Abstract
Systems level investigation of genomic scale information requires the development of truly integrated databases dealing with heterogeneous data, which can be queried for simple properties of genes or other database objects as well as for complex network level properties, for the analysis and modelling of complex biological processes. Towards that goal, we recently constructed PathSys, a data integration platform for systems biology, which provides dynamic integration over a diverse set of databases [Baitaluk et al. (2006) BMC Bioinformatics 7, 55]. Here we describe a server, BiologicalNetworks, which provides visualization, analysis services and an information management framework over PathSys. The server allows easy retrieval, construction and visualization of complex biological networks, including genome-scale integrated networks of protein–protein, protein–DNA and genetic interactions. Most importantly, BiologicalNetworks addresses the need for systematic presentation and analysis of high-throughput expression data by mapping and analysis of expression profiles of genes or proteins simultaneously on to regulatory, metabolic and cellular networks. BiologicalNetworks Server is available at http://brak.sdsc.edu/pub/BiologicalNetworks.
INTRODUCTION
Networks of molecular interactions are widely studied to reveal the complex roles played by genes, gene products and the cellular environments in biological processes. In these networks (or graphs), the nodes represent genes or gene products and the edges represent specific interactions. In a protein–DNA network, an edge may represent the binding of a transcription factor to a promoter region, while in a protein–protein physical interaction network it might represent a recorded evidence of co-immunoprecipitation or a two-hybrid interaction. The nodes of the network are typically associated with additional information about the genes (or gene products), such as positions in the chromosome (or localization sites), or their Gene Ontology (GO) classification. A number of specialized and publicly accessible databases are available, which contain data about the nodes [the SGD database (1)] and the interactions [BIND database (2)], and some databases contain information about both [such as KEGG (3)]. In addition, individual researchers often publish their data as part of their publications or project web sites. Currently, a number of analysis and visualization tools have been developed by different groups for assimilating, visualizing and for the analysis and modeling of these molecular interaction network data, of which the most notable are Cytoscape (4), Osprey (5), PathwayAssist (6), Pathways Database System (7), GeneGO (www.genego.com), VisANT (8,9). Yet another direction of effort is the storage and analysis of large-scale gene and protein expression data (10–14). While some of these store and display expression data, they do not allow query or analysis of such data or integration of novel gene expression data with existing network models.
Ideally, an analysis tool for molecular interaction networks should enable a user to import, efficiently store, effectively retrieve and perform analysis on single genes, gene families, patterns of molecular interactions, as well as on the global structure of the network. The tool needs to be sufficiently flexible for both micro-scale and macro-scale analysis using heterogenous data, and extract the data from a large number of disparate databases; it should allow one to construct interaction networks by curation as well as computation [e.g. using algorithms that convert a time-series microarray dataset into an influence network (15)]; it should enable the users to retrieve different interaction graphs through on-demand queries and construct new graphs by assembling them in a variety of ways. It should also allow the incorporation of novel datasets locally, such as the user's own microarray expression data, and/or overlay these on biological networks to explore novel relationships among genes. These critical needs are the minimal requirements of a systems level analysis of biological pathways.
Previously we reported a general-purpose scalable warehouse of biological information, PathSys (16). PathSys is a comprehensive data warehouse resulting from the integration of molecular interaction data with other graph-structured data, such as ontologies, e.g. GO (17) and taxonomies, e.g. enzyme classification system and functional classification of yeast proteins (3), and state data such as gene expression profiles, from over 20 curated and publicly contributed data sources, biological experimental and PubMed data for the eight representative organisms (Saccharomyces cerevisiae Drosophila melanogaster, etc. for full list see website). It contains more than 100 000 events of regulation, interaction and modification among genes, proteins, cell processes and small molecules. Here we present BiologicalNetworks, the web-based query tool built on top of PathSys, and show how it enables a user to derive novel biological insights at the single gene level and functional relationships at the systems level.
Data integration
PathSys's data integration model achieves the following:
Integration of object and property types from over 20 databases (for a full list see the website), thus creating a controlled vocabulary (ontology) of object/attribute types.
Integration of nomenclature for genes/proteins. Naming conventions between different datasets can be different, and the server-side parser translates between standard nomenclatures and an automated reconciliation procedure assigns multiple names as synonyms to the same ORF.
Integration of different types of networks. BiologicalNetworks supports an arbitrary number of interaction types. Users can upload different types of interactions by specifying different evidence codes that are supported in the BiologicalNetworks (see software documentation for a full list of interaction types).
To illustrate the novelty and capabilities of BiologicalNetworks, in Table 1 we compare BiologicalNetworks against Cytoscape and VisANT. A limitation of Cytoscape and VisANT is that the query capability of these systems is exactly the same as the graph-data manipulation and filtering capability visible on the interface. Thus, the visual integration tools do not have the capacity to take any combination of operations in any order and yet have the system retrieve the specified data in a fashion that optimizes memory and disk operations. We have circumvented this limitation of visual integration by a database-level integration method using a query evaluation engine implementing a query algebra.
Table 1.
Comparison of BiologicalNetworks against Cytoscape and VisANT
| BiologicalNetworks | Cytoscape | VisANT | |
|---|---|---|---|
| Graph manipulation | Developed in house | Based on yFiles package graph engine | Developed in house |
| Project workspace | Project workspace; data sharing, through user/account/user privileges mechanism | Not available | Project workspace could be shared by e-mail |
| Data representation | Generic data model having three types of nodes (primary, connector and graph nodes representing modularity) and Node/Attribute types hierarchies | Ternary relations; no modularity | Ternary relations; modularity presented |
| Input | Local file, database load | .sif formatted file | Database load |
| Output | Local (tab delimited, xml, SBML, BN project) file; database edit/update; image printing | Local file; Image printing | .visML file |
| Data integration | Data integration engine performing data and property types integration, thus creating biological data and properties ontologies | GO database | SGD, KEGG, GO are integrated |
| Filtering | Filtering by any combination of Attribute/Node types from Attribute/Node type hierarchies | Flexible filters with different attributes of node and edge | Several ‘select’ filters available |
| Search | Analytical search tools; | Search node name on the graph | Search by keyword and node name on the graph |
| Keyword search; | |||
| Build/expend pathways; | |||
| Find direct interactions; | |||
| Find covering pathways (all shortest paths); | |||
| Find common targets/regulators; | |||
| Find intersections with curated pathways; | |||
| Network operations | Various layouts, Network intersection/union/subtraction, statistics, search for cycles, Networks comparison (Network BLAST) | Various layouts, several plug-ins for network operations available | Relaxing layout and statistical tool available |
| Microarray data | Import/Export microarray data; | Several plug-ins available | Not available |
| Expression patterns; | |||
| Clustering analysis (different clustering algorithms); | |||
| Visually display (static and dynamic time display) gene expressions on the pathways; | |||
| Building pathways from expression values; | |||
| Building correlation (e.g. Pearson correlation) networks; | |||
| Run GO terms overrepresentation analysis (Fisher's test) on expression clusters, networks or group of genes |
Data representation
Pathways are represented as a graph with three types of nodes. The nodes of the first type are reserved for genes, proteins, small molecules, cellular processes, etc. The nodes of the second type (controls) represent events of functional regulation, chemical reactions or protein–protein interactions; they can have physical meaning, may denote general associations; they can represent shared characteristics between components. The nodes of the third type represent complex objects, such as macromolecular complexes, functional groups, pathways etc. In this case components are made up of subcomponents, being compound or modular, and the connections between modular components (or modules) exist along with interconnections between their subcomponents. Interactions in BiologicalNetworks can also be defined as successively higher-level connections between groups of proteins, complexes, pathways or sub-networks.
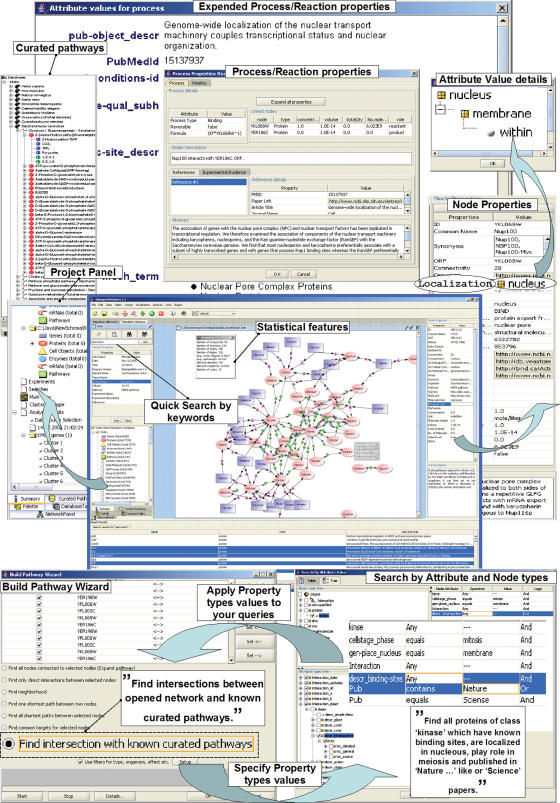
Most importantly, a classification scheme of Property Types representation (about 2000 unique Property Types and 25 000 nodes of the Property Types tree) allows BiologicalNetworks to represent detailed micro-level information. For example, a protein/process is not only localized in the ‘nucleus’, but it is represented as the starting point of finer subdivisions, such as ‘within the nuclear membrane’ and ending in ‘outer surface of the nuclear membrane’ as a ‘component’ or as simply ‘peripherally associated’ (Figure 1). Such a data model and integration environment align well with data representations of existing databases, such as BIND (2), KEGG (3), TransPath (18), eMaze (19) and significantly extends the concepts of other tools, such as Cytoscape and VisANT. To aid biological understanding, interaction networks and protein complexes can be viewed within the context of GO (17) annotations or KEGG (3) pathway assignments.
Figure 1.
BiologicalNetworks data representation and querying.
Interactions have not only information about the relevant literature, but also the experimental system used and a rich array of details on the evidence and classification of the biological properties. For example, a ‘genetic interaction’ between two genes may have information on the wild type/mutant forms, ‘phenotype’ (invasiveness, etc.), mutant ‘allele’, the number of gene copies, etc. Such a rich degree of annotation should play a vital role in understanding the nature of the interaction (see manual for description).
An important goal of systems biology is to generate dynamical models of molecular interaction networks (20). To enable this capability we have stored kinetic parameters as properties of reactions, reactants and products. This has been possible to achieve because of our representation of all three above objects as nodes in the interaction graph. This allows the user to represent the process graphs in SBML (21) format for dynamical simulation using a variety of computational methods.
To accompany BiologicalNetworks, we have developed a preliminary standard for exchanging files that have visual markup and annotation of network layouts. Users of BiologicalNetworks can input several basic data types, including data in standardized network and interaction data exchange formats, such as PSI-MI (22), BioPAX (http://www.biopax.org) and SBML (21).
Data analysis
Once a network dataset has been imported or loaded into BiologicalNetworks, the genes or proteins within it can be queried for other known and predicted interactions from the PathSys's database. Additionally, a repository of curated pathways (∼100 for S.cerevisiae and several hundreds from other organisms) is available for analysis. Imported interactions and components define a network ‘workspace’, which can be annotated and saved for sharing inside user groups working with BiologicalNetworks.
To enable data analysis, the following tools are available:
Search: find and display a list of objects based on a name or a keyword.
Expand: searches the database and displays objects functionally linked to a selected node or a set of nodes. Thus, by alternating expand and filtering options, users can browse through the database building their favorite pathways.
Build pathways: finds a set of links between two or more nodes by searching for the shortest path in the total network of all links in the database. This tool assists in finding regulatory paths between all selected objects.
Find common targets/regulators: searches for common targets or regulators for the group of molecules. This tool as well as Build Pathway can find functional links between proteins in the lists imported from other programs (e.g. gene expression clusters).
Find intersection with curated pathways: searches a group of nodes for other known and predicted interactions from the PathSys's repository of curated pathways.
BiologicalNetworks provides an advanced querying facility for retrieving the data of user's interest by querying Nodes and Properties types. User friendly querying interface allows user to make query with any logical combination of conditions both on Node and Property trees (Figure 1, see User's Tutorial for details).
Networks can also be analyzed for graph topological properties, such as degree distributions, path lengths, shortest paths or clustering coefficients.
Microarray data analysis
Expression data are easily imported through the Import Expression Data Wizard with a minimal amount of data preprocessing. BiologicalNetworks can interpret files of several types, including Tab Delimited (Stanford) Multiple Sample format, the Affymetrix file format, the TIGR format and GenePix file format.
The Expression Experiment Viewer is designed to display a graphical representation of processed gene expression data. It provides a workspace and a suite of algorithms for data analysis, sorting and searching, clustering and normalization, etc. These allow the user the flexibility in creating meaningful views of the expression data.
Results of the clustering analysis are represented in the form of tables and heat maps, and graphically as expression graphs. These viewers appear as a subtree under the Analysis Result within the main Project Properties tree.
Functionalities available from Microarray submenu and Microarray Experiment Manager Menu bar, allows the user to:
Open an expression experiment in a form of a table and heat map;
Sort the experiment by a particular sample;
Expression data can be visually displayed on an existing pathway diagram by showing different shades of green/red depending on the fold change of expression;
Build pathways from expression values;
Build correlation networks (e.g. Pearson correlation);
Run GO terms overrepresentation analysis (Fisher's test) on expression clusters, networks or group of genes.
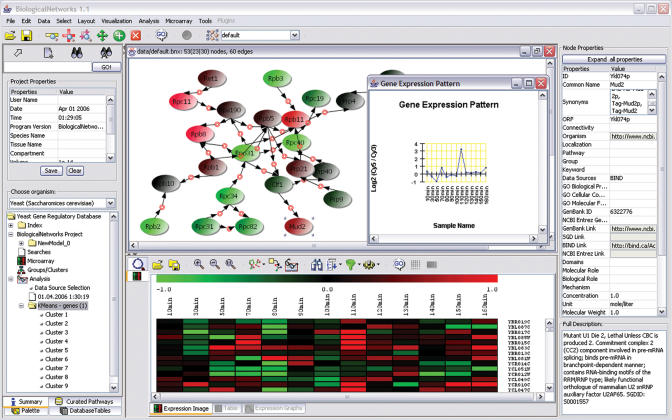
In Figure 2 a sample pathway incorporating expression data from a microarray experiment has been assembled. On the left panel is a hierarchical tree of analysis workspace, where different types of microarray data as well as analysis and associated results can be accessed.
Figure 2.
Microarray data analysis in BiologicalNetworks.
On the pathway diagram genes that are up-regulated in a particular experiment are shown in shades of red, while genes that are down-regulated are shown in shades of green; if no match is found the color gray is used. Using these data it is now possible to provide further annotations of edge property such as positive or negative regulation or to provide new edges between nodes.
In conclusion, the BiologicalNetworks web server allows a systems level analysis of genomic scale information as well as single object queries over a variety of databases for integrative views of biological function and for hypothesis generation.
Acknowledgments
Part of this work was funded by the National Science Foundation grants EIA-0205061 and EIA-0130059 to A.G. and A.R. The authors also thank the National Biomedical Computational Resource for support. Funding to pay the Open Access publication charges for this article was provided by NSF grant FIBR-0527023.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ball C.A., Jin H., Sherlock G., Weng S., Matese J., Andrada R., Binkley G., Dolinski K., Dwight S., Harris M., et al. Saccharomyces Genome Database provides tools to survey gene expression and functional analysis data. Nucleic Acid Res. 2001;29:80–81. doi: 10.1093/nar/29.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bader G., Betel D., Hogue C. BIND--The Biomolecular Interaction Network Database. Nucleic Acid Res. 2001;29:242–245. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acid Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitkreutz B.J., Stark C., Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikitin A., Egorov S., Daraselia N., Mazo I. Pathway Studio--the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- 7.Krishnamurthy L., Nadeau J., Ozsoyoglu G., Ozsoyoglu M., Schaeffer G., Tasan M., Xu W. Pathways database system: an integrated system for biological pathways. Bioinformatics. 2003;19:930–937. doi: 10.1093/bioinformatics/btg113. [DOI] [PubMed] [Google Scholar]
- 8.Hu Z., Mellor J., Wu J., DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z., Mellor J., Wu J., Yamada T., Holloway D., DeLisi C. VisANT: data-integrating visual framework for biological networks and modules. Nucleic Acid Res. 2005;33:W352–W357. doi: 10.1093/nar/gki431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mlecnik B., Scheideler M., Hackl H., Hartler J., Sanchez-Cabo F., Trajanoski Z. PathwayExplorer: web service for visualizing high-throughput expression data on biological pathways. Nucleic Acids Res. 2005;33:W633–W637. doi: 10.1093/nar/gki391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandu R., Guru R.K., Mount D.W. Pathway Miner: extracting gene association networks from molecular pathways for predicting the biological significance of gene expression microarray data. Bioinformatics. 2004;20:2156–2158. doi: 10.1093/bioinformatics/bth215. [DOI] [PubMed] [Google Scholar]
- 12.Chung H.J., Kim M., Park C.H., Kim J., Kim J.H. ArrayXPath: mapping and visualizing microarray gene-expression data with integrated biological pathway resources using Scalable Vector Graphics. Nucleic Acids Res. 2004;32:W460–W464. doi: 10.1093/nar/gkh476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goesmann A., Haubrock M., Meyer F., Kalinowski J., Giegerich R. PathFinder: reconstruction and dynamic visualization of metabolic pathways. Bioinformatics. 2002;18:124–129. doi: 10.1093/bioinformatics/18.1.124. [DOI] [PubMed] [Google Scholar]
- 14.Pan D., Sun N., Cheung K.H., Guan Z., Ma L., Holford M., Deng X., Zhao H. PathMAPA: a tool for displaying gene expression and performing statistical tests on metabolic pathways at multiple levels for Arabidopsis. BMC Bioinformatics. 2003;32:W460–W464. doi: 10.1186/1471-2105-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soinov A., Krestyaninova M., Brazma A. Towards reconstruction of gene networks from expression data by supervised learning. Genome Biol. 2003;4:R6. doi: 10.1186/gb-2003-4-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baitaluk M., Qian X., Godbole S., Raval A., Ray A., Gupta A. PathSys: integrating molecular interaction graphs for systems biology. BMC Bioinformatics. 2006;7:55. doi: 10.1186/1471-2105-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schacherer F., Choi C., Gotze U., Krull M., Pistor S., Wingender E. The TRANSPATH signal transduction database: a knowledge base on signal transduction networks. Bioinformatics. 2001;17:1053–1057. doi: 10.1093/bioinformatics/17.11.1053. [DOI] [PubMed] [Google Scholar]
- 19.van Helden J., Naim A., Lemer C., Mancuso R., Eldridge M., Wodak S.J. From molecular activities and processes to biological function. Brief Bioinform. 2001;2:81–93. doi: 10.1093/bib/2.1.81. [DOI] [PubMed] [Google Scholar]
- 20.Chen K.C., Calzone L., Csikasz-Nagy A., Cross F.R., Novak B., Tyson J.J. Integrative analysis of cell cycle control in budding yeast. Mol. Biol. Cell. 2004;15:3841–3862. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hucka M., Finney A., Sauro H.M., Bolouri H., Doyle J.C., Kitano H., Arkin A.P., Bornstein B.J., Bray D., Cornish-Bowden A., et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;9:24–31. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 22.Hermjakob H., Montecchi-Palazzi L., Bader G., Wojcik J., Salwinski L., Ceol A., Moore S., Orchard S., Sarkans U., von Mering C., et al. The HUPO PSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]