouse secondary antibodies or rhodamine-conjugated anti-rabbit antibodies, protein localization was visualized by fluorescence microscopy (Nikon E800 microscope with G-2E/C and B-2E/C filters). All antibodies were diluted 1:100 in Ca-Tris-buffered saline. Because there are no antibodies available for BALF1, all constructs except where indicated contained N-terminal HA or N-terminal FLAG tags for direct comparison with other BCL-2 family proteins.
Immunoprecipitations were performed with 500 ng of polyclonal HA antibody (Y-11; Santa Cruz Biotechnology) or BAX antibody (13666E; PharMingen) as described previously (10). Immunoblot analyses were performed with monoclonal HA antibody (12CA5; Sigma) or FLAG antibody (M2; Sigma).
RESULTS
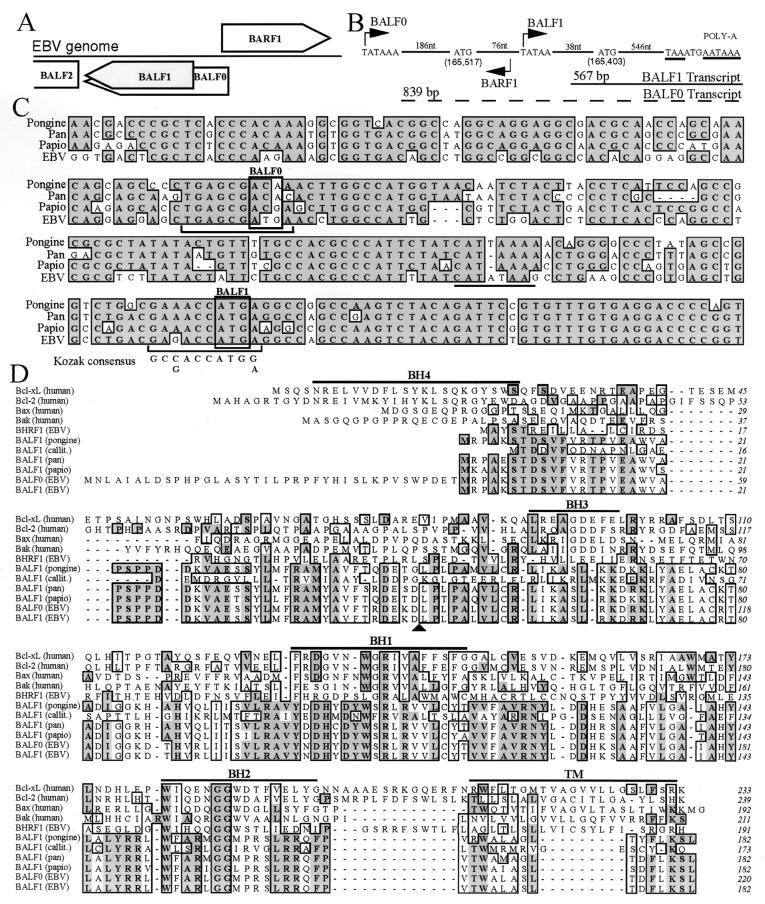
The EBV open reading frame BALF1 is conserved in other related viruses and likely initiates at the second in-frame methionine.
There are two in-frame methionine codons near the beginning of the BALF1 reading frame in EBV, the first of which was originally assumed to be the initiation site of translation at position 165,517 in the EBV genome (Fig. 1A and B). However, sequence analysis of BALF1 homologues from the closely related primate lymphocryptoviruses, pongine herpesvirus 3 (gorilla), pan herpesvirus (pongine herpesvirus 1 of chimpanzee), herpesvirus papio (cercopithecine herpesvirus 12 of baboon), and callitrichine herpesvirus 3 indicates that only the second methionine codon is conserved (Fig. 1C and data not shown). The DNA sequence between the two EBV methionine codons is more divergent in the other viruses (55% homology) than in the remainder of the BALF1 coding sequences (87 to 90% homology), and the EBV reading frame in this stretch is also not preserved in the other viruses (except for pongine herpesvirus 3). Thus, all the primate viruses apparently initiate translation of BALF1 at a site analogous to the second methionine in the EBV genome. Similarly, this internal start codon of EBV BALF1 is likely to be the preferred initiation site of translation, as it has a conserved Kozak initiation sequence that is lacking at the first methionine codon (Fig. 1C). There is also a conserved TATA box 38 bp upstream of the second EBV methionine codon that would produce a transcript of approximately 567 nucleotides plus poly(A) (Fig. 1B). This predicted transcript size is in agreement with the reported BALF1 mRNA size of 0.7 kb (26). In contrast, the calculated size for a polyadenylated transcript that includes the first methionine codon and initiates at the predicted upstream TATA box is approximately 1 kb (Fig. 1B).
FIG. 1.
Human and primate herpesvirus BALF1. (A) Diagram of EBV genome region containing BALF1. Boxes that indicate the direction of transcription represent open reading frames. BALF1 (shaded) results from an internal initiation within the BALF0 reading frame. (B) Diagram of the EBV genome segment containing putative BALF1 transcripts (drawn in reverse of the usual orientation). Nucleotide sequences for putative TATA boxes (bent arrows), initiation codons, stop codon (underlined), and poly(A) signal (underlined) are shown. Numbers of intervening nucleotides (nt) are shown. Calculated transcript sizes [excluding poly(A)] are shown. (C) Alignment of the DNA sequences of BALF1 from EBV (V01555), pan herpesvirus (GenBank accession no. AF306944), herpesvirus papio (GenBank accession no. AF306943), and pongine herpesvirus 3 (GenBank accession no. AY034056). (D) ClustalW alignment of amino acid sequences of the indicated human and viral BCL-2 family members, including callitrichine herpesvirus 3 (GenBank accession no. AF319782). Identical (dark shading) and similar (light shading) amino acids occurring in 6 of 11 entries are marked. Homology domains BH1 to BH4 and the transmembrane domain (TM) of cellular BCL-xL are marked with horizontal lines. The BALF1 consensus caspase cleavage recognition site is marked by an arrowhead.
Without this N-terminal extension in EBV, the BALF1 amino acid sequence is also highly conserved among all of the human and primate viruses, sharing approximately 80 to 90% identity, except with callitrichine herpesvirus 3 which is less well conserved (Fig. 1D). Taking all of this information together, we suggest that the BALF1 protein encoded by EBV corresponds to the smaller reading frame encoding a 182-amino-acid polypeptide beginning at nucleotide 165,403 in the EBV genome. The EBV open reading frame beginning at the first nonconserved methionine is referred to here as BALF0 (Fig. 1A and C). BALF1 proteins encoded by the human and primate lymphocryptoviruses have homology with the BCL-2 family, especially in the BH1 and BH2 homology domains (Fig. 1D) (52) and are more homologous to a BCL-2 homologue of equine herpesvirus 2 than to other BHRF1 proteins. Thus, lymphocryptoviruses contain two BCL-2 homologues, BHRF1 and BALF1.
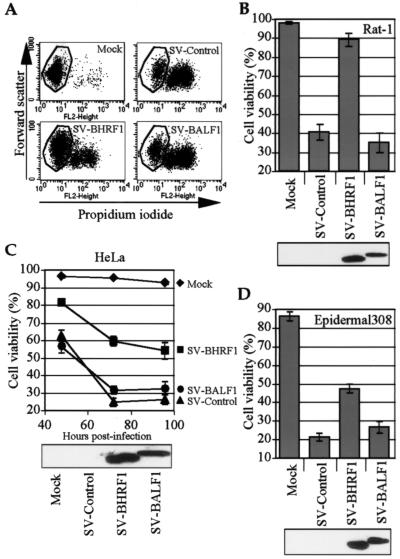
BALF1 fails to protect against Sindbis virus-induced apoptosis.
Many of the gammaherpesvirus BCL-2 homologues have been shown to inhibit apoptosis induced by Sindbis virus and a variety of other cell death stimuli (5, 13, 29, 54). Sindbis virus induces the classic morphological features of apoptosis in many cell types and has been used extensively to study BCL-2 family function in vitro and in vivo (12, 45, 46, 55). To determine if EBV BALF1 functions as an inhibitor of Sindbis virus-induced apoptosis, N-terminal HA-tagged BALF1 was cloned into the Sindbis virus vector, and recombinant virus was used to infect Rat-1 cells. Cells infected with control Sindbis virus underwent apoptosis detected by flow cytometry as a discrete population of shrunken, propidium iodide-positive cells in a two-dimensional analysis (Fig. 2A and B). In contrast to what was observed with the control virus, HA-tagged BHRF1, the known antiapoptotic BCL-2 homologue encoded by EBV (32), dramatically inhibited Sindbis-induced apoptosis. However, BALF1 failed to protect cells in this assay. The recombinant viruses containing BALF1 also did not enhance cell death compared to control viruses (Fig. 2A and B).
FIG. 2.
BALF1 does not inhibit Sindbis virus-induced apoptosis in a variety of cell lines. (A) Rat-1 cells were infected with recombinant Sindbis viruses expressing the indicated constructs and cells were evaluated for apoptosis by flow cytometry to detect cell shrinkage (forward scatter) and membrane permeability (propidium iodide uptake). (B to D) Infected cells were harvested at 24 h post infection for Rat-1 cells (B), the indicated times for HeLa cells (C), and 40 h for 308 epidermal cells (D), treated with propidium iodide, and analyzed by flow cytometry as in the results shown in panel A. Means ± the standard error of the mean (SEM) are shown for three independent experiments. All proteins had N-terminal HA tags. Corresponding immunoblot analyses with anti-HA antibody are shown for each cell type.
BALF1 is expressed as an early lytic cycle transcript (26). Because EBV induces lytic replication in nasopharyngeal epithelium (44) and is shed from the uterine cervix (65), epithelium-derived cell lines were also tested with these recombinant Sindbis viruses. HeLa cells, derived from a human cervical epithelial malignancy, were protected from Sindbis virus-induced apoptosis by BHRF1 but not by BALF1 in a time course experiment (Fig. 2C). BALF1 also failed to protect the murine epidermal 308 cell line (Fig. 2D). In culture, these 308 cells arrange in the classic cobblestone pattern of primary epithelium and express epithelial markers (67). Although BALF1 was sometimes expressed at slightly lower levels than BHRF1 when detected with HA antibody (Fig. 2B to D), this small difference is unlikely to explain the lack of BALF1 activity (see below). N-terminal HA tags do not appear to alter the apoptosis-regulating functions of cellular or viral BCL-2 proteins (data not shown); therefore, we relied on HA-tagged versions because no BALF1 antibody is available.
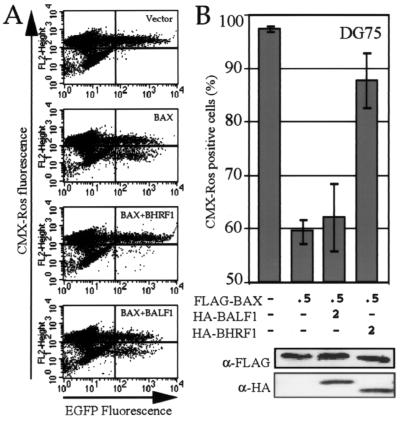
BALF1 failed to protect cells against BAX-induced death in lymphoid cells.
EBV establishes a latent infection in resting B lymphocytes. BALF1 has not been shown to be expressed during the latent phase, but it remains possible that BALF1 functions in a B-cell-specific manner to keep cells alive during reactivation to the lytic phase of the herpesvirus life cycle. Therefore, BALF1 was tested for its ability to inhibit BAX-induced cell death in the DG75 (EBV-negative) B-cell line. One of the functions of the antiapoptotic cellular BCL-2 family members is to inhibit mitochondrial damage and cell death induced by proapoptotic members of the BCL-2 family such as BAX and BAK. BAX overexpression causes release of cytochrome c from mitochondria (42) and loss of membrane potential that can be inhibited by BCL-2, although the mechanisms involved are still unclear (68, 78). DG75 cells were transiently transfected with plasmids containing BAX and BALF1 or controls, and transfected cells were identified by cotransfection with enhanced green fluorescent protein (EGFP). At 18 h after transfection, the number of transfected cells that retained their mitochondrial membrane potential was assessed with the fluorescent dye CMX-Ros. Approximately 30% of cells were killed by the transfection procedure, making a convenient internal control for loss of CMX-Ros staining (Fig. 3A and data not shown). Addition of BAX caused approximately 40% of the transfected GFP-positive cells to lose mitochondrial function, compared to cells lacking overexpressed BAX (Fig. 3A). While BHRF1 protected cells from loss of mitochondrial membrane potential, BALF1 had no ability to overcome BAX activity (Fig. 3A). A graphic representation of the flow cytometry data shows that there is no significant difference in CMX-Ros dye retention of BAX-expressing cells in the presence or absence of BALF1 (Fig. 3B). Immunoblot analysis of transfected cells showed equivalent levels of BAX as well as BALF1 and BHRF1 expression in each sample (Fig. 3B). Mitochondrial integrity in control transfections containing EGFP and either BALF1 or BHRF1 was similar to that of EGFP and the empty vector (data not shown).
FIG. 3.
BALF1 does not protect a lymphocyte cell line against BAX-induced death. (A) DG75 cells were transiently cotransfected with plasmids encoding the indicated proteins with N-terminal HA tags as well as 0.5 μg of EGFP to mark transfected cells. Apoptosis was assessed at 18 h posttransfection by measuring the loss of mitochondrial membrane potential (CMX-Ros fluorescence) in GFP-expressing cells. (B) Graphic presentation of the mean ± SEM for three independent experiments as shown in panel A. Shown below is the amount (in micrograms) of plasmid DNA transfected (with all samples receiving equal amounts of DNA balanced with empty vector) and a corresponding immunoblot analysis.
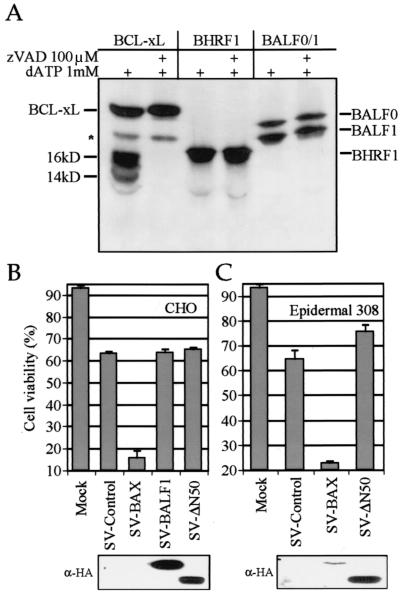
BALF1 and BALF0 are resistant to caspase cleavage by apoptotic cell extracts.
Cellular BCL-2 family members, including BCL-2, BCL-xL, and Bid, can be cleaved in vivo in the loop region by caspases (11, 16, 47). The resulting C-terminal fragments can induce release of cytochrome c and are potently proapoptotic (4, 41, 49). Antiapoptotic herpesvirus BCL-2 homologues have evolved to escape caspase-mediated conversion to proapoptotic molecules by mutation of the cleavage site and by eliminating the proapoptotic activity of their C-terminal fragments (5). Sequence analysis of EBV BALF1 revealed a positionally conserved consensus caspase-3 cleavage site, DEXD, that is also conserved in the pan and papio viruses but not in pongine and callitrichine viruses (Fig. 1D). To explore whether EBV BALF1 protein can be cleaved by caspases during apoptosis, in vitro-translated, 35S-labeled proteins were treated with activated apoptotic extracts prepared from 293 cells. These extracts contain a number of activated caspases, including an abundance of caspase-3, and induce many of the biochemical features of apoptosis (41). As expected, BCL-xL was cleaved into its signature fragments of 16 and 14 kDa (Fig. 4A). Pretreatment of the extracts with the pan-caspase inhibitor zVAD-fmk abolished cleavage, demonstrating that the processing is caspase dependent. BHRF1 was resistant to cleavage by the activated extracts as previously reported (5). Interestingly, when the BALF0 open reading frame was translated in vitro, BALF1 was the more abundant product. Similar to BHRF1, both BALF0 and BALF1 were refractory to caspases and other proteases present in the extracts (Fig. 4A). Identical results were obtained when BALF0 and BALF1 were incubated without 293 extract or with boiled 293 extract or when a construct expressing only BALF1 was tested (data not shown). Resistance to cleavage in 293 cell extracts does not rule out the possibility that BALF1 is cleaved in an EBV-infected cell in vivo. To pursue the possibility that EBV BALF1 has the potential to be converted into a proapoptotic factor by proteolysis, a truncation mutant ΔN50 BALF1 that lacks the N-terminal amino acids 2 to 50 and corresponds approximately to the predicted cleavage fragment was cloned into the Sindbis virus vector and tested for the ability to accelerate Sindbis virus-induced apoptosis. While recombinant virus expressing BAX accelerated the death of Sindbis virus-infected CHO (Fig. 4B) and 308 epidermal (Fig. 4C) cell lines, ΔN50 BALF1 lacked detectable cell killing activity relative to the control. Immunoblot analysis confirmed expression of ΔN50 BALF1 (Fig. 4B and C). The potency of BAX-induced cell death presumably makes detection of BAX protein difficult. Thus, BALF1 lacks latent prodeath activity that is activated by proteolysis or other mechanisms.
FIG. 4.
BALF0-BALF1 protein is resistant to cleavage with apoptotic cell extracts. (A) In vitro-translated, 35S-labeled proteins (without HA tags) were digested with apoptotic 293 cell extracts plus dATP to activate the extract as previously described (5, 24). Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. (B and C) Deletion of the N terminus of BALF1 (ΔN50) does not induce proapoptotic activity. Apoptosis was determined by flow cytometry (forward scatter and propidium iodide staining as described for Fig. 2 at 24 h postinfection of CHO cells (B) and mouse epidermal 308 cells (C) with recombinant Sindbis viruses encoding the indicated proteins or a control virus containing KSBcl-2 in the reverse orientation. Means ± SEM are shown for three independent experiments. All proteins contain an N-terminal HA tag. Corresponding immunoblot analyses with anti-HA antibody are shown for each cell type.
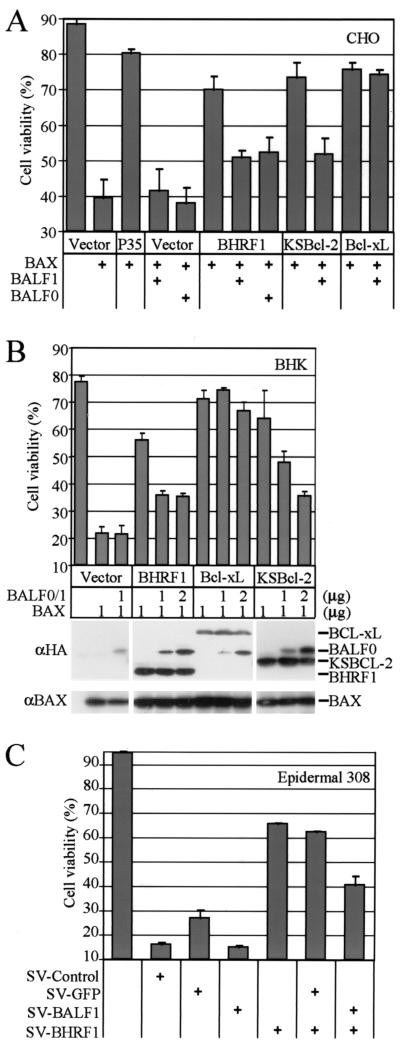
BALF1 and BALF0 antagonize the antiapoptotic activity of BHRF1.
Cellular antiapoptotic BCL-2 family members can antagonize the activity of proapoptotic BCL-2 family members and vice versa, though the mechanisms involved are still debated. Because BHRF1 and BALF1 are apparently both expressed in the early lytic phase of the virus replication cycle, it is possible that one regulates the function of the other. To determine if BALF1 can suppress the antiapoptotic activity of BHRF1, BALF1 was tested for the ability to restore BAX-induced cell death that is otherwise suppressed by BHRF1. BHRF1 suppressed BAX-induced cell death in cotransfected CHO cells (Fig. 5A, compare lanes 2 and 6), while BALF1 did not enhance or suppress the cell-killing function of BAX (compare lane 2 with lanes 4 and 5) as shown above for DG75 cells. However, both BALF1 and BALF0 repressed the protection provided by BHRF1 (Fig. 5A, compare lane 6 with lanes 7 and 8). For this experiment, we used a mutant of BALF0 in which the BALF1 start methionine at amino acid 39 was replaced by a glycine to avoid internal initiation and to force constitutive expression of BALF0. To test whether the effect of BALF1 was specific to EBV, a similar experiment was performed with KSBcl-2, the BCL-2 homologue from Kaposi's sarcoma-associated herpesvirus. Like EBV BHRF1, the antiapoptotic function of KSBcl-2 was also suppressed by BALF1 (Fig. 5A, compare lanes 9 and 10). However, a cellular inhibitor of apoptosis, BCL-xL, was not inhibited by EBV BALF1 (Fig. 5A, compare lanes 11 and 12). Viral BCL-2 proteins provided protection that was almost equivalent to that of the baculovirus caspase inhibitor P35 (compare lanes 2 and 3). BALF0 and BALF1 were also tested for the ability to inhibit BHRF1 and KSBcl-2 in transfected BHK cells. For this experiment, we tested an unaltered BALF0 reading frame that apparently expresses both BALF0 and BALF1 (see Fig. 4), although only BALF0 is detected by the anti-HA antibody. Like the results described for CHO cells, BALF0 and BALF1 had no effect on BAX-induced cell death and no effect on the function of BCL-xL but suppressed the ability of BHRF1 and KSBcl-2 to inhibit BAX-induced cell death (Fig. 5B). Immunoblot analysis verified the expected protein expression levels from the transfected constructs.
FIG. 5.
BALF1 and BALFO antagonize the antiapoptotic activity of BHRF1. (A) Cell viability of CHO cells transfected with the indicated plasmids (plus 0.05 μg of a β-galactosidase plasmid to mark transfected cells) was determined at 18 h posttransfection by scoring the percentage of transfected cells that were live and/or nonapoptotic (counting >200 lacZ-positive cells per sample). All samples received equal amounts of DNA. The data presented are the means ± SEM for at least three independent experiments. (B) BHK cells were transfected with the indicated plasmids and cell viability was determined as described for panel A. A representative immunoblot is shown. (C) Viability of 308 epidermal cells was determined 40 h after coinfection with recombinant Sindbis viruses expressing the indicated proteins (with N-terminal HA tags) as described in the legend to Fig. 2.
This study was extended to include a death stimulus other than BAX. The mouse 308 epidermal cell line was dually infected with recombinant Sindbis viruses expressing BALF1 and BHRF1. Similar to the results with BAX transfection, BALF1 suppressed the protective activity of BHRF1 against Sindbis virus-induced cell death (Fig. 5C). Because Sindbis virus has a superinfection exclusion mechanism (39), care was taken to maximize the number of cells that became dually infected in the first round of infection. To ensure that the observed loss of BHRF1 activity was not due to failure of the two viruses to infect the same cells (in contrast to 50% infection efficiency with each virus), cells were also coinfected with a control virus expressing GFP. GFP failed to suppress the activity of BHRF1, which argued that superinfection exclusion was not an explanation for the observed effects (Fig. 5C). In addition, EGFP fluorescence was detected by flow cytometry, and no differences were observed in the number of fluorescent cells when EGFP virus alone was compared to the combination of EGFP plus BALF1 or EGFP plus BHRF1 viruses (data not shown).
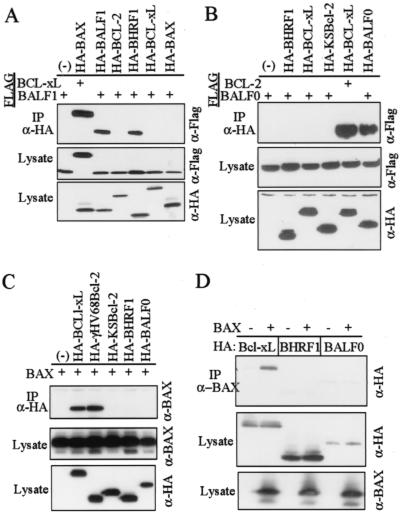
BALF1 but not BALF0 binds to BHRF1 in vitro.
Heterodimerization between proapoptotic and antiapoptotic members of the BCL-2 family has been suggested as a mechanism for titrating their activities (64). However, we and others have challenged this view, suggesting that mechanisms other than direct interaction may contribute to their counteractive functions (12, 36). More recently it has been suggested that the BH3-only members of the BCL-2 family form transient interactions with proapoptotic family members to induce their death-provoking activity, although stable heterodimers may not be formed (14, 57, 74, 81). However, BCL-xL and proapoptotic BAD appear to be bone fide intracellular partners, suggesting that dimerization is limited to specific pairs (19). Nevertheless, dimerization of BALF1 and BHRF1 with concomitant inactivation of BHRF1 offers a simple mechanism to explain BALF1 antagonism of BHRF1 antideath activity. To test this theory, coimmunoprecipitation experiments were performed with transfected COS-1 cells. Precipitation of HA-BAX with anti-HA antibody also coprecipitated FLAG-BCL-xL as expected under these conditions. Coprecipitated FLAG-BCL-xL was detected by immunoblot analysis of the precipitated complexes with anti-FLAG antibody (Fig. 6A, top. In a similar manner, HA-BALF1 and HA-BHRF1 both coprecipitated FLAG-BALF1, but not the FLAG-tagged cellular factors BCL-2, BCL-xL, or BAX. Similarly, BALF1 also coprecipitated with HA-KSBcl-2 (data not shown). This finding is consistent with the model that BALF1 directly binds and inactivates BHRF1 and KSBcl-2. Control immunoblot analyses of the transfected cell lysates with anti-HA and anti-FLAG antibodies verified the expected pattern of protein expression (Fig. 6A). BALF0 was also tested for the ability to coprecipitate other BCL-2 family members. Surprisingly, BALF0 failed to bind BHRF1 and KSBcl-2 as well as the cellular factors, but formed only homodimers (Fig. 6B). This finding suggests that direct binding is not required for BALF0 (and possibly BALF1) to interfere with BHRF1 function. BALF0 was previously reported to interact with BAX (52). Although BAX coprecipitated with BCL-xL and the BCL-2 homologue encoded by murine gammaherpesvirus 68, BALF0 failed to coprecipitate BAX (Fig. 6C), and in the inverse experiment BAX antibody failed to coprecipitate BALF0 (Fig. 6D). BAX also failed to coprecipitate BALF1 (data not shown).
FIG. 6.
BALF1 binds to itself and BHRF1 in vitro. (A) COS-1 cells were transfected with plasmids expressing the indicated HA-tagged BCL-2 family members plus either FLAG-BALF1 or FLAG-BCL-xL as a positive control. Anti-HA immunoprecipitates and total cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with the antibodies indicated on the right of each panel. A representative immunoprecipitation is shown (n = 3). (B, C, and D) Coimmunoprecipitation experiments were performed with lysates of COS-1 cells transfected with the indicated plasmids, as described for panel A.
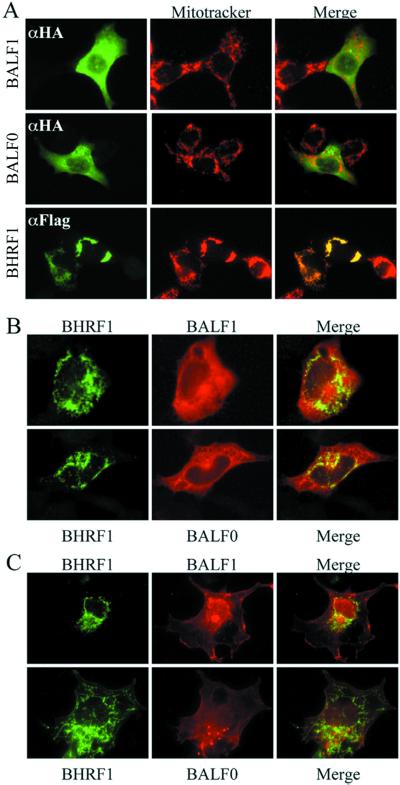
BALF1 and BALF0 have limited colocalization with BHRF1 in cells.
Hsu and Youle (36) reported that coimmunoprecipitation of BAX with other BCL-2 family members is induced by the presence of detergents used in cell lysate preparation and is not an indication of in vivo dimerization events. Therefore, to pursue the possibility that BHRF1 and BALF1 directly interact, they were examined for their ability to colocalize by indirect immunofluorescence microscopy performed with transfected cell lines where antagonistic activity of BALF1 was also observed. Surprisingly, BALF1 and BHRF1 consistently exhibited completely distinct subcellular localizations when transfected separately into CHO cells (Fig. 7A) or COS-1 cells (data not shown). BALF1 was localized diffusely throughout the cytoplasm and apparently excluded from mitochondria that were costained with CMX-Ros (Mitotracker), while BHRF1 displayed a pattern that was identical to the CMX-Ros staining of mitochondria as previously reported (31, 34). Similar results were obtained with BALF0 (Fig. 7A). Thus, BHRF1 has a mitochondrial distribution pattern similar to BCL-2, while BALF1 was cytosolic, similar to BAX prior to the induction of cell death (76). However, BALF1 did not relocalize in cells following a death stimulus (data not shown). We also noted that a large proportion of cells overexpressing BHRF1 display an unusual mitochondrial morphology in which the mitochondria are compressed into a small perinuclear knot in addition to the traditional filigreed mitochondria. Similar perinuclear spots were induced by KSBcl-2 and BCL-xL (data not shown). Examples of both morphologies are shown in the two BHRF1 transfected cells shown in Fig. 7A. This morphology was never seen in untransfected or BALF1-transfected cells. To determine if BHRF1 or BALF1 and BALF0 relocalize when expressed in the same cell, CHO cells were cotransfected with HA-tagged BALF1 and FLAG-tagged BHRF1. Again, BALF1 displayed diffuse cytoplasmic localization even in the presence of BHRF1, which also maintained its mitochondrial localization (Fig. 7B, top row, and data not shown). Similar results were obtained with BALF0 (Fig. 7B, bottom row). Because of the diffuse staining of BALF0 and BALF1, the merged images contained some overlapping signal. However, those regions of the cell that stained most intensively with BALF0 and BALF1 were devoid of BHRF1 (Fig. 7B, right columns). This experiment was repeated with COS-1 cells where BALF0 and BALF1 have a less-diffuse localization. In these merged images, the BALF0-BALF1 and BHRF1 patterns were clearly distinct (Fig. 7C). BALF0 and BALF1 also did not colocalize with a mitochondrial marker in COS-1 cells as expected (data not shown). Colocalization was also not observed in cells induced by die by staurosporine treatment (data not shown). Thus, taking together the immunofluorescence and coimmunoprecipitation experiments. It appears that BHRF1 and BALF1 only form dimers after the cells were broken open, implying that BALF1 inhibits BHRF1 via indirect mechanisms.
FIG. 7.
BALF1 does not colocalize with BHRF1 in cells. (A) Fluorescence microscopy of CHO cells transfected with N-terminal HA-tagged BALF1 (top), HA-tagged BALF0 (middle), or N-terminal FLAG-tagged BHRF1 (bottom). At 24 h posttransfection, cells were treated with CMX-Ros for 1 h (red, center panels) and stained with monoclonal anti-HA or FLAG antibodies (fluorescein isothiocyanate secondary antibody, left panels). The images were merged using SPOT software (yellow, right panels), but the merge failed to detect colocalized proteins. (B) CHO cells were cotransfected with plasmids containing FLAG-tagged BHRF1 (green, fluorescein isothiocyanate) plus either HA-tagged BALF1 (red, top row) or HA-tagged BALF0 (M39G) (red, bottom row). The two images were merged to reveal any colocalization (yellow, third column). (C) The experiment described in panel B was also performed with COS-1 cells.
DISCUSSION
Cellular BCL-2 family members can counteract each other's functions, at least in part due to indirect mechanisms. We now show that BHRF1 is regulated by another BCL-2-related protein, BALF1, also encoded on the EBV genome. Unlike the negative regulatory proteins of the cell, this viral negative regulator lacks overt or latent proapoptotic function while retaining the ability to suppress the antiapoptotic activity of BHRF1. Thus, BALF1 may suppress the antiapoptotic function of BHRF1 in virus-infected cells, perhaps at a later point during infection when the virus no longer requires a viable cell or even benefits from the apoptotic cascade.
How does BALF1 suppress the function of BHRF1? If BALF1 indeed inhibits BHRF1 through an indirect interaction as suggested by the data presented here, it is possible that both BALF1 and BHRF1 target the same downstream cellular machinery and thereby compete with each other to determine cell fate. Consistent with this idea, we have found that both BALF1 and BHRF1 bind to the cellular apoptosis regulator Aven (10) (unpublished data). Other regulatory mechanisms also play key roles in determining BCL-2 family function, including phosphorylation, myristylation, and subcellular localizations (19, 56, 80). We cannot rule out the possibility that a transient interaction between BALF1 and BHRF1 alters protein conformation and/or posttranslational modification to inhibit the protective function of BHRF1. We also cannot rule out the possibility that only a minor fraction of BHRF1 is properly localized or otherwise poised to carry out all of the observed antiapoptotic functions and that this small proportion of the protein is inhibited by direct binding of BALF1. Nevertheless, we favor an indirect mechanism.
The dimerization function of the BCL-2 family is usually attributed to the insertion of an α-helix containing the BH3 domain into a hydrophobic cleft on one side of its partner BCL-2-related protein (63). This BH3 domain of the viral proteins is quite degenerate and lacks the core signature motif (see Fig. 1). Thus, other domains of viral BCL-2 may be involved in binding its partners. Consistent with this ideas, Li et al. (48) recently reported that BHRF1 binds to and is inhibited by the cellular protein PRA1, although the effect of BHRF1 on PRA1 is not known. This interaction requires the BH4 and BH1 domains of BHRF1, and any role for BH3 is not clear. The cleft in cellular BCL-2 molecules apparently can also be occupied by their own C-terminal hydrophobic tail that, when extracted from the cleft, serves to anchor the protein to intracellular membranes (69). Concealment of the hydrophobic tail of BALF1 in its cleft could contribute to our observation that BALF1 is cytosolic. Apparently this is the case for BAX, which is cytosolic in healthy cells, or in cells where BAX functions as an antiapoptotic factor but relocalizes to mitochondria to facilitate cell death by exposing its hydrophobic tail (35, 76). However, we failed to detect a relocalization of BALF1 during cell death.
In contrast to a previous report (52), EBV BALF1 and BALF0 lacked antiapoptotic function. We found that both BAX- and Sindbis virus-induced apoptosis was inhibited by cellular and viral BCL-2 homologues, except BALF1 (5, 12, 54). Even using the same assay previously reported to show antiapoptotic function of BALF0-BALF1, which used camptothecin-treated HeLa cells (52), we were unable to detect protective function (data not shown), although we did not test the GFP-BALF0 fusion used previously. The outcome was unchanged regardless of the strategy employed to assess cell death. BHRF1 but not BALF1 blocked trypan blue uptake, membrane blebbing, cell shrinkage, propidium iodide staining, nuclear chromatin condensation, and mitochondrial membrane depolarization measured with CMX-Ros (see above and data not shown). While it is possible that BALF1 exerts its protective effect downstream of BAX-induced mitochondrial depolarization, parallel experiments measuring exposure of phosphatidyl serine on the plasma membrane, a late step in the apoptotic cascade that occurs after caspase activation, gave similar results and failed to detect protection by BALF1 (data not shown). Taken together, these results suggest that while BALF1 acts to negatively regulate viral antiapoptotic BCL-2 proteins, it has no profound intrinsic death modulatory function on its own. Nevertheless, the study of BALF1 in other model systems may yet reveal new functions, including anti- or proapoptotic activity.
In contrast to our findings with CHO and BHK cells, where BALF1 suppressed the protective function of BHRF1, we were unable to demonstrate BALF1-mediated attenuation of BHRF1 in DG75 cells (data not shown). This could indicate that the suppressive effect of BALF1 is cell type dependent. Conversely, it may reflect limitations of the DG75 system. DG75 cells have at least one genetic lesion related to the BCL-2 family, as they are known to lack detectable BAX protein due to a frameshift mutation (8). In addition, these cells are exceptionally refractory to cell death induced by a variety of stimuli and therefore may also be resistant to the effects of BALF1. This finding in DG75 cells is consistent with the model that BALF1 does not interfere directly with BHRF1 but may also have a cellular target. It is intriguing to speculate that after eliminating some of the negative regulatory functions of cellular BCL-2 in the BHRF1 protein, EBV developed an auxiliary regulatory protein, BALF1, to modulate the death response by the virus.
It had been difficult to reconcile the reported 0.7-kb EBV BALF1 polyadenylated mRNA size (26) with the predicted mRNA size of BALF0 (previously referred to as BALF1). The sequences for BALF1 from several primate lymphocryptoviruses have shed light on this discrepancy. BALF1 from all sequenced viruses appears to initiate at a conserved methionine within BALF0, suggesting that EBV BALF1 protein also initiates at this “internal methionine.” Though BHRF1 and BALF1 are reported to be expressed during the early lytic virus replication cycle, several laboratories have identified spliced BHRF1 transcripts apparently derived from a latent cycle promoter (2, 6, 59). Like BHRF1, BALF1 is positioned downstream of another latency promoter, and others have suggested that BALF1 is also expressed during latency (52). This raises the possibility that BHRF1 and BALF1 also modulate apoptosis during latency by playing additional roles in viral pathogenesis.
Acknowledgments
We thank Y.-R. Fannjiang and C. Gabernet Castello for helpful discussions; S. H. Yuspa, M. Wawersik, and P. Coulombe for 308 epidermal cells; Y. Lazebnik for 293 cell extracts; and S. Mitchell for technical assistance.
This work was supported by NIH grants CA73581 and NS34175 (J.M.H.), BBSRC grant 323/C09500 (M.H.), and Nuffield Bursary grant URB/00020/G (C.P.).
REFERENCES
- 1.Adams, J. M., and S. Cory. 1998. The Bcl-2 protein family: arbiters of cell survival. Science 281:1322-1326. [DOI] [PubMed] [Google Scholar]
- 2.Austin, P. J., E. Flemington, C. N. Yandava, J. L. Strominger, and S. H. Speck. 1988. Complex transcription of the Epstein-Barr virus BamHI fragment H rightward open reading frame 1 (BHRF1) in latently and lytically infected B lymphocytes. Proc. Natl. Acad. Sci. USA 85:3678-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basanez, G., A. Nechushtan, O. Drozhinin, A. Chanturiya, E. Choe, S. Tutt, K. A. Wood, Y.-T. Hsu, J. Zimmerberg, and R. J. Youle. 1999. Bax, but not Bcl-xL, decreases the lifetime of planar phospholipid bilayer membranes at subnanomolar concentrations. Proc. Natl. Acad. Sci. USA 96:5492-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basanez, G., J. Zhang, B. N. Chau, G. I. Maksaev, V. Frolov, T. A. Brandt, J. Burch, J. M. Hardwick, and J. Zimmerberg. 2001. Pro-apoptotic cleavage products of Bcl-xL form cytochrome c-conducting pores in pure lipid bilayers. J. Biol. Chem. 276:31083-31091. [DOI] [PubMed] [Google Scholar]
- 5.Bellows, D. S., B. N. Chau, P. Lee, Y. Lazebnik, W. H. Burns, and J. M. Hardwick. 2000. Antiapoptotic herpesvirus Bcl-2 homologs escape caspase-mediated conversion to proapoptotic proteins. J. Virol. 74:5024-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodescot, M., and M. Perricaudet. 1986. Epstein-Barr virus mRNAs produced by alternative splicing. Nucleic Acids Res. 14:7103-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bossy-Wetzel, E., D. D. Newmeyer, and D. R. Green. 1998. Mitochondrial cytochrome c release in apoptosis occurs upstream of DEVD-specific caspase activation and independently of mitochondrial transmembrane depolarization. EMBO J. 17:37-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brimmell, M., R. Mendiola, J. Mangion, and G. Packham. 1998. BAX frameshift mutations in cell lines derived from human haemopoietic malignancies are associated with resistance to apoptosis and microsatellite instability. Oncogene 16:1803-1812. [DOI] [PubMed] [Google Scholar]
- 9.Budihardjo, I., H. Oliver, M. Lutter, X. Luo, and X. Wang. 1999. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 15:269-290. [DOI] [PubMed] [Google Scholar]
- 10.Chau, B. N., E. H. Y. Cheng, D. A. Kerr, and J. M. Hardwick. 2000. Aven, a novel inhibitor of caspase activation, binds Bcl-xL and Apaf-1. Mol. Cell 6:31-40. [PubMed] [Google Scholar]
- 11.Cheng, E. H. Y., D. G. Kirsch, R. J. Clem, R. Ravi, M. B. Kastan, A. Bedi, K. Ueno, and J. M. Hardwick. 1997. Conversion of Bcl-2 to a Bax-like death effector by caspases. Science 278:1966-1968. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, E. H. Y., B. Levine, L. H. Boise, C. B. Thompson, and J. M. Hardwick. 1996. Bax-independent inhibition of apoptosis by Bcl-xL. Nature (London) 379:554-556. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, E. H. Y., J. Nicholas, D. S. Bellows, G. S. Hayward, H.-G. Guo, M. S. Reitz, and J. M. Hardwick. 1997. A Bcl-2 homolog encoded by Kaposi's sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc. Natl. Acad. Sci. USA 94:690-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng, E. H. Y. A., M. C. Wei, S. Weiler, R. A. Flavell, T. W. Mak, T. Lindsten, and S. J. Korsmeyer. 2001. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell 8:705-711. [DOI] [PubMed] [Google Scholar]
- 15.Chittenden, T., C. Flemington, A. B. Houghton, R. G. Ebb, G. J. Gallo, B. Elangovan, G. Chinnadurai, and R. J. Lutz. 1995. A conserved domain in Bak, distinct from BH1 and BH2, mediates cell death and protein binding functions. EMBO J. 14:5589-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clem, R. J., E. H. Y. Cheng, C. L. Karp, D. G. Kirsch, K. Ueno, A. Takahashi, M. B. Kastan, D. E. Griffin, W. C. Earnshaw, M. A. Veliuona, and J. M. Hardwick. 1998. Modulation of cell death by Bcl-xL through caspase interaction. Proc. Natl. Acad. Sci. USA 95:554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clouston, W. M., and J. F. R. Kerr. 1985. Apoptosis, lymphocytotoxicity and the containment of viral infections. Med. Hypotheses 18:399-404. [DOI] [PubMed] [Google Scholar]
- 18.Crawford, D. 2001. Biology and disease associations of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:461-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datta, S. R., A. Katsov, L. Hu, A. Petros, S. W. Fesik, M. B. Yaffe, and M. E. Greenberg. 2000. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol. Cell 6:41-51. [PubMed] [Google Scholar]
- 20.Earnshaw, W. C., L. M. Martins, and S. H. Kaufmann. 1999. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 68:383-424. [DOI] [PubMed] [Google Scholar]
- 21.Everett, H., and G. McFadden. 1999. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 7:160-165. [DOI] [PubMed] [Google Scholar]
- 22.Fadeel, B., Z. Hassan, E. Hellstrom-Lindberg, J. I. Henter, S. Orrenius, and B. Zhivotovsky. 1999. Cleavage of Bcl-2 is an early event in chemotherapy-induced apoptosis of human myeloid leukemia cells. Leukemia 13:719-728. [DOI] [PubMed] [Google Scholar]
- 23.Fadeel, B., B. Zhivotovsky, and S. Orrenius. 1999. All along the watchtower: on the regulation of apoptosis regulators. FASEB J. 13:1647-1657. [DOI] [PubMed] [Google Scholar]
- 24.Faleiro, L., R. Kobayashi, H. O. Fearnhead, and Y. A. Lazebnik. 1997. Multiple species of CPP32 and Mch2 are the major active caspases present in apoptotic cells. EMBO J. 16:2271-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fanidi, A., D. C. Hancock, and T. D. Littlewood. 1998. Suppression of c-Myc-induced apoptosis by the Epstein-Barr virus gene product BHRF1. J. Virol. 72:8392-8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farrell, P. J. 1989. Epstein-Barr virus genome, p. 103-132. In G. Klein (ed.), Advances in viral oncology; tumorigenic DNA viruses. Raven Press, Ltd., New York, N.Y.
- 27.Foghsgaard, L., and M. Jaattela. 1997. The ability of BHRF1 to inhibit apoptosis is dependent on stimulus and cell type. J. Virol. 71:7509-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grandgirard, D., E. Studer, L. Monney, T. Belser, I. Fellay, C. Borner, and M. R. Michel. 1998. Alphaviruses induce apoptosis in Bcl-2-overexpressing cells: evidence for a caspase-mediated, proteolytic inactivation of Bcl-2. EMBO J. 17:1268-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardwick, J. M. 1998. Comparing and contrasting BHRF1 with human Bcl-2. Epstein-Barr Virus Rep. 5:31-35. [Google Scholar]
- 30.Hardwick, J. M., and B. Levine. 2000. Sindbis virus vector system for functional analysis of apoptosis regulators. Methods Enzymol. 322:492-508. [DOI] [PubMed] [Google Scholar]
- 31.Hardwick, J. M., P. M. Lieberman, and S. D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henderson, S., D. Hue, M. Rowe, C. Dawson, G. Johnson, and A. Rickinson. 1993. Epstein-Barr virus-coded BHRF1 protein, a viral homologue of bcl-2, protects human B cells from programmed cell death. Proc. Natl. Acad. Sci. USA 90:8479-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hengartner, M. O. 2000. The biochemistry of apoptosis. Nature 407:770-776. [DOI] [PubMed] [Google Scholar]
- 34.Hickish, T., D. Robertson, P. Clarke, M. Hill, F. di Stefano, C. Clarke, and D. Cunningham. 1994. Ultrastructural localization of BHRF1: an Epstein-Barr virus gene product which has homology with bcl-2. Cancer Res. 54:2808-2811. [PubMed] [Google Scholar]
- 35.Hsu, Y.-T., K. G. Wolter, and R. J. Youle. 1997. Cytosol-to-membrane redistribution of Bax and Bcl-XL during apotosis. Proc. Natl. Acad. Sci. USA 94:3668-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu, Y.-T., and R. J. Youle. 1997. Nonionic detergents induce dimerization among members of the Bcl-2 family. J. Biol. Chem. 272:13829-13834. [DOI] [PubMed] [Google Scholar]
- 37.Huang, H., J. H. Zhou, S. M. Zhou, J. H. Hu, X. H. Pan, X. T. Kong, L. Yu, X. Y. Sun, and W. Wu. 1997. Epstein-Barr virus BHRF1 prohibits the cells of nasopharyngeal carcinoma from apoptosis. J. Laryngol. Otol. 111:1147-1150. [DOI] [PubMed] [Google Scholar]
- 38.Jurgensmeier, J. M., Z. Xie, Q. Deveraux, L. Ellerby, D. Bredesen, and J. C. Reed. 1998. Bax directly induces release of cytochrome c from isolated mitochondria. Proc. Natl. Acad. Sci. USA 95:4997-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karpf, A. R., E. Lenches, E. G. Strauss, J. H. Strauss, and D. T. Brown. 1997. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J. Virol. 71:7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawanishi, M. 1997. Epstein-Barr virus BHRF1 protein protects intestine 407 epithelial cells from apoptosis induced by tumor necrosis factor alpha and anti-Fas antibody. J. Virol. 71:3319-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirsch, D. G., A. Doseff, B. N. Chau, D.-S. Lin, N. C. de Souza-Pinto, R. Hansford, M. B. Kastan, Y. A. Lazebnik, and J. M. Hardwick. 1999. Caspase-3-dependent cleavage of Bcl-2 promotes release of cytochrome c. J. Biol. Chem. 274:21155-21161. [DOI] [PubMed] [Google Scholar]
- 42.Kluck, R. M., E. Bossy-Wetzel, D. R. Green, and D. D. Newmeyer. 1997. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science 275:1132-1136. [DOI] [PubMed] [Google Scholar]
- 43.Lee, M.-A., and J. L. Yates. 1992. BHRF1 of Epstein-Barr virus, which is homologous to human proto-oncogene bcl-2, is not essential for transformation of B cells or for virus replication in vitro. J. Virol. 66:1899-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemon, S. M., L. M. Hutt, J. E. Shaw, J. L. Li, and J. S. Pagano. 1977. Replication of EBV in epithelial cells during infectious mononucleosis. Nature 268:268-270. [DOI] [PubMed] [Google Scholar]
- 45.Levine, B., J. E. Goldman, H. H. Jiang, D. E. Griffin, and J. M. Hardwick. 1996. Bcl-2 protects mice against fatal alphavirus encephalitis. Proc. Natl. Acad. Sci. USA 93:4810-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis, J., G. A. Oyler, K. Ueno, Y. Fannjiang, B. N. Chau, J. Vornov, S. J. Korsmeyer, S. Zou, and J. M. Hardwick. 1999. Inhibition of virus-induced neuronal apoptosis by Bax. Nat. Med. 5:832-835. [DOI] [PubMed] [Google Scholar]
- 47.Li, H., H. Zhu, C. Xu, and J. Yuan. 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94:491-501. [DOI] [PubMed] [Google Scholar]
- 48.Li, L. Y., H. M. Shih, M. Y. Liu, and J. Y. Chen. 2001. The cellular protein PRA1 modulates the anti-apoptotic activity of Epstein-Barr virus BHRF1, a homologue of Bcl-2, through direct interaction. J. Biol. Chem. 276:27354-27362. [DOI] [PubMed] [Google Scholar]
- 49.Luo, X., I. Budihardjo, H. Zou, C. Slaughter, and X. Wang. 1998. Bid, a Bcl-2 interacting protein, mediates cytochrome c release from mitochondira in response to activation of cell surface death receptors. Cell 94:481-490. [DOI] [PubMed] [Google Scholar]
- 50.Macho, A., D. Decaudin, M. Castedo, T. Hirsch, S. A. Susin, N. Zamzami, and G. Kroemer. 1996. Chloromethyl-X-rosamine is an aldehyde-fixable potential-sensitive fluorochrome for the detection of early apoptosis. Cytometry 25:333-340. [DOI] [PubMed] [Google Scholar]
- 51.Marchini, A., B. Tomkinson, J. I. Cohen, and E. Kieff. 1991. BHRF1, the Epstein-Barr virus gene with homology to Bcl-2, is dispensable for B-lymphocyte transformation and virus replication. J. Virol. 65:5991-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marshall, W. L., C. Yim, E. Gustafson, T. Graf, D. R. Sage, K. Hanify, L. Williams, F. Fingeroth, and R. W. Finberg. 1999. Epstein-Barr virus encodes a novel homolog of the bcl-2 oncogene that inhibits apoptosis and associates with Bax and Bak. J. Virol. 73:5181-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meier, P., A. Finch, and G. Evan. 2000. Apoptosis in development. Nature 407:796-801. [DOI] [PubMed] [Google Scholar]
- 54.Nava, V. E., E. H. Y. Cheng, M. Veliuona, S. Zou, R. J. Clem, M. L. Mayer, and J. M. Hardwick. 1997. Herpesvirus saimiri encodes a functional homolog of the human bcl-2 oncogene. J. Virol. 71:4118-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nava, V. E., A. Rosen, M. A. Veliuona, R. J. Clem, B. Levine, and J. M. Hardwick. 1998. Sindbis virus induces apoptosis through a caspase-dependent, CrmA-sensitive pathway. J. Virol. 72:452-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nechushtan, A., C. L. Smith, Y.-T. Hsu, and R. J. Youle. 1999. Conformation of the Bax C-terminus regulates subcellular location and cell death. EMBO J. 18:2330-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nechushtan, A., C. L. Smith, I. Lamensdorf, S. H. Yoon, and R. J. Youle. 2001. Bax and Bak coalesce into novel mitochondria-associated clusters during apoptosis. J. Cell Biol. 153:1265-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oldstone, M. B. 1997. How viruses escape from cytotoxic T lymphocytes: molecular parameters and players. Virology 234:179-185. [DOI] [PubMed] [Google Scholar]
- 59.Pearson, G. R., J. Luka, L. Petti, J. Sample, M. Birkenbach, D. Braun, and E. Kieff. 1987. Identification of an Epstein-Barr virus early gene encoding a second component of the restricted early antigen complex. Virology 160:151-161. [DOI] [PubMed] [Google Scholar]
- 60.Raff, M. C. 1996. Size control: the regulation of cell numbers in animal development. Cell 86:173-175. [DOI] [PubMed] [Google Scholar]
- 61.Rouabhia, M., L. Germain, F. Belanger, and F. A. Auger. 1993. Cultured epithlium allografts: Langerhans cell and Thy-1+ dendritic epidermal cell depletion effects on allograft rejection. Transplantation 56:259-264. [PubMed] [Google Scholar]
- 62.Sarid, R., T. Sato, R. A. Bohenzky, J. J. Russo, and Y. Chang. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat. Med. 3:293-298. [DOI] [PubMed] [Google Scholar]
- 63.Sattler, M., H. Liang, D. Nettesheim, R. P. Meadows, J. E. Harlan, M. Eberstadt, H. S. Yoon, S. B. Shuker, B. S. Chang, A. J. Minn, C. B. Thompson, and S. W. Fesik. 1997. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 275:983-986. [DOI] [PubMed] [Google Scholar]
- 64.Sedlak, T. W., Z. N. Oltvai, E. Yang, K. Wang, L. H. Boise, C. B. Thompson, and S. J. Korsmeyer. 1995. Multiple Bcl-2 family members demonstrate selective dimerizations with Bax. Proc. Natl. Acad. Sci. USA 92:7834-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sixbey, J. W., and P. Shirley. 1991. Epstein-Barr virus infection of mucosal surfaces: detection of genomic variants with altered pathogenic potential. Springer Semin. Immunopathol. 13:167-179. [DOI] [PubMed] [Google Scholar]
- 66.Strasser, A., L. O'Connor, and V. M. Dixit. 2000. Apoptosis signaling. Annu. Rev. Biochem. 69:217-245. [DOI] [PubMed] [Google Scholar]
- 67.Strickland, J. E., D. A. Greenhalgh, A. Koceva-Chyla, H. Hennings, C. Restrepo, M. Balaschak, and S. H. Yuspa. 1988. Development of murine epidermal cell lines which contain an activated rasHa oncogene and form papillomas in skin grafts on athymic nude mouse hosts. Cancer Res. 48:165-169. [PubMed] [Google Scholar]
- 68.Susin, S. A., N. Zamzami, M. Castedo, T. Hirsch, P. Marchetti, A. Macho, E. Daugas, M. Geuskens, and G. Kroemer. 1996. Bcl-2 inhibits the mitochondrial release of an apoptogenic protease. J. Exp. Med. 184:1331-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suzuki, M., R. J. Youle, and N. Tjandra. 2000. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell 103:645-654. [DOI] [PubMed] [Google Scholar]
- 70.Takizawa, T., R. Fukuda, T. Miyawaki, K. Ohashi, and Y. Nakanishi. 1995. Activation of the apoptotic Fas antigen-encoding gene upon influenza virus infection involving spontaneously produced beta-interferon. Virology 209:288-296. [DOI] [PubMed] [Google Scholar]
- 71.Tarodi, B., T. Subramanian, and G. Chinnadurai. 1994. Epstein-Barr virus BHRF1 protein protects against cell death induced by DNA-damaging agents and heterologous viral infection. Virology 201:404-407. [DOI] [PubMed] [Google Scholar]
- 72.Thompson, C. B. 1995. Apoptosis in the pathogenesis and treatment of disease. Science 267:1456-1462. [DOI] [PubMed] [Google Scholar]
- 73.Wada, N., M. Matsumura, Y. Ohba, N. Kobayashi, T. Takizawa, and Y. Nakanishi. 1995. Transcription stimulation of the Fas-encoded gene by nuclear factor for interleukin-6 expression upon influenza virus infection. J. Biol. Chem. 270:18007-18012. [DOI] [PubMed] [Google Scholar]
- 74.Wei, M. C., W. X. Zong, E. H. Cheng, T. Lindsten, V. Panoutsakopoulou, A. J. Ross, D. A. Roth, G. R. MacGregor, C. B. Thompson, and S. J. Korsmeyer. 2001. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292:272-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Williams, T., D. Sale, and S. A. Hazlewood. 2000. BHRF1 is highly conserved in primate virus analogues of Epstein-Barr virus. Intervirology 44:55-58. [DOI] [PubMed] [Google Scholar]
- 76.Wolter, K. G., Y.-T. Hsu, C. L. Smith, A. Nechushtan, X.-G. Xi, and R. J. Youle. 1997. Movement of Bax from the cytosol to mitochondria during apoptosis. J. Cell. Biol. 139:1281-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin, X.-M., Z. N. Oltvai, and S. J. Korsmeyer. 1993. Bcl-2 functions by counteracting its dimerization partner, Bax, a death accelerator protein. Blood 82:441a.
- 78.Zamzami, N., S. A. Susin, P. Marchetti, T. Hirsch, I. Gomez-Monterrey, M. Castedo, and G. Kroemer. 1996. Mitochondrial control of nuclear apoptosis. J. Exp. Med. 183:1533-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zha, H., and J. C. Reed. 1997. Heterodimerization-independent functions of cell death regulatory proteins Bax and Bcl-2 in yeast and mammalian cells. J. Biol. Chem. 272:31482-31488. [DOI] [PubMed] [Google Scholar]
- 80.Zha, J., S. Weiler, K. J. Oh, M. C. Wei, and S. J. Korsmeyer. 2000. posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science 290:1761-1765. [DOI] [PubMed] [Google Scholar]
- 81.Zong, W. X., T. Lindston, A. J. Ross, G. R. MacGregor, and C. B. Thompson. 2001. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev. 15:1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]