Abstract
CARMAweb (Comprehensive R-based Microarray Analysis web service) is a web application designed for the analysis of microarray data. CARMAweb performs data preprocessing (background correction, quality control and normalization), detection of differentially expressed genes, cluster analysis, dimension reduction and visualization, classification, and Gene Ontology-term analysis. This web application accepts raw data from a variety of imaging software tools for the most widely used microarray platforms: Affymetrix GeneChips, spotted two-color microarrays and Applied Biosystems (ABI) microarrays. R and packages from the Bioconductor project are used as an analytical engine in combination with the R function Sweave, which allows automatic generation of analysis reports. These report files contain all R commands used to perform the analysis and guarantee therefore a maximum transparency and reproducibility for each analysis. The web application is implemented in Java based on the latest J2EE (Java 2 Enterprise Edition) software technology. CARMAweb is freely available at https://carmaweb.genome.tugraz.at.
INTRODUCTION
Expression profiling using microarrays has become a widely used method for the study of gene-expression patterns. Different microarray technologies have become available, including the Affymetrix GeneChip platform (http://www.affymetrix.com), spotted two-color cDNA or oligo microarrays (1), or the ABI single-channel microarrays (Applied Biosystems, http://www.appliedbiosystems.com). All microarray platforms require analytical pipelines with modules for (i) data preprocessing including data normalization, (ii) statistical analysis for identification of differentially expressed genes, (iii) cluster analysis and (iv) Gene Ontology (GO) analysis. The module for normalization and data preprocessing is platform dependent and aims to reduce technical variability without altering the biological variance in the data. After data normalization, the selection of differentially expressed genes is often the main objective of a microarray experiment. Additionally, genes might be grouped into clusters according to the similarity of their expression patterns. Finally, genes can be mapped onto GO (2) terms in order to get an overview of the biological processes, cellular components or molecular functions for which the genes of interest might be involved.
In the past years, Bioconductor (3) (based on the statistical programming language R, http://www.R-project.org) has become the reference tool for the analysis of microarray data because it is based on the most complete set of up-to-date algorithms. However, for scientists without adequate programming experience, the command line usage of R and Bioconductor is too cumbersome. Moreover, the performance of laboratory desktop computers is often insufficient to analyze microarray data with tens of thousands of features. Therefore, many analysis tools with a graphical user interface and powerful computing servers have been developed, including web-based tools like GEPAS (4), ArrayPipe (5), MIDAW (6), RACE (7) or Expression Profiler (8). Of these, only GEPAS and Expression Profiler support both Affymetrix and two-color arrays. To the best of our knowledge, there is currently no web service available for the analysis of the increasingly popular ABI system. MIDAW and RACE use R and Bioconductor packages as analytical engines as well, but these web applications focus either on the analysis of two-color microarrays (MIDAW) or Affymetrix GeneChips (RACE). Presently, only Expression Profiler allows loading microarray data from the ArrayExpress database (9). ExpressionProfiler enables direct handling only for raw data from the Affymetrix platform, whereas for two-color microarrays external manipulation of the raw data files has to be performed. The raw data files derived from the image analysis software are usually large and difficult to handle, especially for inexperienced users. Thus, researchers working with two-color microarray data have to navigate several websites and transfer the data between the servers to complete their analyses.
We have therefore developed the web application CARMAweb (Comprehensive R-based Microarray Analysis web service) based on both the latest Java 2 Enterprise Edition (J2EE) software technology and R in combination with Bioconductor.
CARMAweb provides the following unique features:
Support for Affymetrix, two-color and ABI microarrays,
Import of raw data from a variety of imaging software tools for two-color microarrays (Agilent Feature Extraction, ArrayVision, BlueFuse, GenePix, ImaGene, QuantArray, SPOT or raw data files from the Stanford Microarray Database),
A complete analytical pipeline for Affymetrix, two-color and ABI microarrays including modules for preprocessing, detection of differentially expressed genes, clustering and visualization, as well as GO mapping,
Generation of comprehensive analysis report files.
METHODS
CARMAweb is designed as a multi-tier application based on the J2EE environment, including Java Server Pages and Servlets for the web tier, and Enterprise Java Beans for the middle tier. With the exception of the module for cluster analysis, visualization and classification, all calculations are performed in R using functions of the Bioconductor packages. The connection between Java and R is established through Rserve (http://stats.math.uni-augsburg.de/Rserve/). Each analysis is processed in R using the R function Sweave (http://www.ci.tuwien.ac.at/~leisch/Sweave). Sweave is a tool that allows embedding of R code into LaTeX documents. Sweave executes the R commands from the input file, which is created by the web application. Output from R, R commands and descriptive text are written into a LaTeX file. Thus, code, results and descriptions are presented in a consistent way. After the analysis the LaTeX file generated by Sweave is transformed into a pdf analysis report file. These analysis report files contain all R commands used to perform the analysis, together with descriptions for the various methods used. This guarantees a maximum of transparency and reproducibility of each analysis performed in CARMAweb. The CARMAweb user guide gives a short introduction to the various analysis methods available in the web application. Test datasets are provided for each microarray platform.
The current implementation of CARMAweb runs on a server equipped with two AMD Opteron (64 bit CPU) processors and 4 GB of physical memory. CARMAweb will be updated regularly to the newest R and Bioconductor releases. The current version of CARMAweb uses R version 2.2 and Bioconductor release 1.7.
PROGRAM DESCRIPTION
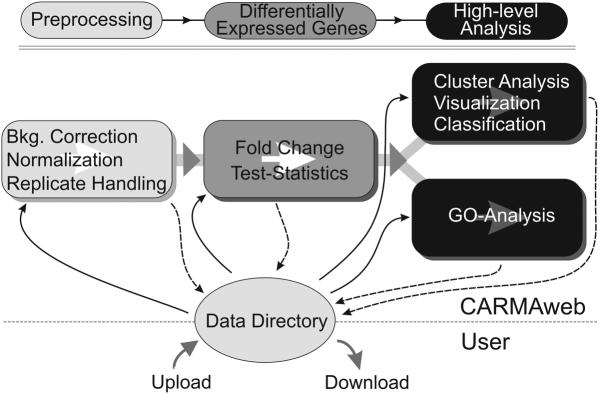
The design and modular conception of CARMAweb allows the use of the different analysis modules either individually or combined into an analytical pipeline (Figure 1). After preprocessing of the raw data and identification of differentially expressed genes, cluster analysis, visualization and GO analysis can be performed. All analysis result files, i.e. tables with normalized expression values, differentially expressed genes or cluster analysis results can be returned to the users' data directory and subsequently used as input files for other analysis modules of CARMAweb or for other applications. Detailed descriptions and help texts for the various processing steps and methods of the different modules are provided as pop-up tool tips.
Figure 1.
CARMAweb analysis workflow. The different modules of CARMAweb can either be used individually or in combination, resulting in an analytical pipeline. Analysis result files can be returned to the user's data directory and then be used as input for the other modules (e.g. the GO analysis module).
Data preprocessing
Data preprocessing is an essential step in the analysis of microarray data. The user has to choose an appropriate method from a wide range of available methods depending on the particularities of the data, i.e. their biological characteristics and the platform used.
Two-color microarrays. A large number of image analysis tools is available for two-color microarrays, and several features essential for the data preprocessing (i.e. flags determination, background estimates) differ between them. CARMAweb allows importing raw data files from Agilent Feature Extraction, ArrayVision, BlueFuse, GenePix, ImaGene, QuantArray, SPOT or raw data files from the Stanford Microarray Database. For background correction, CARMAweb allows several options (i) background subtraction, (ii) background subtraction followed by the minimum method (any intensity which is zero or negative after correction is set to half the minimum of the positive corrected intensities), (iii) the moving minimum method (background estimates are replaced with the minimum of the backgrounds of the spot and its eight neighbors, and are then subtracted from the foreground) or (iv) the method described in (10). After background correction, methods like the median normalization, the loess or print tip loess normalization or the robust spline normalization (normalizes using robustly fitted regression splines and empirical Bayes shrinkage) are provided by CARMAweb to normalize within-array. Afterwards between arrays normalization can be performed using the median scaling or the quantile method. Additionally the variance stabilizing normalization (11), which combines both within and between array normalization, has also been included in CARMAweb. Most of these preprocessing methods are outlined in (12). The preprocessing of two-color microarrays is carried out in CARMAweb using functions from Bioconductors limma and vsn packages.
Affymetrix GeneChips. Preprocessing of Affymetrix GeneChips generally consists of the following steps: (i) background correction, (ii) normalization, (iii) correction for non-specific binding and (iv) summarization, where the measured probe intensities are averaged to one expression value per probe set. CARMAweb uses the affy package from Bioconductor for this purpose, and allows the usage of methods like the Affymetrix MAS5 algorithm or even more sophisticated methods like RMA (robust multi-array average) (13,14) or GCRMA (modified version of RMA that uses probe sequence information for the background correction) to perform the preprocessing. A comparison of the different Affymetrix preprocessing methods is outlined in (15). Additionally it is possible to define custom preprocessing methods by selecting different algorithms for each one of the preprocessing steps. For Affymetrix GeneChip analyses, Affymetrix raw data files (CEL files) are used as input files.
ABI microarrays. The module for the ABI microarray preprocessing supports tabulator-delimited text files, which can be exported from ABIs scanning software. These text files already contain background-corrected expression values from one or more microarrays. CARMAweb permits reading of microarray data from one or more of such exported text files, and allows the adjustment of raw (background-corrected) expression values across all microarrays of one experiment using quantile normalization. Alternatively, the assay-normalized signal provided by ABI might be used for the analysis (see Applied Biosystems 1700 Chemiluminescent Microarray Analyzer User's manual http://www.appliedbiosystems.com). Quality parameters (flags, signal to noise, cv) can be used to filter out poor quality spots.
Following the microarray preprocessing, replicated arrays can be averaged in an optional replicate handling step. This function also allows averaging of the replicated spots within each microarray, and its aim is to increase the quality of the microarray data by reducing the noise.
Detection of differentially expressed genes
The detection of differentially expressed genes can be performed in CARMAweb for microarray experiments with a small number of biological replicates using a simple fold change. Additionally CARMAweb allows ranking of genes according to the number of comparisons in which they were selected as differentially expressed.
In microarray experiments with a sufficient number of arrays, differentially expressed genes can be detected in CARMAweb using statistical tests like the Mann Whitney U test (16), the Student's t-test (16), the permutation (randomization) test (16), the moderated t-statistics (based on an empirical Bayes approach, the Bioconductors limma package) (17) or the significance analysis of microarrays (SAM, Bioconductors siggenes package) (18). Because microarray experiments generate large multiplicity problems in which thousands of hypotheses are tested simultaneously within one experiment (19), an adjustment of the calculated P-values should be performed. Bioconductors multtest package provides suitable methods to adjust P-values regarding this multiple hypothesis testing problem. Available adjustment methods are the procedure introduced by Benjamini and Hochberg (20) for strong control of the FDR (false discovery rate, expected proportion of false positives among the rejected hypotheses) or the method by Westfall and Young (21) to control the FWER (family-wise error rate, probability of at least one false positive). CARMAweb allows the use of all methods described in (19) for the adjustment of raw P-values.
To alleviate the loss of power from the formidable multiplicity of gene-by-gene hypothesis testing within a microarray experiment, a non-specific pre-filtering of the data can also be performed. This pre-filtering consists in the reduction beforehand of the number of genes to be tested, removing those that are either not relevant for the study in question or those expected to be unaltered through the experimental conditions. This can be achieved by focusing the analysis only on those genes for which variance across conditions is in the top x%, where x is a user-defined value.
Cluster analysis, dimension reduction and visualization, and classification
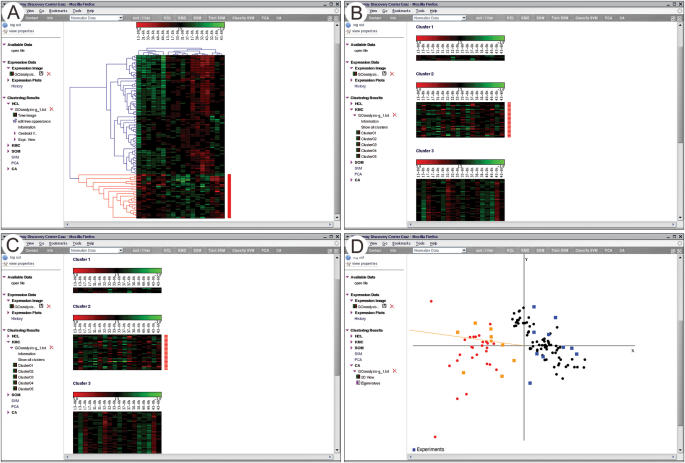
For cluster analysis, dimension reduction and visualization, and for classification of microarray data, the module GenesisWeb can be used (Figure 2). This module is based on the cluster analysis suite Genesis (22), and uses its server (23) to perform the calculations. Cluster analysis and visualization requires intensive graphical user interaction that is not supported by R. The cluster analysis module of CARMAweb supports an interactive selection, coloring and export of clusters, and also displays other important information like the expression values or gene names both as tool tips and in the status bar of the browser when the user moves the mouse over the image.
Figure 2.
The cluster analysis module GenesisWeb offers interactive cluster selection. (A) Result from a hierarchical cluster analysis. (B) Result from k-means cluster analysis of the same dataset. (C) Result from SOM cluster analysis. (D) Visualization of a CA of the same dataset. Clusters interactively selected in any of the cluster analyses can be highlighted in further analyses (shown here as red labeled genes).
Expression data can be adjusted beforehand with methods like mean or median centering, logarithmic transformation or division by the SD across samples or genes. Genes and/or samples can be grouped according to their expression similarity using the hierarchical clustering algorithm (HCL) (24), the k-means clustering method (KMC) (25) or self-organizing maps (SOM) (26). A wide range of distance-measurement methods is available to measure the similarity of gene or sample expression patterns (e.g. Euclidian distance, Pearson correlation, Spearman's rank or Kendall's tau). As mentioned before all result images are interactive, thus allowing the selection, coloring and export of clusters. Additional information, like gene names or expression values, is displayed both as tool tips and in the status bar of the browser when the user moves the mouse over expression or cluster images.
The available dimension reduction and visualization methods are principal component analysis (PCA) (27) and correspondence analysis (CA) (28). Whereas PCA can be used to identify key variables (or combination of variables) in the datasets, CA allows simultaneous detection of dependencies between samples and genes in microarray data. Visualization tools available with the dimension reduction procedure also enable selection, coloring and export of genes that group together in the space spanned by the principal components.
Support vector machines (SVM) (29) can be used for classification of microarray data. The aim of this supervised classification method is to classify genes or samples by using the information gathered from the training on a dataset with known classification. For example, an SVM can learn in the training step what expression features are specific for a given functional group of genes specified by the researcher, and use this information to decide whether any given gene is likely to be a member of the group or not.
The tool can be used as a standalone web application at https://carmaweb.genome.tugraz.at/genesis, or in combination with CARMAweb, where it is possible to return cluster analysis results to the user's data directory. As input files, tabulator-delimited text files containing expression values (e.g. from an earlier analysis that detected differentially expressed genes, or from files uploaded by the user) are supported.
GO analysis
The Gene Ontology project (2) provides three independent ontologies for gene products. The three ontologies refer to the cellular component, biological process and molecular function of a gene product and allow its description in a hierarchical manner. The GO analysis aims to assist in the biological interpretation of the results by finding GO terms that are significantly often associated to genes in a given gene list. CARMAweb uses the GOstats and GO package from Bioconductor for the GO term analysis. The GO term analysis module of CARMAweb supports as input any tabulator-delimited text file that contains one column with EntrezGene (formerly LocusLink) identifiers of the genes of interest. This kind of input file can be either file uploaded by the user or result file from a previous analysis. The result of the GO analysis is a GO graph, and a table with GO terms and P-values calculated for over-representation of the genes in the corresponding GO terms. The GO graph is the collection of unique GO terms that are associated with one or more of the genes of interest. In order to allow calculation of P-values, an additional file containing the EntrezGene identifiers of all genes that can be detected with the microarrays in use needs to be submitted. Affymetrix users can specify the GeneChip used in the analysis instead of submitting a file with all EntrezGene identifiers. Although some correction for multiple testing should be performed on the P-values, such tests are not independent and the sampling distribution is unclear (30), so CARMAweb at present does not perform any correction.
Output and analysis results
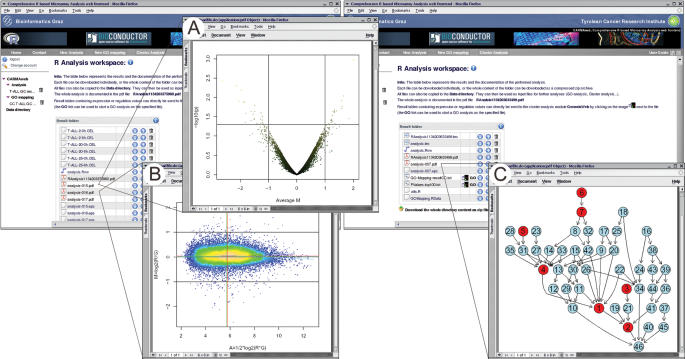
Each analysis is processed in its own workspace, which is accessible only to the user performing the analysis (Figure 3). The analysis result includes all raw data files, the analysis report file containing all commands and descriptions about the methods used, and all result tables and plots created during the analysis. Additionally the R workspace of each analysis step can be exported to an RData file, which can be used to continue the analysis in R on a local workstation. The result tables can comprise tables of normalized expression values for all genes in all arrays, tables with expression values of the subsets of differentially expressed genes, or tables containing the raw P-values and adjusted P-values using the various adjustment methods. In an Affymetrix GeneChip analysis all probe sets are annotated to the identifiers of the publicly available databases (GenBank (31), UniGene, EntrezGene) using the Bioconductor annaffy package. Analysis result files can be returned to the users' data directory and be used as input for further analyses.
Figure 3.
Result workspaces of a differentially expressed genes analysis (left) and a GO analysis (right). (A) Volcano plot displaying the mean differential expression against P-values (−log10 of the P-value) of all genes. (B) MA plot. Points are colored according to local point density with brighter colors coding for higher density. (C) The induced GO graph of the genes of interest. Red nodes represent over-represented GO terms.
Visualizations of the microarray data like MA plots, histograms, box plots or volcano plots are available as single files and are additionally embedded into the analysis report file. The content of each analysis workspace can be downloaded after completion as a single zip archive, or each file can be downloaded separately. The GO term analysis produces a directed acyclic graph of all GO terms to which the genes of interest are associated (Figure 3). Additionally a table containing all GO terms with the corresponding P-value is created. The P-values provide information about the over-representation of the genes of interest to the term compared with the total number of genes associated with it. The table contains the number of genes of interest that are mapped to the specific term and the total number of genes present on the microarray in use that are associated with the GO term.
Future development
The next release of CARMAweb will provide a complete SOAP (Simple Object Access Protocol) interface to its analysis facilities, thus allowing other applications to use the analysis and processing steps available in CARMAweb.
CONCLUSIONS
The web application CARMAweb that we have developed combines the advantages of an intuitive web-based graphical user interface with the wide range of state-of-the-art microarray normalization and analysis methods provided by Bioconductor. Owing to the modular structure of CARMAweb and the standards-based software engineering, extensions or new functionalities can be implemented easily without complex and time-consuming alterations of the source code.
CARMAweb provides several unique features in a modular and flexible system for the analysis of microarray data. First, data from three platforms, namely Affymetrix GeneChip, two-color microarray and the ABI microarray platform can be analyzed. Second, a wide range of file formats for two-color microarray raw data is supported. Third, a complete analytical pipeline for the supported platforms is provided, including preprocessing, detection of differentially expressed genes, cluster analysis, dimensionality reduction and visualization, classification, and GO analysis. Fourth, data exploration is enhanced by analysis report files that include the parameters and commands used. The report files that are generated specifically for each analysis guarantee a maximum of transparency and reproducibility. Furthermore, these report files provide a unique way for the documentation of any analyses that have been performed by recording how and with which methods the analysis results have been derived. In sum, based on its flexibility in selecting different analysis steps, its possibility for customization and its comprehensive web-based graphical user interface, CARMAweb is a powerful tool for microarray data analysis.
Acknowledgments
We thank Reinhard Kofler and Stefan Schmidt from the Tyrolean Cancer Research Institute for discussions and providing test datasets available in CARMAweb. We also thank Hubert Hackl and Robert Molidor from the Institute of Genomics and Bioinformatics for discussions and comments, and James McNally from the National Cancer Institute, National Institutes of Health for comments on the manuscript. This work was supported by the bm:bwk project GEN-AU BIN (Bioinformatic Integration Network) and the Tyrolean Cancer Research Institute. Funding to pay the Open Access publication charges for this article was provided by bm:bwk project GEN-AU BIN.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrero J., Al-Shahrour F., Diaz-Uriarte R., Mateos A., Vaquerizas J.M., Santoyo J., Dopazo J. GEPAS: a web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 2003;31:3461–3467. doi: 10.1093/nar/gkg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hokamp K., Roche F.M., Acab M., Rousseau M.-E., Kuo B., Goode D., Aeschliman D., Bryan J., Babiuk L.A., Hancock R.E.W., et al. ArrayPipe: a flexible processing pipeline for microarray data. Nucleic Acids Res. 2004;32:W457–W459. doi: 10.1093/nar/gkh446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romualdi C., Vitulo N., Del Favero M., Lanfranchi G. MIDAW: a web tool for statistical analysis of microarray data. Nucleic Acids Res. 2005;33:W644–W649. doi: 10.1093/nar/gki497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Psarros M., Heber S., Sick M., Thoppae G., Harshman K., Sick B. RACE: Remote Analysis Computation for gene Expression data. Nucleic Acids Res. 2005;33:W638–W643. doi: 10.1093/nar/gki490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapushesky M., Kemmeren P., Culhane A.C., Durinck S., Ihmels J., Korner C., Kull M., Torrente A., Sarkans U., Vilo J., et al. Expression Profiler: next generation—an online platform for analysis of microarray data. Nucleic Acids Res. 2004;32:W465–W470. doi: 10.1093/nar/gkh470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brazma A., Parkinson H., Sarkans U., Shojatalab M., Vilo J., Abeygunawardena N., Holloway E., Kapushesky M., Kemmeren P., Lara G.G., et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards D.E. Non-linear normalization and background correction in one-channel cDNA microarray studies. Bioinformatics. 2003;19:825–833. doi: 10.1093/bioinformatics/btg083. [DOI] [PubMed] [Google Scholar]
- 11.Huber W., von Heydebreck A., Sultmann H., Poustka A., Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 12.Smyth G.K., Speed T.P. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 13.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 14.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 16.Motulsky H. Intuitive Biostatistics. Oxford: Oxford University Press; 1995. [Google Scholar]
- 17.Smyth G.K., Yang Y.H., Speed T. Statistical issues in cDNA microarray data anlalysis. Methods Mol. Biol. 2003;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- 18.Tusher V., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5124. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudoit S., Shaffer J.P., Boldrick B.J. Multiple hypothesis testing in microarray experiments. Stat. Sci. 2003;18:71–103. [Google Scholar]
- 20.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. 1995;57:289–300. [Google Scholar]
- 21.Westfall P.H., Young S. Resampling-Based Multiple Testing: Examples and Methods for P-Value Adjustment. Wiley, NY: 1993. [Google Scholar]
- 22.Sturn A., Quackenbush J., Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 23.Sturn A., Mlecnik B., Pieler R., Rainer J., Truskaller T., Trajanoski Z. Client-server environment for high-performance gene expression data analysis. Bioinformatics. 2003;19:772–773. doi: 10.1093/bioinformatics/btg074. [DOI] [PubMed] [Google Scholar]
- 24.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavazoie S., Hughes J.D., Campbell M.J., Cho R.J., Church G.M. Systematic determination of genetic network architecture. Nature Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 26.Tamayo P., Slonim D., Mesirov J., Zhu Q., Kitareewan S., Dmitrovsky E., Lander E.S., Golub T.R. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic di erentiation. Proc. Natl Acad. Sci. USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeung K.Y., Ruzzo W.L. Principal component analysis for clustering gene expression data. Bioinformatics. 2001;17:763–774. doi: 10.1093/bioinformatics/17.9.763. [DOI] [PubMed] [Google Scholar]
- 28.Fellenberg K., Hauser N.C., Brors B., Neutzner A., Hoheisel J.D., Vingron M. Correspondence analysis applied to microarray data. Proc. Natl Acad. Sci. USA. 2001;98:10781–10786. doi: 10.1073/pnas.181597298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown M.P., Grundy W.N., Lin D., Cristianini N., Sugnet C.W., Furey T.S., Ares M.J., Haussler D. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl Acad. Sci. USA. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gentleman R., Scholtens D., Ding B., Carey V.J., Huber W. Case Studies Using Graphs on Biological Data. In: Gentleman R., Carey V.J., Huber W., Irizarry R.A., Dudoit S., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. NY: Springer; 2005. pp. 375–378. [Google Scholar]
- 31.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:34–38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]