Abstract
Although the majority of the organisms use the same genetic code to translate DNA, several variants have been described in a wide range of organisms, both in nuclear and organellar systems, many of them corresponding to metazoan mitochondria. These variants are usually found by comparative sequence analyses, either conducted manually or with the computer. Basically, when a particular codon in a query-species is linked to positions for which a specific amino acid is consistently found in other species, then that particular codon is expected to translate as that specific amino acid. Importantly, and despite the simplicity of this approach, there are no available tools to help predicting the genetic code of an organism. We present here GenDecoder, a web server for the characterization and prediction of mitochondrial genetic codes in animals. The analysis of automatic predictions for 681 metazoans aimed us to study some properties of the comparative method, in particular, the relationship among sequence conservation, taxonomic sampling and reliability of assignments. Overall, the method is highly precise (99%), although highly divergent organisms such as platyhelminths are more problematic. The GenDecoder web server is freely available from http://darwin.uvigo.es/software/gendecoder.html.
INTRODUCTION
The genetic code of an organism provides the translation table between the languages in which DNA and proteins are coded by establishing a correspondence between each specific nucleotide triplet (codon) and each amino acid. A relevant property of the genetic code is that it is nearly universal, i.e. distantly related organisms such as Escherichia coli and humans share the same code. Rather than being random or accidental, the form of the genetic code has been shown to be related with stereochemical properties of amino acids and codons, minimization of mutation impact, and with biosynthetic relationships among the different amino acids [reviewed in (1)]. Interestingly, variants of the standard genetic code have been found in several nuclear and organellar systems, in a wide variety of organisms [reviewed in (2)]. Most of these variants, in which some codon has been reassigned to a different amino acid, are found in animal mitochondria, where 11 variants have been already described (3). Pressure towards small size of mitochondrial genomes, and hence towards reducing the total number of tRNAs, might be the cause for the high frequency of codon reassignments in mitochondria (4). At the same time, the small size of mitochondrial genomes makes the effects of codon reassignments less likely to be deleterious.
Genetic code variants are usually found by comparative sequence analyses. By inspecting a multiple alignment, when a codon of a given species appears at homologous positions where a particular amino acid is consistently found in other species, then the query codon is expected to translate as that particular amino acid. The strength of this simple approach depends on several factors. First, we should compare with the appropriate species, i.e. they should not be too distant. Second, to increase statistical power, we should have enough observations (number of appearances of a specific codon). Third, we want to make comparisons at homologous positions that are more or less conserved across species. Such comparative analyses have been applied before either manually (5,6) or with the computer (7), but we lack a bioinformatic tool that automates this process. Here we introduce a web server called GeneDecoder that allows for the automatic prediction of animal mitochondrial genetic codes.
GENDECODER
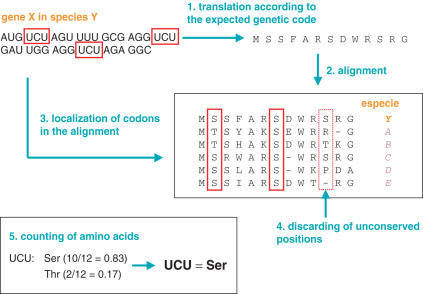
The way GenDecoder operates is depicted in Figure 1. It takes as input an animal mitochondrial genome (the query) and translates each of its 13 protein-coding genes according to the expected, but not necessarily true, translation table. These amino acid sequences are then aligned with a set of appropriate reference sequences for which the genetic code is known. At this stage, variable positions might be discarded according to some conservancy thresholds (see below). Subsequently, the positions at which each query codon appears in the multiple alignments are identified and the frequency of each amino acid at those positions is counted. Finally, each codon is assigned to the amino acid that most frequently appeared at homologous positions. GenDecoder uses the BioPerl library (8) to parse and retrieve mitochondrial genomes from GenBank (9). Sequence alignments are built using Clustalw (10) and inter-conversion between different sequence formats is carried out with ReadSeq (D. Gilbert, http://iubio.bio.indiana.edu/).
Figure 1.
Scheme of GenDecoder's workflow. The example is based on the UCU codon. A similar pipeline is executed for every other codon and using the whole set of 13 mitochondrial protein-coding genes.
Sequence conservation
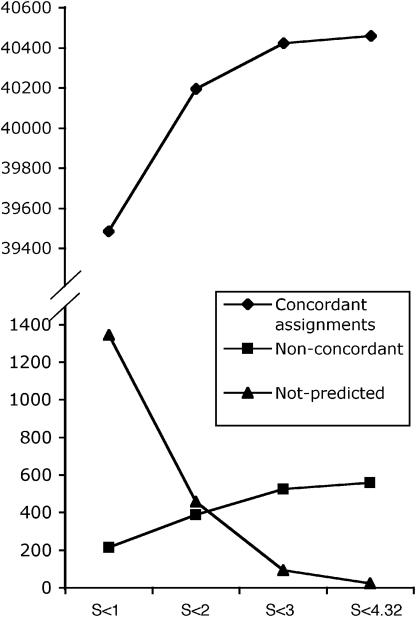
Multiple alignments allow determining to what extent each protein position is conserved. GenDecoder takes advantage of this information to filter out those positions that, because of their high variability, represent a source of noise. Different thresholds based on the percentage of gaps and the Shannon entropy can be selected in order to determine whether an alignment column is included in the analysis. Figure 2 shows the performance of GenDecoder for 681 metazoan species under four different entropy thresholds. By using restrictive thresholds the specificity of the method (fraction of codons successfully predicted) increases but, since fewer observations are available for each codon, there is a decrease in sensitivity (fraction of codons for which a prediction is made), especially for low-frequency codons. In general, GenDecoder is highly accurate (e.g. 99% at entropy threshold of 2).
Figure 2.
Performance of GenDecoder under different entropy thresholds and using the sampling-balanced alignments. The accuracy under different parameters for 41 042 codon assignments corresponding to 681 species is summarized in the graph. In every case columns with >20% of gaps were ignored. Comparison of this figure with the one appearing in (3) indicates that the use of taxonomically balanced alignments displaces the optimal point towards less restrictive entropy thresholds.
The effect of taxonomic sampling
Comparing the appropriate species is also important to obtain trustworthy predictions of the genetic code. If the species being predicted is evolutionary distant from the reference species, then less sites at their protein sequences will be conserved and consequently codon assignment predictions will be less reliable. In addition, if the taxonomic sampling is biased (i.e. species from some lineage are strongly overrepresented) predictions for poorly represented taxa might be less reliable. Our method minimizes these possible pitfalls by comparing query sequences against pre-established 54-taxa multiple alignments that consist of a balanced representation of each metazoan phylum, i.e. a dataset in which no particular phylum is overrepresented. Our subjective selection included 18 vertebrates and 36 invertebrates, comprising 15 arthropods, 5 molluscs, 3 nematodes, 3 platyhelminths, 3 cnidarians, 3 echinoderms, 3 cephalochordates, 1 annelid, 1 hemichordate and 1 branchiopoda. In addition to this metazoan-balanced dataset, two other datasets are available comprising 10 and 12 species of Platyhelminthes and Nematoda, respectively.
By assuming that assignments that were non-concordant with GenBank annotations are wrong [although this is not always true (3)] we were able to estimate the precision of the method for the different lineages of animals (Table 1). We found that prediction is worse for highly divergent lineages like platyhelminths and nematodes (see below). We also analysed the gain in precision that a balanced representation of metazoans provided over using highly biased multiple alignments containing all available metazoan mt-genomes. Results show that overall the performance of the method is better under a balanced representation of metazoan taxa (Table 1). Remarkably, just vertebrates can benefit from using sampling-biased alignments as reference alignments, because those biases are mainly related to the abundance of vertebrate mt-genomes in GenBank. On the other hand, the performance for platyhelminths and nematodes largely increases under a balanced taxa-representation but still a large number of non-concordant predictions (73 and 56, respectively) are obtained for these lineages. Importantly, if platyhelminths and nematodes are analysed using the Platyhelminthes and Nematoda reference datasets, the number of non-concordant assignments is significantly reduced (10 and 21, respectively). Most non-concordant predictions are related with codons appearing at very low frequency and/or codons for which the most frequent amino acid is scarce (data not shown).
Table 1.
Performance of GenDecoder and the importance of using an appropriate taxonomic sampling
| Number of species | 54-Taxa multiple alignments | All-metazoans multiple alignments | |||
|---|---|---|---|---|---|
| #Concordant/total | FP/TP (%) | Number of concordant/total | FP/TP (%) | ||
| Annelida | 4 | 244/247 | 1.2 | 244/248 | 1.6 |
| Arthropoda | 87 | 5116/5222 | 2.1 | 5048/5265 | 4.3 |
| Brachiopoda | 2 | 122/123 | 0.8 | 118/124 | 5.1 |
| Cephalochordata | 5 | 303/303 | 0.0 | 305/306 | 0.3 |
| Cnidaria | 4 | 246/248 | 0.8 | 242/248 | 2.5 |
| Echinodermata | 11 | 671/676 | 0.7 | 672/678 | 0.9 |
| Hemichordata | 1 | 60/60 | 0.0 | 60/60 | 0.0 |
| Mollusca | 15 | 911/924 | 1.4 | 895/926 | 3.5 |
| Nematoda | 12 | 634/690 | 8.8 | 600/703 | 17.2 |
| Platyhelmynthes | 10 | 525/598 | 13.9 | 475/601 | 26.5 |
| Porifera | 3 | 176/178 | 1.1 | 176/178 | 1.1 |
| Vertebrata | 461 | 27 288/27 375 | 0.3 | 27 547/27 498 | 0.2 |
Note: discrepancies in the number of assignments between the two experiments are related with the different behaviour that the conservancy threshold manifests with different alignments (e.g. there were 598 and 601 assignments for platyhelminths in the two experiments).
#Concordant/total, number of assignments concordant with GenBank/total number of assignments. Unassigned codons, i.e. codons that either are not used or do not appear at conserved positions (in this case entropy > 2.0; gaps > 20%), are not considered in this table.
#FP/TP, false-positive rate. Non-concordant/concordant assignments × 100.
WEB SERVER
Using GenDecoder's interface is straightforward. The user must provide an animal mt-genome either by uploading a GenBank formatted file or, if an entry is already available at the Genome section of GenBank, by indicating the corresponding NCBI TaxID for that species (e.g. 7227 for Drosophila melanogaster). Note that if a GenBank-formatted file is submitted, it must follow gene nomenclature standards (e.g. ND1, COX1 or CO1, ATP8). The thresholds used to define a column as ‘noisy’ might be left as default (columns with entropy higher than 2 or with >20% of gaps are ignored) in an initial analysis and then, they can be modified in order to investigate whether a given assignment is consistently predicted across different thresholds. The Metazoa dataset (default) is usually the best reference dataset, except for platyhelminths and nematodes.
Output
The output of GenDecoder provides detailed information about codon-usage, the frequency of the different amino acids associated with each codon, some statistics about the GC content at that species, and the final genetic code prediction (Figure 3). In addition, it offers the possibility of inspecting the corresponding alignments with JalView (11) as well as inspecting which alignment columns support each codon assignment.
Figure 3.
GenDecoder output for the acantocephalan L.thecatus. Codon-imp, number of codons at conserved positions (in this case S < 2.0, gaps < 20%); Codon-num, number of codons in the mt-genome; Freq-aa, first decimal in the frequency of the most frequent amino acid; Diff-freq, difference between the frequency of the predicted and expected amino acids (first decimal).
As a rule of thumb aimed to highlight potentially unreliable predictions in the output, assignments are indicated using lowercase when there are less than four codon observations or when the difference in the frequency of the most frequent amino acid is not sufficiently larger (0.25 different) than the frequency of the expected amino acid (if the predicted and expected amino acids differ from each other). When a codon is not present in an mt-proteome it is indicated by a dash symbol (‘-’). Similarly, if a codon is present but not at alignments columns for which the conservancy threshold holds, then its meaning is not predicted and such occurrence is reported using a question mark (‘?’).
A CASE STUDY
To illustrate how GenDecoder works, the analysis of the acantocephalan Leptorhynchoides thecatus (12) (taxonomic identifier 60532) is described below. The annotation of the genetic code for such a species illustrates the case in which a phylum is sampled for the first time, and potential reference species necessarily belong to different phyla. The result of GenDecoder (Figure 3) indicates that, apart from three predictions, the assignments for L.thecatus are concordant with the invertebrate genetic code, as already annotated in GenBank. The meaning of TGT/TGC codons is predicted as alanine instead of cysteine, and the ATC codon is predicted as leucine instead of isoleucine. The codon TGT appears 68 times in the mt-genome of L.thecatus, and 31 of these occur at alignment positions for which the default conservancy thresholds hold (S < 2.0, <20% gaps). At these 31 positions, alanine and cysteine occur with frequencies 0.21 and 0.12, respectively. The difference in favour of alanine is not large enough to trust this prediction. With respect to the TGC codon, its prediction as alanine is also likely wrong since it is based on just one codon occurrence. Interestingly, cysteine codons are sometimes badly predicted just because this amino acid is seldom used in proteins. The prediction of ATC as leucine instead of isoleucine is based on nine codon occurrences. Since both amino acids are highly similar, and the signal supporting the prediction of ATC as leucine is weak, the prediction is also considered unreliable. Hence we could conclude that L.thecatus mitochondrion has a conventional invertebrate genetic code.
CONCLUSIONS
The comparative approach for the prediction of the genetic code is simple but highly precise. Cases in which the method fails to correctly predict the genetic code are mostly related with taxonomic sampling biases or large evolutionary distances between the predicted and the reference species. We tried to minimize these problems by using a balanced representation of metazoans, as well as by using particular datasets for highly divergent phyla, i.e. for Platyhelminthes and Nematoda.
Recent results (3) suggest that as more animal mitochondrial genomes are sequenced, further new genetic codes are expected to appear, particularly at phyla that are not well sampled yet. Hence, we recommend that every new animal mt-genome be scanned with GenDecoder before its public release. Importantly, results of the method should be interpreted cautiously in order to distinguish between artefacts of the method and real codon reassignments. In this sense, most likely wrong assignments are related with cases in which the assigned codon appears at very low frequency and/or with cases in which the frequency of the most frequent amino acid is low and not very different than the frequency of the expected amino acid.
Even though many of the variant genetic codes occur in metazoan mitochondria, the systematic application of comparative methods to other systems will probably reveal that other variant genetic codes still wait to be unveiled. In this direction, methodologies such as the one presented here would be appropriate for any kind of organism/genome, but we recommend some prior investigation about taxonomic sampling before its application. Alternatively, the development of methods able to take into account sequence weights (13,14) and/or able to weight each amino acid observation at reference species by their evolutionary distance with respect to the query species (15,16) might help solving these questions. Such improvements will surely increase the precision of the method, but they will have the drawback of making interpretation of results less intuitive.
Acknowledgments
We would like to acknowledge the valuable contribution of two anonymous referees. This work was supported by a research grant from the Fundación BBVA (Spain). D.P. is also supported by the Ramón y Cajal programme of the Spanish Government. Funding to pay the Open Access publication charges for this article was provided by Fundación BBVA (Spain).
Conflict of interest statement. None declared.
REFERENCES
- 1.Di Giulio M. The origin of the genetic code: theories and their relationships, a review. Biosystems. 2005;80:175–184. doi: 10.1016/j.biosystems.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Knight R.D., Freeland S.J., Landweber L.F. Rewiring the keyboard: evolvability of the genetic code. Nature Rev. Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- 3.Abascal F., Posada D., Knight R.D., Zardoya R. Parallel evolution of the genetic code in arthropod mitochondrial genomes. PLoS Biol. 2006 doi: 10.1371/journal.pbio.0040127. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knight R.D., Landweber L.F., Yarus M. How mitochondria redefine the code. J. Mol. Evol. 2001;53:299–313. doi: 10.1007/s002390010220. [DOI] [PubMed] [Google Scholar]
- 5.Beagley C.T., Okimoto R., Wolstenholme D.R. The mitochondrial genome of the sea anemone Metridium senile (Cnidaria): introns, a paucity of tRNA genes, and a near-standard genetic code. Genetics. 1998;148:1091–1108. doi: 10.1093/genetics/148.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrell B.G., Bankier A.T., Drouin J. A different genetic code in human mitochondria. Nature. 1979;282:189–194. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- 7.Telford M.J., Herniou E.A., Russell R.B., Littlewood D.T. Changes in mitochondrial genetic codes as phylogenetic characters: two examples from the flatworms. Proc. Natl Acad. Sci. USA. 2000;97:11359–11364. doi: 10.1073/pnas.97.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stajich J.E., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank: update. Nucleic Acids Res. 2004;32:D23–D26. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clamp M., Cuff J., Searle S.M., Barton G.J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 12.Steinauer M.L., Nickol B.B., Broughton R., Orti G. First sequenced mitochondrial genome from the phylum Acanthocephala (Leptorhynchoides thecatus) and its phylogenetic position within Metazoa. J. Mol. Evol. 2005;60:706–715. doi: 10.1007/s00239-004-0159-8. [DOI] [PubMed] [Google Scholar]
- 13.Henikoff S., Henikoff J.G. Position-based sequence weights. J. Mol. Biol. 1994;243:574–578. doi: 10.1016/0022-2836(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 14.Valdar W.S. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 15.Lockhart P.J., Steel M.A., Hendy M.D., Penny D. Recovering evolutionary trees under a more realistic model of sequence evolution. Mol. Biol. Evol. 1994;11:605–612. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- 16.Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]