Abstract
Type B leukemogenic virus (TBLV) is highly related to mouse mammary tumor virus but induces rapidly appearing T-cell lymphomas in mice. Unlike other T-cell tumors induced by retroviruses, only 5 to 10% of TBLV-induced lymphomas have detectable viral integrations near c-myc by Southern blotting, whereas Northern blotting has shown that most tumors have two- to sixfold overexpression of c-myc RNA. In this report, PCR was used to demonstrate that at least 30% of these lymphomas have TBLV insertions near c-myc. Some tumors contained multiple TBLV proviruses in different locations and orientations, suggesting that the tumors are polyclonal. The integrated proviruses near c-myc had different numbers (two to four) of long terminal repeat (LTR) enhancer repeats, although LTRs with three-repeat enhancers dominated the proviral population. Passage of polyclonal tumors in immunocompetent mice and semiquantitative PCR revealed that only cells with particular integrations were selected for growth. In three of six tumors tested, proviruses containing four-repeat enhancers near c-myc were selected during tumor passage. Since tumor cell selection may be accomplished by overexpression of c-myc RNA due to proximity to the unique TBLV LTR enhancer, we inserted LTRs at various locations within a plasmid containing the entire c-myc locus and cellular flanking sequences. To quantitatively measure effects on transcription, the Renilla luciferase gene was substituted for most of c-myc exon 2, and transient transfections were performed with c-myc reporter constructs in two different T-cell lines. As expected, insertion of a TBLV LTR with three-repeat enhancers in either orientation, 5" and 3", of the myc gene elevated reporter activity from 2- to 160-fold, consistent with enhancer function, but four-repeat LTRs had lower levels of expression compared to three-repeat LTRs. Surprisingly, LTR insertions that gave maximal c-myc expression in transient-transfection assays declined in tumor cells selected for growth in vivo. Selection for clonal growth may occur in tumor cells that have modest c-myc overexpression after proviral insertion to prevent apoptosis.
Type B leukemogenic virus (TBLV) is a retrovirus that is highly related to mouse mammary tumor virus (MMTV) (2, 3). However, TBLV induces T-cell lymphomas with a short latency in mice (5). Members of our group and others have shown that TBLV induces immature CD4+ CD8+ or CD4− CD8− T-cell lymphomas after intrathymic inoculation of newborn animals (37, 41). Ball and colleagues have shown that TBLV and MMTV primarily differ with respect to the structure of the long terminal repeat (LTR) U3 region (4). The TBLV LTR has a deletion of 440 bp relative to those of the mammotropic MMTVs; this deletion results in the loss of negative regulatory elements (NREs) that bind to the transcriptional repressor proteins, special AT-rich binding protein 1 (SATB1) and CCAAT displacement protein (CDP) (6, 33). Loss of the binding sites for these proteins alleviates the repression of MMTV expression normally observed in lymphoid tissues (33). In addition to the loss of the NREs, the TBLV LTR also has a triplication of 62 bp that flanks the deletion. Duplication or triplication of regulatory elements within retroviral LTRs is characteristic of viral enhancers (18, 26, 27, 30). All MMTV-like viruses that induce leukemias have a deletion within the U3 region, but only a fraction of them, including TBLV, have multimerization of other LTR sequences (4, 22, 29, 38, 60). Previous studies have suggested that the multimerized sequences in the MMTV LTR form a transcriptional enhancer that is active in T cells (38, 60). Our recent data also indicate that the TBLV LTR contains a cell-type-specific enhancer (36).
Transgenic mice expressing a TBLV LTR-linked c-myc transgene developed T-cell lymphomas after a short latent period with approximately the same kinetics as that observed in mice after virus injections (46). The transgene was expressed primarily in the thymus but was also expressed at lower levels in the spleen, salivary gland, small intestine, and lung. However, tumors were not observed in nonthymic tissues. Recently, members of our group, using Southern blotting, have shown that 5 to 10% of TBLV-induced tumors have integrations near the c-myc proto-oncogene, and in the cases analyzed, the proviruses were integrated downstream and in the transcriptional orientation typical of MMTV enhancer insertions in mammary tumors (10, 43, 44, 48). The majority of tumors tested showed c-myc overexpression (51), suggesting that c-myc transcription is activated indirectly by TBLV infection or that TBLV-induced tumors are polyclonal, precluding the detection by Southern blotting of TBLV insertions near c-myc.
Integrations near c-myc are commonly observed by Southern analysis of retrovirus-induced B- and T-cell lymphomas (11, 17, 21, 42). In Moloney murine leukemia virus (MuLV)- or mink cell focus-forming virus-induced T-cell lymphomas, 20 to 40% of tumors have an insertion near c-myc (11, 32, 54). The proviruses are integrated upstream in the antisense orientation relative to the orientation of c-myc or downstream in the sense orientation, consistent with MuLV enhancer activation of c- myc expression. However, retroviral promoter insertions of c-myc have been observed in reticuloendotheliosis virus- or feline leukemia virus-induced T-cell lymphomas (23, 42), and a promoter insertion mechanism is most commonly observed in B-cell lymphomas induced by avian retroviruses (17, 21).
In this study, PCR analyses showed that 30% of the TBLV-induced tumors have proviruses upstream, downstream, or within the c-myc gene. The majority of the TBLV insertions in c-myc were not detected by Southern blotting, and some tumors had multiple different integrations, indicating the polyclonal nature of these tumors. Semiquantitative PCR analysis showed that only tumor cells with particular integrations near c-myc were selected during passage in immunocompetent mice. To determine the effects of TBLV LTR insertions on upregulation of c-myc expression, we constructed a c-myc promoter-reporter gene construct that contained various insertions of the LTR in cellular flanking sequences. These experiments suggested that proviral insertions that gave the highest levels of c-myc expression were selected against during tumor progression.
MATERIALS AND METHODS
Induction of tumors.
Mice were bred and maintained in the Animal Resources Center at The University of Texas at Austin. Thymomas were induced by intrathymic inoculation of neonatal BALB/cJ or C57BL/6J mice (less than 48 h after birth) with 30 to 50 μl of culture supernatant from the TBLV-producing cell line 485-10 from CFW mice (12) (kindly provided by J. Ball, University of Western Ontario). Virus-containing supernatants were prepared by incubating 485-10 cells at 107 per ml for 12 to 18 h at 37°C. Cells were removed by low-speed centrifugation, and the supernatant was subjected to irradiation (4,500 rads) to kill any remaining cells. Irradiated supernatants were stored in aliquots at −70°C. Inoculated mice were monitored for symptoms that correlated with the onset of thymomas. Tumors were passaged by intraperitoneal injection of 3 × 106 to 30 × 106 cells into weanling BALB/c mice. The mice were monitored for tumor development and harvested within 12 to 30 days.
PCR analysis.
The Expand Long-Template PCR System (Roche Molecular Biochemicals, Mannheim, Germany) was used to detect TBLV integrations. Amplification conditions for TBLV insertion sites within and flanking the c-myc gene included incubation at 94°C, followed by 30 cycles of PCR, with one cycle consisting of 20 s at 94°C, 30 s at 57.5°C, and 4 min at 68°C. The reaction mixture was then incubated at 68°C for 8 min. PCR products were isolated after purification by agarose gel electrophoresis and cloned using pGEMT-Easy Vector System I (Promega, Madison, Wis.). All restriction and modifying enzymes were obtained from New England Biolabs (Beverly, Mass.), except SstI (Life Technologies, Gaithersburg, Md.), and calf intestinal alkaline phosphatase (CIP) (Roche). DNA from clones containing PCR product insertions was sequenced as described previously (59). Semiquantitative PCR analysis was performed as described above, except serial threefold dilutions of tumor DNA were used as indicated in figure legends. DNA concentrations were determined by absorbance readings at 260 nm and were confirmed by Hoechst staining and fluorimetry. Seminested PCR products were obtained in two steps. First, PCR was performed as described above using a c-myc-specific primer and a primer specific for the TBLV LTR. The products were purified using a Qiaquick nucleotide removal kit (Qiagen, Inc., Valencia, Calif.), diluted, and then used in a second PCR with TBLV-specific LTR primers that flank the enhancer repeat region.
Construction and cloning of plasmid vectors.
The vectors pRL-TK (Promega), pGEMT-Easy (Promega), and pBluescript II SK(−) (Stratagene, La Jolla, Calif.) were obtained commercially. The plasmid pTBLV-LUC was engineered by substitution of the ClaI-to-SstI fragment of the C3H MMTV LTR in pLC-LUC (6) with the ClaI-to-SstI fragment of the TBLV LTR; this region of the TBLV LTR includes the triplicated enhancer sequence as well as the deletion of the NREs (4). Construction of the pTBLV-WT-LUC vector has been previously described (36). Briefly, 62-bp enhancer elements corresponding to the sequence published previously (4) were amplified by PCR and ligated. Three copies of the element were cloned into a vector, pd6, at the engineered StuI site that replaces bp 523 to 1024 of the C3H LTR, mimicking the naturally occurring deletion and substitution. The vector pTBLV-4R-LUC was prepared by PCR amplification of a TBLV LTR containing four copies of the enhancer element from T16 tumor DNA. The PCR product was digested with ClaI and SstI and substituted for the corresponding region in pLC-LUC.
The vector containing the Renilla luciferase gene, pc-mycRluc, and derivatives containing the TBLV LTR that were inserted upstream or downstream of c-myc were constructed in several steps. The plasmid pc-mycRluc includes c-myc exon 1, intron 1, bp 1 to 15 of exon 2, bp 61 to 897 of exon 3, and upstream and downstream flanking regions. The Renilla luciferase gene was inserted at the c-myc start codon and replaced all but 15 bp of exon 2 and intron 2. All PCR primers used to construct pc-mycRluc have restriction endonuclease sites (underlined) that were engineered in the 5" end for cloning purposes. The Renilla luc gene [including the stop codon and simian virus 40 poly(A) signal] from pRL-TK was amplified using the sense primer (5" GTC GAC ATG ACT TCG AAA GTT TAT GAT CC 3") and the antisense primer (5" AGA TCT TAC CAC ATT TGT AGA GG 3") and cloned into the pGEMT-Easy vector. Plasmid DNA from a clone containing the PCR product was digested with SalI and BglII, and the ∼1.2-kb fragment was purified by agarose gel electrophoresis prior to ligation. Most of c-myc exon 3 (bp 61 to 897) and 4.7 kb of the downstream flanking region were amplified from BALB/c genomic DNA using the sense primer (5" AGA TCT CTG CCA AGA GGT CGG AGT CGG 3") and the antisense primer (5" GAA TTC GCT TCT ACT CAA CCC TTA CTC 3") and cloned into pGEMT-Easy vector as described above. Plasmid DNA from a clone containing the PCR product was digested with BglII and EcoRI, and the ∼5.5-kb fragment was purified by agarose gel electrophoresis. The 1.2-kb Rluc/poly(A) fragment, the 5.5-kb fragment containing c-myc exon 3 and downstream region, and linearized pBluescript II SK(−) vector (digested with SalI and EcoRI and treated with CIP) were ligated into a 9.7-kb intermediate construct, pPB. Subsequently, ∼5.1 kb of DNA was amplified from BALB/c genomic DNA using the sense primer (5" GGT ACC CGT GAC CTG ATC TCT AGC TTC TCC 3") and the antisense primer (5" GTC GAC CGT CGT GGC TGT CTG CTG GAG GG 3") and cloned into pGEMT-Easy vector as described above. This PCR fragment includes 2.9 kb upstream of c-myc, exon 1, intron 1, and 15 bp of exon 2. Plasmid DNA from a clone containing the PCR product was digested with KpnI and SalI, and the ∼5.1-kb fragment was purified by agarose gel electrophoresis. The intermediate construct, pPB, was linearized by partial digestion with KpnI and complete digestion with SalI, treated with CIP, and ligated to the 5.1-kb fragment to make the parental vector, pc-mycRluc. Plasmid constructs were verified by restriction fragment analysis and direct sequencing. The pc-mycRluc vector was modified by digestion with an enzyme that cuts only in the 3" flanking region (AatII and StuI) or by linearization with an enzyme that cuts once in both the 5" and 3" flanking regions (KpnI and AfeI). In addition, a unique NcoI site was placed ∼1.5 kb upstream of c-myc exon 1 as described previously (36) using the sense primer (5" AAT AAA TCT AGA ACC ATG GCA CAG AGC AAA AGA C 3") and the antisense primer (5" GTC TTT TGC TCT GTG CCA TGG TTC TAG ATT TAT T 3"). Linear pc-mycRluc was dephosphorylated and ligated to the 1.2-kb HindIII fragment of pTBLV-LUC (36), the 1.26-kb HindIII fragment from pTBLV4R-LUC, or the 1.2-kb HindIII fragment from pTBLV-WT-LUC. The Klenow fragment of DNA polymerase or T4 DNA polymerase was used to generate blunt ends after digestion with enzymes that left overhanging 5" or 3" ends. Clones containing the TBLV-LTR HindIII cassette in both forward and reverse orientations were isolated and verified by restriction enzyme analysis and sequencing.
Cell lines and transfections.
Culture of human Jurkat T cells and murine RL-1 cells has been described previously (36). DNA samples for transfection were prepared as described by Bramblett et al. (6). Jurkat T cells were transfected using Superfect transfection reagent (Qiagen) according to the manufacturer. Cells (2.5 × 106) were plated in six-well plates in a volume of 2.5 ml of complete medium on the day of transfection. Samples included 0.5 μg of pc-mycRluc (or a derivative) and 2 μg of pTBLV-LUC to monitor DNA uptake. DNA was mixed with 75 μl of RPMI medium with no additives. Superfect (10 μl) was mixed with the DNA and incubated for 10 min at room temperature. This mixture was added dropwise to the cells and incubated for 48 h at 37°C before harvest for reporter gene assays.
Reporter gene assays.
Methods for reporter gene assays have been described previously (36). Lysates were obtained using passive lysis buffer (Promega), subjected to two or three freeze-thaw cycles, and then clarified by centrifugation. Protein concentrations were determined, and the Dual-Luciferase reporter assay system (Promega) was used to determine both Renilla and firefly luciferase activities. If the test plasmids contained a firefly luciferase reporter gene, then the cotransfected pRL-TK plasmid was used to normalize for DNA uptake. If the test plasmids contained the Renilla luciferase reporter gene, pTBLV-LUC was used to normalize for DNA uptake.
RESULTS
Polyclonal insertions near the c-myc proto-oncogene.
We previously detected TBLV integration by Southern blotting near the c-myc proto-oncogene in a low percentage of virus-induced T-cell tumors (51). However, the majority of tumors showed two- to sixfold c-myc overexpression as judged by Northern blotting (51). These studies also suggested that the tumors induced by TBLV were polyclonal, since the pattern of TBLV proviruses appeared to change during passage of tumors in vivo (51). We reasoned that many TBLV-induced tumors had sustained proviral integrations near c-myc that were not detected by conventional Southern blotting due to the presence of multiple tumor cell clones.
Therefore, a PCR strategy that could detect TBLV integrations upstream, downstream, and within the coding region of the c-myc gene (Fig. 1) was devised. In each PCR, one primer was located within the c-myc gene in the sense or antisense orientation, and a second primer was located within the TBLV LTR in either orientation. The size of the resulting PCR product was an indication of the distance between the TBLV provirus and the proto-oncogene; the orientation of each primer allowed us to determine the transcriptional orientation of the provirus relative to that of the c-myc gene. Tumor integrations were confirmed by using PCR and at least two different primer pairs as well as by cloning and sequencing.
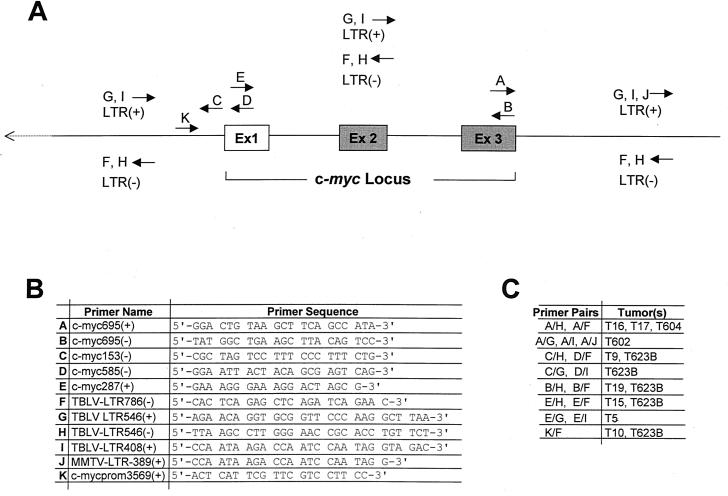
FIG. 1.
Strategy for detection of integrated TBLV proviruses. (A) The forward and reverse arrows indicate the locations of the sense (+) and antisense (−) primers, respectively, used for PCRs. The positions of exons (Ex) are indicated. (B) Names and sequences of the primers shown in panel A used to detect integrated TBLV proviruses. (C) Proviral integrations identified by sets of primer pairs.
Using all of the available primer combinations for each tumor, we were able to construct a map of the myc locus with the locations and orientations of TBLV proviruses within each tumor population (Fig. 2). In the TBLV-induced lymphomas that we tested, 10 of 35 (30%) had detectable integrations in or near the c-myc gene. As previously reported, two of these tumors (T16 and T17) had integrations downstream and in the same transcriptional orientation as that of the oncogene, as determined by Southern blotting (51). However, the PCR analysis revealed that two other tumors (T604 and T602) contained TBLV insertions downstream of c-myc, whereas two additional tumors had TBLV integrations upstream of the oncogene (T9 and T623B). Of the four tumors that had downstream integrations, three tumors had proviruses in the same orientation as that of c-myc (T16, T17, and T604), and one (T602) had a TBLV provirus in the opposite orientation. Of the two tumors with insertions upstream of c-myc, one (T9) had an insertion of TBLV in the opposite orientation, and one tumor (T623B) had multiple proviral integrations in both orientations.
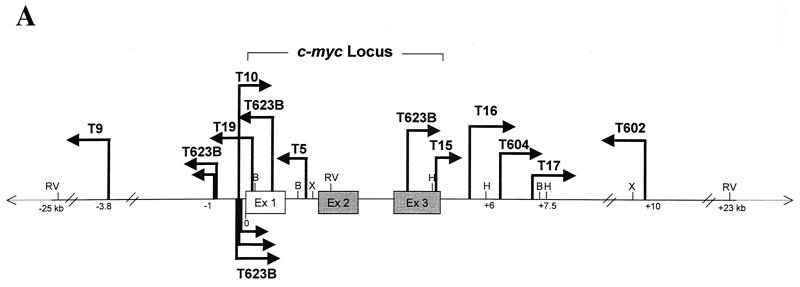
FIG. 2.
Locations of TBLV proviruses within the c-myc locus. (A) Diagram of TBLV insertions into the c-myc region. The arrows indicate the approximate positions of proviral insertions and their transcriptional orientations with respect to that of c-myc. Only proviral integrations that could be confirmed by direct PCR product sequencing or by sequencing of the cloned PCR products have been included on this map. Only proviruses observed in primary TBLV-induced tumors are shown. The positions of some restriction enzyme sites are shown: EcoRV (RV), BamHI (B), XbaI (X), and HindIII (H). Numbers below the line indicate distance from the first base of c-myc exon 1 (Ex 1) in kilobases. The hatch marks indicate that the linear map is not to scale. The proviral insertions in tumors T16 and T17 could be detected by Southern blotting, whereas the other insertions could not. The nontranslated exon 1 of c-myc is depicted as a white box, while the two coding exons (exons 2 and 3) are represented by the solid gray boxes. (B) Representative PCR results used to construct the diagram in panel A. Agarose gel analysis of PCR products obtained with representative tumor DNAs and the primers c-myc695+ and TBLV LTR546−. Lane M contains molecular size standards.
Of the integrations detected within the c-myc coding region, tumors T623B and T19 had insertions within exon 1 in the opposite transcriptional orientation, whereas tumors T623B and T15 had proviruses within exon 3 in the same orientation. Tumor T5 had a proviral integration within intron 1 in the opposite orientation. Only the detected integrations in exon 3 would interrupt the c-myc coding region. Nevertheless, Northern blot analysis of total RNA from these tumors did not reveal hybrid transcripts between c-myc and the TBLV provirus (51) (data not shown). This suggests that such disruptive integrations are not abundant in the tumor cell populations.
As noted above, the T623B tumor had multiple integrations near c-myc. PCR analysis of primary tumor DNA showed a large number of bands that were not resolved by agarose gel electrophoresis (data not shown). Cloning and sequencing of eight PCR products from the T623B tumor showed that most of the TBLV integrations were clustered within 1 kb upstream of the first exon of c-myc. There were two tight clusters (within 45 to 70 bp of each other) of integrations upstream of c-myc and in both orientations separated by about 400 bp. T10 also had an insertion in the same region.
Together, these results indicate that TBLV frequently targets the c-myc proto-oncogene in primary T-cell lymphomas and that the majority of tumors are polyclonal. The high frequencies of these insertions and the detection of multiple proviruses near c-myc in the same tumor suggest that proviral insertion near the c-myc gene is a powerful selective factor for tumor cell growth.
Characterization of TBLV enhancer repeats in proviruses integrated near the c-myc gene.
Because previous experiments indicated that the nature of the retroviral enhancer was critical for proviral oncogenicity in T cells (8, 31, 55), we characterized the LTRs from integrated TBLV proviruses near the oncogene. PCR was performed with tumor DNAs and primers that would flank the copy of the TBLV enhancer closest to the c-myc proto-oncogene, and the resulting products were subjected to sequencing analysis. To our surprise, some of the TBLV proviruses had a different number of enhancer repeats than that previously reported for an LTR cloned from a TBLV-induced tumor (4). In two proviruses that were integrated approximately 0.5 and 2 kb downstream of the c-myc gene in the T16 and T17 tumors, respectively (Fig. 2), there were four repeats within the TBLV enhancer (Table 1). Interestingly, the T16 and T17 tumors were the most clonal of the characterized tumors based on our ability to detect TBLV integrations by Southern blotting (51). All of the other proviruses from polyclonal tumors contained two or three enhancer repeats within the LTR closest to the c-myc oncogene (Table 1).
TABLE 1.
Analysis of the enhancer repeats in TBLV LTRs near the c-myc gene
| TBLV-induced tumor | Proviral locationa | Proviral orientationb | No. of LTR enhancer repeatsc
|
|
|---|---|---|---|---|
| 5" LTR | 3" LTR | |||
| T9 | Upstream | Opposite | 3 | |
| T623B | Upstream | Opposite | 3, 3d | |
| T623B | Upstream | Same | 2, 2, 3d | |
| T623B | Exon 1 | Opposite | 3 | |
| T5 | Intron 1 | Opposite | 3 | |
| T623B | Exon 3 | Same | 2 | |
| T16 | Downstream | Same | 4 | |
| T604 | Downstream | Same | 3 | |
| T17 | Downstream | Same | 4 | |
| T602 | Downstream | Opposite | 3 | |
Location of the TBLV provirus relative to the c-myc proto-oncogene.
Transcriptional orientation of the TBLV provirus relative to that of the c-myc gene.
Number of copies of the 62-bp repeat element in the TBLV LTR.
The numbers of TBLV LTR enhancer repeats found in different clustered proviruses in this orientation as verified by cloning and sequencing.
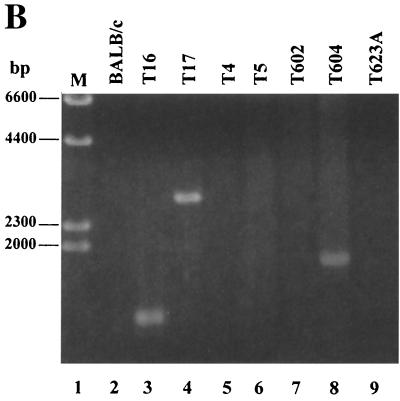
To determine if our starting TBLV population was a mixture of viruses, we performed PCR on high-molecular-weight DNA obtained from the cell line used for virus preparation (485-10). Using LTR primers localized outside the enhancer repeats, we obtained a predominant PCR product consistent with three-repeat enhancers as reported by Ball et al. (4). This PCR product was cloned, and 10 independent clones were sequenced (data not shown). All clones had three-repeat enhancers, and the sequence was identical to that previously reported (4) except that the first two repeats had G residues instead of A residues in a portion of the first two repeats (5" TAA GTA GGT TTA TGG 3"). Both the G and A residues resulted in a termination codon within the sag gene.
We also performed PCR on DNA derived from the 485-10 tumor used for virus preparation using TBLV-specific LTR primers that flanked the enhancer repeats (Fig. 3A). As expected, gel analysis of these PCR mixtures showed that the three-repeat enhancer was dominant, although fragment sizes consistent with one- or two-repeat enhancers could be detected. Similar analysis of DNAs from the primary tumors T5, T9, T16, and T17 also revealed that the three-repeat enhancer is dominant in proviruses integrated into tumor DNA. However, the four-repeat enhancer was detected in tumors T5, T16, and T17 (Fig. 3A); quantitation indicated that the three-repeat enhancer was three- to sevenfold more abundant than the four-repeat enhancer in these tumors (data not shown). Since the semiclonal tumors T16 and T17 both had TBLV proviruses with four-repeat enhancers downstream of c-myc, as detected by Southern blotting (51) (Fig. 2), these results suggested that four-repeat enhancers at particular insertion sites near c-myc have a selective advantage over three-repeat enhancers during tumor progression.
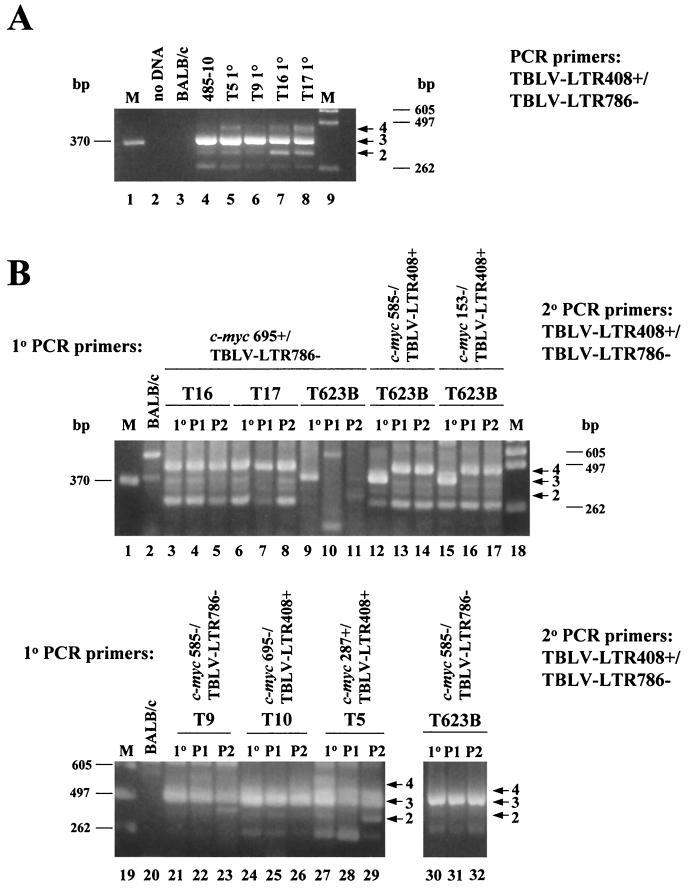
FIG. 3.
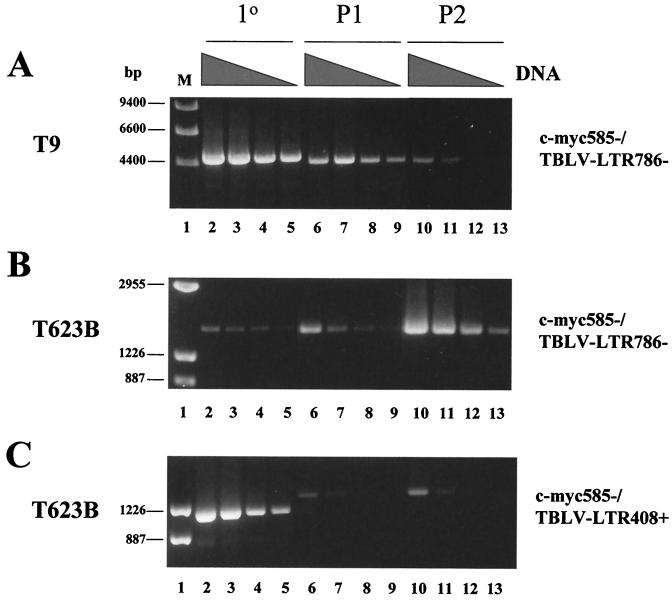
PCR analysis of tumor DNAs to determine the number of 62-bp repeats in the TBLV LTR enhancer. (A) PCR analysis of DNA from primary (1°) TBLV-induced tumors (T5, T9, T16, and T17) using sense and antisense primers that flank the LTR enhancer repeats (LTR408+ and LTR786−). Lanes M contain molecular size markers, and lane 2 is blank. The tumor cell line 485-10 used for virus production is shown in lane 4. Use of DNA from uninfected BALB/c mice shows the specificity of the PCRs. The numbered arrows to the right of the gel indicate the positions of fragments with different numbers (two to four) of enhancer repeats. (B) Seminested PCR analysis of tumor DNA. Primary PCRs were performed using DNAs from unpassaged (1°) or passaged (P1 or P2) tumors and the indicated c-myc and TBLV LTR primers. The c-myc-LTR junction fragments were then purified, diluted, and used for secondary (2°) PCR with primers that flank the LTR enhancer repeats (LTR408+ and LTR786−). The numbered arrows to the right of the gels indicate the positions of fragments with different numbers (two to four) of enhancer repeats. The strong band at ca. 260 bp in some lanes represents one copy of the 62-bp enhancer region, and this has been confirmed by sequencing. All PCR mixtures were analyzed by agarose gel electrophoresis.
Selection for TBLV enhancer repeats during tumor passage.
To determine if there was selection for particular integration sites or enhancer composition during tumor progression, we passaged the semiclonal tumors T16 and T17 and the polyclonal tumors T5, T9, T10, and T623B in immunocompetent syngeneic mice. DNA extracted from the primary or passaged tumors was then subjected to nested PCR. Primary PCR products obtained using primers that would detect a TBLV LTR-c-myc junction fragment were purified, diluted, and used for secondary PCR with primers that flanked the LTR enhancer repeats. Gel analysis of the secondary PCRs allowed us to determine if there was a change in the number of enhancer repeats at particular TBLV integration sites found near c-myc after tumor passage (Fig. 3B). Cloning and sequencing verified the identities of fragments composed of two, three, or four copies of the 62-bp repeats. As anticipated, PCR results generated from the semiclonal tumors T16 and T17 showed that the four-repeat enhancer was dominant using primers that detected insertions downstream of c-myc (lanes 3 and 6). The polyclonal tumor T623B had no detectable product in primary PCRs (data not shown), but LTRs with three-repeat enhancers were detectable in the primary tumor (lane 9). However, such insertions did not appear to be selected during tumor passage (lanes 10 and 11). TBLV insertions containing the three-repeat enhancer were detected predominantly in primary T623B DNA upstream and in the same orientation as c-myc (lane 12). However, both passages of this primary tumor selected for the appearance of LTRs with the four-repeat enhancer (lanes 13 and 14). Similar results were obtained using a different c-myc primer for the primary PCRs (lanes 15 to 17).
Selection for TBLV insertions with four-repeat LTRs was not apparent at every integration site analyzed. For example, insertions into the T9 site about 4 kb upstream and in the transcriptional orientation opposite that of c-myc had predominantly three-repeat enhancers (Fig. 3B, lanes 21 to 23). Insertions into intron 1 in the T5 tumor predominantly had three-repeat enhancers, although two-repeat enhancers were detectable (lanes 27 to 29). Insertions within exon 3 in T623B also had predominantly three-repeat enhancers (lanes 30 to 32).
To determine the relative abundance of tumor cells carrying different TBLV insertions near c-myc, we performed semiquantitative PCRs with DNAs derived from primary and passaged tumors. In the T9 tumor where we detected an insertion ca. 4 kb upstream and in the transcriptional orientation opposite that of c-myc (see Fig. 2 for map position), we observed selection against tumor cells carrying TBLV insertions with a three-repeat enhancer during tumor passage (Fig. 4A). Alternatively, tumor cells in T623B carrying a TBLV insertion with a three-repeat enhancer upstream and in the transcriptional orientation opposite that of c-myc became more abundant during in vivo passage (Fig. 4B). However, in the same tumor, cells carrying a retroviral insertion with a three-repeat enhancer upstream and in the same transcriptional orientation as the oncogene disappeared from the population (Fig. 4C, compare lanes 2 to 5 with lanes 6 to 9), yet cells having a similar insertion with a four-repeat enhancer appeared after one tumor passage (lanes 6 to 9) and persisted after a second passage (lanes 10 to 13). The new insertion was confirmed to be ca. 300 bp upstream of c-myc in the same orientation by cloning and sequencing. Sequencing also indicated that this insertion consisted of the 5" LTR and gag leader sequences (data not shown).
FIG. 4.
Semiquantitative PCR analysis to determine the relative abundance of TBLV integrations near c-myc after tumor passage. (A) DNA (200 ng) from the primary (1°) or passaged (P1 or P2) T9 tumor was subjected to threefold serial dilutions (the amount of DNA indicated by the height of the black triangles over the lanes) and used for PCRs with an antisense primer for c-myc exon 1 (c-myc585−) and an antisense TBLV LTR primer (LTR786−). (B) DNA (70 ng) from the primary or passaged T623B tumor was subjected to threefold serial dilutions and used for PCRs with an antisense primer in c-myc exon 1 (c-myc585−) and an antisense primer in the TBLV LTR (LTR786−). (C) DNA (600 ng) from the primary or passaged T623B was subjected to threefold serial dilutions and used for PCRs with an antisense primer in c-myc exon 1 (c-myc585−) and a sense primer in the TBLV LTR (LTR408+). All PCR mixtures were analyzed by agarose gel electrophoresis.
Effects of the enhancer repeats on TBLV promoter activity.
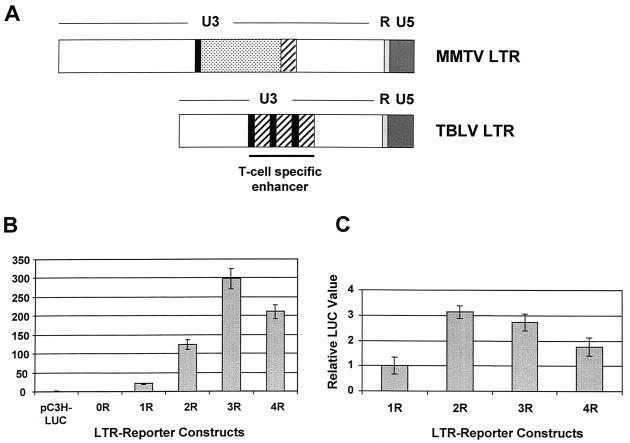
Comparison of the published sequences of the TBLV and MMTV LTRs shows that the TBLV LTR has a deletion of NREs as well as a triplication of the sequences flanking the deletion (Fig. 5A). We assumed that the selection for a four-repeat enhancer in proviral LTRs would serve to further upregulate c-myc expression compared to LTRs with three-repeat enhancers. To test directly the effects of the repeats in CD4+ Jurkat T cells, we performed transient-transfection assays with TBLV LTRs that contained no enhancer or various numbers of repeats (Fig. 5B). These experiments showed that three-repeat enhancers reproducibly gave the highest level of reporter gene expression from the TBLV promoter compared to LTRs with other repeat numbers up to four. Since TBLV induces tumors in immature thymocytes, we also tested these constructs in the RL1 CD4+ CD8+ cell line (Fig. 5C). Results showed that two- and three-repeat enhancers gave greater expression than those with one or four repeats. Therefore, these experiments indicated that the TBLV enhancers with four repeats were less effective in promoting TBLV transcription in mature or immature transformed thymocytes than enhancers with two or three copies of the 62-bp sequence.
FIG. 5.
Enhancer activity of TBLV LTRs with different numbers of enhancer repeats. (A) Comparison of the structures of the MMTV and TBLV LTRs. The NRE-containing region of MMTV that is deleted in TBLV is indicated (dotted box). The sequences flanking this region (solid black boxes or hatched boxes) are multimerized in the TBLV LTR to form the enhancer. (B) Transient-transfection assays with luciferase reporter plasmids containing TBLV LTRs with different numbers of enhancer repeats (from zero [0R] to four [4R]) in Jurkat T cells. (C) Transient-transfection assays with luciferase reporter plasmids containing TBLV LTRs with different numbers of enhancer repeats in RL1 T cells. Relative luciferase activity was determined after normalization for DNA uptake by cotransfection with the pRL-TK plasmid. Luciferase activity was determined in Jurkat cells relative to the activity of the C3H LTR-luciferase reporter plasmid that contains the NREs (pC3H-LUC). For transfections of RL1 cells, luciferase activity was determined relative to the activity of the pTBLV-1R-LUC plasmid, since pC3H-LUC activity was undetectable in these cells. The C3H LTR-luciferase reporter was engineered to delete the NREs including the sequences that constitute the 62-bp repeat (0R) or retain one, two, three, or four copies of the repeat (1R, 2R, 3R, and 4R, respectively). There was no difference between the values obtained for the engineered pTBLV-3R-luciferase plasmid and a reporter plasmid containing the naturally occurring alteration from the TBLV LTR (pTBLV-LUC) (data not shown). The means of triplicate assays (± standard deviations) are shown.
Effects of TBLV enhancers on c-myc expression.
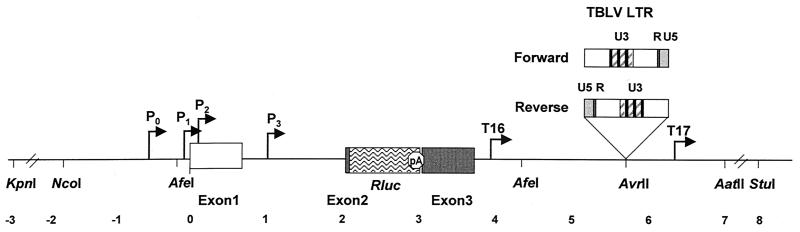
To determine the effects of different TBLV enhancers on the ability to affect c-myc transcription, we developed a novel plasmid vector containing the entire murine c-myc coding region, including 3.5 and 5 kb of upstream and downstream flanking sequences, respectively (Fig. 6). In addition, we substituted the majority of c-myc exon 2 for the Renilla luciferase gene at the transcription start site to directly measure effects on oncogene transcription and to avoid potential secondary effects of c-myc overexpression on apoptosis (47, 49). This vector then was used for insertion of TBLV LTRs containing three-repeat enhancers in both orientations relative to the c-myc promoters at various distances in cellular flanking DNA. Previous data suggest that expression of c-myc promoters is relatively normal if there is an appropriate enhancer within the construct (28).
FIG. 6.
Strategy for detection of TBLV enhancer effects on c-myc transcription. The locations of different c-myc promoters (P0 to P3) in the pc-mycRluc vector are indicated. The stop codon following the Renilla luciferase gene should prevent expression of sequences from the third exon of c-myc. TBLV LTRs were inserted in both orientations at the restriction sites shown in cellular DNA flanking the c-myc locus. The TBLV LTR insertions at the AvrII site represent examples of the constructs that were made. The numbers below the map indicate distance (in kilobases) relative to the beginning of exon 1.
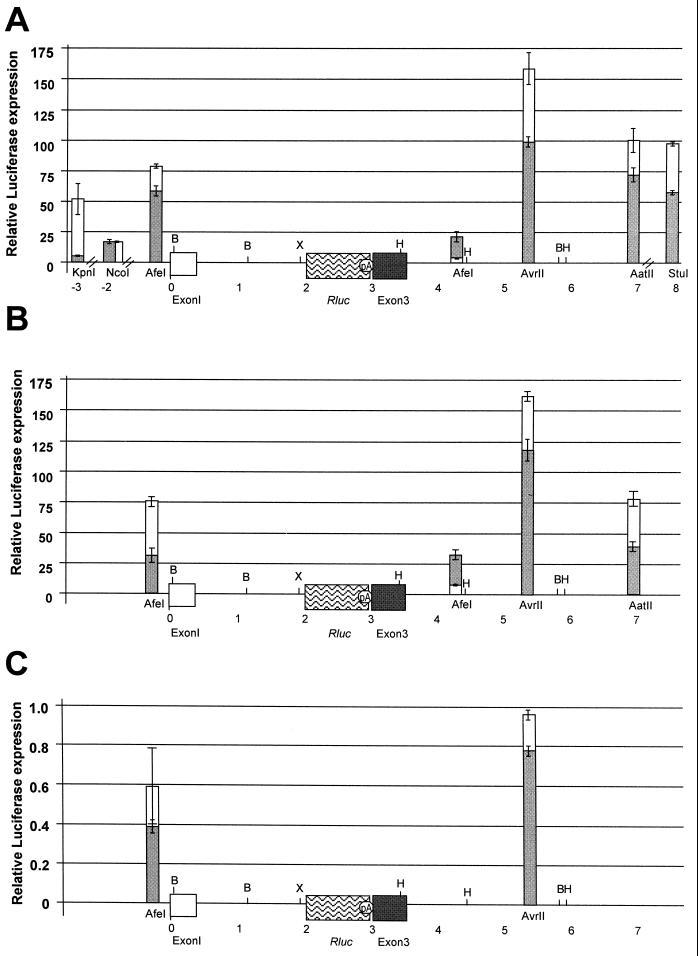
The LTR-containing constructs were transfected into triplicate cultures of Jurkat T cells, and the luciferase activity was determined relative to that of the parent construct lacking the TBLV LTR (Fig. 7A). Values were normalized for DNA uptake by cotransfection of a plasmid expressing firefly luciferase and displayed on a map according to their position within the c-myc locus. These experiments revealed that LTRs inserted at virtually any location were capable of stimulating transcription of the c-myc promoters between 2- and 160-fold, consistent with the enhancer function of the TBLV LTR. As expected, in many instances, enhancer activity was relatively independent of the orientation. Insertion of a TBLV LTR missing the enhancer repeats in the forward or reverse orientation in either the AatII or AvrII site downstream of c-myc gave no elevation of reporter gene activity relative to that of the pc-mycRluc vector alone (data not shown), demonstrating the specific effect of the 62-bp repeats.
FIG. 7.
Transcriptional activity of the c-myc reporter plasmids containing TBLV LTRs with three-repeat enhancers. (A) Activity of the c-myc reporter plasmid after transient transfections in Jurkat T cells. Relative luciferase activity was determined after normalization for DNA uptake. Luciferase activity was determined relative to the activity of the c-myc reporter plasmid in the absence of LTR insertion. The means of triplicate assays (± standard deviations) are shown. The positions of LTR insertions are shown relative to the first exon of c-myc. The hatch marks on the x axis indicate that the graph is not drawn to scale. Results of assays with plasmids containing LTRs either in the same transcriptional orientation as the c-myc gene (gray bars) or in the transcriptional orientation opposite that of the c-myc gene (white bars) are shown. Restriction enzyme site abbreviations: B, BamHI; X, XbaI; H, HindIII. (B) Activity of the c-myc reporter plasmid after transient transfections in RL1 cells. Results are expressed as in panel A. (C) Activity of the c-myc reporter plasmid after transient transfections in XC fibroblast cells. Results are expressed as in panel A. Note the differences in the scales of relative luciferase expression for each panel.
Transfections of LTR-containing pc-mycRluc plasmids were also performed in RL1 immature T cells (Fig. 7B). Although we tested fewer constructs in this line, the trend for LTR enhancer activity on the c-myc promoter was the same as that observed using Jurkat T cells. As expected from previous results (36), transfections of these plasmids into XC rat fibroblasts showed that the insertion of the TBLV LTR had no effect on c-myc expression in non-T-cell lines (Fig. 7C). In fact, when these plasmids were transfected into XC cells, expression from the c-myc promoters was slightly repressed when the TBLV LTR was present.
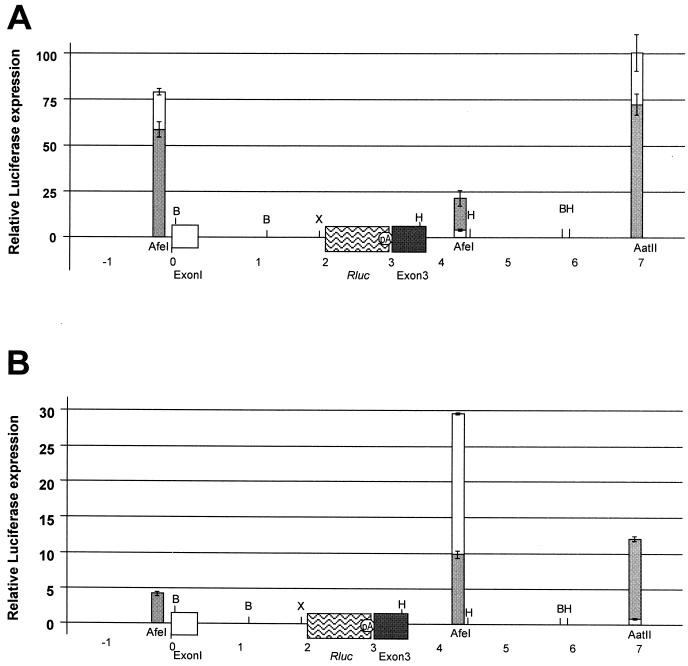
Because the most clonal tumors had TBLV proviruses with four-repeat enhancers in the LTR, we also transfected plasmids that had four-repeat LTRs at several different locations relative to c-myc (Fig. 8). Surprisingly, the presence of four repeats lowered overall expression 2- to 100-fold from c-myc promoters relative to that obtained with LTRs with three repeats. An exception was insertion of the four-repeat LTR in the AfeI site downstream and in the reverse orientation, a proviral insertion site that was not observed in tumor cells (Fig. 2). Nevertheless, in most instances, the presence of four 62-bp repeats in the enhancer appeared to lower transcriptional activity from both the TBLV and c-myc promoters (Fig. 2 and 5).
FIG. 8.
Comparison of the enhancer activity of LTRs containing three- or four-repeat enhancers on the c-myc promoters. Insertions in the same transcriptional orientation as the c-myc gene are shown as gray bars, whereas insertions in the opposite orientation are shown as white bars. Restriction enzyme site abbreviations: B, BamHI; X, XbaI; H, HindIII. (A) Transient-transfection assays in Jurkat cells using c-mycRluc plasmids containing three-repeat enhancers. These results are duplicated from Fig. 7 using a different scale for ease of comparison to results in panel B. (B) Transient-transfection assays in Jurkat cells using c-mycRluc plasmids containing the four-repeat enhancer. Results are reported as described in the legend to Fig. 7. For unknown reasons, the LTR with the four-repeat enhancer could not be cloned in the reverse orientation in the AfeI site upstream of c-myc exon 1; therefore, only the forward orientation is shown.
DISCUSSION
Polyclonal nature of TBLV-induced tumors.
Selection for TBLV proviruses near the c-myc oncogene occurs more frequently in T-cell tumors than originally detected by Southern blotting experiments (51). PCR-based assay results described here indicate that at least 30% of TBLV-induced tumors contain a provirus within 5 kb of c-myc. Since the majority of the proviral integrations could be detected only by PCR, these results suggest that TBLV-induced tumors are polyclonal, in agreement with Southern blotting results that showed both loss and gain of proviruses after tumor passage in vivo (51). Furthermore, the T623B tumor had multiple proviruses that were detected by PCR in or near c-myc (Fig. 2); since these proviruses were not detected by Southern analysis, these results are consistent with a tumor comprised of a heterogeneous population of cells, each with an independent TBLV insertion near the oncogene.
Leukemias induced by other retroviruses containing proviral insertions near c-myc appear to be clonal or oligoclonal (56, 58). Why are tumors induced by TBLV polyclonal? One of the differences between TBLV and other retroviruses is the presence of the superantigen or Sag reading frame. Because of the deletion within the U3 region of TBLV LTR, the 3" half of the sag gene is lost due to the deletion of the NREs (4). The C terminus of the Sag protein is known to interact with the β chain of the T-cell receptor (TCR), leading to proliferation of Sag-reactive T cells (1, 9, 24). Therefore, the Sag protein encoded by TBLV would lack the amino acid sequence necessary for TCR interactions.
Could the truncated TBLV Sag protein perform some other function important for T-cell proliferation? Evidence from leukemias induced by other thymotropic MMTVs suggests that both the truncated sag transcript and the gene product can be detected in infected cells (14, 39, 50). Previous reports have postulated that the full-length Sag product, in addition to its properties as a superantigen, is either a transcriptional activator or suppressor (52, 57). Thus, it is possible that the expression of the truncated Sag encoded by TBLV acts by a transcriptional mechanism to give a polyclonal proliferation of T cells. Such a proliferating T-cell population may provide ample opportunities for additional TBLV infection events and proviral integration near the c-myc proto-oncogene.
Enhancer function of the TBLV LTR.
Proviruses integrated near c-myc generally are inserted upstream in the opposite orientation or downstream in the same transcriptional orientation as MMTV proviruses found near Wnt-1 or fgf-3 in virus-induced mammary carcinomas (10, 43, 44, 48). Previous experiments have shown that the levels of c-myc RNA are elevated two- to sixfold in TBLV-induced tumors compared to healthy mouse thymus or radiation-induced tumors (51). This level of c-myc elevation may be an underestimate in cells with TBLV insertions near the oncogene because of tumor heterogeneity. Nevertheless, these results are compatible with the enhancer insertion model for TBLV-induced tumors. TBLV proviruses were also detected in the c-myc coding region using primary tumor DNA and PCR primers within the c-myc coding regions (Fig. 2). Thus far, we have not detected hybrid transcripts between the TBLV LTR and c-myc coding sequences in RNA extracted from these T-cell tumors (51), suggesting that most cells in the population overexpress RNA from the normal c-myc promoters (13, 34).
Prior results in transgenic mice indicate that the TBLV LTR can direct reporter gene expression preferentially in CD4+ CD8+ T cells in vivo (46). The TBLV U3 region has a deletion of sequences shown to suppress MMTV transcription in lymphoid tissues (6, 22, 33) as well as a triplication of sequences flanking the deletion (4). Our current data indicate that the TBLV triplicated region acts as a cell-type-specific enhancer from both the MMTV promoter and the thymidine kinase promoter (36). In addition, the cell-type-specific enhancer combined with the deletion of NREs, is sufficient to change the pathogenicity of MMTV from mammary tumors to T-cell lymphomas (F. Mustafa and J. Dudley, unpublished observations). In agreement with these results, we show here that the TBLV LTR has enhancer activity for the c-myc promoters in T cells. As expected, enhancement of c-myc expression by the TBLV LTR was not observed when pc-mycRluc constructs were analyzed in XC rat fibroblast cells (Fig. 5C) and enhancement was eliminated after removal of the 62-bp LTR repeats (not shown).
Evidence for modulation of c-myc overexpression.
Several types of evidence argue that there is selection for particular TBLV insertion sites near c-myc. First, although viruses containing four-repeat LTR enhancers appeared to be less than 10% of the injected population, both of the most clonal tumors (as judged by Southern blotting) had proviral insertions with four copies of the 62-bp enhancer repeat immediately downstream of c-myc. Second, seminested PCR analysis showed that the polyclonal tumor 623B population lost proviral insertions that had three-repeat enhancers just upstream and in the same orientation as the c-myc promoters after growth in immunocompetent mice, consistent with selection against such insertions during tumor passage. In contrast, the same tumor showed an apparent increase in the number of cells carrying proviruses with four-repeat enhancers near c-myc using the same primers and PCR conditions. This insertion was confirmed by sequencing. Thus, there appeared to be selection during passage in mice (three of six tumors tested) for proviruses carrying four-repeat enhancers near c-myc. Third, semiquantitative PCR showed that there was selection for TBLV proviruses containing a three-repeat enhancer at a position approximately 1 kb upstream and in the opposite orientation in tumor 623B. Interestingly, this is similar to the orientation and position of MuLV proviruses integrated near c-myc in many murine T-cell lymphomas (11, 25, 32, 40, 45). However, another TBLV integration with three-repeat enhancers in the LTR located approximately 4 kb upstream and in the orientation opposite that of c-myc appeared to decrease in the tumor population after passage (Fig. 4A). Such results indicate that only particular insertions near c-myc are selected during tumor progression.
What is the nature of the selection? Previous experiments have suggested that MuLV LTR enhancers have been selected for optimal activity during T-cell leukemogenesis (8, 20, 55). Mutations in c-Myb, Ets, or AML-1 (RUNX1) binding sites that compromise enhancer activity of proviral LTRs were shown to revert following characterization of proviruses found in T-cell tumors (35, 61). Recent experiments indicate that proviruses with two-repeat enhancers predominate early during infections, yet lymphomas predominantly had proviruses with three- or four-repeat enhancers (40). Our data suggest that TBLV proviruses that have been clonally selected during lymphoma growth frequently have insertions that activate c-myc transcription through the enhancer activity of the LTR, but this activity may be modulated during tumor cell selection. Possible mechanisms for modulation of c-myc expression include (i) selection for tumor cells carrying TBLV insertions with other than the optimal number of enhancer repeats, (ii) selective growth of cells with a TBLV proviral orientation relative to c-myc that minimizes oncogene overexpression, and (iii) selection for increasing distance between c-myc and the TBLV provirus.
The following data support the assumption that TBLV proviruses near c-myc may be selected for suboptimal oncogene expression. First, proviruses located near c-myc in the most clonal tumors, T16 and T17, had four-repeat enhancers in the LTR, rather than the three repeats observed in proviruses derived from the tumor population used to prepare the inoculated virus. Our experiments indicated that four-repeat enhancers reproducibly gave lower transcriptional activity from the TBLV promoter than the three-repeat enhancers in CD4+ Jurkat and CD4+ CD8+ RL1 T-cell lines (Fig. 5). Using constructs where TBLV enhancer activity was assessed on the c-myc promoters (Fig. 7 and 8), four-repeat LTRs gave up to 100-fold-lower expression than did three-repeat LTRs (insertions into the AatII site in the reverse orientation). An exception to this paradigm was insertion of the LTR with a four-repeat enhancer into the AfeI site about 1 kb downstream and in the reverse orientation from c-myc. In this case, four-repeat LTRs gave approximately 30-fold elevation of oncogene expression, whereas three-repeat LTRs had approximately twofold elevation. However, proviruses with four-repeat enhancers in the LTR (from the T16 and T17 tumors) were detected only in the forward orientation downstream of c-myc. If we compare insertions at the AfeI site downstream and in the same orientation as that of c-myc (the site closest to that of the T16 tumor integration), LTRs with four-repeat enhancers gave 10-fold elevation of oncogene expression compared to 20-fold for LTRs with three repeats. Second, the placement of LTRs relative to c-myc did not appear to be optimal for oncogene overexpression. For example, the highest expression with three-repeat LTRs was observed with insertions into the AvrII site 1.75 kb downstream and in the orientation opposite that of c-myc. All of the proviruses located in this region (T16, T604, and T17) were oriented in the forward orientation (Fig. 2). Only the T602 tumor had a provirus that was located downstream and in the reverse orientation and had three-enhancer repeats in the LTR. This provirus was located approximately 2.5 kb further downstream than insertions tested at the AvrII site, and the trend for constructs tested was a decrease in c-myc expression in sites 3" to AvrII. Third, semiquantitative PCR indicated that cells carrying proviruses with three-repeat enhancers and in optimal insertion sites for c-myc overexpression (for example, some of the T623B insertions upstream and in the same orientation as the oncogene and the T9 insertion [Fig. 2]) were diluted within the population after tumor passage. In the case of T623B, cells carrying a solo LTR with a four-repeat enhancer quite similar to the TBLV LTR that we tested at the AfeI site upstream of c-myc (Fig. 8) were selected after tumor passage. Such results are consistent with selection for suboptimal c-myc expression, since four-repeat enhancers at this position decreased expression ca. 15-fold relative to expression with three-repeat enhancers.
Why would there be selection against the highest levels of c-myc expression? One explanation is that there is a threshold level of c-myc RNA that is required to drive tumor cell growth, but above this threshold, oncogene expression becomes cytotoxic or leads to apoptosis. Recent experiments using a tetracycline-regulated c-myc oncogene indicate that continued expression is required for maintenance of leukemia cell growth (16). However, several pieces of evidence also suggest that certain levels of c-myc lead to tumor cell apoptosis. For example, in mice that express a hybrid protein that brings c-Myc under the control of the ligand-binding domain of a modified estrogen receptor, viability of the tumor cells was decreased in the presence of an estrogen derivative (where c-myc expression was increased) (7). Moreover, other studies have shown that upregulation of antiapoptotic genes, such as Gfi-1/pal-1 and Bcl-2, can complement upregulation of c-myc during leukemogenesis (15, 19, 53). Thus, these data lead to the intriguing hypothesis that transient increases in c-myc levels or decreases in antiapoptotic gene expression could be used therapeutically to induce tumor cell destruction.
Acknowledgments
This work was supported in part by grants R01 CA34780 and P01 CA77760 from the National Institutes of Health.
We thank members of the Dudley lab for useful comments on the manuscript.
REFERENCES
- 1.Acha-Orbea, H. 1992. Retroviral superantigens. Chem. Immunol. 55:65-86. [PubMed] [Google Scholar]
- 2.Ball, J. K., L. O. Arthur, and G. A. Dekaban. 1985. The involvement of a type-B retrovirus in the induction of thymic lymphomas. Virology 140:159-172. [DOI] [PubMed] [Google Scholar]
- 3.Ball, J. K., G. A. Dekaban, J. A. McCarter, and S. M. Loosmore. 1983. Molecular biological characterization of a highly leukaemogenic virus isolated from the mouse. III. Identity with mouse mammary tumour virus. J. Gen. Virol. 64:2177-2190. [DOI] [PubMed] [Google Scholar]
- 4.Ball, J. K., H. Diggelmann, G. A. Dekaban, G. F. Grossi, R. Semmler, P. A. Waight, and R. F. Fletcher. 1988. Alterations in the U3 region of the long terminal repeat of an infectious thymotropic type B retrovirus. J. Virol. 62:2985-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball, J. K., and J. A. McCarter. 1979. Biological characterization of a leukemogenic virus isolated from the CFW mouse. Cancer Res. 39:3080-3088. [PubMed] [Google Scholar]
- 6.Bramblett, D., C. L. Hsu, M. Lozano, K. Earnest, C. Fabritius, and J. Dudley. 1995. A redundant nuclear protein binding site contributes to negative regulation of the mouse mammary tumor virus long terminal repeat. J. Virol. 69:7868-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron, E. R., J. Morton, C. J. Johnston, J. Irvine, M. Bell, D. E. Onions, J. C. Neil, M. Campbell, and K. Blyth. 2000. Fas-independent apoptosis in T-cell tumours induced by the CD2-myc transgene. Cell Death Differ. 7:80-88. [DOI] [PubMed] [Google Scholar]
- 8.Chatis, P. A., C. A. Holland, J. W. Hartley, W. P. Rowe, and N. Hopkins. 1983. Role for the 3" end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc. Natl. Acad. Sci. USA 80:4408-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, Y., J. W. Kappler, and P. Marrack. 1991. A superantigen encoded in the open reading frame of the 3" long terminal repeat of mouse mammary tumour virus. Nature 350:203-207. [DOI] [PubMed] [Google Scholar]
- 10.Clausse, N., D. Baines, R. Moore, S. Brookes, C. Dickson, and G. Peters. 1993. Activation of both Wnt-1 and Fgf-3 by insertion of mouse mammary tumor virus downstream in the reverse orientation: a reappraisal of the enhancer insertion model. Virology 194:157-165. [DOI] [PubMed] [Google Scholar]
- 11.Corcoran, L. M., J. M. Adams, A. R. Dunn, and S. Cory. 1984. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell 37:113-122. [DOI] [PubMed] [Google Scholar]
- 12.Dekaban, G. A., and J. K. Ball. 1984. Integration of type B retroviral DNA in virus-induced primary murine thymic lymphomas. J. Virol. 52:784-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eick, D., A. Polack, E. Kofler, G. M. Lenoir, A. B. Rickinson, and G. W. Bornkamm. 1990. Expression of P0- and P3-RNA from the normal and translocated c-myc allele in Burkitt's lymphoma cells. Oncogene 5:1397-1402. [PubMed] [Google Scholar]
- 14.Elliott, J. F., B. Pohajdak, D. J. Talbot, J. Shaw, and V. Paetkau. 1988. Phorbol diester-inducible, cyclosporine-suppressible transcription from a novel promoter within the mouse mammary tumor virus env gene. J. Virol. 62:1373-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fanidi, A., E. A. Harrington, and G. I. Evan. 1992. Cooperative interaction between c-myc and bcl-2 proto-oncogenes. Nature 359:554-556. [DOI] [PubMed] [Google Scholar]
- 16.Felsher, D. W., and J. M. Bishop. 1999. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell 4:199-207. [DOI] [PubMed] [Google Scholar]
- 17.Fung, Y. K., A. M. Fadly, L. B. Crittenden, and H. J. Kung. 1981. On the mechanism of retrovirus-induced avian lymphoid leukosis: deletion and integration of the proviruses. Proc. Natl. Acad. Sci. USA 78:3418-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graves, B. J., R. N. Eisenman, and S. L. McKnight. 1985. Delineation of transcriptional control signals within the Moloney murine sarcoma virus long terminal repeat. Mol. Cell. Biol. 5:1948-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimes, H. L., C. B. Gilks, T. O. Chan, S. Porter, and P. N. Tsichlis. 1996. The Gfi-1 protooncoprotein represses Bax expression and inhibits T-cell death. Proc. Natl. Acad. Sci. USA 93:14569-14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallberg, B., J. Schmidt, A. Luz, F. S. Pedersen, and T. Grundstrom. 1991. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J. Virol. 65:4177-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayward, W. S., B. G. Neel, and S. M. Astrin. 1981. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 290:475-480. [DOI] [PubMed] [Google Scholar]
- 22.Hsu, C. L., C. Fabritius, and J. Dudley. 1988. Mouse mammary tumor virus proviruses in T-cell lymphomas lack a negative regulatory element in the long terminal repeat. J. Virol. 62:4644-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isfort, R., R. L. Witter, and H. J. Kung. 1987. c-myc activation in an unusual retrovirus-induced avian T-lymphoma resembling Marek's disease: proviral insertion 5" of exon one enhances the expression of an intron promoter. Oncogene Res. 2:81-94. [PubMed] [Google Scholar]
- 24.Janeway, C. A., Jr. 1991. Selective elements for the Vβ region of the T cell receptor: Mls and the bacterial toxic mitogens. Adv. Immunol. 50:1-53. [DOI] [PubMed] [Google Scholar]
- 25.Kung, H. J., C. Boerkoel, and T. H. Carter. 1991. Retroviral mutagenesis of cellular oncogenes: a review with insights into the mechanisms of insertional activation. Curr. Top. Microbiol. Immunol. 171:1-25. [DOI] [PubMed] [Google Scholar]
- 26.Laimins, L. A., P. Gruss, R. Pozzatti, and G. Khoury. 1984. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J. Virol. 49:183-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laimins, L. A., P. Tsichlis, and G. Khoury. 1984. Multiple enhancer domains in the 3" terminus of the Prague strain of Rous sarcoma virus. Nucleic Acids Res. 12:6427-6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavenu, A., S. Pournin, C. Babinet, and D. Morello. 1994. The cis-acting elements known to regulate c-myc expression ex vivo are not sufficient for correct transcription in vivo. Oncogene 9:527-536. [PubMed] [Google Scholar]
- 29.Lee, W. T., O. Prakash, D. Klein, and N. H. Sarkar. 1987. Structural alterations in the long terminal repeat of an acquired mouse mammary tumor virus provirus in a T-cell leukemia of DBA/2 mice. Virology 159:39-48. [DOI] [PubMed] [Google Scholar]
- 30.Levinson, B., G. Khoury, G. V. Woude, and P. Gruss. 1982. Activation of SV40 genome by 72-base pair tandem repeats of Moloney sarcoma virus. Nature 295:568-572. [DOI] [PubMed] [Google Scholar]
- 31.Li, Y., E. Golemis, J. W. Hartley, and N. Hopkins. 1987. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J. Virol. 61:693-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, Y., C. A. Holland, J. W. Hartley, and N. Hopkins. 1984. Viral integration near c-myc in 10-20% of mcf 247-induced AKR lymphomas. Proc. Natl. Acad. Sci. USA 81:6808-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, J., D. Bramblett, Q. Zhu, M. Lozano, R. Kobayashi, S. R. Ross, and J. P. Dudley. 1997. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol. Cell. Biol. 17:5275-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcu, K. B., S. A. Bossone, and A. J. Patel. 1992. myc function and regulation. Annu. Rev. Biochem. 61:809-860. [DOI] [PubMed] [Google Scholar]
- 35.Martiney, M. J., K. Rulli, R. Beaty, L. S. Levy, and J. Lenz. 1999. Selection of reversions and suppressors of a mutation in the CBF binding site of a lymphomagenic retrovirus. J. Virol. 73:7599-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mertz, J. A., F. Mustafa, S. Meyers, and J. P. Dudley. 2001. Type B leukemogenic virus has a T-cell-specific enhancer that binds AML-1. J. Virol. 75:2174-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyers, S., P. D. Gottlieb, and J. P. Dudley. 1989. Lymphomas with acquired mouse mammary tumor virus proviruses resemble distinct prethymic and intrathymic phenotypes defined in vivo. J. Immunol. 142:3342-3350. [PubMed] [Google Scholar]
- 38.Michalides, R., and E. Wagenaar. 1986. Site-specific rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in murine T-cell leukemias. Virology 154:76-84. [DOI] [PubMed] [Google Scholar]
- 39.Miller, C. L., R. Garner, and V. Paetkau. 1992. An activation-dependent, T-lymphocyte-specific transcriptional activator in the mouse mammary tumor virus env gene. Mol. Cell. Biol. 12:3262-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morrison, H. L., B. Soni, and J. Lenz. 1995. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. J. Virol. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller, R. E., J. K. Ball, and F. P. Chan. 1989. Characterization of cell markers in type B retroviral-induced thymic lymphomas. I. Surface antigen phenotype and karyotype in developing and primary lymphomas. Leukemia Res. 13:553-559. [DOI] [PubMed] [Google Scholar]
- 42.Neil, J. C., D. Hughes, R. McFarlane, N. M. Wilkie, D. E. Onions, G. Lees, and O. Jarrett. 1984. Transduction and rearrangement of the myc gene by feline leukaemia virus in naturally occurring T-cell leukaemias. Nature 308:814-820. [DOI] [PubMed] [Google Scholar]
- 43.Nusse, R. 1991. Insertional mutagenesis in mouse mammary tumorigenesis. Curr. Top. Microbiol. Immunol. 171:43-65. [DOI] [PubMed] [Google Scholar]
- 44.Nusse, R., and H. E. Varmus. 1982. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 31:99-109. [DOI] [PubMed] [Google Scholar]
- 45.O'Donnell, P. V., E. Fleissner, H. Lonial, C. F. Koehne, and A. Reicin. 1985. Early clonality and high-frequency proviral integration into the c-myc locus in AKR leukemias. J. Virol. 55:500-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paquette, Y., L. Doyon, A. Laperriere, Z. Hanna, J. Ball, R. P. Sekaly, and P. Jolicoeur. 1992. A viral long terminal repeat expressed in CD4+ CD8+ precursors is downregulated in mature peripheral CD4− CD8+ or CD4+ CD8− T cells. Mol. Cell. Biol. 12:3522-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelengaris, S., B. Rudolph, and T. Littlewood. 2000. Action of Myc in vivo—proliferation and apoptosis. Curr. Opin. Genet. Dev. 10:100-105. [DOI] [PubMed] [Google Scholar]
- 48.Peters, G., S. Brookes, R. Smith, and C. Dickson. 1983. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell 33:369-377. [DOI] [PubMed] [Google Scholar]
- 49.Prendergast, G. C. 1999. Mechanisms of apoptosis by c-Myc. Oncogene 18:2967-2987. [DOI] [PubMed] [Google Scholar]
- 50.Racevskis, J. 1986. Expression of the protein product of the mouse mammary tumor virus long terminal repeat gene in phorbol ester-treated mouse T-cell-leukemia cells. J. Virol. 58:441-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajan, L., D. Broussard, M. Lozano, C. G. Lee, C. A. Kozak, and J. P. Dudley. 2000. The c-myc locus is a common integration site in type B retrovirus-induced T-cell lymphomas. J. Virol. 74:2466-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salmons, B., V. Erfle, G. Brem, and W. H. Günzburg. 1990. naf, a trans-regulating negative-acting factor encoded within the mouse mammary tumor virus open reading frame region. J. Virol. 64:6355-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheijen, B., J. Jonkers, D. Acton, and A. Berns. 1997. Characterization of pal-1, a common proviral insertion site in murine leukemia virus-induced lymphomas of c-myc and Pim-1 transgenic mice. J. Virol. 71:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selten, G., H. T. Cuypers, M. Zijlstra, C. Melief, and A. Berns. 1984. Involvement of c-myc in MuLV-induced T cell lymphomas in mice: frequency and mechanisms of activation. EMBO J. 3:3215-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Speck, N. A., B. Renjifo, E. Golemis, T. N. Fredrickson, J. W. Hartley, and N. Hopkins. 1990. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 4:233-242. [DOI] [PubMed] [Google Scholar]
- 56.Tremblay, P. J., C. A. Kozak, and P. Jolicoeur. 1992. Identification of a novel gene, Vin-1, in murine leukemia virus-induced T-cell leukemias by provirus insertional mutagenesis. J. Virol. 66:1344-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Klaveren, P., and P. Bentvelzen. 1988. Transactivating potential of the 3" open reading frame of murine mammary tumor virus. J. Virol. 62:4410-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villemur, R., Y. Monczak, E. Rassart, C. Kozak, and P. Jolicoeur. 1987. Identification of a new common provirus integration site in Gross passage A murine leukemia virus-induced mouse thymoma DNA. Mol. Cell. Biol. 7:512-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wrona, T. J., M. Lozano, A. A. Binhazim, and J. P. Dudley. 1998. Mutational and functional analysis of the C-terminal region of the C3H mouse mammary tumor virus superantigen. J. Virol. 72:4746-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanagawa, S., A. Murakami, and H. Tanaka. 1990. Extra mouse mammary tumor proviruses in DBA/2 mouse lymphomas acquire a selective advantage in lymphocytes by alteration in the U3 region of the long terminal repeat. J. Virol. 64:2474-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zaiman, A. L., A. Nieves, and J. Lenz. 1998. CBF, Myb, and Ets binding sites are important for activity of the core I element of the murine retrovirus SL3-3 in T lymphocytes. J. Virol. 72:3129-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]