Abstract
Nucleosomes, a basic structural unit of eukaryotic chromatin, play a significant role in regulating gene expression. We have developed a web tool based on DNA sequences known from empirical and theoretical studies to influence DNA bending and flexibility, and to exclude nucleosomes. NXSensor (available at http://www.sfu.ca/~ibajic/NXSensor/) finds nucleosome exclusion sequences, evaluates their length and spacing, and computes an ‘accessibility score’ giving the proportion of base pairs likely to be nucleosome-free. Application of NXSensor to the promoter regions of housekeeping (HK) genes and those of tissue-specific (TS) genes revealed a significant difference between the two classes of gene, the former being significantly more open, on average, particularly near transcription start sites (TSSs). NXSensor should be a useful tool in assessing the likelihood of nucleosome formation in regions involved in gene regulation and other aspects of chromatin function.
INTRODUCTION
Transcription factors, enhancer-binding proteins and chromatin-remodeling factors all play a role in regulating gene expression. In addition, nucleosomes, the basic structural units of chromatin, are now thought to also be involved in this process [(1,2); see also (3,4)]. The dissociation or displacement of nucleosomes from DNA by the action of other protein factors, such as DNA- and histone-modifying enzymes (methylases, kinases, acetylases and deacetylases) likely gives the basal transcription apparatus access to the promoter regions of genes (5–7). A means for assessing the distribution of nucleosomes along stretches of regulatory DNA would therefore be a useful addition to our understanding of gene regulation.
In eukaryotic cells, nucleosomes are formed by the binding of DNA to histones (8). The nucleosome consists of 147 bp of DNA wound around a histone-octamer core (9–11). In human DNA, adjacent nucleosomes are separated on average by about 50 bp of ‘linker’ DNA, so that successive nucleosomes occur about every 200 bp. Most DNA sequences are neutral or favorable to nucleosome formation, but some sequences, by virtue of their influence on the curvature and flexibility of the DNA double helix (12,13), are unfavorable to nucleosome formation; we have called these ‘nucleosome exclusion sequences’ (NXSs). Thus, nucleosomes are not uniformly or randomly spaced along DNA, allowing for variation in access to promoter and other regulatory regions by the basal transcription apparatus, transcription factors and other proteins.
We have developed a program, NXSensor (available at http://www.sfu.ca/~ibajic/NXSensor/), that locates NXSs in DNA and defines regions where nucleosomes are unlikely to be formed because of the presence of such sequences and their spacing at distances less than the number of base pairs in the nucleosome. We have applied NXSensor to sets of promoter sequences whose genes are known to differ in their expression patterns among different tissues, and were able to show, in agreement with recent reports, significant differences among promoter regions in the number and position of such sequences.
BACKGROUND AND DESCRIPTION OF NXSensor
DNA sequences chosen as NXSs were the following, based on experimental observations (14–16):
Other studies lend support to the unfavorability of these sequences for nucleosome formation because of their hindering of DNA bending or flexibility (12,13,17).
A NXS was defined as one of the DNA sequences above, a contiguous non-overlapping DNA sequence long enough to comprise one full turn of the DNA double helix. The NXSensor program annotates individual DNA regions by marking NXSs and then examining their spacing to find sequences between NXSs that are <147 bp in length, on the assumption that nucleosomes are unlikely to form in such regions. The remaining segments of DNA are those where nucleosomes may be located.
VALIDATION AND APPLICATION
To test the general validity of the NXSs chosen, and to test the ability of the NXSensor program to find and annotate such sequences, the DNA sequences of positioned nucleosomes listed in the nucleosome positioning region database (NPRD) were examined (18). Ideally, in these nucleosome-associated DNA sequences there should be no NXSs as defined here. We found that only nine (8%) of the 112 positioned nucleosomes contained any NXSs. These sequences (a total of 12 NXSs) comprised 199 bp in a total of 16 829 nucleosome-associated base pairs (1.18%), which represents reasonable agreement with expectation. Exceptions in which a NXS is accomodated within a nucleosome may be the result of the presence of additional protein factors associated with nucleosomal DNA.
As a further test of NXSensor, the SV40 viral sequence was examined because it is known to contain a nucleosome-free segment of about 400–500 bp within its 5243 bp genome (19–21). The results showed a close correspondence between the experimentally-determined location of the nucleosome-free region and the nucleosome-free region predicted by NXSensor's analysis of DNA sequence (Supplementary Figure 1).
In a final comprehensive test, NXSensor was used to analyze the promoter regions of two sets of human genes, housekeeping (HK) and tissue-specific (TS) genes. These two sets of genes were chosen in order to assess the potential distribution of nucleosomes in promoters of genes that are used differently in different tissues, to substantiate the conclusions of other researchers who have recently investigated this question using different approaches, and to demonstrate further the usefulness of the NXSensor tool.
One hundred genes from each set, HK and TS, were selected on the basis of the tissue expression patterns given in the Novartis Research Foundation's Genomic Institute ‘SymAtlas’ (http://symatlas.gnf.org/) (22). The genes selected were at the two extremes of tissue expression patterns: HK genes showing significant expression in all 73 normal tissues of the SymAtlas, and TS genes showing, according to gene-specific non-cross-hybridizing probe sets (those with ‘_at’ suffixes), significant expression in only one or two tissues. To avoid possible bias in selecting HK genes, we included as wide a variety of basic cell functions as possible, and in selecting TS genes, as wide a variety of cell- and tissue types as possible. Genes with only one region of transcription initiation were used instead of those with widely-spaced alternative transcription start sites (TSSs), as indicated in the UCSC Genome Browser (http://genome.ucsc.edu/), and double-checked in the Database of Transcriptional Start Sites (DBTSS, http://dbtss.hgc.jp/). The list of genes is available at http://www.sfu.ca/~ibajic/NXSensor/200genes.xls.
To examine the region around the TSS of each gene, we used the sequence from 1000 bases upstream (which would include the complete promoter of most genes) to 1000 bases downstream from the TSS (−1000 to +1000), and called this 2000 bp stretch the ‘promoter region,’ centered on the TSS. The TSS was either the predominant site in, or a site near the middle of, the cluster of oligo-capped cDNA clones shown in the DBTSS (23). In cases where no oligo-capped cDNA clones were available from the DBTSS (21 of 100 HK genes, 36 of 100 TS genes), the RefSeq (NCBI) annotated TSSs were used. All sequences tested were datamined from the UCSC Genome Browser.
The basic ‘nucleosome segments’ option of NXSensor annotates the submitted sequence by highlighting in gray the segments available for nucleosome binding, leaving open the segments unlikely to be bound to nucleosomes by virtue of the number and positions of NXSs (Figure 1 and Supplementary Figure 1). The annotated promoter region sequences were used to construct two basic measures. The ‘NXScore’ defined as the number of NXSs in a window of 147 bp at a given position within the 2000 base promoter region, estimates the likelihood that nucleosomes are excluded from promoter regions at different distances from the TSS. This measure is the obverse of the ‘nucleosome formation potential’ based on nucleosome binding sequences used by Levitsky et al. (1,24). The NXScore is averaged over all promoter regions of each class of genes, HK and TS, to generate a graph of average NXScore versus position, centered on a given window (Figure 2).
Figure 1.
NXSensor output for two promoter regions, −1000 to +1000 relative to the major TSS, which is highlighted in green. Left column: HK gene RBPMS, RNA-binding protein; right column: TS gene F12, coagulation factor XII. Top, nucleosome-segment output: shaded regions show where nucleosomes are more likely to be located, unshaded regions are more likely to be nucleosome-free; bottom row, locations of NXSs.
Figure 2.
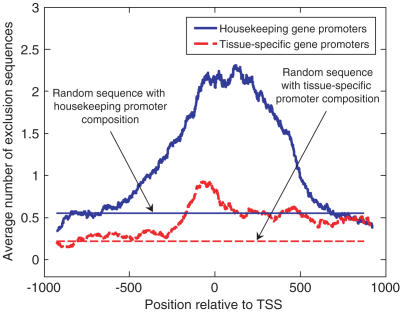
Average number of exclusion sequences per window of 147 bases in 100 HK (solid blue line) and 100 TS (dashed red line) promoter regions centered on the TSS. Also shown as thinner horizontal straight lines are the expected numbers of exclusion sequences in a random-sequence of the same base composition as that of the sum of the promoter regions in each class. The average number of observed exclusion sequences in a window centered at TSS is 2.10 for HK promoters, which is significantly higher (P = 0.0014) than the expected number of 0.53 in a random-sequence of the same composition. Similarly, for TS promoters, the average number of observed sequences in a window centered at TSS is 0.82, which is higher than the expected number of 0.22 in a random-sequence of the same composition, at the significance level of P = 0.06.
The second measure was the ‘accessibility score,’ measuring the overall accessibility of the 2000 base promoter region to protein factors. The accessibility score is defined as the proportion of base pairs in the region likely to be free of nucleosomes, according to the number and spacing of the NXSs. Accessibility scores can vary from 0.0 (all sequences potentially occupied by nucleosomes) to 1.0 (no sequences likely to be occupied by nucleosomes). In calculating the accessibility score ACC(10), short exclusion sequences of 10 bp or less flanked by two segments having the potential for nucleosome binding were not considered ‘open’ because of the unlikelihood that any protein would be able to bind effectively such a short sequence between two nucleosomes. Accessibility scores for two examples are given in Figure 1.
A direct comparison of the average number of NXSs, per window of 147 bases, in HK and TS promoter regions is shown in Figure 2. In both sets of promoter regions there were more NXSs nearer the TSS. There was a significant difference between HK-gene promoters and TS-gene promoters in the mean number of NXSs in the specific 147 bp window centered on the TSS (HK, 2.10 and TS, 0.82; P < 0.01, Kolmogorov–Smirnov test). Supplementary Figure 2 gives an illustrative example of 10 000 bases around the TSS of a HK gene, in which NXSensor analysis suggests that essentially only the TSS region is free of nucleosomes.
Figure 2 also shows that the mean number of NXSs in this window for the HK genes was significantly higher than that expected for random-sequence DNA with the same base composition as the HK-gene promoter regions (2.10 for HK genes, 0.53 for the corresponding random-sequence DNA, P = 0.0014). In contrast, the mean number of NXSs in this window for the TS genes was hardly different from the NXSs expected in random-sequence DNA having the TS promoter base composition (0.82 for TS genes, 0.22 for the corresponding random-sequence DNA, P = 0.06).
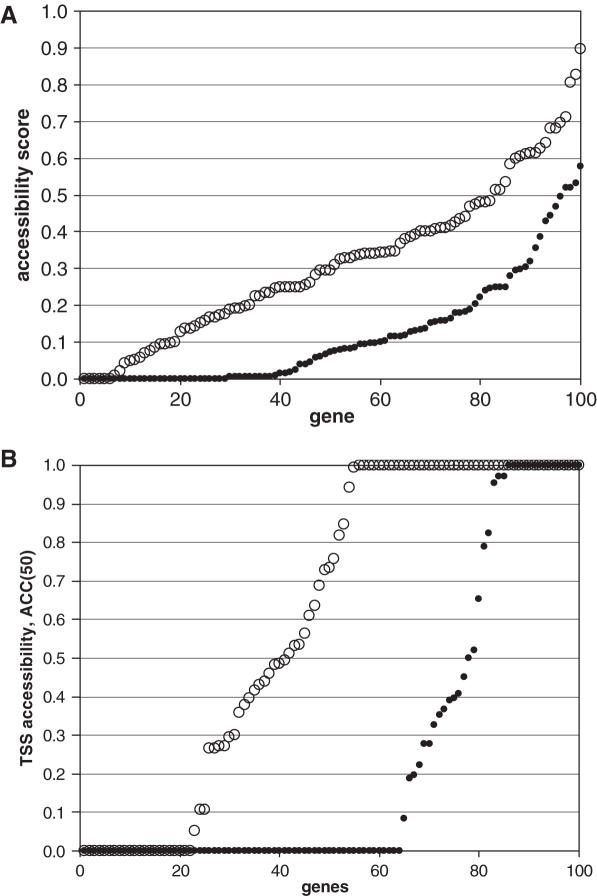
From NXSensor output we obtained accessibility scores, ACC(10), for the promoter regions in our dataset. As shown in Figure 3A, there was a significant difference between the distributions of accessibility scores of HK genes and of TS genes, the HK genes having significantly higher accessibility scores than the TS genes (P ≤ 0.01, Kolmogorov–Smirnov test).
Figure 3.
(A) Accessibility scores ACC(10) for promoter regions, −1000 to +1000 relative to the TSS. Open circles, HK-gene promoter regions; dots, TS-gene promoter regions. The gene promoter regions are ordered on the abscissa according to increasing accessibility score. (B) Accessibility scores ACC(50) in the vicinity of the TSS segment, −110 to +60 (accessibility to the basal transcription apparatus). Open circles, HK gene promoters; dots, TS-gene promoters. The gene promoters are ordered on the abscissa according to increasing accessibility score.
Further analysis was conducted using a modified accessibility score, ACC(50), which includes sequences at least 50 bp in length likely to be nucleosome-free. This is the length of sequence estimated to be occupied by the basal transcription apparatus (25,26). ACC(50) was examined for the DNA region from −110 to +60, the region in which the basal transcription apparatus would have to bind in order to initiate transcription at the majority of TSSs clustered around the modal or central TSS at +1 (27). The comparison of this TSS region (Figure 3B) showed that 58% of the HK genes, but only 22% of the TS genes, had an ACC(50) of 0.5 or higher. Of the HK genes, 45% had an ACC(50) of 1.0, while only 15% of the TS genes did. The overall distributions of HK gene and TS gene ACC(50) scores were significantly different by the Kolmogorov–Smirnov test (P ≤ 0.01).
DISCUSSION
Work in recent years has shown that the nucleosomes are neither uniformly nor randomly positioned along the DNA double helix. In a number of instances the location of nucleosomes in control regions of DNA was found to be an important feature of gene regulation. The validity of this statement was recognized explicitly by Levitsky et al. (1), and is the basis for the nucleosome positioning region database (NPRD), (http://srs6.bionet.nsc.ru/srs6/) (18). The ‘nucleosome formation potential’ scores have recently been used to demonstrate differences in the intergenic regions (28) and more specifically in the promoter regions (29) of human HK and TS genes.
We have taken a different approach by utilizing a combination of published experimental and observational data to identify DNA sequences unfavorable for nucleosome formation because of their influence on DNA bending and flexibility. Other sequences not used here may also be unfavorable for nucleosome formation. One example is TGGA repeats (30), but these occurred too infrequently in our sample of 200 promoter regions to have any influence on the results.
Using our NXSensor program (http://www.sfu.ca/~ibajic/NXSensor/), we have shown that, as expected, these NXSs are rare in DNA sequences occupied by nucleosomes listed in the NPRD. NXSensor also accurately predicts the location of the nucleosome-free zone in viral SV40 DNA. In a set of HK and TS promoter region DNA sequences, NXSensor analysis has demonstrated that the promoter regions of HK genes are likely to be relatively nucleosome-free, a finding that may help to explain the wide tissue distribution of their expression. This is in contrast to those of TS genes, whose promoter regions contain sequences more favorable for nucleosome formation (28,29), and which are therefore likely to require additional TS transcription factors to modify or displace nucleosomes before the genes can be expressed.
The promoter regions of the two sets of genes differed in base composition and in how many of them had CpG islands (31,32). The GC content of the promoter regions of HK genes was 56.86%, compared with 50.44% for TS genes. CpG islands were characteristic of 92 of 100 HK genes, but only 19 of 100 TS genes, in agreement with the observations of others (32). The greater frequency of NXSs of the [(G/C)3N2]≥3 type in HK-gene promoter regions compared with TS-gene promoter regions is partly a reflection of the higher GC content of the former (56.86% G + C in HK, 50.44% G + C in TS). However, the average number of [(G/C)3N2]3 sequences in HK-gene promoter regions, 15.4, was even higher than the 7.8 ± 2*2.7 (S.D.) expected in random sequences of the same length and base composition (P < 0.05). In addition, in spite of their higher GC content, HK-gene promoter regions had almost as many nucleosome-unfriendly polyA and polyT tracts as did the TS-gene promoter regions (50 in HK, as compared with 52 in TS). These two observations show that the frequency of both types of NXS in HK-gene promoters is higher than would be expected from their base composition, suggesting a functional significance, possibly related to nucleosome exclusion.
Promoter regions of HK genes are typically GC-rich. Presumably this is the consequence of selection pressure for certain kinds of promoter sequences. It may be speculated that one component of such selection is for sequences that tend to exclude nucleosomes. Another possible component is selection for GC-rich sequences that bind certain ubiquitous transcription factors, such as Sp-1. These two components of selection might be related, in that the factors that regulate expression of HK genes may have evolved to bind to nucleosome-free regions.
We were unable to find any correlation between accessibility score and expression levels of these genes as given in SymAtlas (22) (see Supplementary Figure 3). It is likely that the lower nucleosome occupancy of promoter regions of HK genes is related not to their expression levels but primarily to their ubiquity of expression in different tissues.
The NXSensor web tool is flexible enough to allow for different definitions of regions likely to be free of nucleosomes and more accessible to other protein factors. NXSensor can be set for more stringent exclusion criteria by increasing the number of tandem NXSs required for nucleosome exclusion. The minimum length of ‘nucleosome-free’ sequence can be increased to accommodate the space required for larger protein complexes, an approach we took to show that the region immediately surrounding TSSs of HK genes is likely to be more accessible to the basal transcription apparatus than is the corresponding region of TS genes.
Here we have applied NXSensor to the promoter regions of individual genes and classes of genes. NXSensor may also be used to investigate other control regions farther from coding sequences, such as enhancer and inhibitor regions, as well as sites of methylation, imprinting, recombination, repair, pre-mRNA splicing and indeed any DNA sequences where nucleosome location is likely to be a factor in overall chromatin organization and function (33–38).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Supplementary Material
Acknowledgments
The authors thank Steven Green (Biology Department, University of Miami) for statistical advice, John Walker and Andrew Su (Genomics Institute of the Novartis Research Foundation) for advice on the interpretation of SymAtlas gene expression data, Terace Fletcher (Department of Biochemistry and Molecular Biology, University of Miami) for helpful discussions, and anonymous reviewers for many cogent comments. Part of the work performed while I.V.B. was with the Electrical and Computer Engineering Department, University of Miami, Coral Gables, Florida 33124, USA. Funding to pay the Open Access publication charges for this article was provided by the Dr. John T. Macdonald Foundation Center for Medical Genetics.
Conflict of interest statement. None declared.
REFERENCES
- 1.Levitsky V.G., Podkolodnaya O.A., Kolchanov N.A., Podkolodny N.L. Nucleosome formation potential of eukaryotic DNA: calculation and promoters analysis. Bioinformatics. 2001;17:998–1010. doi: 10.1093/bioinformatics/17.11.998. [DOI] [PubMed] [Google Scholar]
- 2.Weaver R.F. Molecular Biology, 3rd edn. Boston: McGraw-Hill; 2005. pp. 342–423. Ch. 12, 13. [Google Scholar]
- 3.Gao J., Benyajati C. Specific local histone-DNA sequence contacts facilitate high-affinity, non-cooperative nucleosome binding of both adf-1 and GAGA factor. Nucleic Acids Res. 1998;26:5394–5401. doi: 10.1093/nar/26.23.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vicent G.P., Nacht A.S., Smith C.L., Peterson C.L., Dimitrov S., Beato M. DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol. Cell. 2004;16:439–452. doi: 10.1016/j.molcel.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 5.Schrem H., Klempnauer J., Borlak J. Liver-enriched transcription factors in liver function and development. Part I: the hepatocyte nuclear factor network and liver-specific gene expression. Pharmacol. Rev. 2002;54:129–158. doi: 10.1124/pr.54.1.129. [DOI] [PubMed] [Google Scholar]
- 6.Morse R.H. Getting into chromatin: how do transcription factors get past the histones? Biochem. Cell. Biol. 2003;81:101–112. doi: 10.1139/o03-039. [DOI] [PubMed] [Google Scholar]
- 7.Adkins M.W., Tyler J.K. Transcriptional activators are dispensable for transcription in the absence of spt6-mediated chromatin reassembly of promoter regions. Mol. Cell. 2006;21:405–416. doi: 10.1016/j.molcel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Chakravarthy S., Park Y.J., Chodaparambil J., Edayathumangalam R.S., Luger K. Structure and dynamic properties of nucleosome core particles. FEBS Lett. 2005;579:895–898. doi: 10.1016/j.febslet.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 9.Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. X-ray structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 10.Davey C.A., Sargent D.F., Luger K., Maeder A.W., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 11.Richmond T.J., Davey C.A. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 12.Drew H.R., McCall M.J., Calladine C.R. New approaches to DNA in the crystal and in solution. In: Cozzarelli N.R., Wang J.C., editors. DNA Topology and its Biological Effects. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. pp. 1–56. Monograph 20. [Google Scholar]
- 13.Travers A.A., Klug A. Bending of DNA in nucleoprotein complexes. In: Cozzarelli N.R., Wang J.C., editors. DNA Topology and its Biological Effects. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. pp. 57–106. Monograph 20. [Google Scholar]
- 14.Wang Y.H., Griffith J.D. The [(G/C)3NN]n motif: a common DNA repeat that excludes nucleosomes. Proc. Natl Acad. Sci. USA. 1996;93:8863–8867. doi: 10.1073/pnas.93.17.8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satchwell S.C., Drew H.R., Travers A.A. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 16.Suter B., Schnappauf G., Thoma F. Poly(dA.dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo. Nucleic Acids Res. 2000;28:4083–4089. doi: 10.1093/nar/28.21.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McConnell K.J., Beveridge D.L. Molecular dynamics simulations of B′-DNA: sequence effects on A-tract-induced bending and flexibility. J. Mol. Biol. 2001;314:23–40. doi: 10.1006/jmbi.2001.4926. [DOI] [PubMed] [Google Scholar]
- 18.Levitsky V.G., Katokhin A.V., Podkolodnaya O.A., Furman D.P., Kolchanov N.A. NPRD: Nucleosome Positioning Region Database. Nucleic Acids Res. 2005;33:D67–D70. doi: 10.1093/nar/gki049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson G.W., Hallick L.M. Mapping the in vivo arrangement of nucleosomes on simian virus 40 chromatin by the photoaddition of radioactive hydroxymethyltrimethylpsoralen. J. Virol. 1982;41:78–87. doi: 10.1128/jvi.41.1.78-87.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choder M., Bratosin S., Aloni Y. A direct analysis of transcribed minichromosomes: all transcribed SV40 minichromosomes have a nuclease-hypersensitive region within a nucleosome-free domain. EMBO J. 1984;3:2929–2936. doi: 10.1002/j.1460-2075.1984.tb02234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batson S.C., Rimsky S., Sundseth R., Hansen U. Association of nucleosome-free regions and basal transcription factors with in vivo-assembled chromatin templates active in vitro. Nucleic Acids Res. 1993;21:3459–3468. doi: 10.1093/nar/21.15.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki Y., Yamashita R., Sugano S., Nakai K. DBTSS, DataBase of Transcriptional Start Sites: progress report 2004. Nucleic Acids Res. 2004;32:D78–D81. doi: 10.1093/nar/gkh076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levitsky V.G., Katokhin A.V., Podkolodnaya O.A., Furman D.P. Nucleosomal DNA organization: an integrated information system. In: Kolchanov N., Hofestaedt R., editors. Bioinformatics of Genome Regulation and Structure. Kluwer Academic Publishers, Boston; 2004. pp. 3–12. [Google Scholar]
- 25.Roeder R.G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 26.Woychik N.A., Hampsey M. The RNA polymerase II machinery: structure illuminates function. Cell. 2002;108:453–463. doi: 10.1016/s0092-8674(02)00646-3. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki Y., Taira H., Tsunoda T., Mizushima-Sugano J., Sese J., Hata H., Ota T., Isogai T., Tanaka T., Morishita S., et al. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2001;2:388–393. doi: 10.1093/embo-reports/kve085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinogradov A.E. Noncoding DNA, isochores and gene expression: nucleosome formation potential. Nucleic Acids Res. 2005;33:559–563. doi: 10.1093/nar/gki184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganapathi M., Srivastava P., Kumar S., Sutar S.K.D., Kumar K., Dasgupta D., Singh G.P., Brahmachari V., Brahmachari S.K. Comparative analysis of chromatin landscape in regulatory regions of human housekeeping and tissue specific genes. BMC Bioinformatics. 2005;6:126. doi: 10.1186/1471-2105-6-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao H., Widlund H.R., Simonsson T., Kubista M. TGGA repeats impair nucleosome formation. J. Mol. Biol. 1998;281:253–260. doi: 10.1006/jmbi.1998.1925. [DOI] [PubMed] [Google Scholar]
- 31.Takai D., Jones P.A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antequera F. Structure, function and evolution of CpG island promoters. Cell. Mol. Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C., Yang T.P. Nucleosomes are translationally positioned on the active allele and rotationally positioned on the inactive allele of the HPRT promoter. Mol. Cell. Biol. 2001;21:7682–7695. doi: 10.1128/MCB.21.22.7682-7695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baumann M., Mamais A., McBlane F., Xiao H., Boyes J. Regulation of V(D)J recombination by nucleosome positioning at recombination signal sequences. EMBO J. 2003;22:5197–5207. doi: 10.1093/emboj/cdg487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powell N.G., Ferreiro J., Karabetsou N., Mellor J., Waters R. Transcription, nucleosome positioning and protein binding modulate nucleotide excision repair of the Saccharomyces cerevisiae MET17 promoter. DNA Repair. 2003;2:375–386. doi: 10.1016/s1568-7864(02)00239-2. [DOI] [PubMed] [Google Scholar]
- 36.Davey C., Fraser R., Smolle M., Simmen M.W., Allan J. Nucleosome positioning signals in the DNA sequence of the human and mouse H19 imprinting control regions. J. Mol. Biol. 2003;325:873–887. doi: 10.1016/s0022-2836(02)01340-2. [DOI] [PubMed] [Google Scholar]
- 37.Kogan S., Trifonov E.N. Gene splice sites correlate with nucleosome positions. Gene. 2005;352:57–62. doi: 10.1016/j.gene.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Pennings S., Allan J., Davey C.S. DNA methylation, nucleosome formation and positioning. Brief Funct. Genomic Proteomic. 2005;3:351–361. doi: 10.1093/bfgp/3.4.351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.