Abstract
Marine unicellular coccolithophore algae produce species-specific calcite scales otherwise known as coccoliths. While the coccoliths and their elaborate architecture have attracted the attention of investigators from various scientific disciplines, our knowledge of the underpinnings of the process of biomineralization in this alga is still in its infancy. The processes of calcification and coccolithogenesis are highly regulated and likely to be complex, requiring coordinated expression of many genes and pathways. In this study, we have employed cDNA microarrays to investigate changes in gene expression associated with biomineralization in the most abundant coccolithophorid, Emiliania huxleyi. Expression profiling of cultures grown under calcifying and noncalcifying conditions has been carried out using cDNA microarrays corresponding to approximately 2,300 expressed sequence tags. A total of 127 significantly up- or down-regulated transcripts were identified using a P value of 0.01 and a change of >2.0-fold. Real-time reverse transcriptase PCR was used to test the overall validity of the microarray data, as well as the relevance of many of the proteins predicted to be associated with biomineralization, including a novel gamma-class carbonic anhydrase (A. R. Soto, H. Zheng, D. Shoemaker, J. Rodriguez, B. A. Read, and T. M. Wahlund, Appl. Environ. Microbiol. 72:5500-5511, 2006). Differentially regulated genes include those related to cellular metabolism, ion channels, transport proteins, vesicular trafficking, and cell signaling. The putative function of the vast majority of candidate transcripts could not be defined. Nonetheless, the data described herein represent profiles of the transcription changes associated with biomineralization-related pathways in E. huxleyi and have identified novel and potentially useful targets for more detailed analysis.
Coccolithophores are a diverse group of calcifying unicellular marine algae, which includes the extremely abundant species Emiliania huxleyi. This group of phytoplankton has captured the attention of scientists from diverse disciplines, particularly because of the important roles it plays in the cycling of carbon and sulfur in the marine environment. The fixation of CO2 via photosynthesis and the production of CO2 in the calcification process affect global carbon cycling and thus link coccolithophores with climate and the chemical balance between atmosphere, hydrosphere, and geosphere (20). Furthermore, the striking morphological diversity of the more than 200 species of extant coccolithophores (13) and the seemingly endless range of fossil coccolith types (40) provide a unique resource with which to study biodiversity and speciation in the marine environment (23, 24). The biomineralized skeletons or “coccoliths” (Fig. 1A) produced by these microscopic organisms have also captured the attention of materials scientists interested in the microscale and nanoscale fabrication of three-dimensional biomineralized structures for novel applications related to telecommunications, optoelectronics, and medicine (31, 37).
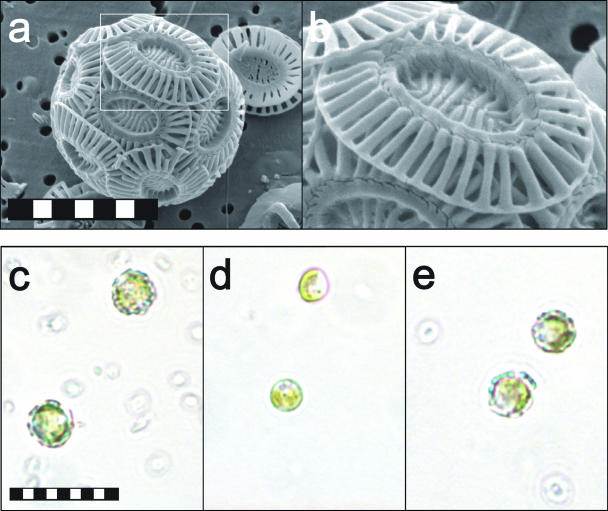
FIG. 1.
Scanning electron microscopy (SEM) and light micrographs of E. huxleyi. (a) SEM micrograph of E. huxleyi “coccosphere” in filtered plankton sample. Scale bar, 4 μm. (b) Detail of E. huxleyi coccolith in SEM micrograph. (c) Light micrograph of calcifying E. huxleyi 1516 cells and isolated coccoliths in phosphate-limiting (f/2) media. Scale bar, 5 μm. (d) Noncalcifying E. huxleyi 1516 cells in phosphate-replete (f/2) media. (e) Calcifying B39 E. huxleyi cells and isolated coccoliths in phosphate-limiting (f/2) media.
Although hypotheses continue to be debated in the literature, the function of the calcite skeletons and the role of biomineralization in coccolithophores remain unresolved. Knowledge of the mechanisms of calcification in coccolithophores is equally poor. Coccolithogenesis takes place within the coccolith vesicle, a specialized apparatus derived from the Golgi body, in a highly regulated and reproducible manner. The molecules involved in the acquisition and transport of calcium ions and inorganic carbon within the cell and those involved in the precipitation of calcite and crystal growth have not been identified or adequately characterized. While acidic polysaccharides and a single protein, “GPA,” have been implicated, the exact roles of these molecules (if any) in calcification have not been experimentally demonstrated (6, 7, 15).
The inability to define the function and to resolve the mechanisms of calcification and coccolithogenesis with previous experimental approaches obviates the need for employing novel molecular genetic tools to investigate these processes. Molecular approaches, however, have been complicated by the absence of a viable transformation system. Further complicating matters is the fact that coccolithophores grow as diploid, vegetative cells whose sexual life cycle has yet to be controlled in culture. Genomic and functional genomic strategies have been applied recently and together with proteomics methods offer great potential for imparting new insight into the fundamental biology of coccolithophores and for the development of molecular tools and techniques. Studies describing the sequencing and analysis of expressed sequence tags (ESTs) from the model coccolithophore species E. huxleyi under calcifying and noncalcifying conditions were the first reports of gene expression in coccolithophores (35, 36). More recently, serial analysis of gene expression (SAGE) and suppressive subtractive hybridization, coupled with real-time reverse transcriptase PCR (RT-PCR), have demonstrated the power of genomic approaches for identifying genes involved in phosphate limitation and/or calcification in E. huxleyi (9, 17). With the goal of further defining genes potentially involved in biomineralization in E. huxleyi, we have constructed the first coccolithophore cDNA microarrays and tested their capability as a tool for high-throughput gene expression analysis of biomineralization and coccolithogenesis processes.
Microarrays emerged during the dawn of the genome-sequencing era and are arguably the most promising new laboratory technique of this decade. In marine systems, microarray analysis has recently been applied to characterize populations of marine bacterioplankton (22), analyze redox regulation of genes in a dinoflagellate (18), evaluate functional gene diversity and distribution in microbial communities (38), identify patterns of differential gene expression associated with hypoxic stress in fish (12), and study the innate immune response in the liver of Atlantic salmon (34). While microarrays have been used to address a number of important questions in marine biology, they have not yet been applied to investigate biomineralization in any organism, including the coccolithophorids. This is largely because of the lack of cDNA libraries and/or available sequence information from these organisms.
Using a cDNA library constructed from the RNA of calcifying cells of E. huxleyi strain CCMP 1516 grown in the laboratory (35), we printed cDNA microarray slides containing some 2,298 elements. These microarrays were used to examine differential gene expression patterns in calcifying and noncalcifying E. huxleyi cultures grown in phosphate-limiting and phosphate-replete media, respectively. Among the transcripts analyzed, 79 were found to be up-regulated and 48 were found to be down-regulated in calcifying cells maintained in phosphate-limiting media relative to noncalcifying cells maintained in phosphate-replete media. These 127 transcripts were then subjected to full-length sequencing and subjected to BLASTX homology searches. To validate microarray analysis results, we also analyzed the relative gene expression patterns of a small subset of differentially expressed transcripts by real-time RT-PCR. In order to distinguish genes involved in biomineralization from those associated with phosphate limitation, we analyzed the expression of differentially expressed transcripts in another strain of E. huxleyi (strain MBA B39), which in contrast to strain 1516 was found to calcify in phosphate-replete media. The combined microarray and real-time RT-PCR analysis results enabled us to identify 46 genes that may be directly or indirectly involved in coccolith biomineralization and 36 genes that either were related to phosphate limitation or were false positives.
With the completion of the genome sequencing of E. huxleyi strain 1516 (a collaborative effort between our laboratory and the U.S. Department of Energy), microarrays are likely to play an important role in the functional characterization of new genes in this important organism. The prospect of printing whole-genome arrays or genome chips (8, 32) of E. huxleyi will open up exciting possibilities for the understanding of coccolithophore cell biology and physiology at the molecular level. The relatively small cDNA microarrays presented here highlight the potential of this approach as a means of studying gene expression patterns in coccolithophores.
MATERIALS AND METHODS
Emiliania huxleyi cultures and construction of cDNA library.
Cultures of E. huxleyi strain 1516, obtained from the Provasoli Guillard National Center for Culture of Marine Phytoplankton (CCMP), were found to calcify when grown in phosphate-limiting (1.67 μM) f/2 medium and to produce mostly noncalcifying cells when grown in phosphate-replete (41.7 μM) f/2 medium (Fig. 1) (35). Using RNA extracted from calcifying cultures of strain 1516, a cDNA library was constructed by Wahlund et al. (35), from which several thousand clones were randomly selected.
Experimental design.
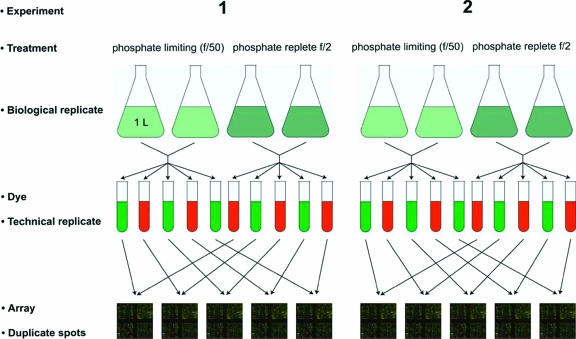
In this study, two biological replicate experiments were performed whereby RNA was extracted from pooled 1-liter batch cultures of calcifying and noncalcifying cells grown in phosphate-limiting and phosphate-replete media, respectively (Fig. 2). Each replicate experiment consisted of five replicated microarray hybridizations for a total of 10 technical replicates. Technical replicates were hybridized with reciprocally labeled cDNA. In the forward reaction, the cDNA from calcifying cells was labeled with Cy5 and cDNA from noncalcifying cells was labeled with Cy3, whereas in the reverse reaction, cDNA from noncalcifying cells was labeled with Cy3 and cDNA from calcifying cells was labeled with Cy5. Competitive hybridizations were carried out by combining 20 μg of extracted and differentially labeled total RNA from calcifying and noncalcifying samples and applying the mixture to the array printed on a glass slide. All array experiments were conducted under the same hybridization and wash conditions with the same batch of arrays.
FIG. 2.
Experimental design employed in this study. In each case, the starting material consists of RNA samples collected from calcifying and noncalcifying cultures of E. huxleyi. Samples from experimental replicates 1 and 2 were fluorescently labeled. Reciprocal dye swap experiments were performed and assayed using 10 microarrays indicated by connecting lines in each diagram.
Construction of microarrays.
Microarrays were printed using a subset of 2,298 clones from the cDNA library derived from calcifying cells grown under phosphate-limiting conditions (35). A total of 3,000 ESTs from the library were generated from the 5′ ends. These sequences were assembled into the 2,298 unique contigs and singletons that were used to construct the microarray. A panel of control elements was also printed and included gene sequences from Bacillus (diaminopimelate decarboxylase and dehydrodipicolinate reductase genes and pheB) and Escherichia coli (biotin synthesis protein gene) as negative controls; the multiple cloning site of the parent vector (pMAB58), deoxynucleoside triphosphates (dNTPs), and poly(A) sequences as blocking controls; genomic DNA from E. huxleyi as a positive control; and a site-specific recombinase gene from P1 bacteriophage (cre) as a doping control.
cDNA clones were picked and amplified in triplicate in 96-well plates containing the following: 20 ng/ml template DNA, 1 μM forward primer (5′-TATAACCGCTTTGGAATCACT-3′), 1 μM reverse primer (5′-GTAAATTTCTGGCAAGGTAGAC-3′); 200 μM dNTPs (TaKaRa Biomedicals, Tokyo, Japan), Taq polymerase buffer with MgCl2 (Promega Corporation, Madison, Wis.), and 2 to 4 U Taq polymerase. The reaction mixture was incubated at 94°C for 3 min; cycled 29 times at 94°C for 45 s, 55°C for 1 min, and 72°C for 3 min; and then held at 4°C before storage at −20°C. The three PCRs were pooled and purified using QIAGEN QIAquick 96 PCR purification kit (QIAGEN Inc., Valencia, Calif.). To check the efficiency of the amplification, 3 μl of each PCR product was run on a Ready-To-Run 96-well gel (Amersham Biosciences, Little Chalfont, United Kingdom) stained with ethidium bromide. Ten random samples were selected from each plate, and the concentration of amplified DNA was determined spectrophotometrically at 260 nm. Finally, purified products were dried down and resuspended to a concentration of 500 μg/ml.
Microarrays were printed using poly-lysine-coated slides. Prior to printing, the cDNA samples were diluted to a concentration of 250 μg/ml in 50% dimethyl sulfoxide. The 2,298 sequences and the 8 controls (total of 2,306 elements) were then printed in duplicate on 68 slides using a Roboarrayer (RoboDesign, Carlsbad, Calif.) with a 4-by-4 pin configuration. Array spots were ∼150 μm in diameter, and the distance between spots was ∼200 μm. The printed slides were baked at 80°C for 4 h and blocked for 15 min with a succinic anhydride-NaBO4 solution. This solution was freshly prepared by dissolving 5 g of succinic anhydride (Sigma, St. Louis, Mo.) in 315 ml of N-methyl-pyrrolidinone and adding 35 ml of 0.2 M NaBO4. After blocking, the DNA was denatured in 95°C double-distilled H2O (ddH2O) for 2 min, dehydrated by immersion in 95% ethanol for 1 min, and dried by centrifugation for 5 min at 600 × g. Printed arrays were stored at room temperature in a desiccator until they were used. The quality of the printed arrays was assessed by random 9-mer staining using a Cy3-labeled Poly-N 9-mer (QIAGEN).
RNA isolation and microarray hybridization.
E. huxleyi strain 1516 was grown at 20°C in phosphate-limiting and phosphate-replete f/2 artificial seawater supplemented with kanamycin (100 μg/ml) under a 12-h-light/12-h-dark cycle with cool white florescent light at 660 μmol m−2 s−2. Cells of a second, more robust E. huxleyi strain, B39, which calcifies under both phosphate-replete and phosphate-limiting conditions (Fig. 1c), were obtained from the Marine Biological Association (MBA) Marine Algae Culture Collection, Plymouth, United Kingdom, and grown in f/2 media as described above. Cells were harvested from all cultures on the seventh day after inoculation, during late log phase. Prior to harvesting, the cultures were observed under the light microscope to check their degree of calcification (Fig. 1c to e).
Total RNA extraction was performed using a modified version of the standard guanidinium isothiocyanate procedure (30) with a decalcification step as described by Wahlund et al. (35). The integrity of the isolated RNA was assessed by denaturing agarose gel electrophoresis, and its concentration was measured spectrophotometrically. Twenty micrograms of total RNA from the calcifying and noncalcifying cultures of E. huxleyi 1516 was used to produce fluorescently labeled cDNA with the Superscript III indirect labeling system (Invitrogen, Carlsbad, Calif.), whereby amino-modified dNTPs incorporated during first-strand cDNA synthesis are coupled to NHS ester-containing Cy3 or Cy5 dyes (Amersham Pharmacia, Little Chalfont, United Kingdom). Unincorporated dye molecules were removed using PureLink spin columns, and the purified fluorescently labeled probes were dried by centrifugation in a SpeedVac. The dye labeling was reversed in separate experiments to eliminate the possibility of dye incorporation bias.
Microarray slides were prehybridized in a solution of 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1% bovine serum albumin, and 0.1% sodium dodecyl sulfate (SDS) at 45°C for 30 min. After being rinsed twice in dH2O and submersion in isopropanol, the slides were dried by centrifugation (2 min, 697 × g). Labeled cDNA probes from the calcifying and noncalcifying E. huxleyi cells were resuspended and combined in a total of 50 μl hybridization solution [25% formamide, 5× SSC, 0.1% SDS, 27.5 μg poly(A), 100 ng Cy5-labeled cre, 5 μg human Cot-1 DNA]. The probe solution was incubated at 95°C for 3 min and cooled to room temperature before being applied to a prehybridized microarray under a coverslip. Arrays were hybridized for 18 h at 45°C. The slides were washed for 5 min each as follows: (i) 1× SSC plus 0.1% SDS at 45°C, (ii) 0.2× SSC plus 0.1% SDS at 30°C, (iii) 0.1× SSC plus 0.1% SDS at 30°C, and (iv) 0.1× SSC at 22°C before being rinsed briefly in dH2O and dried by centrifugation (2 min, 697 × g). A total of 10 independent hybridizations were performed. The fluorescence of the hybridized microarrays was measured with the GenePix 4000A Scanner (Axon Instruments, Union City, Calif.).
Microarray data analysis.
Fluorescent images were captured, and the Cy3 and Cy5 fluorescence signal at each target spot was measured using the GenePix Pro 4.1 software (Axon). A grid based on the predefined microarray layout was manually aligned to spot signals to optimize the subsequent automated spot recognition and signal quantitation. Poor-quality spots were automatically excluded from the analysis when the spot intensity was not greater than the background threshold, was irregular in size, or overlapped an adjacent spot. Images were also inspected manually, and spots from low-quality areas of the array were flagged and excluded from further analysis. The local median background signal was subtracted from the median hybridization signal of each separate spot. After background subtraction, quality spots exhibiting an intensity/background ratio of >3 in either channel were subjected to global robust locally weighted regression (LOWESS) normalization (5) designed to equalize the fluorescence ratio of the medians for all quality spots. Prior to statistical analysis, median fluorescence measurements were logarithm transformed (log2). For each gene, a paired t test (P value of <0.01) was conducted using log2-transformed median fluorescent intensity values to assess the significance of the relative transcript abundance levels. A Bonferroni multiple comparisons procedure was also performed, and an adjusted P value of 1.36E−5 was applied to reduce the probability of a type I error. Transcripts exhibiting a significant P value and an expression ratio greater than twofold were subjected to full-length sequencing.
Because some of the EST clones printed were similar in sequence and likely to represent the same gene, full-length sequences generated were assembled using Phrap for Linux (Codon Code Corporation). After assembly, an attempt to putatively identify the function of transcripts was made using BLASTX homology searches of GenBank with a probability cutoff of 1 in 1,000 (E value, ≤1.0E−3).
Real-time RT-PCR.
Open reading frames (ORFs) within the full-length sequences of each of the significantly up- and down-regulated cDNA clones revealed by microarray were identified using ORF Finder (NCBI: www.ncbi.nlm.nih.gov/gorf). Real-time RT-PCR primers were designed to ORFs exhibiting significant BLASTX homology results or to the longest ORF of transcripts returning no significant GenBank hits, using Primer Express Software version 1.0 (Applied Biosystems, Foster City, Calif.). Primers were designed to have a G+C content of 50 to 60%, a melting temperature of >50°C, C's > G's, ≤3 identical dNTPs in a row, and to amplify a target sequence between 75 and 100 bp. When suitable primers could not be found using these criteria, the parameters C's > G's and having ≤3 identical dNTPs in a row were relaxed. Forward and reverse primers were synthesized by San Diego State University Microchemical Core Facility (SDSU, MCF).
cDNA synthesis was performed with the Omniscript reverse transcriptase kit (QIAGEN, Valencia, Calif.) using total RNA as template from calcifying and noncalcifying cultures of E. huxleyi 1516 maintained in phosphate-limiting and phosphate-replete media, respectively, as well as RNA from calcifying cultures of B39 maintained in phosphate-replete media. Each 20-μl reaction mixture contained 2 μg template RNA, 1× RT buffer, 0.5 mM deoxynucleoside triphosphate, 1 μM oligo(dT) primer, 10 U of RNase inhibitor, and 4 U of Omniscript reverse transcriptase. The reaction mixture was incubated for 60 min at 37°C.
Real-time RT-PCR was performed with the iCycler IQ (Bio-Rad, Richmond, Calif.) using SYBR green chemistry. The thermal profile was 95°C for 10 min followed by 40 cycles of 60°C for 30 s and 82°C for 30 s. Reactions were carried out in a 96-well plate in a 25-μl reaction volume containing 7.5 μl SYBR green Supermix (Bio-Rad), 11.9 μl dH2O, 0.27 μl of each forward and reverse primer (20 nm/ml), and 5 μl of cDNA mixture (1.5 μl cDNA, 148.5 μl dH2O). Each sample was run in triplicate using cDNA synthesized from E. huxleyi RNA and a negative, no-template control. Because of the absence of established control genes for E. huxleyi, normalization of the real-time RT-PCR data was made to accurately quantified total RNA (33).
Relative gene expression was calculated using the ΔCT method. A mean CT value for the expression of each gene in the E. huxleyi cultures was obtained from the three replicates. Pairwise comparisons were made between the mean CT value in different E. huxleyi cultures using the following equation: relative expression = 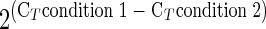 . The relative expression values were converted to changes (fold [FC]) by taking the additive inverse of all values less than 1 (i.e., negative FC). As with the cDNA microarray, only results with an FC of 2 or more were considered significant.
. The relative expression values were converted to changes (fold [FC]) by taking the additive inverse of all values less than 1 (i.e., negative FC). As with the cDNA microarray, only results with an FC of 2 or more were considered significant.
In order to independently test the results of the cDNA microarray, we randomly selected a subset of 20 differentially expressed clones for real-time RT-PCR analysis. The relative expression of these transcripts in calcifying versus noncalcifying E. huxleyi 1516 cultures 7 days postinoculation was analyzed by real-time RT-PCR and compared to the results of the competitive microarray hybridization. Those clones for which the direction of real-time RT-PCR FC was in agreement with the cDNA microarray were considered to be successfully validated.
To distinguish potential biomineralization genes from those associated with phosphate limitation, we ran all working real-time RT-PCR primer sets using cDNA from the noncalcifying E. huxleyi 1516 cultures and cDNA synthesized from RNA derived from calcifying cultures of E. huxleyi strain B39. Both cultures were grown in phosphate-replete f/2 media. Real-time RT-PCR results were then compared to microarray analysis results.
Nucleotide sequence acccession numbers.
The sequences of the E. huxleyi genes have been deposited in the GenBank database under accession no. DQ658243 to DQ658369.
RESULTS
Microarray hybridization.
The microarray hybridizations exhibited a high degree of specificity to the fluorescently labeled E. huxleyi probe sequences. There was little or no detectable signal from the blocking controls or the panel of negative controls representing bacterial gene sequences. Strong hybridization signals were recorded by the positive and doping controls. The high specificity of the E. huxleyi microarrays was indicated by the considerable variation in signal intensity across the elements; intensities that spanned 4 orders of magnitude (Fig. 3a and b). The correlation of the log ratio expression profiles was considered to be satisfactory across the 10 technical replicates, ranging from 0.55 to 0.98 with mean of 0.76. Expression profiles within experimental replicates exhibited mean correlation coefficients of 0.87 and 0.86, while correlation coefficients across experimental replicates were much lower. The median coefficient of variation across arrays was found to be 31% and also reflects acceptable but less than optimal reproducibility. After filtering, however, the median coefficient of variation dropped to 14%. Factors influencing hybridization efficiency such as probe sequence complexity, secondary structure of the target, and the relative position of probe binding sites to the target interfere with the simple correlation of probe signal ratios and relative abundances of the corresponding target molecules. For these reasons, attempts were made herein to independently validate microarray results using real-time RT-PCR.
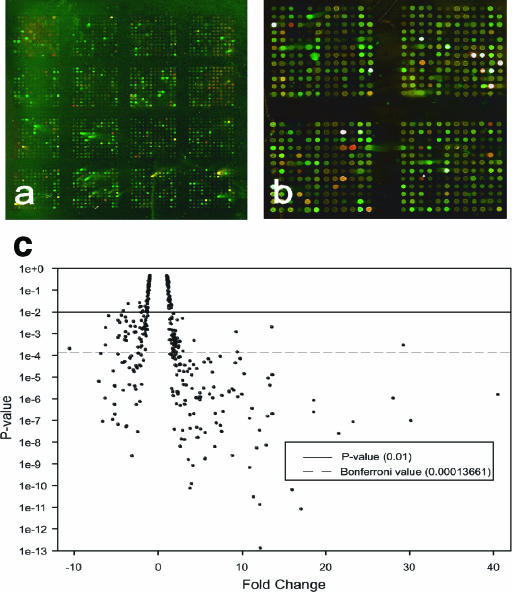
FIG. 3.
Images and statistical analysis of competitively hybridized cDNA microarrays. (a) Image of the entire cDNA microarray after dye swap experiment. Phosphate-limited f/2 1516 cultures are green, and phosphate-replete f/2 1516 cultures are red). (b) Upper right quadrant of the microarray with standard labeling. Phosphate-replete 1516 cultures are green, and phosphate-limited 1516 cultures are red). (c) Statistical analysis of interrogated genes whereby the P value derived from a paired t test is plotted the microarray FC determined by the ratio of the fluorescence medians (phosphate limited versus phosphate replete).
Of the 2,298 EST sequences analyzed by microarray analysis, 62% of the elements registered a signal that was significantly greater than that of the background in one of the two channels. Differential expression of greater than twofold in the calcifying cells grown in low phosphate, relative to noncalcifying cells grown in phosphate-replete media, was exhibited by 187 transcripts at the 95% confidence level; a total of 133 of these transcripts exhibited a P value below the Bonferroni value (P < 1.366 × 10−5). A plot of P value against FC of the genes included in the statistical analysis is shown in Fig. 3c. Among the differentially expressed transcripts, 26 exhibited a change of greater than 10-fold, with the maximum change being 30-fold. The full-length sequences for all but six of the corresponding differentially-expressed cDNA clones were obtained. Because several of the clones shared a high degree of sequence similarity, sequences were assembled using Phrap. Sixty of the sequences were found to be redundant, bringing the total number of unique differentially expressed sequences to 127. Of these sequences, 79 were found to be up-regulated in calcifying cells grown in low-phosphate media and 48 were found to be down-regulated under these same conditions (Tables 1 and 2). An attempt to putatively identify the function of each of the 127 transcripts was made using BLASTX homology searches against known proteins in GenBank (Table 1). The cellular function of the 45 transcripts returning significant hits varied and included vesicular trafficking, cell signaling, lipid biosynthesis, transport, and general metabolism. The function of the majority (a total of 82) of the transcripts, however, could not be predicted.
TABLE 1.
Results of microarray analysis showing transcripts significantly up- or down-regulated in calcifying phosphate-limited relative to noncalcifying phosphate-replete E. huxleyi 1516 culturesa
| Gene identification no. | Homolog description | E value | P value | FCb |
|---|---|---|---|---|
| 77-2-5-E02.r.5 | No hit | 3.00E−04 | 29.19 | |
| 77-2-35-G03.r.1 | No hit | 1.09E−06 | 27.99 | |
| 77-2-9-C04.r.1 | No hit | 2.58E−08 | 21.52 | |
| Contig 22 | Hypothetical protein | 4.82E−24 | 3.21E−07 | 20.60 |
| 77-2-41-G02.r.1 | No hit | 8.63E−07 | 18.49 | |
| 77-2-8-C07.r.1 | No hit | 2.46E−07 | 18.48 | |
| 77-2-36-H06.r.1 | No hit | 2.07E−03 | 13.53 | |
| 77-2-36-H03.r.2 | No hit | 9.05E−06 | 13.10 | |
| 77-2-9-E03.r.5 | No hit | 1.58E−07 | 13.05 | |
| Contig 15 | Hypothetical protein | 4.95E−03 | 1.80E−08 | 12.07 |
| 77-2-6-H01.r.2 | No hit | 3.63E−07 | 11.18 | |
| 77-2-32-E05.r.1 | No hit | 1.26E−07 | 10.85 | |
| Contig 9 | No hit | 2.11E−06 | 10.43 | |
| 77-2-41-F03.r.1 | Hypothetical protein | 3.26E−03 | 1.70E−06 | 9.93 |
| Contig 24 | Alpha-soluble NSF attachment protein | 6.91E−23 | 2.87E−07 | 9.51 |
| 77-2-4-E06.r.1 | RabGAP/TBC domain-containing protein | 9.61E−22 | 1.22E−03 | 9.27 |
| Contig 20 | No hit | 3.10E−06 | 9.06 | |
| 77-2-4-G05.r.1 | No hit | 2.38E−09 | 8.85 | |
| Contig 18 | No hit | 2.46E−05 | 8.52 | |
| 77-2-31-F12.r.1 | No hit | 2.62E−07 | 7.70 | |
| Contig 14 | No hit | 2.38E−06 | 7.54 | |
| 77-2-5-B05.r.2 | No hit | 9.21E−05 | 7.36 | |
| 77-2-39-G04.r.1 | No hit | 4.51E−05 | 6.84 | |
| 77-2-8-G04.r.1 | No hit | 6.38E−09 | 6.74 | |
| 77-2-41-E04.r.1 | No hit | 1.68E−05 | 6.43 | |
| 77-2-9-C12.r.1 | Surface antigen BspA, Bacteroides forsythus | 9.03E−37 | 4.99E−05 | 6.23 |
| 77-2-39-E03.r.1 | Predicted dinucleotide-binding enzymes | 7.34E−05 | 7.03E−05 | 5.96 |
| 77-2-8-A07.r.1 | LigA | 2.31E−56 | 1.78E−09 | 5.57 |
| 77-2-15-F10.r.2 | No hit | 1.63E−09 | 5.54 | |
| 77-2-12-C02.r.1 | Guanylate cyclase | 6.27E−08 | 4.98E−05 | 5.44 |
| 77-2-40-B09.r.1 | No hit | 3.14E−06 | 5.39 | |
| Contig 10 | Predicted esterase | 1.83E−05 | 7.11E−06 | 5.03 |
| 77-2-36-F03.r.2 | l-3-Phosphoserine phosphatase | 3.80E−50 | 1.23E−06 | 5.0 |
| Contig 21 | No hit | 7.53E−04 | 4.91 | |
| 77-2-11-H03.r.2 | No hit | 1.24E−06 | 4.88 | |
| Contig 12 | No hit | 9.41E−06 | 4.77 | |
| 77-2-8-C12.r.1 | Outer mitochondrial membrane protein porin | 4.21E−22 | 1.65E−08 | 4.65 |
| 77-2-7-D01.r.1 | No hit | 2.43E−05 | 4.59 | |
| Contig 19 | No hit | 2.42E−06 | 4.38 | |
| 77-2-38-B11.r.1 | No hit | 2.05E−07 | 4.25 | |
| Contig 16 | No hit | 4.93E−05 | 4.23 | |
| 77-2-8-C05.r.1 | No hit | 3.82E−05 | 4.19 | |
| Contig 6 | Hypothetical protein | 2.99E−30 | 1.87E−05 | 4.03 |
| Contig 7 | No hit | 2.55E−04 | 3.98 | |
| 77-2-16-D04.r.1 | No hit | 1.84E−07 | 3.76 | |
| 77-2-4-B06.r.1 | Possible elongation of very-long-chain fatty acid protein | 3.44E−19 | 5.72E−05 | 3.76 |
| 77-2-15-E10.r.2 | No hit | 1.78E−05 | 3.47 | |
| 77-2-9-D02.r.5 | Putative serine/threonine kinase | 1.38E−39 | 4.28E−06 | 3.30 |
| 77-2-41-E05.r.1 | No hit | 5.55E−07 | 3.14 | |
| 77-2-7-A10.r.5 | IMP dehydrogenase/GMP reductase | 4.03E−10 | 3.56E−05 | 3.13 |
| 77-2-8-E04.r.1 | No hit | 4.47E−06 | 3.0 | |
| Contig 3 | No hit | 3.15E−07 | 2.97 | |
| 77-2-24-F03.r.1 | Cytochrome b5 domain-containing protein-like | 8.30E−14 | 3.57E−08 | 2.9 |
| 77-2-19-E12.r.1 | Small nuclear ribonucleoprotein polypeptide D2 | 2.47E−35 | 1.40E−08 | 2.74 |
| 77-2-34-B09.r.1 | No hit | 4.98E−07 | 2.61 | |
| 77-2-14-G05.r.1 | No hit | 3.98E−05 | 2.60 | |
| 77-2-24-H06.r.1 | No hit | 6.53E−09 | 2.60 | |
| 77-2-35-H12.r.1 | Hypothetical protein | 2.17E−03 | 8.76E−05 | 2.60 |
| 77-2-7-B09.r.4 | Proton/amino acid transporter 2 | 5.19E−09 | 4.40E−04 | 2.57 |
| 77-2-9-C08.r.3 | Expressed protein | 7.82E−15 | 8.60E−05 | 2.53 |
| 77-2-18-E09.r.1 | Asparaginyl endopeptidase | 7.83E−70 | 1.94E−05 | 2.49 |
| 77-2-19-F06.r.1 | Phosphate-repressible phosphate permease | 8.44E−04 | 4.22E−04 | 2.41 |
| 77-2-8-B04.r.1 | 4 Fe-S ferridoxin | 1.19E−03 | 2.70E−04 | 2.40 |
| 77-2-14-C05.r.1 | No hit | 8.24E−05 | 2.39 | |
| Contig 13 | No hit | 1.18E−04 | 2.33 | |
| 77-2-38-H09.r.4 | No hit | 2.08E−06 | 2.33 | |
| 77-2-7-A09.r.1 | Putative ammonium transporter | 2.55E−53 | 6.02E−04 | 2.28 |
| 77-2-41-E09.r.1 | Arachidonate 15-lipoxygenase, second type | 5.34E−05 | 4.29E−03 | 2.26 |
| 77-2-40-E03.r.1 | No hit | 2.14E−06 | 2.22 | |
| 77-2-7-H02.r.2 | 4-Aminobutyrate aminotransferase | 9.05E−103 | 1.45E−04 | 2.13 |
| 77-2-19-C01.r.1 | Putative gamma-carbonic anhydrase | 1.73E−48 | 3.31E−05 | 2.10 |
| 77-2-16-F04.r.1 | Hypothetical protein | 8.29E−14 | 4.17E−04 | 2.09 |
| 77-2-1-A06.r.1 | Hypothetical protein | 8.30E−13 | 4.00E−04 | 2.06 |
| 77-2-7-B11.r.5 | Hypothetical protein | 1.85E−14 | 2.73E−04 | 2.06 |
| 77-2-35-D05.r.1 | No hit | 3.70E−04 | 2.05 | |
| Contig 17 | No hit | 2.10E−04 | 2.04 | |
| 77-2-7-G03.r.2 | Similar to ankyrin 2 | 1.44E−18 | 1.38E−03 | 2.04 |
| 77-2-30-H04.r.1 | Aspartate aminotransferase | 5.02E−95 | 6.91E−05 | 2.02 |
| 77-2-15-F09.r.2 | No hit | 2.46E−04 | 2.01 | |
| 77-2-11-H04.r.3 | No hit | 4.24E−04 | −2.02 | |
| 77-2-11-G02.r.1 | No hit | 4.05E−03 | −2.03 | |
| 77-2-11-A09.r.1 | No hit | 1.04E−04 | −2.05 | |
| 77-2-11-A05.r.1 | No hit | 9.82E−03 | −2.07 | |
| 77-2-19-H01.r.1 | N-Acetylneuraminic acid phosphate synthase | 3.92E−17 | 5.36E−04 | −2.11 |
| 77-2-33-C08.r.1 | Hypothetical protein | 1.59E−03 | 3.28E−05 | −2.13 |
| 77-2-11-C12.r.5 | No hit | 2.39E−04 | −2.14 | |
| 77-2-19-B04.r.1 | Conserved hypothetical protein | 7.24E−09 | 7.55E−07 | −2.20 |
| 77-2-10-C04.r.2 | No hit | 3.64E−05 | −2.26 | |
| 77-2-19-G10.r.1 | No hit | 6.05E−04 | −2.27 | |
| Contig 5 | Fucoxanthin-chlorophyll a to c binding protein F | 6.58E−09 | 1.02E−03 | −2.31 |
| 77-2-11-A12.r.1 | No hit | 2.39E−04 | −2.45 | |
| 77-2-10-D09.r.3 | No hit | 2.85E−05 | −2.57 | |
| 77-2-14-F02.r.1 | No hit | 5.35E−07 | −2.64 | |
| 77-2-14-B10.r.1 | No hit | 2.78E−06 | −2.72 | |
| 77-2-16-A09.r.1 | No hit | 1.20E−03 | −2.74 | |
| 77-2-12-F03.r.2 | Proteophosphoglycan | 2.35E−03 | 2.19E−03 | −2.80 |
| 77-2-5-F11.r.4 | Proteasome alpha subunit | 4.13E−77 | 3.90E−06 | −2.84 |
| 77-2-5-H09.r.3 | No hit | 1.16E−03 | −2.87 | |
| 77-2-33-D03.r.1 | No hit | 1.76E−05 | −2.91 | |
| 77-2-24-F09.r.1 | No hit | 3.73E−03 | −2.97 | |
| Contig 23 | Ac1147 | 4.34E−25 | 5.13E−05 | −3.04 |
| 77-2-30-E06.r.1 | No hit | 2.28E−03 | −3.32 | |
| 77-2-38-F06.r.1 | No hit | 7.67E−05 | −3.40 | |
| 77-2-39-H02.r.2 | GA12046-PA | 7.51E−13 | 9.79E−04 | −3.57 |
| 77-2-3-B03.r.2 | Cysteine protease | 1.01E−60 | 4.77E−04 | −3.82 |
| 77-2-30-E07.r.1 | Hypothetical protein | 2.12E−149 | 3.79E−03 | −3.83 |
| 77-2-12-D03.r.1 | Novel protein | 2.00E−10 | 1.24E−03 | −3.93 |
| 77-2-30-G06.r.1 | Phosphoglycerate/bisphosphoglycerate mutase family protein | 4.60E−18 | 7.24E−04 | −3.94 |
| 77-2-12-E01.r.2 | No hit | 5.87E−03 | −3.94 | |
| 77-2-24-E01.r.1 | No hit | 5.20E−04 | −4.03 | |
| 77-2-4-H01.r.2 | No hit | 4.71E−05 | −4.30 | |
| Contig 4 | Fucoxanthin chlorophyll a/c protein | 5.65E−24 | 5.90E−08 | −4.34 |
| 77-2-12-F04.r.1 | Predicted permeases | 2.93E−39 | 7.18E−03 | −4.43 |
| Contig 25 | No hit | 2.12E−05 | −4.58 | |
| 77-2-12-B04.r.1 | No hit | 3.34E−03 | −4.59 | |
| 77-2-24-D02.r.1 | Sulfatase 1 precursor | 8.29E−71 | 8.16E−04 | −4.66 |
| 77-2-24-H08.r.1 | Peptidyl-prolyl cis-trans-isomerase | 1.23E−19 | 4.09E−03 | −4.80 |
| 77-2-24-E02.r.1 | No hit | 1.14E−04 | −4.80 | |
| 77-2-11-A10.r.5 | Trypsin | 5.78E−31 | 6.48E−04 | −4.82 |
| 77-2-31-F09.r.1 | No hit | 5.47E−07 | −5.23 | |
| 77-2-31-C11.r.1 | No hit | 1.92E−05 | −5.38 | |
| 77-2-29-H11.r.1 | No hit | 1.09E−06 | −5.76 | |
| 77-2-12-C05.r.1 | Proteophosphoglycan | 6.16E−04 | 6.84E−03 | −5.93 |
| 77-2-2-B09.r.1 | Similar to kinesin light chain | 1.93E−03 | −6.29 | |
| 77-2-19-E11.r.1 | No hit | 3.02E−04 | −6.32 | |
| 77-2-7-A05.r.1 | Unnamed protein product | 1.88E−12 | 9.02E−08 | −6.66 |
| 77-2-16-G05.r.1 | No hit | 2.08E−04 | −10.59 |
The results of a translated query to the NCBI peptide database are shown, including homologues with an E value of <10−2.
Relative gene expression is expressed as FC with the corresponding P value. For assembled contigs, a mean FC value is given.
TABLE 2.
Real-time RT-PCR validation of cDNA microarray relative gene expression
| Gene identification no.a | FC byb:
|
|
|---|---|---|
| Microarray | Real-time RT-PCR | |
| Contig 24 | 9.51 | 134.05 |
| Contig 20 | 9.06 | 53.20 |
| 77-2-19-F06.r.1 | 2.41 | 35.10 |
| Contig 18 | 8.52 | 34.30 |
| Contig 7 | 3.98 | 28.51 |
| 77-2-7-B09.r.4 | 2.57 | 18.38 |
| Contig 16 | 4.23 | 16.37 |
| Contig 21 | 4.91 | 9.40 |
| 77-2-7-D01.r.1 | 4.59 | 3.17 |
| 77-2-18-E09.r.1 | 2.49 | 3.17 |
| 77-2-7-G03.r.2 | 2.04 | 2.64 |
| 77-2-12-C02.r.1 | 5.44 | 2.05 |
| 77-2-12-E01.r.2 | −3.94 | 1.87* |
| Contig 5 | −2.31 | 1.74* |
| 77-2-41-E04.r.1 | 6.43 | 1.66 |
| 77-2-19-C01.r.1 | 2.10 | 1.66 |
| 77-2-19-E12.r.1 | 2.74 | 1.66 |
| 77-2-40-B09.r.1 | 5.39 | 1.02 |
| 77-2-12-F04.r.1 | −4.43 | −1.23* |
| 77-2-24-H08.r.1 | −4.80 | −3.03 |
Twenty transcripts up- and down-regulated in cDNA microarray randomly selected for real-time RT-PCR analysis.
Relative gene expression in phosphate-limiting relative to phosphate-replete E. huxleyi cultures expressed as FC. Asterisks indicate transcripts that were not validated by real-time RT-PCR.
Real-time RT-PCR validation.
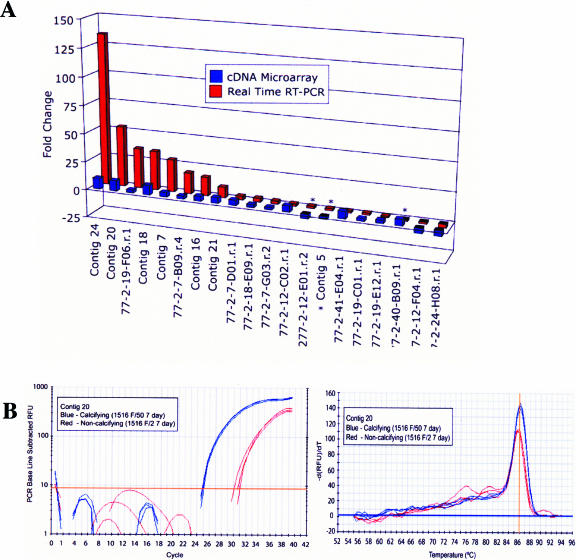
Working real-time RT-PCR primer sets were designed for a total of 82 of the 127 differentially expressed transcripts identified by microarray analysis. Primer sets designed to 25 additional transcripts either failed to yield an amplification signal or did not elicit a single amplification product; viable primer sets could not be found for the remaining 20 transcripts. Twenty differentially expressed transcripts were randomly selected from the 82 working primer sets. Their relative gene expression in calcifying and noncalcifying E. huxleyi 1516 cultures was determined by real-time RT-PCR and compared to cDNA microarray analysis results. Of this subset, a total of 17 (85%) were successfully validated (Table 3 and Fig. 4). The real-time RT-PCR FC of these 17 transcripts, calculated using the ΔCT method (Fig. 4B) was in the same direction recorded in the cDNA microarray (Fig. 4A). FCs for individual transcripts, however, differed in magnitude, highlighting differences in the sensitivity and specificity of the two techniques (Fig. 4A). The FC recorded by real-time RT-PCR was higher for 10 of the transcripts, reflecting the greater sensitivity of real-time RT-PCR analysis. For example, the relative expression of contig 20 was measured and found to be 53-fold higher in calcifying versus noncalcifying cells by real-time RT-PCR (Fig. 4B) and only 9-fold higher when measured by microarray analysis (Fig. 4A). The relative FC was lower in eight cases when measured by real-time RT-PCR as compared to microarray analysis, which may in part be due to the greater specificity afforded by real-time RT-PCR.
TABLE 3.
Transcripts most likely to be involved in biomineralization based upon microarray and real-time RT-PCR analyses
| Gene identification no.a | Homolog descriptionb | FC by:
|
|
|---|---|---|---|
| Microarray | Real-time RT PCR | ||
| 77-2-14-G05.r.1 | No hit | 2.60 | 64.00 |
| 77-2-5-E02.r.5 | No hit | 29.19 | 48.50 |
| Contig 16 | No hit | 4.23 | 42.22 |
| 77-2-15-F09.r.2 | No hit | 2.01 | 32.75 |
| Contig 7 | No hit | 3.98 | 30.55 |
| 77-2-7-G03.r.2 | Similar to ankyrin 2 | 2.04 | 28.51 |
| 77-2-35-D05.r.1 | No hit | 2.05 | 24.82 |
| 77-2-19-C01.r.1 | Gamma-carbonic anhydrase | 24.25 | 2.10 |
| Contig 20 | No hit | 9.06 | 19.70 |
| 77-2-1-A06.r.1 | Hypothetical protein | 2.06 | 18.38 |
| 77-2-6-H01.r.2 | No hit | 11.18 | 16.76 |
| Contig 3 | No hit | 2.97 | 16.76 |
| 77-2-7-A10.r.5 | IMP dehydrogenas/GMP reductase e | 3.13 | 16.37 |
| Contig 14 | No hit | 7.54 | 16.00 |
| 77-2-32-E05.r.1 | No hit | 10.85 | 12.41 |
| Contig 18 | No hit | 8.52 | 12.13 |
| 77-2-41-E04.r.1 | No hit | 6.43 | 11.85 |
| 77-2-8-C05.r.1 | No hit | 4.19 | 11.31 |
| Contig 17 | No hit | 2.04 | 9.85 |
| 77-2-40-E03.r.1 | No hit | 2.22 | 8.98 |
| Contig 10 | Predicted esterase | 5.03 | 8.67 |
| Contig 24 | Alpha-soluble NSF attachment protein | 9.51 | 7.46 |
| 77-2-39-G04.r.1 | No hit | 6.84 | 7.46 |
| 77-2-24-F03.r.1 | Cytochrome b5 domain-containing protein like | 2.90 | 6.96 |
| Contig 22 | Hypothetical protein | 20.6 | 6.81 |
| 77-2-9-C12.r.1 | Surface antigen BspA, Bacteroides forsythus | 6.23 | 5.66 |
| 77-2-36-F03.r.2 | L-3-phosphoserine phosphatase | 5.00 | 5.53 |
| Contig 21 | No hit | 4.91 | 4.19 |
| 77-2-4-B06.r.1 | Possible elongation of very-long-chain fatty acid protein | 3.76 | 3.82 |
| 77-2-41-E09.r.1 | Arachidonate 15-lipoxygenase, second type | 2.26 | 3.25 |
| 77-2-19-F06.r.1 | Phosphate-repressible phosphate permease | 2.41 | 3.17 |
| 77-2-24-H06.r.1 | No hit | 2.60 | 3.17 |
| Contig 15 | Hypothetical protein | 12.07 | 2.76 |
| 77-2-4-G05.r.1 | No hit | 8.85 | 2.24 |
| 77-2-19-E12.r.1 | Small nuclear ribonucleoprotein polypeptide D2 | 2.74 | 2.19 |
| 77-2-8-B04.r.1 | 4 Fe-S ferridoxin | 2.40 | 2.19 |
| 77-2-7-B11.r.5 | Hypothetical protein | 2.06 | 2.05 |
| 77-2-3-B03.r.2 | Cysteine protease | −3.82 | −2.27 |
| 77-2-5-H09.r.3 | No hit | −2.87 | −5.26 |
| 77-2-10-C04.r.2 | No hit | −2.26 | −5.26 |
| 77-2-14-F02.r.1 | No hit | −2.64 | −6.25 |
| Contig 4 | Fucoxanthin chlorophyll a/c protein | −4.34 | −10.00 |
| Contig 23 | Ac1147 | −3.04 | −20.00 |
| 77-2-29-H11.r.1 | No hit | −5.76 | −25.00 |
| 77-2-30-E06.r.1 | No hit | −3.32 | −33.30 |
Transcript identification number based upon differential expression in calcifying B39 relative to noncalcifying E. huxleyi 1516 cultures grown in phosphate-replete media and their differential relative gene expression profiles in calcifying versus noncalcifying 1516 cultures as determined by microarray analysis.
The results of a translated query to the NCBI peptide database are shown (E value, <10−2).
FIG. 4.
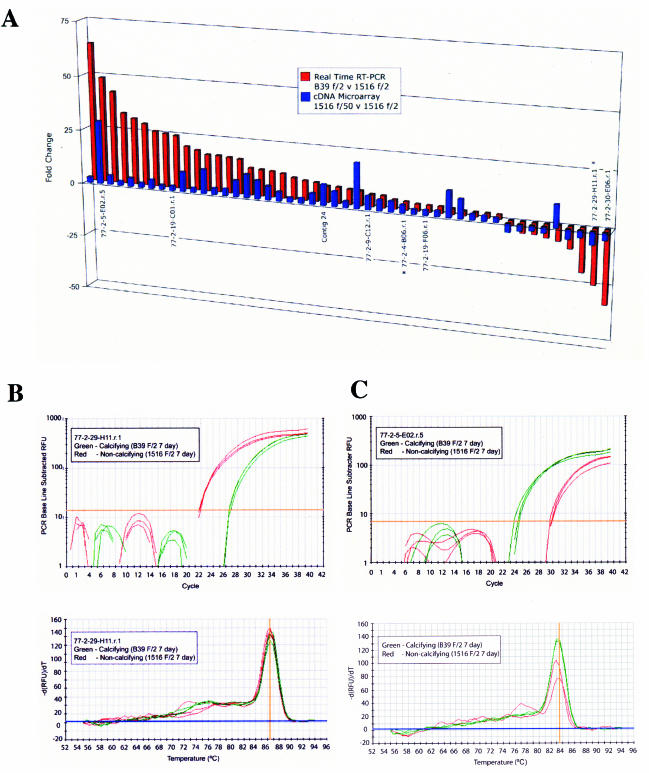
Real-time RT-PCR validation of cDNA microarray. (A) Comparison of relative expression (FC) for 20 randomly chosen transcripts in phosphate-limiting relative to phosphate-replete E. huxleyi cultures determined by cDNA microarray and real-time RT-PCR. Three transcripts that were not validated are indicated by asterisks. (B) Amplification plot and melt curve of a validated transcript (Contig 20), up-regulated in phosphate-limiting relative to phosphate-replete E. huxleyi cultures.
Calcification versus phosphate limitation.
Among the 82 differentially expressed sequences for which we could design working primer sets yielding a single real-time RT-PCR amplification product, 46 exhibited concordant microarray and real-time RT-PCR analysis results in terms of the direction of differential expression. These transcripts, of which 38 were significantly up-regulated and eight were significantly down-regulated, are likely to be involved in some aspect of calcification and coccolithogenesis in E. huxleyi (Table 3 and Fig. 5A). Figure 5C shows the real-time RT-PCR amplification plot and corresponding melt curve of a transcript whose function remains to be determined (77-2-2-E05) that yielded concordant up-regulation in calcifying cells over noncalcifying cells. This particular transcript was found to be up-regulated 28.2-fold in calcifying 1516 cells as determined by microarray analysis and was up-regulated 48.5-fold in calcifying B39 cells over noncalcifying 1516 cells by real-time RT-PCR analysis. The amplification plot and corresponding melt curve of a transcript exhibiting concordant down-regulation in calcifying cells (77-2-29-H11) is shown in Fig. 5B. While the function of 77-2-29-H11 also remains to be defined, it was found to be down-regulated 5.8-fold by microarray analysis and 25-fold by real-time RT-PCR analysis in calcifying cells.
FIG. 5.
Real-time RT-PCR relative gene expression for 63 transcripts significantly up- or down-regulated in calcifying B39 relative to noncalcifying E. huxleyi 1516 cultures grown in phosphate-replete media (red bars). The corresponding cDNA microarray FC data are included for comparison (blue bars). Shown are real-time RT-PCR amplification plots (A) and corresponding melt curves of candidate biomineralization transcripts showing the differential expression of transcripts that are highly down-regulated, 77-2-29-H11 (B), and highly up-regulated, 77-2-5-E02 (C), in calcifying cells. RFU, relative fluorescence units.
A total of 19 transcripts appear to be involved in phosphate limitation. These transcripts were found to be significantly up- or down-regulated by microarray analysis when comparing calcifying and noncalcifying 1516 cultures in phosphate-limited and phosphate-replete media, respectively, but were not “significantly” different in terms of a twofold threshold when comparing their expression in calcifying B39 and noncalcifying cultures of 1516 under identical phosphate-replete growth conditions. This suggests that the differential expression observed when comparing calcifying and noncalcifying cells under phosphate-limiting and phosphate-replete conditions, respectively, is a function of phosphate levels rather than calcification. Transcripts encoding transport proteins, predicted permeases, and proteins involved in intermediary metabolism were among those whose expression appears to be regulated to some degree by phosphate availability. The remaining 17 sequences exhibited discordant microarray and real-time RT-PCR results and most likely represent either false positives or strain-specific expression differences that have little to do with calcification. These transcripts are listed along with those likely to be involved in phosphate limitation in Table 4.
TABLE 4.
Transcripts most likely to be involved in phosphate limitation and/or microarray false positivesa
| Gene identification no. | Homolog descriptionb | FC by:
|
|
|---|---|---|---|
| Microarray for 1516 P− vs 1516 P+ | Real-time RT-PCR for B39 P+ vs 1516 P+ | ||
| 77-2-24-D02.r.1 | Sulfatase 1 precursor | −4.66 | 106.40 |
| 77-2-11-H04.r.3 | No hit | −2.02 | 23.16 |
| 77-2-24-E02.r.1 | No hit | −4.80 | 18.38 |
| 77-2-11-C12.r.5 | No hit | −2.14 | 18.38 |
| 77-2-12-E01.r.2 | No hit | −3.94 | 10.08 |
| Contig 25 | No hit | −4.58 | 7.29 |
| 77-2-12-F03.r.2 | Proteophosphoglycan | −2.80 | 6.50 |
| 77-2-11-A12.r.1 | No hit | −2.45 | 2.83 |
| 77-2-30-E07.r.1 | EH domain protein | −3.83 | 1.87 |
| 77-2-19-G10.r.1 | No hit | −2.27 | 1.59 |
| Contig 5 | Fucoxanthin-chlorophyll A-to-C binding protein F | −2.31 | 1.52 |
| 77-2-14-C05.r.1 | No hit | 2.39 | 1.35 |
| 77-2-24-E01.r.1 | No hit | −4.03 | 1.18 |
| 77-2-11-A10.r.5 | Similar to transmembrane serine protease 9 | −4.82 | 1.15 |
| 77-2-11-H03.r.2 | No hit | 4.88 | 1.07 |
| 77-2-30-G06.r.1 | Phosphoglycerate/bisphosphoglycerate mutase | −3.94 | 1.02 |
| 77-2-12-C02.r.1 | Adenylate cyclase, family 3 | 5.44 | 1.00 |
| 77-2-24-H07.r.1 | Peptidyl-prolyl cis-trans-isomerase | −4.08 | −1.05 |
| 77-2-24-F09.r.1 | No hit | −2.97 | −1.08 |
| 77-2-7-B09.r.4 | Proton/amino acid transporter | 2.57 | −1.12 |
| 77-2-16-A09.r.1 | No hit | −2.74 | −1.12 |
| Contig 12 | No hit | 4.77 | −1.15 |
| 77-2-7-A09.r.1 | Putative ammonium transporter | 2.28 | −1.23 |
| 77-2-12-F04.r.1 | Predicted permeases | −4.43 | −1.23 |
| 77-2-33-C08.r.1 | hypothetical protein | −2.13 | −1.28 |
| 77-2-31-F12.r.1 | No hit | 7.70 | |
| 77-2-8-A07.r.1. | Putative calcium binding protein | 5.57 | −1.96 |
| 77-2-8-C12.r.1 | Outer mitochondrial membrane protein porin | 4.65 | −2.50 |
| 77-2-40-B09.r.1 | No hit | 5.39 | −2.56 |
| 77-2-16-D04.r.1 | No hit | 3.76 | −2.78 |
| 77-2-8-E04.r.1 | No hit | 3.00 | −5.26 |
| Contig 9 | No hit | 10.43 | −7.14 |
| 77-2-7-D01.r.1 | No hit | 4.59 | −14.29 |
| 77-2-18-E09.r.1 | Asparaginyl endopeptidase | 2.49 | −14.29 |
| 77-2-41-F03.r.1 | Hypothetical protein | 9.93 | −20.00 |
| 77-2-36-H06.r1 | No hit | 13.53 | −25.00 |
Shown are results for transcripts that exhibited differential expression in calcifying 1516 cultures relative to noncalcifying 1516 E. huxleyi cultures grown in phosphate-limited media (P−) relative to noncalcifying cultures of 1516 grown in phosphate-replete media (P+). These transcripts also showed no differential expression or discordant relative expression in calcifying B39 cultures compared to noncalcifying 1516 cultures when grown in identical phosphate-replete media as determined by real-time PCR analysis.
The results of a translated query to the NCBI peptide database are shown (E value of <10−2).
In a recent study, Dyhram and colleagues (9) applied long SAGE to identify E. huxleyi transcripts responsive to nitrogen or phosphate starvation. Upon sequencing 11,578 tags, Dyhram and coinvestigators identified 74 unique or differentially expressed sequences from their phosphate-starved library. While 69 of our differentially expressed transcripts (59%) were represented by one or more of their sequence tags, only 8 of these sequences were identified as being uniquely present or up-regulated twofold or greater in their phosphate-starved library. Five of these gene sequences appear to be more likely associated with calcification as opposed to phosphate limitation or starvation. Transcripts identified in using both microarray and SAGE analysis included a phosphate-repressible phosphate permease, two different fucoxanthin chlorophyll a/c binding proteins, a probable 18S rRNA transcript, and four other sequences with no meaningful BLASTX hits. Of these transcripts, the phosphate-repressible permease, one of the fucoxanthin chlorophyll a/c binding proteins, and three transcripts of unknown function appear to be expressed in response to calcification rather than phosphate starvation. The similarities and differences noted when comparing SAGE and microarray analysis highlight the importance of employing complementary approaches to address and help resolve complex metabolic processes.
DISCUSSION
The power of microarrays coupled with real-time RT-PCR analysis as a tool for novel gene discovery in a non-model organism has been demonstrated in this study, wherein the gene expression profiles of E. huxleyi cells under calcifying and noncalcifying conditions have been compared. Fluorescently labeled cDNA was hybridized to an array of 2,298 putative unique sequences, and some 105 differentially expressed genes were subsequently analyzed by real-time RT-PCR analysis. Because some of these ESTs were very similar and are likely to represent the same gene, the number of unique sequences interrogated was less than 2,298. In addition, because the cDNA elements were obtained from a recombinant cDNA library originating from calcifying cells grown under phosphate-limiting conditions, the arrays are not representative of the entire transcriptome. Nevertheless, the data obtained provide an important and novel description of the expression of a large number of E. huxleyi genes.
Microarray validation.
The validity of the microarray results was substantiated by real-time RT-PCR analysis performed on a subset of 20 differentially expressed gene sequences and revealed a relatively low false-positive rate (15%). This suggested that the majority of the 127 differentially expressed clones identified in the microarray experiments were indeed associated with the presence/absence of calcification and/or the level of phosphate, in our experimental cultures. The integrity of the microarray data, moreover, is reflected in the putative function of several of the differentially expressed transcripts. The up-regulation of both a putative phosphate-repressible phosphate permease and the putative l-3-phosphoserine phosphatase evidenced herein represents a logical adaptive response to cope with growth under conditions of limited phosphate availability. The induction of these gene sequences in calcifying E. huxleyi cells is also consistent with the overwhelming body of literature linking phosphate limitation with the processes of calcification and coccolith formation in E. huxleyi (2, 9, 21, 29). The reliability of the microarray data is further substantiated by the increased expression of the gene transcript encoding the calcium binding protein in strain 1516 under calcifying conditions. This particular protein was originally isolated from an E. huxleyi cDNA library by screening with antibody raised to intracellular polysaccharide fractions isolated from cells harboring B-type coccoliths (7). Further immunolocalization of the protein (otherwise known as “GPA” because of its high constituent percentage of glutamic acid, proline, and alanine) revealed high concentrations on the surface of calcifying and noncalcifying cells. While its function has yet to be determined, researchers have hypothesized that it may in conjunction with coccolith polysaccharides be involved in either the nucleation and growth of CaCO3 crystals or the transport of Ca2+ from the cell exterior to the coccolith-forming vesicle.
Phosphate limitation versus calcification.
Our microarray analysis did not allow us to unambiguously differentiate between changes in gene expression due to biomineralization and changes due to phosphate limitation. The physiological relevance of phosphate limitation and the specificity of microarray results in narrowing the search for biomineralization transcripts were demonstrated by further analysis performed using real-time RT-PCR to compare differentially expressed transcripts in strain 1516 and strain B39. Strain B39 calcifies in phosphate-replete media, while strain 1516 does not, and hence comparisons of transcripts across strains were made under phosphate-replete conditions and results obtained were weighed against initial microarray results. A total of 46 differentially expressed transcripts identified by microarray analysis were also significantly differentially expressed when comparing abundance in strains 1516 and B39 under phosphate-replete conditions. These transcripts are most likely involved in biomineralization. Another 19 transcripts were differentially expressed in microarray experiments but showed no differential expression when transcripts were compared in strains 1516 and B39 under phosphate-replete conditions, and hence the logical conclusion is that these sequences are more likely to have a role in phosphate limitation. The 17 sequences exhibiting opposing relative expression levels may represent false positives or strain-specific differences in gene expression. Unfortunately, we were unable to probe 45 transcripts by real-time RT-PCR, either because suitable primer sets could not be designed or primers failed to elicit product during real-time RT-PCR analysis or yielded multiple amplification products. Suitable primer sets could not be designed for 20 of these sequences, and the other 25 either failed to elicit a signal when subjected to real-time RT-PCR analysis or yielded multiple amplification products. The high G+C content of E. huxleyi gene sequences makes primer design and the amplification of sequences using real-time RT-PCR problematic.
Calcification.
The transcripts identified herein as important to calcification and coccolithogenesis were dominated by sequences with no GenBank equivalents, and as expected, those whose function could be predicted were linked to a variety of cellular processes. Calcification and coccolithogenesis in coccolithophores occur intracellularly and as such are expected to involve proteins controlling the nucleation and growth of calcium carbonate crystals and crystalline templating as well as proteins responsible for carbon acquisition and ion transport, coccolithosome biogenesis, vesicle-mediated secretory processes, and protein sorting. Hence, the presence of transcripts showing significant homology to proteins such as carbonic anhydrase, the phosphate-repressible phosphate permease, an α-soluble N-ethylmaleimide-sensitive attachment protein (SNAP) among candidate sequences makes biological sense and is of particular interest.
A carbonic anhydrase identified in both our microarray and real-time RT-PCR screens was identified as a candidate biomineralization transcript. Carbonic anhydrase is in fact known to be associated with biomineralization in other organisms. While the protein plays an important role in many physiological processes, facilitating the transport of inorganic carbon and catalyzing carboxylation/decarboxylation reactions essential to photosynthesis and/or respiration, a gamma carbonic anhydrase from E. huxleyi was found to be linked to biomineralization in E. huxleyi by both microarray and real-time RT-PCR analysis. Microarray analysis showed the transcript to be up-regulated twofold in calcifying 1516 cells under phosphate-limiting conditions, while real-time RT-PCR results revealed up-regulation of 25-fold in calcifying B39 cells under phosphate-replete conditions as compared to the noncalcifying cells of strain 1516. Carbonic anhydrase-like proteins are involved in calcium carbonate biomineralization in mollusks (25) as well as avian eggshell formation (10). In the latter study, researchers found that carbonic anhydrase increases the rate of crystal growth in avian eggshells and works together with two sulfated extracellular matrix proteins. While its role in the process of calcification is hypothesized to be related to carbonate availability rather than nucleation or the regulation of crystal growth (25), its importance is underscored by the presence of functional domains from carbonic anhydrase that have been identified in the two prominent organic matrix proteins of the mollusk shell, Nacrein and N66 (14, 26). This gamma class carbonic anhydrase cDNA has been expressed in E. coli, and the recombinant protein has been purified to generate antisera for localization experiments (29a).
As described above, the up-regulation of the phosphate-repressible phosphate permease in calcifying cells of both strain 1516 and B39 is also in accord with much of the literature. Increases in both coccolith production and calcium content have been observed under phosphate limitation in batch and chemostat-maintained cultures of E. huxleyi (21, 29). According to the study by Shiraiwa (29), when nutrients such as phosphate and nitrate are adequate, photosynthesis and cell growth is enhanced, cell volume is decreased and calcification is suppressed. When nutrients are restricted, photosynthesis and cell growth are suppressed, resulting in increases in cell volume and calcification. Details describing the functional significance and mechanism governing enhanced coccolith production as a consequence of nutrient limitation and, in particular, phosphate depletion have not been elucidated. The overexpression of the phosphate-repressible phosphate permease in calcifying cells of strain B39 under phosphate-replete conditions, however, is somewhat puzzling but nonetheless intriguing. Perhaps the phosphate permease serves as a symporter driving the transport of ions critical to biomineralization. Alternatively, strain B39 may be genotypically aberrant in this regard.
The down-regulation of the GPA-encoding transcript seen in the present investigation when comparing by real-time RT-PCR analysis levels in calcifying cells of B39 relative to noncalcifying cells of 1516 was also unexpected and seems to contradict the notion that GPA plays a central role in biomineralization. While GPA may participate in some aspect of biomineralization in E. huxleyi, compelling evidence for its direct involvement in calcification has not been substantiated in this study. More detailed time-scale experiments comparing both transcript and protein levels in calcifying and noncalcifying cells may help to clarify this issue. It is important to note, however, that microarrays are only a first approximation of protein abundance and as such may not always accurately reflect the functional significance of particular proteins.
It is tempting to speculate on the role that some of the other candidate transcripts may play in biomineralization and coccolithogenesis. Of particular interest was the induction of a couple of genes that code for protein components of the vesicle trafficking machinery; a putative α-soluble N-ethylmaleimide-sensitive factor attachment protein (SNAP), and a RabGAP/TBC-containing protein. The fusion of vesicles in the secretory pathway in most eucaryotes is mediated by the interaction of t-soluble N-ethylmaleimide-sensitive factor attachment protein receptors (t-SNAREs) on the target membrane and v-SNAREs on the vesicle membrane (27, 28). While Rab proteins contribute to the specificity of vesicular transport controlling the rate of vesicle docking and matching v-SNAREs and t-SNARES for membrane fusion, SNAP and NSF disassemble the complex after fusion. In coccolithophores, coccolith formation is known to occur within a “calcifying vesicle” that is initially juxtaposed to the Golgi apparatus but later moves to cell membrane, where it fuses to extrude the completed coccolith (4). The enhanced level of expression shown here for putative SNAP and RabGAP transcripts in E. huxleyi cells under calcifying conditions when secretion of coccoliths is increased suggests SNAREs could potentially govern coccolith vesicle transfer and docking processes.
That the 29 transcripts that we have identified herein as being candidate biomineralization transcripts in E. huxleyi showed no significant homology to proteins in GenBank or showed hits to hypothetical proteins or proteins of unknown function is not surprising, given the proportionately small number of phytoplankton sequences in GenBank and the lack of knowledge about proteins involved in processes of biomineralization across species. Further complicating the situation is the fact that of the biomineralization proteins described so far from marine organisms, few show any sequence similarity to one another (16); prominent biomineralization domains and/or motifs have yet to emerge. Many of our candidate transcripts encode proteins that share some of the biochemical and biophysical properties that are characteristic of biomineralization proteins such as being small acidic proteins or glycoproteins that are elastomeric, rich in Asp and Glu, and contain repeat sequences (1, 11, 39), but these characteristics are not mutually exclusive. More detailed cellular localization studies coupled with in vitro mineralization assays or gene knockouts are required if we are to better define their role in biomineralization processes. We are particularly interested in one of the most prominent up-regulated transcripts, 77-2-5-E02.r.5, which was expressed at levels of 48-fold and 29-fold in calcifying relative to noncalcifying E. huxleyi cultures as determined by real-time RT-PCR analysis and microarray analysis, respectively (Table 4 and Fig. 5C). The longest open reading frame of this particular transcript encodes a protein of 168 amino acids in length and is estimated to be 20,315 Da. It is an alkaline (9.24 pI) protein and is rich in Ala (15.5%) and Gly (10%) residues, but has no obvious sequence motifs or domains. This particular transcript has also been successfully cloned and expressed in E. coli (data not shown) for more detailed cellular localization studies.
Three transcripts implicated in coccolith biomineralization may encode glycoproteins. Although full-length transcripts of 77-2-32-E05, 77-2-39-G04, and 77-2-14-G05 returned no significant BLASTX hits, individual open reading frames identified by ORFinder within these transcripts returned significant BLAST hits to glycoproteins. Many research scientists have focused their attention on the properties of polysaccharides from coccolithophores and their potential role in biomineralization and coccolithogenesis (15, 20). Indeed, acidic polysaccharides (APSs) have been isolated from calcifying strains of E. huxleyi, Gephyrocapsa oceanica, and Pleurochriysis caterae that cannot be recovered from noncalcifying strains (3, 19). Experiments in vitro further show the APSs have high affinity for calcium and can inhibit the deposition of calcium carbonate crystals in solution. Although the relationship between the ability of APSs to inhibit calcification in solution in vitro and the function of coccolith formation in vivo is unclear, Westbroek et al. (37a) suggest polysaccharides anchored to the inner membrane of the coccolithosome form a meshwork that fills the vesicle and governs the orientation and growth of the crystalline units once nucleation has been promoted. Thus, further characterization of the putative glycoprotein transcripts identified herein and determination of their cellular location may be warranted.
It is also unclear whether APSs, proteins, and/or both govern biomineralization processes in coccolithophores. To date most of the studies aimed at identifying macromolecules involved in coccolith formation have attempted to extract and purify compounds by dissolving coccoliths (15). While soluble components of E. huxleyi coccoliths have been analyzed in this manner, the insoluble organic fractions have not. In addition, the organic macromolecules that are responsible for tight control of crystal growth may be varied and present only in minute amounts. Biochemical extraction procedures are prejudiced and favor the recovery of only the most abundant macromolecular species: hence, the need for alternative techniques such as microarray analysis and real-time RT-PCR, which together enable gene transcripts to be interrogated in a comprehensive and quantitative manner.
Conclusions.
The results from this investigation represent preliminary information that will be invaluable in future functional genomics work aimed at defining the role of the many transcripts present on the array whose identity and function are unknown. For example, the existence of a large number of genes responsive to phosphate starvation, a subset of which appear to be biomineralization specific, suggests that E. huxleyi is endowed with a phosphate starvation regulon and possibly a biomineralization regulon as well. Genes specific to phosphate limitation and/or biomineralization can be further analyzed by studying the promoter regions. Gene sequences of this nature have not been previously available for this type of analysis. The data set described here also includes genes whose expression levels vary significantly. Bioinformatics analysis of several hundred such promoters may offer insights into cis-activation elements responsible for regulating the expression of genes involved in biomineralization. Promoters can be used to identify and clone corresponding trans-activating elements using yeast one-hybrid screening or other similar approaches.
Coccolithogenesis in E. huxleyi is a complicated process that is regulated both temporally and spatially. Our knowledge of the molecular and cellular underpinnings of this important biological process has been limited. However, tremendous efforts have been made to understand the molecular basis of biomineralization on a genome-wide scale, and significant progress has been made (9, 17, 35, 36). This microarray-based study represents an important first step in understanding the mechanisms governing biomineralization in coccolithophores and establishing microarray analysis methods for E. huxleyi. A specific set of differentially expressed genes under growth conditions of phosphate deficiency and enhanced calcification has been identified and further partitioned into subsets of transcripts: (i) those likely to be involved in phosphate limitation and (ii) those likely to be involved in biomineralization. While the putative function of the majority of candidate biomineralization transcripts we identified herein is unknown, this was not surprising given the low number of genes identified in other biomineralizing organisms. However, the data presented herein will allow for cellular localization and in vitro biomineralization analysis of some of these genes and will thus provide clues to key aspects governing coccolithogenesis in E. huxleyi.
Acknowledgments
This work has been supported by MBRS-SCORE grant GM 05983 from the National Institutes of Health.
REFERENCES
- 1.Addadi, L., and S. Weiner. 1985. Interactions between acidic proteins and crystals: stereochemical requirements in biomineralization. Proc. Natl. Acad. Sci. USA 82:4110-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, O. K. 1981. Coccolith formation and calcification in an N-cell culture of Emiliania huxleyi during phosphorus-limited growth in batch and chemostat cultures. Ph.D. thesis. University of Oslo, Oslo, Norway.
- 3.Borman, A. H., E. W. de Vrind de Jong, P. W. R. Thierry, and L. Bosch. 1987. Coccolith-associated polysaccharides from cells of Emiliania huxleyi (Haptophyceae). J. Phycol. 23:118-134. [Google Scholar]
- 4.Brownlee, C. W., and A. Taylor. 2004. Calcification in coccolithophores: a cellular perspective. Springer Verlag, Berlin, Germany.
- 5.Cleveland, H. C., J. Quackenbush, and A. Brazma. 2003. Microarray gene expression data analysis: a beginner's guide. Blackwell Science Ltd., Oxford, United Kingdom.
- 6.Corstjens, P. L. A. M., Y. Araki, and E. L. Gonzalez. 2001. A coccolithophorid calcifying vesicle with a vacuolar-type ATPase proton pump: cloning and immunolocalization of the V0 subunit c. J. Phycol. 37:71-78. [Google Scholar]
- 7.Corstjens, P. L. A. M., A. van der Kooij, C. Linschooten, G.-J. Brouwers, P. Westbroek, and E. W. de Vrind-de Jong. 1998. GPA, a calcium-binding protein in the coccolithophorid Emiliania huxleyi (Prymnesiophyceae). J. Phycol. 34:622-630. [Google Scholar]
- 8.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 9.Dyhrman, S. T., S. T. Haley, S. R. Birkeland, L. L. Wurch, M. J. Cipriano, and A. G. McArthur. 2006. Long serial analysis of gene expression for gene discovery and transcriptome profiling in the widespread marine coccolithophore Emiliania huxleyi. Appl. Environ. Microbiol. 72:252-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez, M. S., K. Passalacqua, J. I. Arias, and J. L. Arias. 2004. Partial biomimetic reconstruction of avian eggshell formation. J. Struct. Biol. 148:1-10. [DOI] [PubMed] [Google Scholar]
- 11.Gotliv, B.-A., L. Addadi, and S. Weiner. 2003. Mollusk shell acidic proteins: in search of individual functions. Chem. Bio. Chem. 4:522-529. [DOI] [PubMed] [Google Scholar]
- 12.Gracey, A. Y., J. V. Troll, and G. N. Somero. 2001. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc. Natl. Acad. Sci. USA 98:1993-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordon, R. W., L. Cros, and J. R. Young. 2004. A revised classification scheme for living haptophytes. Micropaleontology 50:55-79. [Google Scholar]
- 14.Kono, M., N. Hayashi, and T. Samata. 2000. Molecular mechanism of the nacreous layer formation in Pinctada maxima. Biochem. Biophys. Res. Commun. 269:213-218. [DOI] [PubMed] [Google Scholar]
- 15.Marsh, M. E. 2003. Regulation of CaCO3 formation in coccolithophores. Comp. Biochem. Physiol. 136:743-754. [DOI] [PubMed] [Google Scholar]
- 16.Nagasawa, H. 2004. Macromolecules in biominerals of aquatic organisms. Thalassas 20:15-24. [Google Scholar]
- 17.Nguyen, B., R. M. Bowers, T. M. Wahlund, and B. A. Read. 2005. Suppressive subtractive hybridization of and differences in gene expression content of calcifying and noncalcifying cultures of Emiliania huxleyi strain 1516. Appl. Environ. Microbiol. 71:2564-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto, O. K., and J. W. Hastings. 2003. Genome-wide analysis of redox-regulated genes in a dinoflagellate. Gene 4:73-81. [DOI] [PubMed] [Google Scholar]
- 19.Ozaki, N., S. Sakuda, and H. Nagasawa. 2001. Isolation and some characterization of an acidic polysaccharide with anti-calcification activity from coccoliths of a marine alga, Pleurochrysis carterae. Biosci. Biotechnol. Biochem. 65:2330-2333. [DOI] [PubMed] [Google Scholar]
- 20.Paasche, E. 2002. A review of the coccolithophorid Emiliania huxleyi (Prymnesiophyceae), with particular reference to growth, coccolith formation, and calcification-photosynthesis interactions. Phycol. Rev. 40:503-529. [Google Scholar]
- 21.Paasche, E. 1998. Roles of nitrogen and phosphorus in coccolith formation in Emiliania huxleyi (Prymnesiophyceae). Eur. J. Phycol. 33:33-42. [Google Scholar]
- 22.Peplies, J., S. C. Lau, J. Pernthaler, R. Amann, and F. O. Glockner. 2004. Application and validation of DNA microarrays for the 16S rRNA-based analysis of marine bacterioplankton. Environ. Microbiol. 6:638-645. [DOI] [PubMed] [Google Scholar]
- 23.Quinn, P. S., A. G. Saez, K-H. Baumann, B. A. Steel, C. Sprengel, and L. K. Medlin. 2004. Coccolithophorid biodiversity: evidence from the cosmopolitan species Calcidiscus leptoporus, p. 299-326. In H. R. Thierstein and J. R. Young (ed.), Coccolithophores: from molecular processes to global impact. Springer, Berlin, Germany.
- 24.Saez, A. G., I. Probert, M. Geisen, P. Quinn, and L. K. Medlin. 2003. Pseudo-cryptic speciation in coccolithophores. Proc. Natl. Acad. Sci. USA 100:7163-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samata, T. 2004. Recent advances in studies on nacreous layer biomineralization. Molecular and cellular aspects. Thalassas 20:25-44. [Google Scholar]
- 26.Samata, T., and M. Kono. 2000. Relationship between the structure of protein in shells and aragonite formation. Kaiyo Monthly 32:373-378. [Google Scholar]
- 27.Sanderfoot, A. A., F. F. Assaad, and N. V. Raikhel. 2000. The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 124:1558-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanderfoot, A. A., and N. V. Raikhel. 1999. The specificity of vesicle trafficking: coat proteins and SNAREs. Plant Cell 11:629-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiraiwa, Y. 2003. Physiological regulation of carbon fixation in the photosynthesis and calcification of coccolithophorids. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 136:775-783. [DOI] [PubMed] [Google Scholar]
- 29a.Soto, A. R., H. Zheng, D. Shoemaker, J. Rodriguez, B. A. Read, and T. M. Wahlund. 2006. Identification and preliminary characterization of two cDNAs encoding unique carbonic anhydrases from the marine alga Emiliania huxleyi. Appl. Environ. Microbiol. 72:5500-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strommer, J., R. Gregerson, and M. Vayda. 1993. Isolation and characterization of plant mRNA, p. 49-66. In B. R. Glick and J. E. Thompson (ed.), Methods in plant molecular biology and biotechnology. CRC Press, Boca Raton, Fla.
- 31.Stupp, S. I., and P. V. Braun. 1997. Molecular manipulation of microstructures: biomaterials, ceramics, semiconductors. Science 277:1242-1248. [DOI] [PubMed] [Google Scholar]
- 32.Sudarsanam, P., V. R. Iyer, V. R. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tricarico, C., P. Pinzani, S. Bianchi, M. Paglierani, V. Distante, M. Pazzagli, S. S. Bustin, and C. Orlando. 2002. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human biopsies. Anal. Biochem. 309:293-300. [DOI] [PubMed] [Google Scholar]
- 34.Tsoi, S. C. M., J. M. Cale, I. M. Bird, K. V. Ewart, L. L. Brown, and S. E. Douglas. 2003. Use of human cDNA arrays for the identification of differentially expressed genes in Atlantic salmon liver during Aeromonas salmonicida infection. Mar. Biotechnol. 5:545-554. [DOI] [PubMed] [Google Scholar]
- 35.Wahlund, T. M., A. R. Hadaegh, R. Clark, B. Nguyen, M. Fanelli, and B. A. Read. 2004. Analysis of expressed sequence tags from calcifying cells of the marine coccolithophorid, Emiliania huxleyi. Mar. Biotechnol. 6:278-290. [DOI] [PubMed] [Google Scholar]
- 36.Wahlund, T. M., X. Zhang, and B. A. Read. 2005. EST expression profiles from calcifying and non-calcifying cultures of Emiliania huxleyi. J. Micropaleontol. 51(Suppl. 1):145-155. [Google Scholar]
- 37.Weiner, S., and L. Addadi. 1997. Design strategies in mineralized biological materials. J. Mater. Chem. 7:689-702. [Google Scholar]
- 37a.Westbroek, P., E. W. de Jong, P. van der Wal, A. H. Borman, J. P. M. de Vrind, D. Kok, W. C. de Bruijin, and S. B. Parker. 1984. Mechanism of calcification in the marine alga Emiliania huxleyi. Philos. Trans. R. Soc. Lond. B 304:435-444. [Google Scholar]
- 38.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wustman, B. A., R. Santos, B. Zhang, and J. S. Evans. 2002. Identification of a “glycine-loop”-like coiled structure in the 34 AA Pro, Gly, Met repeat domain of the biomineral-associated protein, PM27. Biopolymers 65:362-372. [DOI] [PubMed] [Google Scholar]
- 40.Young, J. R., and K. Henriksen. 2003. Biomineralization within vesicles: the calcite of coccoliths, p 189-215. In P. M. Dove, J. J. De Yoreo, and S. Weiner (ed.), Biomineralization, vol. 54. Mineralogical Society of America, Washington, D.C.