Abstract
The human cytomegalovirus (HCMV) virion is a complex structure that contains at least 30 proteins, many of which have been identified. We determined that the HCMV UL35 gene encodes two proteins, including a previously unidentified virion protein. A 22-kDa phosphoprotein (ppUL35A) was translated from a 1.2-kb UL35 transcript by 4 h postinfection; a second phosphoprotein of 75 kDa (ppUL35) was translated from a 2.2-kb transcript predominantly late in infection. The 22-kDa protein localized to the nucleus, while the 75-kDa protein localized to the juxtanuclear compartment and was packaged into virion particles. The 22-kDa protein was identical to the COOH-terminal end of the 75-kDa protein but was not found in virions, thus defining the NH2-terminal portion of the 75-kDa protein as essential for packaging. Expression of the 22-kDa protein inhibited activation of the major immediate-early promoter by ppUL82 (pp71), suggesting that the UL35 22-kDa protein may modulate expression of the major immediate-early gene.
Human cytomegalovirus (HCMV) is a ubiquitous, opportunistic human pathogen that causes severe infection in immunocompromised individuals. It also infects the developing fetus and is a major cause of congenital malformation (see reference 12 for a review). Like the other herpesviruses, HCMV has a complex virion structure. The icosahedral capsid contains the packaged viral genome; the capsid is surrounded by an amorphous proteinaceous region called the tegument or matrix, and a lipid bilayer envelopes the capsid and tegument (see reference 46 for a review). More than 30 viral proteins are found in the complete infectious particle (2, 19, 20, 32; see references 46 and 54 for reviews).
Among the known HCMV virion components, four proteins (pUL46, pUL48/49, pUL85, and pUL86) comprise the mature viral capsid (2, 15, 21, 22, 29). Six glycoproteins have been identified in the viral envelope and include gpUL55 (gB), gpUL100 (gM), gpUL73 (gN), gpUL75 (gH), gpUL115 (gL), and gpUL74 (gO) (13, 28, 31, 38, 42). In addition, gpUL4 (gp48) and pUL33 are structural glycoproteins that are likely localized in the envelope (14, 44).
The remaining 20 to 25 structural proteins of HCMV are believed to reside in the tegument or the matrix, a poorly characterized amorphous region between the capsid and the envelope of the virion. Among the tegument constituents, six phosphoproteins have been identified and include ppUL32 (pp150), ppUL83 (pp65), ppUL82 (pp71), ppUL69, ppUL99 (pp28), and ppUL25 (3, 30, 36, 45, 49, 52, 57, 61). Two of the tegument proteins, ppUL82 (pp71) and ppUL69, are transactivators of viral gene expression (1, 11, 25, 26, 39, 56). In addition, ppUL69 arrests infected cells in the G1 phase of the cell cycle (25, 41). Gene products encoded by IRS1/TRS1, UL98a (pp58), UL48 (p212), UL56 (p130), UL65 (pp67), and UL36 are also found in virions, although their location in viral particles has not yet been determined (9, 10, 17, 23, 34, 35, 47, 51).
The HCMV UL25 gene is a member of the UL25 gene family, which consists of UL25 and UL35 (15). The UL25 gene family is conserved among members of the betaherpesviruses (40, 48, 55). The second member of the UL25 gene family, UL35, is conserved among the betaherpesviruses, with the amino acid similarities spread throughout the predicted protein coding region (40). UL25 encodes a tegument phosphoprotein (3, 61), suggesting that the UL35 open reading frame would also encode a structural protein.
In this report, we demonstrate that the UL35 open reading frame encodes two proteins, one which is found in the viral particle and one that localizes to the nucleus and decreases ppUL82 activation of the major immediate-early promoter.
MATERIALS AND METHODS
Cells, virus, and transfections.
Human diploid fibroblasts (HDFs) were propagated as described (37). HCMV (strain Towne) was obtained from Adam Geballe (Fred Hutchinson Cancer Research Center, Seattle, Wash.). Viral infections were performed using 80 to 90% confluent HDFs at a multiplicity of infection of 10 PFU per cell for 60 min. HCMV virions were concentrated from infected-cell culture medium by ultracentrifugation and purified by banding in potassium tartrate-glycerol gradients as described (29).
To block protein synthesis and restrict transcription to immediate-early genes, cycloheximide was added to the medium 1 h prior to infection at a concentration of 50 μg/ml. Viral DNA replication was inhibited by adding phosphonoformic acid to the medium at a final concentration of 200 μg/ml at the time of infection. For visualization of enhanced green fluorescent protein (EGFP)-tagged fusion proteins, plasmids were transiently transfected into HDFs using Effectene transfection reagent (Qiagen, Valencia, Calif.). Transient transfection assays measuring the level of viral gene expression were performed using DEAE-dextran and 4-methylumbelliferyl-β-d-galactoside (MUG) as described (4).
Plasmids.
pEQ3 and pEQ276 were provided by Adam Geballe; pEQ3 is a promoterless plasmid containing the lacZ gene, and pEQ276 expresses the major immediate-early proteins IE1 and IE2 (7, 8). pBJ176, pBJ201, and pBJ203 have been described previously; pBJ176 expresses the lacZ gene under the control of the major immediate-early promoter and crs (4), pBJ201 contains the major immediate-early promoter (5), and pBJ203 expresses ppUL82 (6).
pBJ352 expresses a portion of the UL35 open reading frame as a His-tagged protein in bacterial cells and was constructed by amplifying nucleotides 47482 to 47855 of HCMV strain AD169 genomic DNA using oligonucleotides 187 (5"-CGGGATCCTTCATGGAACTCCTCGAC-3") and 188 (5"-CCCAAGCTTCTGTAGGCTACCGAGTAGG-3") and Taq polymerase. The amplimer was inserted into pET30a (Novagen, Madison, Wis.).
pBJ397 and pBJ399 served as templates for in vitro transcription-translation reactions used to synthesize UL35. pBJ397 was constructed by amplifying a DNA fragment (nucleotides 46089 to 48214, which contain the entire UL35 open reading frame) from pRL109 (kindly provided by G. Hayward) using Vent polymerase (New England Biolabs, Beverly, Mass.) and oligonucleotides 235 (5"-CCCAAGCTTCATCATGGCCCAAGGATC-3") and 236 (5"-CCCAAGCTTCCCGTTTGTTTATGTGCATG-3"); the amplimer was inserted into pBluescript SK(−) (Stratagene, La Jolla, Calif.). pBJ399 contains the 3" half of the UL35 open reading frame and was generated by digesting pBJ397 with XhoI and BglII, followed by Klenow treatment and religation.
pBJ505 and pBJ506 express EGFP-UL35 fusion proteins. pBJ505 was constructed by inserting the DNA fragment from pBJ397 into pEGFP-C2 (Clontech, Palo Alto, Calif.) so that the UL35 open reading frame was in-frame with EGFP. pBJ506 was constructed by digestion of pBJ505 with BglII and religation, resulting in a plasmid that expresses the 3" half of the UL35 open reading frame as an EGFP fusion protein.
pBJ511 expresses the 75-kDa UL35 protein under the control of the HCMV major immediate-early promoter and was constructed by inserting a HindIII fragment isolated from pBJ397 and containing HCMV sequences from 46089 to 48214 into pBJ201 (5). pBJ511 expresses the 22-kDa UL35 protein under the control of the HCMV major immediate-early promoter and was constructed by digesting pBJ511 with XbaI and BglII, followed by Klenow treatment and ligation.
RNA analyses.
Total cellular RNA was extracted from mock-infected and HCMV-infected HDFs using acid phenol-guanidinium isothiocyanate (16). For Northern blot analyses, 5 μg of total cellular RNA was fractionated in 1.5% agarose-formaldehyde gels in morpholinepropanesulfonic acid (MOPS) buffer and transferred onto nitrocellulose membranes (43). Hybridizations were performed as described, using riboprobes to detect UL35 transcripts and the cellular mRNA for glyceraldehyde phosphate dehydrogenase (GAPDH) (27).
The template for in vitro transcription of an antisense UL35 RNA was generated by amplifying HCMV sequences (15) from HCMV strain Towne genomic DNA using oligonucleotides 253 (5"-ACATTTCAGCGCGTACAAGC-3") and 254 (5"-TAATACGACTCACTATAGGTGCCGTACAGGTTCTTGGAG-3") and Vent polymerase (New England Biolabs). The antisense UL35 riboprobes were synthesized using the PCR-generated template, the Maxiscript in vitro transcription kit (Ambion, Austin, Tex.), and [32P]uracil. The GAPDH probe was generated from the pTRI-GAPDH-Human template (Ambion).
RNase protection assays were performed using the RPAII kit (Ambion) according to the manufacturer's instructions. Protected fragments were analyzed by electrophoresis in urea-polyacrylamide gels.
5"RACE.
For amplification of the 5" ends of the UL35 transcripts, polyadenylated RNA was isolated from HDF cells 96 h after HCMV infection using the Poly(A)Pure kit (Ambion). 5"Rapid amplification of cDNA ends (RACE) was performed with the Marathon cDNA amplification kit (Clontech, Palo Alto, Calif.) as directed. A 28-base oligonucleotide complementary to nucleotides 47383 to 47410 (179, 5"-ACGTCTCGGTGGTGATCTTGCCTTCGTG-3") was used as the UL35-specific primer. Amplification products were inserted into the pCR2.1/TOPO TA cloning vector (Invitrogen, Carlsbad, Calif.). The ends of the amplimers were sequenced using the T7 Sequenase version 2.0 DNA sequencing kit (USB).
Protein analysis.
pBJ397 and pBJ399, which contain the entire and 3" half of the UL35 open reading frame, respectively, were used as templates in in vitro transcription-translation assays using the TNT T7 Quick Coupled System (Promega) and [35S]methionine (New England Nuclear, Boston, Mass.).
Antiserum to UL35 proteins was obtained by immunizing rabbits with purified, His-tagged UL35 peptides from bacteria. Recombinant UL35 protein was produced in Escherichia coli strain BL21(DE3)plysS cells (Novagen, Madison, Wis.) following transformation with pBJ352. Induction and affinity purification of the recombinant protein were performed as directed. Briefly, cells were grown in Luria-Bertani (LB) broth containing 30 μg of kanamycin and 34 μg of chloramphenicol per ml to an optical density at 600 nm of 0.6 at 37°C. Then 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture. After a further 2.5 h of growth, cells were pelleted and lysed by sonication.
Inclusion bodies were dissolved in binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9], 6 M urea), and loaded onto an NiSO4 resin column. After washing with 20 mM imidazole, the recombinant protein was eluted with an elution buffer (1 M imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9], 6 M urea) and dialyzed against 20 mM Tris-HCl (pH 9.0)-100 mM NaCl to a final concentration of 0.75 M urea. Rabbit immunizations were conducted by Spring Valley Laboratories, Inc., in Woodbine, Md., according to standard protocols (24). The antisera were characterized for reactivity against the immunogen.
Western blot analyses and immunoprecipitation experiments were used to characterize the UL35 proteins. For Western blot analyses, mock-infected and HCMV-infected HDFs were lysed with Laemmli sample buffer. The proteins were separated by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis (SDS-12.5% PAGE) and transferred onto Optitran BA-S 85 nitrocellulose membranes (Midwest Scientific). Filters were then incubated with anti-UL35 rabbit serum BIE-2 diluted 1:5,000. Antibody binding was detected with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (IgG; Fisher Scientific) and visualized with bromochloroindolyl phosphate-nitro blue tetrazolium (Gibco-BRL, Rockville, Md.).
Immunoprecipitation of 35S- or 32P-labeled proteins was performed (W. Liu, Y. Zhao, and B. J. Biegalke, unpublished data), lysing cells in radioimmunoprecipitation assay (RIPA) buffer that contained the complete protease inhibitor cocktail (Boehringer Mannheim, Mannheim, Germany). Lysates were precleared with preimmune rabbit serum and protein A-Sepharose beads. UL35 proteins were immunoprecipitated with anti-UL35 rabbit antiserum and protein A-Sepharose beads; antigen-antibody complexes were washed once with RIPA buffer, twice with buffer II (50 mM Tris-Cl, pH 7.5, 500 mM NaCl, 0.1% NP-40, and 0.05% sodium deoxycholate), with a final wash using buffer III (50 mM Tris-Cl [pH 7.5], 0.1% NP-40, 0.05% sodium deoxycholate). Antigen-antibody complexes were eluted from the protein A-Sepharose beads by boiling in 1× Laemmli sample buffer; proteins were size fractionated on 12.5% SDS-polyacrylamide gels. For 35S labeling, cells were labeled with 200 μCi of EasyTag EXPRE35S35S protein labeling mix (NEN) in methionine- and cysteine-free Dulbecco's modified Eagle's medium (DMEM) for 30 min. For 32P labeling, cells were incubated with [32P]orthophosphate in phosphate-free DMEM for 120 min.
Fluorescence microscopy.
EGFP fusion proteins were visualized ≈24 h after transfection, following fixation with 3.5% formaldehyde and counterstaining with 4",6"-diamidino-2-phenylindole (DAPI; Molecular Probes, Eugene, Oreg.).
Mock-infected and HCMV-infected HDF cells were fixed with paraformaldehyde (4%) for 30 min at 4°C and permeabilized with 0.2% Triton X-100 for 2 min. Cells were then incubated with 10% normal human serum at 37°C for 1 h to block HCMV-induced Fc receptors. Subsequently, cells were incubated with rabbit anti-UL35 (BIE-1) or preimmune rabbit serum, both diluted 1:200, in cold phosphate-buffered saline (PBS) containing 10% normal human serum for 2 h on ice. After three washes with PBS, Texas red-labeled goat anti-rabbit IgG (1:800) was added to the cells and incubated for 2 h on ice.
For double staining, cells were incubated with a mixture of rabbit anti-UL35 (1:200) and a mouse monoclonal anti-gB (Virusys, North Berwick, Maine) diluted 1:400. Oregon green-labeled goat anti-mouse IgG (Molecular Probes, Eugene, Oreg.) diluted 1:1,000 was used to detect mouse anti-gB. To rule out cross-reactions between the primary and secondary antibodies, infected cells were stained with rabbit anti-UL35 plus fluorescent goat anti-mouse IgG or mouse anti-gB plus fluorescent goat anti-rabbit IgG as described above. Cells were observed with a Nikon Quantix fluorescence microscope and the appropriate fluorescence filters.
RESULTS
Transcriptional analysis of UL35.
Transcripts originating from the region of the HCMV genome containing the UL35 open reading frame were characterized for size and time of expression using Northern blot analyses, RNase protection assays, and 5"RACE.
Northern blot analyses were performed using total cellular RNA harvested from mock-infected or from HCMV (Towne)-infected human fibroblasts at 4, 24, 48, 72, and 96 h postinfection (hpi). RNA was also harvested from cells infected in the presence of the protein synthesis inhibitor cycloheximide, which limits viral transcription to immediate-early genes. Harvesting of infected-cell RNA from cells treated with the viral DNA synthesis inhibitor phosphonoformic acid was used to prevent true late-gene transcription, which is dependent on viral DNA replication. RNAs were hybridized to a riboprobe complementary to the predicted UL35 open reading frame (nucleotides 47094 to 47384) (15).
Ethidium bromide staining of rRNA demonstrated that equal amounts of RNA were present in all samples (data not shown); the level of the cellular GAPDH transcripts varied with time postinfection, as described (Fig. 1B) (59). Three predominant transcripts of 1.2, 2.2, and 3.6 kb were detected with the UL35 riboprobe (Fig. 1B). The 1.2-kb RNA was synthesized as early as 4 hpi and continued to be transcribed throughout infection. Cycloheximide treatment prevented the synthesis of the 1.2-kb RNA, classifying the transcript as an early RNA (Fig. 1B). The 2.2-kb and 3.6-kb transcripts were detected predominantly at later times of infection (48 to 96 hpi, Fig. 1B), although RNase protection assays demonstrated that a low level of the larger transcripts was present as early as 4 hpi (Fig. 1C). Phosphonoformic acid treatment significantly inhibited the expression of all three transcripts, demonstrating that the level of UL35 transcripts is influenced by replication of the viral genome. In addition to the three predominant transcripts, a minor transcript of 1.8 kb was detected at 8 hpi.
FIG. 1.
Transcript mapping of the UL35 gene. (A) Schematic diagram of the UL35 gene. The UL34 and UL35 open reading frames (ORF) are labeled; the gray rectangle represents the part of UL35 expressed as a His-tagged fusion protein and used as the antigen in the generation of the anti-UL35 antiserum. Horizontal thin arrows, UL35 transcripts of 2.2 and 1.2 kb; horizontal thick arrow, riboprobe used for Northern blot hybridization and RNase protection assays; horizontal arrowhead, oligonucleotide primer used for 5"RACE; vertical arrows, predicted polyadenylation sites; bold T, TA-rich regions; A, AUG codons present in the 5" end of the 1.2-kb RNA. (B) Northern blot analysis of mock-infected cell RNA and RNA from HCMV-infected cells harvested at the indicated times postinfection. The probe is indicated, as are the positions of the rRNAs. M, mock-infected cell RNA; 4, 8, 48, 72, and 96 refer to the time (in hours) postinfection of RNA harvest; 4c, RNA harvested 4 hpi with infection occurring in the presence of cycloheximide; 96p, RNA harvested 96 hpi from cells infected in the presence of phosphonoformic acid. (C) RNase protection analysis of UL35 transcripts. Lane 1, probes for the cellular transcript GAPDH and the UL35 transcripts (−); lane 2, probes treated with RNase mixture (+); lane 3, RNA harvested from mock-infected cells (M); lane 4, RNA harvested from infected cells 4 hpi; lane 5, RNA harvested from infected cells 72 hpi. The positions of the RNA molecular size markers are indicated (in nucleotides). The fragments protected with the GAPDH and the UL35 probes are labeled.
Radiolabeled probes corresponding to the 5" end of the UL35 open reading frame, the UL34 open reading frame, or the UL36 open reading frame were used to further map the UL35 transcripts shown in Fig. 1B. The 2.2- and the 3.6-kb RNAs hybridized to a 5" UL35 probe, while the 1.2-kb transcript did not (data not shown), suggesting that the 1.2- and 2.2-kb RNAs have similar 3" but different 5" ends. The 3.6-kb transcript also hybridized to the UL34 probe but not the UL36 probe (data not shown), suggesting that the 3.6-kb RNA is a readthrough transcript that contains both UL34 and UL35 sequences.
5"RACE was performed to further characterize the UL35 transcripts. Polyadenylated RNA isolated from HCMV-infected fibroblasts 96 hpi served as the template for cDNA amplification. The predominant amplification products of 1.0 and 0.3 kb were cloned, and the ends of the amplimers were sequenced (data not shown). The 5" end of the 1.0-kb RACE product corresponded to nucleotide 46061, which is 32 bases upstream of the predicted UL35 open reading frame (Fig. 1A). The 5" end of the second smaller 0.3-kb RACE product corresponded to nucleotide 47142, which is located within the UL35 open reading frame. These data demonstrated that the UL35 transcripts initiate both at the 5" end and internally in the UL35 open reading frame.
Transcript processing using the polyadenylation signal located at nucleotide 48070 and subsequent polyadenylation would generate RNAs of the observed sizes, with the 2.2-kb transcript corresponding to the entire UL35 open reading frame and the 1.2-kb RNA containing sequences from the 3" end of the open reading frame (Fig. 1A). In addition to the 1.0- and 0.3-kb RACE products, a third RACE amplimer of 3.0 kb was also detected. The 3-kb RACE product was not analyzed, but the size of the amplimer suggested that it was generated by using the 3.6-kb transcript as the template.
RNase protection analyses revealed two UL35-specific protected fragments, one of 186 nucleotides that corresponds to the larger UL35 RNAs and the other (164 nucleotides) corresponding to the smaller UL35 transcript (Fig. 1C). As expected, the cellular GAPDH transcript was detected in RNA harvested from mock- and HCMV-infected cells (Fig. 1C). These data confirmed that the 1.2-kb UL35 transcript initiates internally in the UL35 open reading frame. The smaller protected fragment corresponding to the 1.2-kb RNA was 164 nucleotides in length, indicating a transcription initiation site around nucleotide 47120 (Fig. 1C). This predicted start site is approximately 20 nucleotides 5" of the start site identified by RACE analysis, suggesting that the cDNA used for amplification was an incomplete product of the reverse transcription and amplification reactions.
The UL35 transcript mapping studies demonstrated that at least two mRNAs initiate in or around the UL35 open reading frame. Analysis of the DNA sequence for UL35 and the flanking regions identified potential promoter elements 50 and 30 nucleotides 5" of the initiation sites for the 2.2- and 1.2-kb transcripts, respectively. The potential promoter element for the 2.2-kb transcript is located 5" of the predicted UL34 polyadenylation site (Fig. 1A). The overlap between transcript processing elements and the putative UL35 promoter element may contribute to the generation of a UL34-UL35 readthrough transcript.
Characterization of UL35 proteins.
Two monocistronic mRNAs of 1.2 and 2.2 kb originate from the UL35 open reading frame. The 2.2-kb RNA contains the entire UL35 open reading frame and was predicted to encode a protein of 69 kDa. The open reading frame contained in the 1.2-kb mRNA corresponds to the carboxyl-terminal end of the UL35 open reading frame. The initiation codons present in the 1.2-kb transcript are all in frame with the UL35 initiation codon; the first methionine is located at nucleotide 47338 and is in a poor context for initiation of translation (33). The next methionine, at nucleotide 47434, is in a good context for initiation of translation, suggesting that the second AUG of the 1.2-kb transcript will be used to initiate protein synthesis. The size of the protein encoded by the 1.2-kb RNA was predicted to be either 25 or 21 kDa, depending on the translation initiation start site.
To identify UL35-encoded proteins and to analyze their expression, polyclonal antiserum was generated to the carboxyl-terminal end of the UL35 open reading frame (Fig. 1A). Two different immune sera obtained from two different rabbits were used in Western blot analyses and immunoprecipitation experiments and yielded identical results. For Western blot analyses, lysates of mock-infected cells, lysates of HCMV-infected human fibroblasts harvested at different times postinfection, and purified virions were size fractionated on SDS-polyacrylamide gels, transferred to filter paper, and incubated with the anti-UL35 antiserum or with the preimmune antiserum. Antigen-antibody interactions were detected enzymatically.
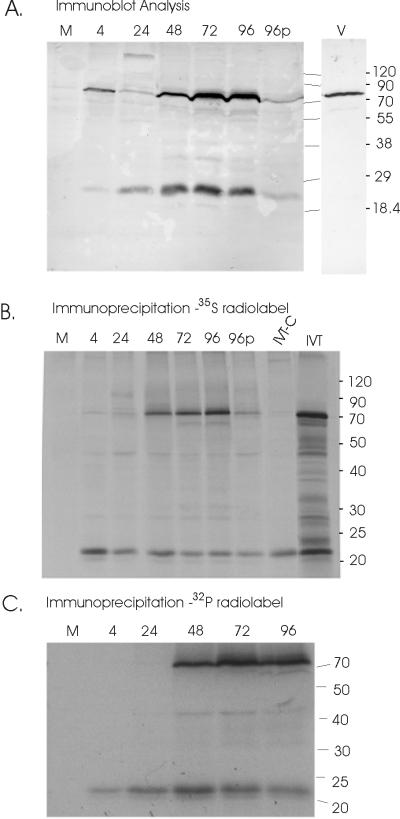
No predominant proteins were detected using the preimmune antiserum (data not shown). The anti-UL35 antiserum detected two proteins in infected cells at 4 hpi, proteins of 75 and 22 kDa (Fig. 2A). By 24 hpi, the 75-kDa protein was significantly decreased in abundance, suggesting that the 75-kDa protein seen at 4 hpi was a component of the infecting virions. This was confirmed by detection of the 75-kDa protein in gradient-purified virion particles (Fig. 2A). The 75-kDa protein was most abundant at 96 hpi, while the 22-kDa protein accumulated throughout the course of infection (Fig. 2A). Treatment with phosphonoformic acid inhibited expression of both UL35 proteins, corresponding to the decrease seen in transcript levels (Fig. 1B). A cross-reactive protein of 135 kDa was detected at 24 hpi; the origin of this protein is unknown. The 75-kDa protein was detected in dense bodies as well as virions (data not shown), suggesting that the protein is a component of the tegument.
FIG. 2.
Characterization of UL35 proteins. (A) Immunoblotting of cell lysates harvested following mock infection (M) or harvested at the indicated time (in hours) postinfection. V, purified virions. (B) Immunoprecipitation of [35S]methionine-labeled proteins using the rabbit anti-UL35 antiserum from mock-infected cells (M) or HCMV-infected cells at the indicated times (in hours) postinfection. IVT-C, in vitro transcription-translation products of pBJ399, which contains the open reading frame present on the short UL35 transcript and beginning with the second AUG (see Fig. 1A); IVT, in vitro transcription-translation products of pBJ397, which contains the entire UL35 open reading frame. (C) Immunoprecipitation of 32P-labeled proteins from mock-infected (M) and HCMV-infected cells at the indicated times postinfection with the rabbit anti-UL35 antiserum. The positions of the protein molecular size markers are indicated (in kilodaltons).
Immunoprecipitation assays allowed us to examine the levels of de novo-synthesized UL35 proteins. Immunoprecipitation experiments were performed using mock-infected-cell or infected-cell lysates from cells pulse-labeled with [35S]methionine or with [32P]orthophosphate. Lysates were harvested from infected cells at periodic intervals throughout the viral life cycle. After clearing the cell lysates with preimmune antiserum, radiolabeled proteins of 75 and 22 kDa were precipitated from infected cells with the anti-UL35 antiserum (Fig. 2B and C). The 22-kDa protein was synthesized throughout infection. The 75-kDa protein was first detectable at 48 hpi, with an increase in protein levels at 72 and 96 hpi (Fig. 2B). Again, phosphonoformic acid treatment of infected cells decreased the levels of the UL35 proteins.
The size of the 22-kDa protein suggested that the second AUG on the 1.2-kb RNA was utilized as the translation start site. To examine this hypothesis, plasmids were constructed so that the entire UL35 open reading frame (pBJ397) or the open reading frame initiating with the second AUG of the 1.2-kb transcript (pBJ399) could be transcribed and translated in vitro. The protein encoded by the COOH end of UL35, initiating with methionine 2 of the 1.2-kb RNA, was 22 kDa in size, comigrating with the immunoprecipitation product seen in infected cells (Fig. 2B). The full-length open reading frame yielded a 75-kDa protein as well as some protein fragments or incomplete translation products. The incomplete translation products included a 22-kDa protein; synthesis of the 22-kDa protein from the entire UL35 open reading frame is presumably an artifact of the in vitro translation system. The proteins synthesized by the in vitro translation reactions were immunoprecipitated by the anti-UL35 antiserum, confirming their identify (data not shown).
Many of the virion proteins, particularly those located in the tegument, are phosphoproteins. The two proteins encoded by the UL35 transcripts have a number of potential phosphorylation sites, suggesting that they, like the related protein ppUL25, would be modified by phosphorylation. Immunoprecipitation of 32P-labeled proteins was used to determine if the UL35 proteins were phosphorylated. The 75-kDa and the 22-kDa UL35 proteins were both phosphorylated, as evidenced by the precipitation of 32P-labeled proteins (Fig. 2C). Specific amino acid residues that are phosphorylated on the two proteins have yet to be identified.
Intracellular location of UL35 proteins.
Two overlapping proteins are synthesized at different times of infection from the UL35 open reading frame. The 75-kDa UL35 protein is synthesized most abundantly late in infection, is packaged into virion particles, and enters the cell upon infection. We examined the intracellular distribution of the UL35 proteins during infection by immunofluorescence. HCMV-infected cells were fixed and stained with the anti-UL35 antiserum and the Texas red-conjugated secondary antibody (goat anti-mouse IgG) at different times postinfection.
Very early in infection (1 hpi), UL35 proteins are broadly distributed throughout the cell (Fig. 3). At 4 hpi, the nuclei were fluorescent and there was an additional punctate perinuclear pattern of fluorescence in some of the cells. By 9 hpi, the nuclear fluorescence became more predominant and intensified by 24 hpi, with infected cells exhibiting nuclear fluorescence during the remainder of the viral replication cycle. In addition to the nuclear fluorescence pattern, a diffuse pattern of cytoplasmic fluorescence was also detected, with the cytoplasmic fluorescence intensity markedly increasing by 72 hpi. At 96 hpi, although the nuclei continued to fluoresce following antibody staining, the cytoplasmic fluorescence became more intense and was concentrated around the nucleus.
FIG. 3.
Localization of UL35 proteins in HCMV-infected fibroblasts. HCMV-infected cells and mock-infected cells were fixed and incubated with the anti-UL35 antiserum or with the preimmune rabbit antiserum (96 hpi -- preimmune). Antibody binding was detected with a secondary antibody, Texas red-conjugated goat anti-rabbit IgG. Cells were photographed using phase contrast microscopy (phase) and fluorescence microscopy (fluorescence) at 400× magnification.
Mock-infected cells incubated with the anti-UL35 antiserum and infected cells incubated with the preimmune antiserum had little fluorescence, demonstrating the specificity of the immunofluorescence assays (Fig. 3). Comparison of the immunofluorescence data with the protein synthesis data (Fig. 2B) suggests that the nuclear fluorescence seen early in infection, i.e., 24 hpi, was due to the 22-kDa protein, while the cytoplasmic fluorescence seen at 1 hpi and late in infection reflected the presence of the 75-kDa protein.
The intracellular localization patterns were examined for each of the UL35 proteins in the absence of other viral proteins by utilizing EGFP-tagged UL35 fusion proteins. The entire UL35 open reading frame and the open reading frame encoding the 22-kDa protein were fused in-frame to the C terminus of EGFP in the eukaryotic expression plasmid pEGFG-C2. The resulting plasmids were transfected into human fibroblasts and observed for fluorescence following overnight incubation. The 75-kDa UL35 protein localized in the cytoplasm as punctate perinuclear fluorescence (Fig. 4A); the 22-kDa protein was concentrated in the nucleus (Fig. 4A). EGFP alone exhibited bright green fluorescence in both the cytoplasm and nucleus (data not shown).
FIG. 4.
(A) Localization of UL35-EGFP fusion proteins in transfected fibroblasts. Cells were transfected with pBJ505, which expresses the UL35 75-kDa protein as an EGFP fusion (75-kDa EGFP), or with pBJ506, which expresses the UL35 22-kDa protein as an EGFP fusion (22-kDa EGFP). Cells were fixed and stained with DAPI; cells were observed for green (EGFP fusions) or blue (DAPI staining) fluorescence. Transfected cells are indicated with arrows. (B) Colocalization of UL35 and gB. HCMV-infected cells (96 hpi) were incubated with rabbit antiserum to UL35 and with mouse antiserum to gB. The primary antibody reactions were detected with Texas red-conjugated goat anti-rabbit IgG or with Oregon green-conjugated rabbit anti-mouse IgG. Cells were visualized by phase contrast microscopy and fluorescence microscopy, using red and green filters. The merged panel is the combination of the green and red fluorescence. The magnification for all pictures was 400×.
The patterns of localization seen for the EGFP fusion proteins support the immunofluorescence studies that suggested the 75-kDa protein localized to the cytoplasm and the 22-kDa protein localized to the nucleus during viral infection. These results are somewhat unexpected, as the 22-kDa protein is contained entirely within the 75-kDa protein. Thus, the dominant signal(s) for intracytoplasmic localization and packaging into virion particles is contained within the NH2-terminal region of the protein.
The perinuclear pattern of fluorescence seen with the anti-UL35 antiserum at 96 hpi has been associated with assembly of viral particles (3, 53). We compared the pattern of UL35 localization with that of the structural glycoprotein gB. The rabbit polyclonal anti-UL35 antiserum and a mouse monoclonal antibody to gB, and goat anti-rabbit IgG-Texas red conjugate or rabbit anti-mouse IgG-Oregon green conjugate secondary antibodies were used to stain HCMV-infected cells at 96 hpi. The cytoplasmic and perinuclear pattern of UL35 staining colocalized with antibody staining of gB (Fig. 4B). Staining with preimmune serum or secondary antibodies alone resulted in little fluorescence (data not shown). These data suggest that incorporation of the 75-kDa UL35 protein into the virion occurs in the juxtanuclear locale.
Regulation of gene expression by UL35.
The nuclear localization pattern of the 22-kDa UL35 protein suggested that it could potentially have a role in regulating gene expression. The UL35 proteins were screened for potential effects on viral gene expression using transient transfection assays. A plasmid expressing the entire UL35 open reading frame (pBJ511) or a plasmid expressing the 22-kDa UL35 protein (pBJ525) was transiently transfected into HDFs along with reporter gene constructs in which expression of the reporter gene, lacZ, was regulated by viral promoters. Several viral promoters were screened for effects of the 75- and 22-kDa UL35 proteins on viral gene expression in the presence and absence of other viral transactivators, including IE1 and IE2. The levels of gene expression were determined by measuring the fluorescence of the medium containing the cleaved substrate MUG.
The results of the transient transfection assays demonstrated that the 22-kDa UL35 protein had a slight repressive effect on expression from the major immediate-early promoter and, more significantly, caused a fourfold decrease in pp71-mediated activation of the major immediate-early promoter (Fig. 5). The 22-kDa UL35 protein did not inhibit IE1 activation of the major immediate-early promoter or enhance IE2-mediated repression of the promoter. The 75-kDa protein had a slight but reproducible activating effect on the major immediate-early promoter. Expression from other viral promoters, including the US3, UL35, UL119, and TRL4/IRL4 promoters, was not influenced by the expression of the 22-kDa protein either alone or in the presence of the major immediate-early proteins IE1 and IE2 (data not shown).
FIG. 5.
Repression of major immediate-early promoter expression by the 22-kDa UL35 protein. pBJ176, which expresses the lacZ gene under the control of the major immediate-early enhancer, promoter, and crs, was transfected into HDFs along with a control plasmid (pBJ201), a plasmid expressing the 22-kDa UL35 protein (UL35A; pBJ525), or a plasmid expressing the 75-kDa UL35 protein (UL35; pBJ511) in combination with a plasmid expressing the major immediate-early proteins IE1 and IE2 (pEQ276) or a plasmid expressing ppUL82 (pBJ203). Reporter gene activity was determined ≈36 h after transfection by measuring the fluorescence of the cleavage product of the β-galactosidase substrate MUG. The amount of plasmid DNA was kept constant by adding the appropriate amount of the control plasmid pBJ201. Background levels of β-galactosidase activity obtained with the promoterless lacZ plasmid pEQ3 were subtracted from the test values. The data presented here are the averages of duplicate transfections with 1 standard deviation. The transfections were repeated more than six times, with similar activities obtained.
DISCUSSION
Sequence analysis of the genome of HCMV strain AD169 (15) predicted that the virus encoded over 200 proteins. Analyses of different regions of the viral genome have identified a number of additional proteins synthesized by the virus. Synthesis of related and overlapping proteins is one strategy used by the virus to generate additional proteins without an increase in genome size. The UL35 gene is an example of one locus encoding two overlapping and related proteins. A 75-kDa UL35 protein (ppUL35) is encoded by the entire UL35 open reading frame; a second protein of 22 kDa (which we named ppUL35A) is encoded by the COOH end of the UL35 open reading frame. The two proteins are differentially localized and packaged, suggesting that they have different functions during the viral life cycle.
ppUL35A is synthesized throughout infection, appearing by 4 hpi. ppUL35A localized to the nuclei of transfected cells; nuclear staining during infection suggested that ppUL35A also localizes to the nucleus in infected cells. The nuclear location of ppUL35A protein suggested a potential role in regulating gene expression. Indeed, expression of ppUL35A inhibited activation of the major immediate-early promoter by ppUL82, suggesting that ppUL35A modulates the activity of the major immediate-early promoter early in infection.
ppUL35 was synthesized relatively late in infection and was packaged into virions. Despite the presence of a nuclear localization signal, in the absence of other viral proteins, ppUL35 localized to a cytoplasmic, perinuclear region of transfected fibroblasts. This suggests that the additional amino acids found in ppUL35 obscure the nuclear localization signal present in the COOH end of the protein and in ppUL35A. Immunofluorescence demonstrated that UL35 proteins, most likely the cytoplasmic 75-kDa protein, colocalized with the envelope glycoprotein, gB. Several structural proteins of HCMV, including ppUL25, UL99, gB, gH, ppUL32 (pp150), ppUL99 (pp28), and ppUL83 (pp65), also localize to a juxtanuclear structure (3, 36, 53), which has been suggested to be a viral assembly site (18, 59, 60). ppUL35 is another member in this congregation of structural proteins and may contribute to packaging or assembly of the viral particle.
Expression of the UL35 gene has a striking similarity to that observed for IRS1 (51). The proteins encoded by IRS1 and the related gene TRS1 are packaged within the virion and cooperate with the major immediate-early proteins IE1 and IE2 in activating viral promoters (50, 51). A second protein, pIRS1263, which is encoded by the 3" end of the IRS1 gene, localizes to the nucleus and inhibits transcriptional activation of an early and a late gene promoter (51).
The nuclear localization of ppUL35A and pIRS1263 and the packaging of the larger UL35 and IRS1 proteins into the virion suggested that the proteins might have regions of similar amino acids. Both open reading frames contain similar nuclear localization signals in the carboxy-terminal portion of the coding regions; however, no other significant similarities or regions of similar amino acid composition were identified by Clustal W analysis.
Synthesis of two related proteins from one open reading frame, proteins with predicted differences in function as for UL35, extends the protein-coding capacity of the viral genome. Given the size of the HCMV genome (235 kb), it is somewhat surprising that the virus goes to such lengths to synthesize additional proteins. The complexities of gene expression and protein synthesis undoubtedly contribute to the long replication cycle of HCMV in tissue culture and its success as an opportunistic human pathogen.
Acknowledgments
We thank Mark Berryman for assistance with microscopy, Adam Geballe for pEQ plasmids, helpful suggestions, and critical reading of the manuscript, and Frank Horodyski for critical reading of the manuscript.
This work was supported in part by Council of Tobacco Research grant 4740 and an Ohio University Baker Award to B.J.B.
REFERENCES
- 1.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battista, M. C., G. Bergamini, M. C. Boccuni, F. Campanini, A. Ripalti, and M. P. Landini. 1999. Expression and characterization of a novel structural protein of human cytomegalovirus, pUL25. J. Virol. 73:3800-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biegalke, B. J. 1995. Regulation of human cytomegalovirus US3 gene transcription by a cis-repressive sequence. J. Virol. 69:5362-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biegalke, B. J. 1997. IE2 protein is insufficient for transcriptional repression of the human cytomegalovirus US3 promoter. J. Virol. 71:8056-8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biegalke, B. J. 1999. Human cytomegalovirus US3 gene expression is regulated by a complex network of positive and negative regulators. Virology 261:155-164. [DOI] [PubMed] [Google Scholar]
- 7.Biegalke, B. J., and A. P. Geballe. 1990. Translational inhibition by cytomegalovirus transcript leaders. Virology 177:657-667. [DOI] [PubMed] [Google Scholar]
- 8.Biegalke, B. J., and A. P. Geballe. 1991. Sequence requirements for activation of the HIV-1 LTR by human cytomegalovirus. Virology 183:381-385. [DOI] [PubMed] [Google Scholar]
- 9.Bogner, E., M. Reschke, B. Reis, T. Mockenhaupt, and K. Radsak. 1993. Identification of the gene product encoded by open reading frame UL56 of the human cytomegalovirus genome. Virology 196:290-293. [DOI] [PubMed] [Google Scholar]
- 10.Bradshaw, P. A., M. R. Duran-Guarino, S. Perkins, J. I. Rowe, J. Fernandez, K. E. Fry, G. R. Reyes, L. Young, and S. K. H. Foung. 1994. Localization of antigenic sites on human cytomegalovirus virion structural proteins encoded by UL48 and UL56. Virology 205:321-328. [DOI] [PubMed] [Google Scholar]
- 11.Bresnahan, W. A., and T. Shenk. 2000. UL82 virion protein activates expression of immediate-early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 97:14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britt, W. J., and C. A. Alford. 1996. Cytomegalovirus, p. 2493-2524. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 13.Britt, W. J., and D. Auger. 1986. Synthesis and processing of the envelope gp55-116 complex of human cytomegalovirus. J. Virol. 58:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, C. P., D. H. Vesole, J. Nelson, M. B. Oldstone, and M. F. Stinski. 1989. Identification and expression of a human cytomegalovirus early glycoprotein. J. Virol. 63:3330-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, I. I. I., T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-170. [DOI] [PubMed] [Google Scholar]
- 16.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 17.Davis, M. G., and E. S. Huang. 1985. Nucleotide sequence of a human cytomegalovirus DNA fragment encoding a 67-kilodalton phosphorylated viral protein. J. Virol. 56:7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gershon, A. A., D. L. Sherman, A. Zhu, C. A. Gabel, R. T. Ambron, and M. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68:6372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson, W. 1981. Structural and nonstructural proteins of strain Colburn cytomegalovirus. Virology 111:516-537. [DOI] [PubMed] [Google Scholar]
- 20.Gibson, W. 1983. Protein counterparts of human and simian cytomegaloviruses. Virology 128:391-406. [DOI] [PubMed] [Google Scholar]
- 21.Gibson, W., M. K. Baxter, and K. S. Clopper. 1996. Cytomegalovirus “missing” capsid protein identified as heat-aggregable product of human cytomegalovirus UL46. J. Virol. 70:7454-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson, W., K. S. Clopper, W. J. Britt, and M. K. Baxter. 1996. Human cytomegalovirus (HCMV) smallest capsid protein identified as product of short open reading frame located between HCMV UL48 and UL49. J. Virol. 70:5680-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goins, W. F., and M. F. Stinski. 1986. Expression of a human cytomegalovirus late gene is posttranslationally regulated by a 3"-end processing event occurring exclusively late after infection. Mol. Cell. Biol. 6:4202-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. USA 97:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homer, E. G., A. Rinaldi, M. J. Nicholl, and C. M. Preston. 1999. Activation of herpesvirus gene expression by the human cytomegalovirus protein pp71. J. Virol. 73:8512-8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horodyski, F. M., L. M. Riddiford, and J. W. Truman. 1989. Isolation and expression of the eclosion hormone gene from the tobacco hornworm, Manduca sexta. Proc. Natl. Acad. Sci. USA 86:8123-8127. [DOI] [PMC free article] [PubMed]
- 28.Huber, M. T., and T. Compton. 1997. Characterization of a novel third member of the human cytomegalovirus glycoprotein H glycoprotein L complex. J. Virol. 71:5391-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of noninfectious virion-like particles released from cells infected with human strains of cytomegalovirus. Virology 130:118-133. [DOI] [PubMed] [Google Scholar]
- 30.Jahn, G., T. Kouzarides, M. Mach, B.-C. Scholl, B. Plachter, B. Traupe, E. Preddie, S. C. Satchwell, B. Fleckenstein, and B. G. Barrell. 1987. Map position and nucleotide sequence of the gene for the large structural phosphoprotein of human cytomegalovirus. J. Virol. 61:1358-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kari, B., W. Li, J. Cooper, R. Goertz, and B. Radeke. 1994. The human cytomegalovirus UL100 gene encodes the gC-II glycoproteins recognized by group 2 monoclonal antibodies. J. Gen. Virol. 75:3081-3086. [DOI] [PubMed] [Google Scholar]
- 32.Kim, K. S., V. J. Sapieza, R. I. Carp, and H. M. Moon. 1976. Analysis of structural polypeptides of purified human cytomegalovirus. J. Virol. 20:604-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lahijani, R. S., E. W. Otteson, and S. C. St. Jeor. 1992. A possible role for nonsense suppression in the synthesis of a human cytomegalovirus 58-kilodalton protein. Virology 186:309-312. [DOI] [PubMed] [Google Scholar]
- 35.Lahijani, R. S., E. W. Otteson, and S. C. St. Jeor. 1991. Characterization of a human cytomegalovirus 1.6-kilobase late mRNA and identification of its putative protein product. J. Virol. 65:373-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landini, M. P., B. Severi, G. Furlini, and L. Baldiali. 1987. Human cytomegalovirus components: intracellular and intraviral localization of p28 and p65-69 by immunoelectron microscopy. Virus Res. 8:15-23. [DOI] [PubMed] [Google Scholar]
- 37.LaPierre, L., and B. J. Biegalke. 2001. Identification of a novel transcriptional repressor encoded by human cytomegalovirus. J. Virol. 75:6062-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, L., J. A. Nelson, and W. J. Britt. 1997. Glycoprotein H-related complexes of human cytomegalovirus: identification of a third protein in the gCIII complex. J. Virol. 71:3090-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, B., and M. F. Stinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and Ap-1 cis-acting elements. J. Virol. 66:4434-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, Y., and B. J. Biegalke. 2001. Characterization of a cluster of late genes of guinea pig cytomegalovirus. Virus Genes 23:247-256. [DOI] [PubMed]
- 41.Lu, M. S., and T. Shenk. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mach, M., B. Kropff, P. Dal Monte, and W. Britt. 2000. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 74:11881-11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual, p. 89-94. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 44.Margulies, B. J., H. Browne, and W. Gibson. 1996. Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology 225:111-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer, H., A. T. Bankier, M. P. Landini, B. Ruger, and M. Mach. 1988. Identification and prokaryotic expression of the gene coding for the highly immunogenic 28-kilodalton structural phosphoprotein (pp28) of human cytomegalovirus. J. Virol. 62:2243-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mocarski, E. S., Jr. 1996. Cytomegaloviruses and their replication, p. 2447-2492. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven, Philadephia, Pa.
- 47.Patterson, C. E., and T. Shenk. 1999. Human cytomegalovirus UL36 protein is dispensable for viral replication in cultured cells. J. Virol. 73:7126-7131. [DOI] [PMC free article] [PubMed]
- 48.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roby, C., and W. Gibson. 1986. Characterization of phosphoproteins and protein kinase activity of virions, noninfectious enveloped particles, and dense bodies of human cytomegalovirus. J. Virol. 59:714-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romanowski, M. J., E. Garrido-Guerrero, and T. Shenk. 1997. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J. Virol. 71:5703-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romanowski, M. J., and T. Shenk. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J. Virol. 71:1485-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruger, B., S. Klages, B. Walla, T. Albrecht, F. Fleckenstein, P. Tomlinson, and B. Barrell. 1987. Primary structure and transcription of the genes coding for the two virion phosphoproteins pp61 and pp71 of human cytomegalovirus. J. Virol. 61:446-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spaete, R., R. Gehrz, H. Land, and M. P. Landini. 1994. Human cytomegalovirus structural proteins. J. Gen. Virol. 75:3287-3308. [DOI] [PubMed] [Google Scholar]
- 55.Vink, C., E. Beuken, and C. A. Bruggeman. 2000. Complete DNA sequence of the rat cytomegalovirus genome. J. Virol. 74:7656-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winkler, M., S. A. Rice, and T. Stamminger. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68:3943-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winkler, M., and T. Stamminger. 1996. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J. Virol. 70:8984-8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, Z., M. D. Gershon, Y. Hao, R. T. Ambron, D. A. Gabel, and A. A. Gershon. 1995. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J. Virol. 69:7951-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu, Z., Y. Hao, M. D. Gershon, R. T. Ambron, and A. A. Gershon. 1996. Targeting of glycoprotein I (gE) of varicella-zoster virus to the trans-Golgi network by an AYRV sequence and an acidic amino acid-rich patch in the cytosolic domain of the molecule. J. Virol. 70:6563-6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zini, N., M. C. Battista, S. Santi, M. Riccio, G. Bergamini, M. P. Landini, and N. M. Maraldi. 1999. The novel structural protein of human cytomegalovirus, pUL25, is localized in the viral tegument. J. Virol. 73:6073-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]