Abstract
Lactic acid bacteria (LAB) are generally sensitive to H2O2, a compound that they can paradoxically produce themselves, as is the case for Lactobacillus bulgaricus. Lactobacillus plantarum ATCC 14431 is one of the very few LAB strains able to degrade H2O2 through the action of a nonheme, manganese-dependent catalase (hereafter called MnKat). The MnKat gene was expressed in three catalase-deficient LAB species: L. bulgaricus ATCC 11842, Lactobacillus casei BL23, and Lactococcus lactis MG1363. While the protein could be detected in all heterologous hosts, enzyme activity was observed only in L. casei. This is probably due to the differences in the Mn contents of the cells, which are reportedly similar in L. plantarum and L. casei but at least 10- and 100-fold lower in Lactococcus lactis and L. bulgaricus, respectively. The expression of the MnKat gene in L. casei conferred enhanced oxidative stress resistance, as measured by an increase in the survival rate after exposure to H2O2, and improved long-term survival in aerated cultures. In mixtures of L. casei producing MnKat and L. bulgaricus, L. casei can eliminate H2O2 from the culture medium, thereby protecting both L. casei and L. bulgaricus from its deleterious effects.
Lactobacillus bulgaricus is a representative of the group of lactic acid bacteria (LAB) that are widely used in dairy fermentations. It is one of the economically most important LAB, as its main application is found in the worldwide production of yogurt. Moreover, a new market for derived dairy fermentation products that often contain additional probiotic bacteria such as Lactobacillus casei is rapidly emerging.
In industrial processes, these bacteria can be exposed to oxidative stress when reactive oxygen species (ROS) with a high oxidizing potential accumulate in the cell. In addition to damage repair mechanisms, detoxification capacity is an important factor of resistance. Bacterial species are more or less sensitive to oxygen according to their enzymatic equipment to prevent or repair ROS damage. An analysis of the recently completed genome sequence of L. bulgaricus (24) confirmed previous observations suggesting that this bacterium, like the majority of LAB, is very poorly equipped with enzymes to detoxify oxygen-derived compounds. Under microaerobic conditions, it produces H2O2, which eventually causes growth arrest (15, 23). It does not contain catalase, peroxidase, or superoxide dismutase activity to eliminate H2O2 or other ROS. We therefore decided to investigate whether the addition of a gene coding for a catalase in L. bulgaricus or in its partner in mixed fermentations could improve its survival.
Two distinct families of catalases are known, differing in both the conformation of the folded protein and the nature of the catalytic redox cofactor, heme iron or nonheme manganese. In order to avoid the addition of heme (not synthesized by LAB) in the fermentation medium, a heme-independent pseudocatalase was chosen. Such a catalase has been purified from Lactobacillus plantarum ATCC 14431 (11), and the corresponding gene has been cloned (8). The enzyme, hereafter called MnKat, was shown to be a hexamer with two manganese ions per subunit (5) and is therefore also referred to as manganese catalase.
Intracellular Mn content varies greatly among LAB (4), where higher Mn concentrations have been implicated in oxidative stress resistance, acting as an O2− scavenger that could replace superoxide dismutase. L. plantarum ATCC 14431 is situated at the high end of the spectrum of intracellular Mn concentrations, with a reported value of 10.8 mM (4). L. bulgaricus ATCC 11842 is situated at the other extreme, with a reported Mn content of only 0.06 mM. As the concentration needed to support in vivo MnKat activity is not known, we decided to express the MnKat gene not only in L. bulgaricus but also in L. casei, where a value of 10.8 mM Mn was reported for strain YIT-00010 (4). For comparison, the gene was expressed in Lactococcus lactis, where an intermediate Mn content of 1.0 mM was measured for strain ML-3 (4). We show in a model system that L. casei that expresses active MnKat can serve as a tool to remove H2O2, protecting L. bulgaricus, its partner, in mixed dairy fermentations.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Plasmids were constructed using Escherichia coli as a host in the transformation and were subsequently transferred to lactobacilli or lactococci by electroporation using a Gene-Pulser apparatus (Bio-Rad) (13, 17, 20). A BamHI-SacI fragment containing the L. plantarum manganese catalase gene (hereafter called MnKat) under the control of its native transcription and translation signals was isolated from pMN115 and cloned into pLEM415 to produce pLEM415MnKat, which was subsequently used to transform L. bulgaricus Vi104 (20) and L. casei. pMN115 was linearized with BamHI and fused to pIL253 linearized with the same enzyme to obtain pVE3874, which was subsequently used to transform Lactococcus lactis and L. casei.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli TG1 | supE hsd Δ5 thi Δ(lac-proAB) F′ (traD36 proAB-lacZ ΔM15) | 7 |
| L. plantarum ATCC 14431 | Endogenous production of the manganese catalase MnKat | 11 |
| L. casei BL23 | 2 | |
| L. bulgaricus Vi104 | Derivative of strain ATCC 11842 | 20 |
| L. bulgaricus Vi1038 | Smr derivative of strain ATCC 11842 | M. van de Guchte, unpublished |
| Lactococcus lactis NZ9000 | Derivative of MG1363 carrying nisR/K genes on the chromosome | 12 |
| Plasmids | ||
| pMN115 | Amr; vector carrying the L. plantarum MnKat gene | 8 |
| pLEM415 | Emr | 6 |
| pLEM415mnkat | Emr; vector carrying the L. plantarum ATCC 14431 MnKat gene | This work |
| pIL253 | Emr | 21 |
| pVE3874 | Emr Amr; cointegrate between pIL253 and pMN115 | This work |
Media and growth conditions.
L. plantarum was grown in APT medium (Difco) at 37°C. L. casei and L. bulgaricus were grown in MRS medium (Difco) at 37°C or 42°C, respectively, or in 1% reconstituted skim milk (Compagnie laitière Food Service, Le Pecq, France). Lactococcus lactis subsp. cremoris was grown in M17 medium (Difco) containing 0.5% glucose (GM17) at 30°C. E. coli was grown in LB medium with stirring (200 rpm) at 37°C. When necessary, erythromycin (Em) was added at 5 μg/ml (for LAB) or 100 μg/ml (for E. coli), and ampicillin was added at 100 μg/ml. Aerated cultures (45 ml) were grown in 250-ml Erlenmeyer flasks with shaking (200 rpm for L. casei and 150 rpm for L. bulgaricus), and nonaerated cultures (10 ml) were grown in 25-ml closed tubes. CFU of LAB strains were counted as follows: appropriate sample dilutions were prepared in cold peptone (1 g/liter; Difco), plated onto GM17 (for Lactococcus lactis) or MRS medium (for lactobacilli), and incubated for 48 h at optimal temperature before counting was done. L. bulgaricus was incubated in airtight jars with GENbox (Biomérieux) to generate an anaerobic atmosphere. Kinetics of acidification and growth were determined by pH measurements and plating of L. bulgaricus milk cultures. These cultures were obtained after dilution (1/100 in milk) of a stationary-phase culture (grown for 24 h in MRS medium at 42°C) and were incubated at 39°C with or without stirring (150 rpm). After mixed incubations at 39°C in milk with H2O2, viable cells of each strain were counted after plating onto MRS medium with Em at 37°C for L. casei (pLEM415 or pLEM415MnKat) or onto MRS medium at 42°C under anaerobic conditions for L. bulgaricus.
DNA manipulations.
General molecular biology techniques were used as previously described (19). An incubation of 30 min at 37°C with lysozyme (10 mg/ml) in TES buffer (50 mM Tris-HCl, pH 8.0, 1 mM EDTA, 25% sucrose) was performed before plasmid extraction from LAB strains. Restriction enzymes were used as recommended by the suppliers. PCR amplifications were performed using Taq DNA polymerase (Invitrogen) and a thermocycler (Perkin-Elmer).
RNA preparation and reverse transcription-PCR.
RNA of LAB was purified from 25 ml of a fresh culture (optical density at 600 nm [OD600] of ∼0.6). After cell rupture (Fast Prep FP120; ThermoSavant), RNA was extracted with an aqua phenol (Appligène)-chloroform mix (1:1 [vol/vol]) and quantified. RNA extracts were treated with the DNA-free kit (Ambion) to remove all DNA. The total absence of DNA was checked by PCR using primers against the Em resistance gene of pAMβ1 (5′-GACAGTCATCTATTCAACTTA and 5′-GACGATATTCTCGATTGACC). cDNA of the MnKat gene was obtained with the primer 5′-GGAATACCACCCATTGCTTCTGGG using Moloney murine leukemia virus reverse transcriptase (Promega) as recommended by the supplier. cDNA of the MnKat gene was detected by PCR using the primers 5′-GTTGGGCGTCAACTGGTGCTG and 5′-GGAATACCACCCATTGCTTCTGGG.
Preparation of protein extracts and Western blotting.
Cellular protein extracts were prepared from exponentially growing (OD600 of ∼0.5) Lactococcus lactis cultures as previously described (14). For lactobacilli, the same procedure was applied, with minor modifications: cells were incubated for 1 h at 37°C in a buffer containing lysozyme (10 mg/ml) and mutanolysine (0.1 mg/ml) to obtain efficient lysis. The protein concentration of extracts was determined by the Bradford method (5a) with bovine serum albumin as the standard. Protein extracts (5 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) (16% acrylamide) and transferred onto a polyvinylidene difluoride membrane (Millipore) for immunodetection (19). Monoclonal antibodies against MnKat were used at 1:10,000 dilution. G-horseradish peroxidase conjugate (Bio-Rad) and a chemiluminescence kit (Perkin-Elmer) were used as recommended by the suppliers.
Preparation of protein extracts for nondenaturing polyacrylamide gel analysis.
Proteins from exponential-phase (OD600 of ∼0.5) or stationary-phase (24 h) L. plantarum and L. casei cultures were extracted without denaturation by mechanical disruption (using a Fast Prep FP120; ThermoSavant) in 1 ml of buffer (50 mM potassium phosphate-0.1 mM EDTA, pH 7.8). The protein concentration was assayed by the Bradford method (5a) with bovine serum albumin as the standard. Total protein extracts (5 μg) were separated by nondenaturing PAGE (10% acrylamide). Catalase activity was visualized using the method of Woodbury et al. (25). This method is based on the reduction (by H2O2) of potassium ferricyanide(III) to potassium ferrocyanide(II), which reacts with ferric chloride to form a stable green precipitate. The presence of catalase, which degrades H2O2 in protein extracts, becomes visible as an uncolored area.
Detection of catalase activity.
Cultures of LAB strains grown overnight were inoculated at a 1:50 dilution in fresh medium supplemented with erythromycin. Control strains harboring pLEM415 or pIL253 were used as negative controls. At an OD600 of ∼0.5, 2 ml of culture was centrifuged, and cells were resuspended in 30 μl of TES buffer. TES-resuspended cells (20 μl) were mixed with 10 μl of 8 M H2O2. The presence of catalase activity leads to bubble formation resulting from the transformation of H2O2 to H2O and O2. A quantitative assay of catalase activity was performed according to the method of Sinha (22). Briefly, exponentially growing cells (OD600 of ∼0.5) were concentrated 25-fold in phosphate buffer (0.1 M, pH 7), and total cellular proteins were extracted without denaturation by mechanical disruption (using a Fast Prep FP120; ThermoSavant). The protein concentration was assayed by the Bradford method (5a) with bovine serum albumin as the standard. Total protein extracts (0.1 μg) were mixed with 0.8 mM H2O2 in phosphate buffer. To measure the H2O2 concentration, every minute, an aliquot was mixed with 3 volumes of a solution of dichromate in acetic acid (1/3 dipotassium chromate [50 g/liter], 2/3 glacial acetic acid). The samples were boiled, and the OD570 was measured. The quantity of H2O2 was determined using a reference curve established with different H2O2 solutions (from 2 mM to 80 mM H2O2). Catalase activity is expressed as micromoles of H2O2 degraded per minute per milligram of protein.
Survival after short-term H2O2 exposure.
To test the level of H2O2 resistance of LAB strains producing or not producing MnKat, exponential-phase (OD600 of ∼0.5) or stationary-phase (10-fold dilution of culture grown overnight) cultures were incubated with 8 or 10 mM H2O2, respectively, for 1 h at the growth temperature. For assays performed using milk, cultures were grown in MRS medium until an OD600 of ∼0.5 was reached. Cells were then washed in cold peptone (1%), resuspended in a volume of milk equivalent to the volume of MRS medium removed, and incubated with H2O2. To test the H2O2 resistance in mixtures of L. casei and L. bulgaricus, each partner was prepared separately as described above and mixed in equal quantities in milk before the addition of H2O2. After 1 h, H2O2 was eliminated by the addition of bovine catalase (10 U/ml; Sigma), and viable bacterial counts were determined by plating onto MRS medium. Cells incubated for 1 h without H2O2 were used as a reference to calculate the survival of each strain.
Long-term survival of aerated cultures.
A culture grown overnight was diluted 1,000-fold in MRS medium. Aeration was performed using 250-ml Erlenmeyer flasks containing 45 ml of culture with stirring at 200 rpm. Viable cells were counted at 24-h intervals during 4 days.
RESULTS
Production and activity of manganese catalase in different LAB strains.
The gene encoding the manganese-dependent catalase (MnKat) of Lactobacillus plantarum ATCC 14431 was introduced into two lactobacilli, L. bulgaricus and L. casei, and into Lactococcus lactis. The heterologous expression of MnKat, under the control of its native transcription and translation signals, was examined at three levels: mRNA, protein, and enzyme activity. MnKat mRNA was detected after specific PCR, suggesting that transcription signals are recognized in both lactobacilli and Lactococcus lactis (data not shown). In these bacteria, Western blots using MnKat-specific antibodies revealed a single protein of the same size as the protein in L. plantarum ATCC 14431 (∼30 kDa) (Fig. 1). Catalase activity was checked by using three different methods. A first indication of the presence or absence of activity was obtained by the detection of gas (O2) formation upon the addition of H2O2. Activity was observed only in L. plantarum ATCC 14431 and in the MnKat-producing L. casei strain (Fig. 2A, lanes a and b). When MnKat-containing plasmids were isolated from L. bulgaricus or Lactococcus lactis and introduced into L. casei, the latter acquired catalase activity, showing that these plasmids encode an intact catalase (see Fig 2A, lane e, for plasmid from Lactococcus lactis; results are not shown for plasmid from L. bulgaricus). The absence of catalase activity in L. bulgaricus and Lactococcus lactis (Fig. 2A, lanes c and d) is therefore not due to mutations in the gene encoding the protein detected in Western blots. The addition of MnSO4 (50 mM) to L. bulgaricus or Lactococcus lactis cultures did not restore catalase activity (results not shown). For L. casei, specific catalase activity was then visualized by nondenaturing protein PAGE followed by activity staining (Fig. 2B). A signal was obtained for L. casei (pLEM415MnKat) at the same migration distance as that for L. plantarum ATCC 14431, while no signal was detected for L. casei (pLEM415) (Fig. 2B). Finally, a quantitative assay of catalase activity directly showed that L. casei acquired the capacity to degrade H2O2 upon the introduction of pLEM415MnKat. For the same amount of cellular proteins, the activity in the L. plantarum extracts was about four times higher than that in the L. casei extracts (Fig. 2C). This difference corresponds to the difference in the amount of enzyme produced in the two bacteria (Fig. 1), indicating that the specific activity of the enzyme (i.e., the activity per milligram of enzyme) is comparable in L. casei and L. plantarum.
FIG. 1.
Detection of MnKat by Western blotting. Proteins were extracted from exponentially growing LAB cultures expressing (+) or not expressing (−) MnKat, and total protein extracts (5 μg) were separated by sodium dodecyl sulfate-PAGE and revealed using MnKat-specific antibodies. (a) L. plantarum ATCC 14431; (b) L. casei BL23; (c) L. bulgaricus ATCC 11842; (d) Lactococcus lactis MG1363.
FIG. 2.
(A) Catalase activity in LAB strains expressing MnKat. H2O2 was added to bacterial cells resuspended in TES buffer. The presence of catalase activity leads to bubble formation resulting from the transformation of H2O2 to H2O and O2. Lanes: a, L. plantarum ATCC 14431 (wild type [wt]); b, L. casei (pLEM415 [−] or pLEM415MnKat [+]); c, L. bulgaricus (pLEM415 [−] or pLEM415MnKat [+]); d, Lactococcus lactis (pIL253 [−] or pVE3874 [+]); e, L. casei (pIL253 [−] or pVE3874 [+]). (B) MnKat activity staining in protein extracts separated by PAGE. Proteins from exponentially growing (E) or stationary-phase (S) cultures of L. plantarum and L. casei expressing (+) or not expressing (−) MnKat were extracted and separated by nondenaturing PAGE. Catalase activity was visualized using the method of Woodbury (22). Lanes: a, L. plantarum ATCC 14431; b, L. casei (pLEM415) (−); c, L. casei (pLEM415MnKat) (+). B.C., purified bovine catalase (Sigma); numbers indicate the numbers of units of enzyme. (C) Catalase activity in protein extracts. The degradation of H2O2 was measured in crude protein extracts. a, L. plantarum ATCC 14431; b, L. casei (pLEM415); c, L. casei (pLEM415MnKat). Catalase activity is calculated as micromoles H2O2 degraded/minute/milligram of protein. The results presented correspond to the averages of three different assays. Error bars correspond to the values of standard errors of the means (SEM).
Effect of MnKat on the physiology of L. casei.
The production of MnKat by L. casei had no incidence on its growth kinetics in standing MRS cultures (results not shown). Its effect under oxidative stress conditions was evaluated by oxidative stress induced by either H2O2 or aeration.
(i) Survival after short-term exposure to H2O2.
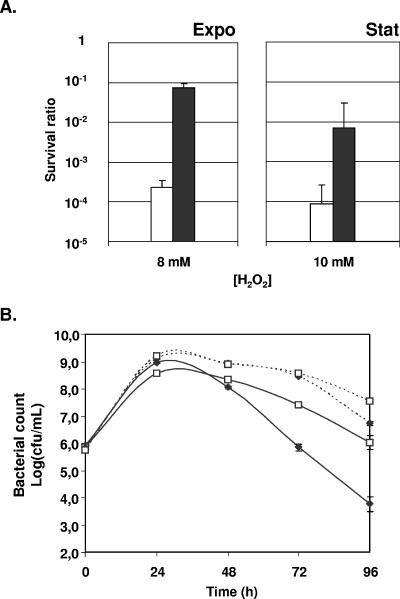
L. casei was incubated for 1 h in MRS medium in the presence of various H2O2 concentrations to determine discriminating conditions that would allow the evaluation of the effect of MnKat on the survival of exponentially growing cells or stationary-phase cells (data not shown). Exponentially growing cells were more sensitive to H2O2 than stationary-phase cells. The production of MnKat in L. casei conferred improved survival rates under these conditions: the survival of exponentially growing cells in response to 8 mM H2O2 was increased 320-fold, and the survival of stationary-phase cells in response to 10 mM H2O2 was increased 80-fold (Fig. 3A).
FIG. 3.
Survival of L. casei under oxidative stress conditions. (A) Survival after short-term exposure to H2O2. Survival after exposure to H2O2 (8 or 10 mM in exponential or stationary phase, respectively) is expressed as the ratio of viable cells measured in the presence over the count measured in the absence of added H2O2. Open bars, L. casei (pLEM415); closed bars, L. casei (pLEM415MnKat). (B) Long-term survival in aerated MRS cultures. Survival in aerated (plain lines) or nonaerated (dotted lines) stationary-phase cultures was determined by CFU counting at 24-h intervals. ⧫, L. casei (pLEM415); □, L. casei (pLEM415MnKat). The results presented correspond to the averages of three independent experiments. Error bars correspond to the SEM.
(ii) Aerated cultures.
As H2O2 is only one of the numerous ROS produced in aerated cultures, we evaluated the capacity of MnKat-producing L. casei isolates to grow and survive in stirred cultures. In MRS medium, aeration induced slower growth of L. casei and an earlier entry into stationary phase, leading to significant differences in the final population densities (∼2 × 109 CFU/ml and ∼4 × 108CFU/ml for nonaerated and aerated cultures, respectively). The production of MnKat had no effect on this difference (data not shown). The survival of aerated or nonaerated cultures of L. casei producing or not producing MnKat was determined by viable counts at 24-h intervals during stationary phase. The results show a clear effect of aeration on long-term survival. After 3 days in stationary phase, viable counts of L. casei (pLEM415) in aerated cultures were 800-fold lower than those found in nonaerated cultures (Fig. 3B). After 3 days in aerated cultures, survival of L. casei (pLEM415MnKat) was 160-fold higher than survival of L. casei (pLEM415) (Fig. 3B). The production of MnKat significantly reduced mortality in both nonaerated and, more importantly, aerated cultures.
Protection of L. bulgaricus from H2O2 by L. casei producing MnKat. (i) Behavior of L. bulgaricus in aerated milk cultures.
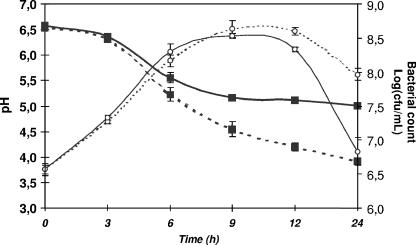
L. bulgaricus has been reported to produce H2O2, which eventually causes growth arrest under (micro)aerobic conditions in MRS medium (15, 23). To evaluate the impact of oxygen on L. bulgaricus in milk, the industrially important culture medium for these bacteria, the growth of aerated and nonaerated cultures were first compared. During the first 6 h, growth was similar for both cultures (Fig. 4). However, in the presence of oxygen, we observed (a) an earlier shift into stationary phase, leading to slightly lower maximal CFU counts, and (b) an important decrease in the survival rates after 24 h (6 × 106 CFU/ml versus 8 × 107 CFU/ml for aerated and nonaerated cultures, respectively). Furthermore, acidification of aerated milk (Fig. 4) was significantly slower (acidification rate of 0.36 ± 0.05 and 0.27 ± 0.02 pH units · h−1 for nonaerated and aerated cultures, respectively), and the final pH was higher (pH 5 versus pH 4 for aerated and nonaerated cultures, respectively). These results clearly illustrate the toxic effects of aeration on L. bulgaricus in milk cultures that notably affect acidification and survival.
FIG. 4.
Impact of aeration on growth and acidification kinetics of L. bulgaricus in milk. After inoculation (1/100) with a stationary-phase culture of L. bulgaricus, milk was incubated at 39°C and aerated by stirring at 150 rpm (plain lines) or not aerated (dotted lines). Kinetics of acidification (▪) and growth (○) were determined by pH measurements and plating. The results presented correspond to the averages of three different assays. Error bars correspond to the SEM.
(ii) Survival of L. bulgaricus and L. casei after mixed incubation with H2O2 in milk.
The most probable cause of the above-mentioned effects of aeration in milk cultures is the formation of H2O2, similar to previous observations described for MRS cultures (15, 23). Therefore, to determine whether L. casei could improve the survival of L. bulgaricus through the production of catalase, we directly measured the survival of both lactobacilli in milk with known concentrations of added H2O2. Exponential-phase cells or stationary-phase cells of both species were coincubated in the presence of 8 or 10 mM H2O2, respectively. The beneficial effect of MnKat on L. casei was significant in both cases: the survival of the MnKat-producing L. casei strain was 63-fold and 2,500-fold higher than that of the control strain for exponential- and stationary-phase cells, respectively (Fig. 5). The production of MnKat by L. casei markedly affected the survival of L. bulgaricus. When exponential-phase cells of the two species were mixed, L. bulgaricus survived 10-fold better in the presence of the catalase-producing L. casei strain than in the presence of the control strain. During coincubation of stationary-phase cells, the survival of L. bulgaricus reached 63% in the presence of the MnKat-producing L. casei strain, while in the presence of the control strain, only 0.1% of the L. bulgaricus cells survived (Fig. 5). These results clearly show that an MnKat-producing L. casei strain can efficiently protect bacteria in its environment from the deleterious effects of H2O2 via detoxification of the medium.
FIG. 5.
Survival of L. bulgaricus and L. casei MnKat in a mixture in milk after H2O2 exposure. L. bulgaricus and L. casei (pLEM415 [□] or pLEM415MnKat [▪]) were resuspended and mixed in milk (volume/volume) before the addition of 8 or 10 mM of H2O2 for cells in exponential phase (A) or stationary phase (B), respectively. Enumeration of each partner in mixtures was performed after 1 h of exposure. Survival after exposure to H2O2 is expressed relative to survival in the absence of added H2O2. The results presented correspond to the averages of three different assays. Error bars correspond to the SEM.
DISCUSSION
Dairy LAB are facultative anaerobic microorganisms that reduce pyruvate to lactate to regenerate NAD+. Oxygen is generally associated with toxicity in these bacteria that cannot use oxygen as a terminal electron acceptor. However, they are often exposed to oxygen during industrial fermentation, product handling, and storage. L. bulgaricus can eliminate oxygen in a reaction that produces H2O2, thereby preventing the formation of extremely damaging ROS like O2− (superoxide) and OH. (hydroxyl radical) (15). L. bulgaricus does not, however, produce catalase or peroxidase to eliminate H2O2, which is toxic itself. Therefore, we investigated how a heterologous catalase might be used to protect L. bulgaricus from the H2O2 it produces.
It is generally believed that LAB are not able to synthesize heme. Two previous studies reported the introduction of heme-dependent catalases in lactobacilli (10, 16). Both cases concerned species that are used for the fermentation of meat, where heme is amply available. So far, no LAB that are able to synthesize heme themselves have been identified, and the use of this type of catalase would thus be limited to environments where heme is naturally available or is added for this purpose (18). As the addition of heme to industrial-scale dairy fermentations may not be feasible or desirable, we present here the potential of MnKat, a heme-independent catalase. MnKat is the only manganese-dependent catalase isolated from LAB. This enzyme disrupts H2O2 using Mn as a catalytic redox cofactor, thereby avoiding the need for heme. The subunits of the homohexameric MnKat are smaller than those of regular homotetrameric catalases (27 and ∼60 kDa, respectively), and the enzyme is more resistant to elevated temperatures (11).
The MnKat gene, encoding a nonheme manganese-dependent catalase from L. plantarum, was successfully expressed in L. casei, L. bulgaricus, and Lactococcus lactis under the control of its native expression signals. Enzyme activity was detected only in L. casei. The possibility that the absence of activity in L. bulgaricus and Lactococcus lactis was due to mutations in the gene encoding the protein was excluded by the introduction of the corresponding plasmids isolated from these bacteria into L. casei, to which they conferred a catalase-positive phenotype. The quantities of protein produced, although lower than that produced in its original host, were comparable in the three bacteria. Therefore, the most likely explanation for the observed absence of enzyme activity can be found in the Mn content of the different bacteria, reportedly similar in L. casei and L. plantarum but at least 10- and 100-fold lower in Lactococcus lactis and L. bulgaricus, respectively. The scarce information available in the literature gives us no reason to believe that within a bacterial species, the Mn content would be strain dependent (4), and it therefore does not seem unreasonable to extrapolate these data to the strains used in the present study.
As observed in several pathogenic bacteria (9), in LAB, the cellular Mn content is tightly regulated to maintain a constant level that can highly differ between species (4). In L. plantarum, a high intracellular Mn concentration was shown to be maintained by active transport, independent of the extracellular Mn concentration (3). This tight regulation of the intracellular Mn concentration would explain why catalase activity in L. bulgaricus and Lactococcus lactis could not be restored by the addition of MnSO4 to the growth medium. In vitro, the extent of the Mn dependency of the catalase has not been established, as even prolonged dialysis was not sufficient to dislodge the native Mn ions from the purified protein (11). Our results may therefore be interpreted as a first, indirect, appreciation of the Mn concentration required to support activity of this enzyme.
Expression of the MnKat gene in L. casei conferred a marked increase in the survival rate after exposure of exponentially growing as well as stationary-phase cells to H2O2. In addition, an important improvement in long-term survival was observed in both aerated and, to a lesser extent, nonaerated cultures, while exponential growth under these conditions was not significantly affected. Taken together, these results indicate that (i) L. casei produces H2O2 in amounts that have no detectable effect on exponentially growing cultures and (ii) deleterious effects observed in stationary phase could be due to either the amounts of H2O2 reached or the accumulation of H2O2 damage.
We then investigated whether L. casei (pLEM415MnKat) could protect L. bulgaricus from H2O2 in a model system in milk. In a mixture of stationary-phase cells of L. bulgaricus and L. casei (pLEM415MnKat), 63% of the L. bulgaricus cells survived a 1-h exposure to 10 mM H2O2, compared to only 0.1% when the mixture contained L. casei without catalase. In these mixtures, L. casei (pLEM415MnKat) survived 2,500-fold better than L. casei without catalase (10% and 0.004%, respectively). The major requirement for probiotic products is the delivery of sufficient numbers of live bacteria. To meet this criterion while prolonging shelf life, bacterial survival in the products is of high importance. Our results should open the way to improve these products by introducing antioxidative LAB strains.
Although heterologous catalases have been expressed in several other lactobacilli and Lactococcus lactis (1, 10, 16, 18), this work constitutes the first report of heterologous expression of a nonheme catalase in bacteria relevant to dairy industries, offering the advantage that no heme has to be added to the culture medium for enzyme activity. In addition, this is the first demonstration of the protection of a bacterial species from H2O2 damage by the production of a heterologous catalase by its partner in mixed fermentations.
It is important to note that MnKat is a cytoplasmic enzyme that is not secreted into the culture medium. Therefore, in this system, L. casei should be considered an efficient tool to remove H2O2 from the medium and thereby protect L. bulgaricus by lowering its H2O2 exposure. One could envisage the utilization of such a system for the production of starter cultures with improved viability from bacteria that produce H2O2 by growing the starter strain and the catalase producer in separate compartments of a membrane reactor.
Acknowledgments
Tatiana Rochat is the recipient of a MENRT grant from the French government.
We thank T. Igarashi for the gift of plasmid pMN115, containing the cloned Mn-catalase gene from L. plantarum ATCC 14431. We thank J. W. Whittaker (Department of Environmental and Biomolecular Systems, OGI School of Science and Engineering at OHSU, Oregon) for monoclonal antibodies against L. plantarum MnKat.
REFERENCES
- 1.Abriouel, H., A. Herrmann, J. Starke, N. M. Yousif, A. Wijaya, B. Tauscher, W. Holzapfel, and C. M. Franz. 2004. Cloning and heterologous expression of hematin-dependent catalase produced by Lactobacillus plantarum CNRZ 1228. Appl. Environ Microbiol. 70:603-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acedo-Felix, E., and G. Perez-Martinez. 2003. Significant differences between Lactobacillus casei subsp. casei ATCC 393T and a commonly used plasmid-cured derivative revealed by a polyphasic study. Int. J. Syst. Evol. Microbiol. 53:67-75. [DOI] [PubMed] [Google Scholar]
- 3.Archibald, F. S., and M. N. Duong. 1984. Manganese acquisition by Lactobacillus plantarum. J. Bacteriol. 158:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archibald, F. S., and I. Fridovich. 1981. Manganese, superoxide dismutase, and oxygen tolerance in some lactic acid bacteria. J. Bacteriol. 146:928-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barynin, V. V., M. M. Whittaker, S. V. Antonyuk, V. S. Lamzin, P. M. Harrison, P. J. Artymiuk, and J. W. Whittaker. 2001. Crystal structure of manganese catalase from Lactobacillus plantarum. Structure (Cambridge) 9:725-738. [DOI] [PubMed] [Google Scholar]
- 5a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Fons, M., T. Hege, M. Ladire, P. Raibaud, R. Ducluzeau, and E. Maguin. 1997. Isolation and characterization of a plasmid from Lactobacillus fermentum conferring erythromycin resistance. Plasmid 37:199-203. [DOI] [PubMed] [Google Scholar]
- 7.Gibson, T. J. 1984. PhD thesis. University of Cambridge, Cambridge, England.
- 8.Igarashi, T., Y. Kono, and K. Tanaka. 1996. Molecular cloning of manganese catalase from Lactobacillus plantarum. J. Biol. Chem. 271:29521-29524. [DOI] [PubMed] [Google Scholar]
- 9.Kehres, D. G., and M. E. Maguire. 2003. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol. Rev. 27:263-290. [DOI] [PubMed] [Google Scholar]
- 10.Knauf, H. J., R. F. Vogel, and W. P. Hammes. 1992. Cloning, sequence, and phenotypic expression of katA, which encodes the catalase of Lactobacillus sake LTH677. Appl. Environ. Microbiol. 58:832-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kono, Y., and I. Fridovich. 1983. Isolation and characterization of the pseudocatalase of Lactobacillus plantarum. J. Biol. Chem. 258:6015-6019. [PubMed] [Google Scholar]
- 12.Kuipers, O. P., P. G. de Ruyters, M. Kleerezen, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 13.Langella, P., Y. Le Loir, S. D. Ehrlich, and A. Gruss. 1993. Efficient plasmid mobilization by pIP501 in Lactococcus lactis subsp. lactis. J. Bacteriol. 175:5806-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Loir, Y., A. Gruss, S. D. Ehrlich, and P. Langella. 1998. A nine-residue synthetic propeptide enhances secretion efficiency of heterologous proteins in Lactococcus lactis. J. Bacteriol. 180:1895-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marty-Teysset, C., F. de la Torre, and J. Garel. 2000. Increased production of hydrogen peroxide by Lactobacillus delbrueckii subsp. bulgaricus upon aeration: involvement of an NADH oxidase in oxidative stress. Appl. Environ. Microbiol. 66:262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noonpakdee, W., S. Sitthimonchai, S. Panyim, and S. Lertsiri. 2004. Expression of the catalase gene katA in starter culture Lactobacillus plantarum TISTR850 tolerates oxidative stress and reduces lipid oxidation in fermented meat product. Int. J. Food Microbiol. 95:127-135. [DOI] [PubMed] [Google Scholar]
- 17.Posno, M., R. J. Leer, N. van Luijk, M. J. F. van Giezen, P. T. H. M. Heuvelmans, B. C. Lokman, and P. H. Pouwels. 1991. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl. Environ. Microbiol. 57:1822-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rochat, T., A. Miyoshi, J. J. Gratadoux, P. Duwat, S. Sourice, V. Azevedo, and P. Langella. 2005. High-level resistance to oxidative stress in Lactococcus lactis conferred by Bacillus subtilis catalase KatE. Microbiology 151:3011-3018. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Serror, P., T. Sasaki, S. D. Ehrlich, and E. Maguin. 2002. Electrotransformation of Lactobacillus delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis with various plasmids. Appl. Environ. Microbiol. 68:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 22.Sinha, A. K. 1972. Colorimetric assay of catalase. Anal. Biochem. 47:389-394. [DOI] [PubMed] [Google Scholar]
- 23.van de Guchte, M., S. D. Ehrlich, and E. Maguin. 2001. Production of growth-inhibiting factors by Lactobacillus delbrueckii. J. Appl. Microbiol. 91:147-153. [DOI] [PubMed] [Google Scholar]
- 24.van de Guchte, M., S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J. M. Batto, T. Walunas, J. F. Gibrat, P. Bessieres, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA 103:9274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodbury, W., A. K. Spencer, and M. A. Stahman. 1971. An improved procedure using ferricyanide for detecting catalase isozymes. Anal. Biochem. 44:301-305. [DOI] [PubMed] [Google Scholar]