Abstract
We report here a functional analysis of spo5+(mug12+) of Schizosaccharomyces pombe, which encodes a putative RNA-binding protein. The disruption of spo5+ caused abnormal sporulation, generating inviable spores due to failed forespore membrane formation and the absence of a spore wall, as determined by electron microscopy. Spo5 regulates the progression of meiosis I because spo5 mutant cells display normal premeiotic DNA synthesis and the timely initiation of meiosis I but they show a delay in the peaking of cells with two nuclei, abnormal tyrosine 15 dephosphorylation of Cdc2, incomplete degradation of Cdc13, retarded formation and repair of double strand breaks, and a reduced frequency of intragenic recombination. Immunostaining showed that Spo5-green fluorescent protein (GFP) appeared in the cytoplasm at the horsetail phase, peaked around the metaphase I to anaphase I transition, and suddenly disappeared after anaphase II. Images of Spo5-GFP in living cells revealed that Spo5 forms a dot in the nucleus at prophase I that colocalized with the Mei2 dot. Unlike the Mei2 dot, however, the Spo5 dot was observed even in sme2Δ cells. Taken together, we conclude that Spo5 is a novel regulator of meiosis I and that it may function in the vicinity of the Mei2 dot.
The generation of inheritable haploid gametes from diploid parental cells by meiosis is an essential event for sexually reproducing eukaryotic organisms. The fission yeast Schizosaccharomyces pombe proceeds to meiosis upon nutritional starvation when two cells with opposite mating types conjugate and the two haploid nuclei fuse, thereby producing a zygote with a diploid nucleus. Genome-wide expression profiles of S. pombe successfully identified a large number of genes whose expressions are specifically upregulated during meiosis and appear to support a variety of meiosis-specific events (13). Large-scale deletion analysis based on the S. pombe transcriptome data has identified novel genes that are important for meiotic chromosome segregation, meiosis-specific DNA double-strand breakage, telomere clustering, and homologous pairing (3, 12). We also took advantage of the transcriptome data to comprehensively identify meiosis-specific genes that generate proteins with coiled-coil motifs and found that four genes, named meu13+ (17), mcp7+ (23), mcp6+ (24), and mcp5+ (25), play pivotal roles in homologous pairing and meiotic recombination.
Recent large-scale cDNA sequencing projects in mammals have revealed that eukaryotic cells contain numerous mRNA-like noncoding RNAs (ncRNAs) that are expected to play physiological roles, particularly in gene expression (2). In S. pombe, we previously isolated five kinds of meiosis-specific transcripts (32) as well as 68 mRNA-like ncRNAs (33) that appear to be antisense and multiply overlapping mRNA-like transcripts. We also identified overlapping transcripts, one of which appears to be a mRNA-like ncRNA (10), and three kinds of meiosis-specific transcripts that are derived from the complementary strand of rec7+ (14). Bidirectional transcripts named spo6-S and spo6-L have been found to derive from the coding region of spo6+, which expresses a meiosis-specific protein with sequence similarity to Dbf4, a regulator of DNA replication (18, 20). While the direction of spo6-S encodes Spo6 protein, the direction of spo6-L is reversed and generates an antisense ncRNA. Although these ncRNAs may play important roles in the progression of meiosis, neither their functions nor their putative association partners (RNA-binding proteins) are clear.
An mRNA-like ncRNA named meiRNA of S. pombe is expressed only during meiosis from the sme2 gene. It is essential for meiosis I (MI) but not for either cell growth or premeiotic DNA synthesis (34). meiRNA is a cofactor of an RNA-binding protein called Mei2 that is required at two distinct stages of meiosis, namely, once prior to premeiotic DNA synthesis and then prior to meiosis I (36). Mei2-like protein is also found in other organisms (9). Mei2 forms a dot in meiotic prophase nuclei, and meiRNA is required for this nuclear localization of Mei2 (35). While localization of the Mei2 dot coincides with the sme2 locus, it is the transcripts of sme2 rather than the DNA sequence of the gene that determine this localization of the Mei2 dot (28). These results do not appear to simply reflect the attachment of Mei2 to meiRNA that is undergoing transcription; rather, various observations suggest that this localization involves a specialized platform structure that permits a large number of proteins to assemble and thereby mediate the proper progression of meiosis I. However, little is known about this structure and its functions.
As an initial step to understanding the putative physiological roles of these mRNA-like ncRNAs, we searched for meiosis-specific proteins that harbor putative RNA-binding motifs and may associate with these ncRNAs. Based on the S. pombe transcriptome data, we identified three candidate genes. We here report our detailed analysis of one of these, mrb1+, which was named after meiotic RNA-binding protein. In the course of our analysis, we noted that mrb1+ is equal to spo5+ (C. Shimoda, personal communication;6), although the DNA sequence of spo5+ has not been registered in the DNA bank. We recently found that it is also named mug12+ (12). Hereafter, we will call it spo5+ because this was the original name. Of note is that Spo5/Mug12/Mrb1 localized at prophase of meiosis I as nuclear dots that colocalized with the Mei2 dot.
MATERIALS AND METHODS
GFP tagging of the spo5+ gene.
To construct the Spo5-green fluorescent protein (GFP) strain, we performed PCR as described previously (23) and obtained a DNA fragment carrying the open reading frame and 3′ downstream region of the spo5+ gene. For this purpose, we synthesized the following two oligonucleotides and used them as primers: spo5-F (5′-GCGTCGACGGCGCGCCGATGAATGGAATAATTACGCCTC-3′) and spo5-R (5′-GCGCGGCCGCCCATTAGCAGAATGAGCGGG-3′). The underlined sequences denote the artificially introduced restriction enzyme sites for SalI and AscI and for NotI, respectively. We used the same primers (spo5-3F and spo5-3R) as described above. The 3′ downstream PCR product of the spo5+ gene was inserted into the pRGT1 vector via the XhoI-SacI sites. The PCR product of the spo5+ open reading frame, the 1.8-kb HindIII fragment containing the ura4+ cassette (4), and the pRGT1-spo5-3′ vector were inserted into the pBluescriptII KS(+) vector via the EcoRI-NotI, HindIII, and NotI-SacI sites, respectively. This plasmid construct was then digested with SphI, and the resulting construct was introduced into the TP4-5A strain. The Ura+ transformants were then screened by PCR to identify the spo5::spo5-GFP-ura4+ strain.
Construction of Spo5 mutants.
To construct the Spo5N and Spo5C mutant-expressing strains, which bear only the N- and C-terminal, respectively, portions of Spo5, we performed PCR and obtained DNA fragments carrying the desired Spo5 regions. For this purpose, we synthesized the following oligonucleotides and used them as primers: spo5-SalI-F (5′-GGGTAACAAAGTAAACACTGGCAGTCGAC-3′), spo5-EcoRI-R 5′-ACTGAATTCGTAGGCACAGTCGCTGAAGG-3′), spo5-D-SalI-F (GCGTCGACTTCATTGCACTTCAATAATTAAGGCG), spo5-DI-R (5′-GTGCGGCCGCACATTCCGAGTACGAGAAGTGCTTTCCATG-3′), and spo5-DV-R 5′-CGAGGAATTCGTAGACATTAATTGTTTTTGTTTTTGAGGCG-3′). The underlined sequences denote the artificially introduced restriction enzyme site for SalI, NotI, or EcoRI.
We also performed PCR to create the Spo5FA3-, Spo5FA4-, and Spo5FAFA-expressing strains by using the following oligonucleotides as primers: spo5-F341A-F (5′-CGAATTTATGTAAAGGATATGGCGCCGCATGCTTTGAAGAAGAGAAATCTGC-3′), spo5-F341A-R (5′-CAGATTTCTCTTCTTCAAAGCATGCGGCGCCATATCCTTTACATAAATTCG-3′), spo5-F427A-F (5′-CGTGACTCTAAGGAACAATCCCGCGGTGTTGGGGCTGCTCGTATGCAAGATCG-3′), and spo5-F427A-R (5′-CGATCTTGCATACGAGCAGCCCCAACACCGCGGGATTGTTCCTTAGAGTCACG-3′). The underlined sequences denote the artificially introduced nucleotides that serve to replace phenylalanine 341 and/or phenylalanine 427 with alanine or that introduce restriction enzyme sites for NarI or SacII. These Spo5-FA3, Spo5-FA4, and Spo5-FAFA constructs were then created by PCR using the Spo5 construct as the template. These plasmid constructs were digested with NruI. The resulting construct was introduced into AS18 (h90 ade6-M216 leu1-32 ura4-D18 spo5::ura4+) (Table 1). We then screened the Leu+ transformants and confirmed the precise integration of the constructs by PCR and digestion by the relevant restriction enzyme.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| CD16-1 | h+/h−ade6-M210/ade6-M216 cyh1/++/lys5-391 | 19 |
| CD16-5 | h−/h−ade6-M210/ade6-M216 cyh1/++/lys5-391 | 19 |
| TP4-5A | h−ade6-M210 leu1-32 ura4-D18 | 34 |
| JZ670 | h−/h−ade6-M210/ade6-M216 leu1-32/leu1-32 pat1-114/pat1-114 | 29 |
| AS29 | h−/h−ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 pat1-114/pat1-114 spo5::ura4+/spo5::ura4+ | This study |
| AS6 | h90ade6-M210 leu1-32 ura4-D18 | This study |
| CRL266 | h90leu1-32 | Y. Hiraoka |
| AS17 | h90ade6-M210 leu1-32 ura4-D18 spo5::ura4+ | This study |
| AS18 | h90ade6-M216 leu1-32 ura4-D18 spo5::ura4+ | This study |
| YN68 | h90leu1-32::[GFP-psy1+-leu1+] | C. Shimoda |
| AS44 | h90ade6-M216 ura4-D18 spo5::ura4+leu1-32::[GFP-psy1+-leu1+] | This study |
| AS24 | h90ade6-M210 leu1-32 ura4-D18 spo5::[spo5-GFP-3′UTR-ura4+] mei2::[mei2-CFP-kanr] | This study |
| AS47 | h90ade6-M210 leu1-32 ura4-D18 spo5::[spo5-GFP-3′UTR-ura4+] sme2::ura4+ | This study |
| AS7 | h90ade6-M210 leu1-32 ura4-D18 spo5::[spo5-GFP-3′UTR-ura4+] | This study |
| TK155 | h90ura4-D18 spo5::ura4+leu1-32::[spo5-GFP-3′UTR-leu1+] | This study |
| TK161-FA3 | h90ade6-M216 ura4-D18 spo5::ura4+leu1-32::[spo5-F341A- GFP-3′UTR-leu1+] | This study |
| TK162-FA4 | h90ura4-D18 spo5::ura4+leu1-32::[spo5-F427A-GFP-3′UTR-leu1+] | This study |
| TK163-FAFA | h90ade6-M216 ura4-D18 spo5::ura4+leu1-32::[spo5-F341A/F427A- GFP-3′UTR-leu1+] | This study |
| TK164-N | h90ade6-M216 ura4-D18 spo5::ura4+leu1-32::[spo5N-GFP-3′UTR-leu1+] | This study |
| TK164-C | h90ade6-M216 ura4-D18 spo5::ura4+leu1-32::[spo5C-GFP-3′UTR-leu1+] | This study |
| TK24-M26 | h−ade6-M26 ura4-D18 | This study |
| TK24-469 | h+ade6-469 his2 leu1-32 ura4-D18 | This study |
| TK171-M26 | h−ade6-M26 ura4-D18 spo5::ura4+leu1-32::[spo5-F341A/F427A-GFP-3′UTR-leu1+] | This study |
| TK171-469 | h+ade6-469 his2 ura4-D18 spo5::ura4+leu1-32::[spo5-F341A/F427A-GFP-3′UTR-leu1+] | This study |
Fluorescence microscopy.
To detect Spo5-GFP in living or fixed meiotic cells, mid-log-phase spo5+-GFP cells (AS7) were cultured in EMM2-N medium to induce meiosis as described previously (24). The living cells were then stained with 3.0 μg/ml Hoechst 33342 for 5 min and mounted on a coverslip by spotting. To produce fixed cells, meiotic cells were fixed according to the procedure of Hagan and Hyams (5) that uses glutaraldehyde and paraformaldehyde. In indirect immunofluorescence microscopy, the spindle pole bodies (SPBs) were stained with anti-Sad1 antibody (a gift from M. Yanagida, University of Kyoto). The samples were then stained with 0.5 mg/ml Hoechst 33342 in phosphate-buffered saline for 1 min and mounted with antifade-containing Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Fluorescence images of these cells were observed by using a fluorescence microscope (BX51; Olympus, Tokyo, Japan) with a Cool SNAP charge-coupled-device camera (Roper Scientific, San Diego, CA). Immunofluorescence images were acquired using Adobe Photoshop 7.0.
Antibody and Western blotting.
To generate an anti-Spo5 polyclonal antibody, rabbits were injected with a recombinant glutathione S-transferase (GST)-fused full-length Spo5 protein. The antiserum was then affinity purified against the Spo5 protein. For Western blot analysis, we extracted the proteins by using two different methods. With the first method (see Fig. 5C), 2.8 × 108 S. pombe cells were suspended in 0.4 ml of HB buffer (25 mM MOPS [morpholinepropanesulfonic acid], pH 7.2, 15 mM MgCl2, 15 mM EGTA, 60 mM β-glycerophosphate, 15 mM p-nitrophenylphosphate, 0.1 mM sodium vanadate, and 1% Triton X-100) and boiled for 10 min. The cells were then disrupted with acid-washed glass beads by using a vortex mixer, and the glass beads were removed by centrifugation, thus generating the whole-cell extract. With the second method (see Fig. S4 in the supplemental material), 2.8 × 108 S. pombe cells were suspended in 0.4 ml of 20% trichloracetic acid (TCA) solution. The cells were disrupted with acid-washed glass beads by using a vortex mixer, and then a 5% TCA solution was added and the total protein precipitates were obtained by centrifugation. The whole-cell extract (HB buffer) or protein precipitate (TCA) was then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane (Immobilon; Millipore, Bedford, MA). These blots were probed with rabbit anti-Spo5 polyclonal antibody, and the bands were visualized by using the Renaissance system (NEN Life Science, Boston, MA).
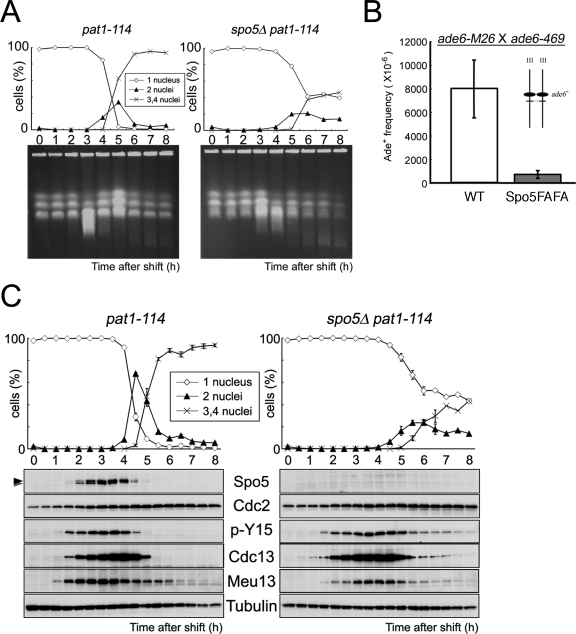
FIG. 5.
Analysis of the meiotic progression and Spo5 protein expression in spo5Δ cells. (A) The timing of DSBs repair was analyzed by PFGE using DNA from pat1-114 and spo5Δ pat1-114 cells at the indicated time points of meiotic progression. Chr I, Chr II, and Chr III indicate the positions of chromosomes 1, 2, and 3, respectively, that were detected by staining DNA with ethidium bromide. The smear bands represent the DSBs that appear during meiosis. An asterisk indicates the position of degraded DNA derived from dead cells. (B) Intragenic recombination between ade6-M26 and ade6-469 on chromosome III was examined in WT (TK24-M26 × TK24-469) or Spo5FAFA (TK171-M26 × TK171-469) strains. The data show the average of three independent experiments with standard errors (error bars). (C) Determination of the kinetics of meiosis using pat1-114 strains and Western blot analysis of Spo5 protein expression. JZ670 (pat1-114) and AS29 (spo5Δ pat1-114) strains were induced to enter synchronous meiosis, and cell extracts were prepared every 30 min. These extracts were dissolved in HB buffer (see Materials and Methods) and subjected to Western blot analysis using antibodies against Spo5, Cdc2, phosphor Tyr15 of Cdc2 (p-Y15), Cdc13, or Meu13. The Cdc2 and α-tubulin levels acted as loading controls. Meu13 is presented as a meiotic timing control.
Electron microscopy (EM) observations.
Cells were incubated on ME plates at 28°C for 18 h, collected, mounted on a copper grid to form a thin layer, and immersed in liquid propane cooled with liquid nitrogen (Leica EM CPC; Leica Microsystems, Vienna, Austria). The frozen cells were transferred to 2% OsO4 in dry acetone and kept at −80°C for 3 days. Thereafter, they were warmed gradually from −80°C to 0°C over 5 h, held for 1 h at 0°C, and then warmed from 0°C to 23°C (room temperature) for 2 h (Leica EM AFS; Leica Microsystems). After being washed with dry acetone three times, the samples were infiltrated with increasing concentrations of Spurr's resin in dry acetone and finally with 100% Spurr's resin. After polymerization, ultrathin sections were cut by an ultramicrotome (Leica Ultracut UCT; Leica Microsystems) and stained with uranyl acetate and lead citrate. The sections were viewed on an electron microscope (H-7600; Hitachi Co., Tokyo, Japan) at 100 kV.
Meiotic double strand break (DSB) assays.
For pulsed-field gel electrophoresis (PFGE), meiosis was induced in pat1-114 and pat1-114 spo5Δ cells by shifting the temperature and ca. 2.5 × 108 cultured cells were collected at the indicated times (27). DNA plug preparation and cells lysis were performed as described previously (16). PFGE was conducted in a 0.5% chromosomal grade agarose gel (Bio-Rad) in a Bio-Rad CHEF Mapper system at 14°C for 48 h with a 2 V/cm, 120°C-induced angle in 1× TAE buffer (40 mM Tris-acetate, pH 8.0, 1 mM EDTA), with a switch time of 30 to 45 min.
Recombination assay.
The intragenic recombination rates were determined as described previously (27). Briefly, haploid parental strains were grown on yeast extract-peptone-dextrose plates at 33°C. Cells were mated and sporulated on ME plates at 28°C. After 3 days of incubation, the spores were treated with 1% glusulase (DuPont NEN, Boston, MA) for about 3 h at room temperature and checked under a microscope for the complete digestion of contaminating vegetative cells. The glusulase-treated spores were washed with water and then used to measure the intragenic recombination rates. To examine the frequency of intragenic recombination, we used two ade6 alleles (ade6-M26 and ade6-469) since intragenic recombination between these alleles produces the ade6+ allele.
RESULTS
Spo5 is a putative meiosis-specific RNA-binding protein.
We previously reported the existence of many mRNA-like ncRNAs in S. pombe (32, 33). To identify meiosis-specific RNA-binding proteins that may associate with these ncRNAs, we searched the S. pombe genome database for uncharacterized genes that both harbor RNA-binding motifs and show enhanced expression during meiosis (http://www.genedb.org/genedb/pombe/index.jsp) (13). We screened 275 genes in the genome database AmiGO (http://www.genedb.org/amigo/perl/go.cgi) using “RNA binding” as a query word. In addition, we selected genes that display more enhanced expression in meiotic prophase I than in other meiotic phases based on the cDNA microarray data (13). This selection yielded three candidate genes, SPBC29A10.02, SPCC1682.08c, and SPAC4G9.05.
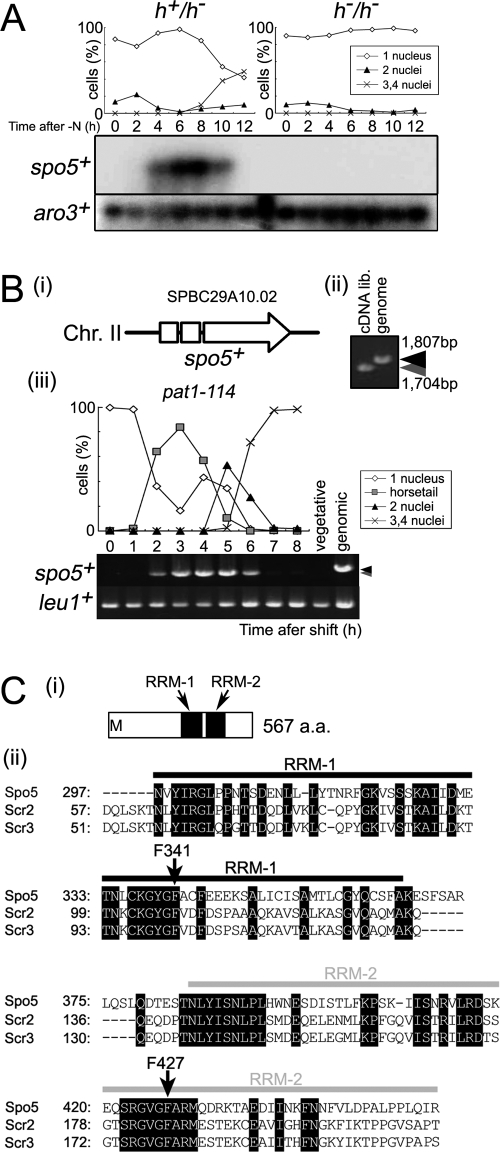
We then obtained DNA fragments from each of these genes and used them as probes for time course Northern blot analysis (Fig. 1A) or reverse transcription-PCR (RT-PCR) (Fig. 1B) to examine the RNA that was obtained from CD16-1 (h+/h−) and CD16-5 (h−/h−) cells harvested at various times after the induction of meiosis by nitrogen starvation. This analysis identified three novel genes that show dramatically induced transcription in only CD16-1 (h+/h−), the strain that can enter meiosis. We denoted the genes mrb1+ (AB248101; SPBC29A10.02), which is equal to spo5+ (Fig. 1A and B), mcp2+, which was named after meiotic coiled-coil protein (24) (AB189990; SPCC1682.08c) because it harbors a coiled-coil motif, and mpf1+, which was named after meiotic PUF family protein (AB248102; SPAC4G9.05) because it belongs to the PUF family (see Fig. S1 in the supplemental material). Hereafter, we will concentrate on our functional analysis of spo5+.
FIG. 1.
spo5+ is a meiosis-specific gene that encodes a protein with two RNA-binding motifs. (A) Northern blot analysis of spo5+ expression during meiosis. Total RNA was extracted from diploid h+/h− (CD16-1) and h−/h− (CD16-5) cells at the indicated times after they were subjected to nitrogen starvation, which induces CD16-1 but not CD16-5 cells to enter meiosis. Total RNA of these cells was blotted and probed with the protein-coding regions of the spo5+ and aro3+ (loading control) genes. The graph indicates the meiotic profiles of the cells used for RNA extraction. The frequencies of Hoechst 33342-stained cells with one, two, three, or four nuclei were assessed by counting at least 200 cells under a microscope. (B) PCR analysis of spo5+ transcripts during meiosis. (i) The structure of the spo5+ gene. The spo5+ gene consists of three exons which are shown by two open boxes and an open arrow. Chr., chromosome. (ii) The JZ670 (pat1-114) strain was induced to enter meiosis synchronously, and the total RNA from cells harvested at the indicated times was extracted and reverse transcribed to cDNA. PCR amplification of spo5+ was performed by using alternative cDNA from meiotic cells that were 3 h into meiosis, cDNA from vegetative cells, and genomic DNA as the template. PCR products were resolved on 1% agarose gels and stained by ethidium bromide. Black and gray arrowheads indicate the sizes of genomic spo5+ and the spliced spo5+ cDNA, respectively. lib., library. (iii) The difference in size of the spo5+ fragments amplified from genomic DNA or cDNA obtained 4 h into meiosis. (C) Schematic representation of the RRM in the RRM-bearing protein family. (i) The Spo5 protein has two RRMs in its C-terminal region as determined by SMART (http://smart.embl-heidelberg.de/). These motifs are designated as RRM-1 and RRM-2. a.a., amino acid. (ii) Alignment of RRM-1 and RRM-2 sequences with the RRM motif of Spo5, Scr2, and Scr3.
Spo5 consists of 567 amino acids and harbors two putative RNA-recognition motifs (RRMs) (Fig. 1C). Spo5 and Mei2 differ in structure since Mei2 has three RRMs; moreover, the amino acid sequence of these RRMs lacks significant similarity with those of the Spo5 RRMs. Homology searches using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST/) revealed no overall homologs of Spo5 in other organisms. Nonetheless, RRM-1 and RRM-2 of Spo5 displayed significant homologies to sequences in the human RNA-binding proteins Scr2 and Scr3 (Fig. 1C), which we have previously isolated as multicopy suppressors of the cdc2 and cdc13 mutants of S. pombe (11).
Northern blot analysis of RNA obtained from CD16-1 (h+/h−) cells indicated that spo5+ transcription during meiosis peaked around prophase I (horsetail phase) and meiosis I (8 h after induction) (Fig. 1A). RT-PCR using RNA from spo5+ cells in the genetic background of a pat1-114 strain, a temperature-sensitive strain used to attain synchronous meiosis, confirmed this timing of spo5+ transcription, as spo5+ transcripts appeared after the horsetail phase (2 h) and disappeared after meiosis II (6 h) (Fig. 1B). Western blot analysis using an anti-Spo5 polyclonal antibody that we prepared (see Fig. S2A in the supplemental material) indicated that Spo5 protein displays meiosis-specific expression, the details of which will be discussed later (see Fig. 5C). These results suggest that Spo5 may play a role in meiosis I. Notably, unlike some meiotic genes of S. pombe that display regulated splicing in meiosis (1), the two introns of spo5+ appear to be simultaneously spliced without showing regulated splicing (Fig. 1B).
Spo5 is essential for sporulation.
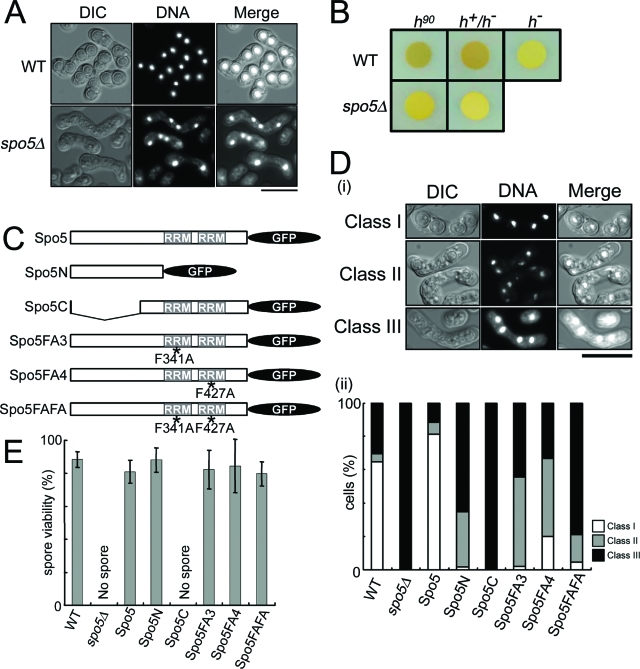
To assess the physiological role of spo5+, a deletion mutant that does not express Spo5 protein was constructed by one-step gene replacement. Diploid cells in which one of the spo5+ genes had been replaced by ura4+ were sporulated and germinated. The segregation ratio compared to that of the wild type (WT) was 1:1. All of the resulting spores were viable, indicating that the spo5+ gene is not essential for vegetative growth. The growth properties, cell sizes, and morphologies of spo5Δ cells were also indistinguishable from those of the WT cells. However, when the diploid h90 spo5Δ cells were observed 16 h after being induced to enter meiosis, many appeared to fail meiotic progression, as they carried incomplete numbers of nuclei (i.e., 1, 2, or 3). Moreover, even the cells containing four nuclei appeared to lack spore walls, as shown in the differential interference contrast image of Fig. 2A, whereas most WT cells display normal spores (Fig. 2A). This spo5Δ phenotype is not due to a delay in the progression of sporulation because spore walls were not observed even after 28 h or 48 h (data not shown). Staining of the spores by iodine vapor also confirmed that spo5Δ cells are defective in spore-wall formation (Fig. 2B).
FIG. 2.
The spo5Δ cells are defective in spore wall formation. (A) AS6 (h90) and AS17 (h90 spo5Δ) cells were induced to enter meiosis by nitrogen starvation, and their sporulation was examined after 16 h. Shown are differential interference contrast (DIC) fluorescence photographs of cellular DNA stained by Hoechst 33342 and their merged images. The bar indicates 10 μm. (B) The spore wall is absent in spo5Δ cells that are induced to sporulate. WT and spo5Δ cells were spotted onto sporulation plates (SPA), incubated at 28°C for 3 days, and then exposed for 1 min to iodine vapor, which stains sporulated cells dark brown. (C) Schematic representation of the truncated forms of spo5+ that are expressed as C-terminal GFP fusion proteins (see Fig. S7 in the supplemental material). The plasmids carrying these constructs were put into the leu1+ locus of spo5Δ cells. The Spo5 (strain TK155; h90 spo5Δ spo5+-GFP) drawn on top is the full-length Spo5 to which the GFP was added at the C terminus. Spo5N (TK164-N; h90 spo5Δ spo5N-GFP) and Spo5C (TK164-C; h90 spo5Δ spo5C-GFP) consist only of the Spo5 N- or C-terminal regions, respectively. The Spo5FA3-, Spo5FA4-, or Spo5FAFA-expressing strain (TK163-FAFA; h90 spo5Δ spo5-F341A/F427A-GFP) expresses full-length Spo5 proteins with point mutations that replace F341 and/or F427 with alanines (F341A and/or F427A) (Fig. 1C). Asterisks indicate point mutations where F341 and/or F427 were replaced with alanines. (D) The morphology of asci in various mutants. (i) The ascus phenotypes are classified into three classes. Class I contains the cells that form normal spores, and class II contains the cells that form abnormal spore walls, while class III contains the cells that did not form spore walls. WT (AS6), spo5Δ (AS17), Spo5 (TK155; this is an spo5Δ strain that has been engineered to express Spo5), Spo5N (TK164-N), Spo5C (TK164-C), Spo5-FA3 (TK161-FA3), Spo5-FA4 (TK162-FA4), and Spo5-FAFA (TK163-FAFA) cells at mid-log phase were cultured in EMM2, transferred to EMM2-N medium, and cultured further at 28°C for 16 h. At least 250 cells with three, four, or more nuclei were counted under a microscope. (ii) The graph shows the frequency with which the spores of each strain fell into each of these three classes. DIC, differential interference contrast. (E) Spore viability of each strain. The data show the averages of three independent experiments with standard errors (error bars).
To determine which part of the Spo5 molecule is important for sporulation, we prepared h90 spo5 strains that expressed Spo5 mutants; these mutants consisted of either N- or C-terminal portions of Spo5 only or expressed full-length proteins with point mutations that substituted alanines for F341 and/or F427, which are residues in RRM-1 and -2 (Fig. 2C). We then induced these cells to enter meiosis by nitrogen starvation. First, we demonstrated that the Spo5 strain in which the native Spo5 was designed to be expressed from its native promoter causes the phenotype to recover to the WT level, which confirms that Spo5 plays an essential role in spore wall formation (Fig. 2D). The cells observed under a microscope are grouped into three classes according to their ascus phenotypes; class I cells formed normal spores, class II cells formed abnormal spore walls, and class III cells did not form spore walls at all. The Spo5C mutant showed the most severe phenotype, which was similar to that of spo5Δ; namely, all cells lacked spore walls (Fig. 2D) and no viable spore was observed (Fig. 2E). This indicates that the N terminus of Spo5 plays a key role in spore wall formation. The three strains expressing Spo5 containing a point mutation in F341 and/or F427 failed to form spore walls similar to those of the Spo5N mutant that lacks both RRMs. However, the spore viability of these mutants was almost normal (Fig. 2E). These results indicate that the F341 and F427 residues of Spo5 in RRM-1 and RRM-2 also play important roles in the full function of Spo5, but the N-terminal portion of Spo5 that does not harbor RRMs is more important for sporulation.
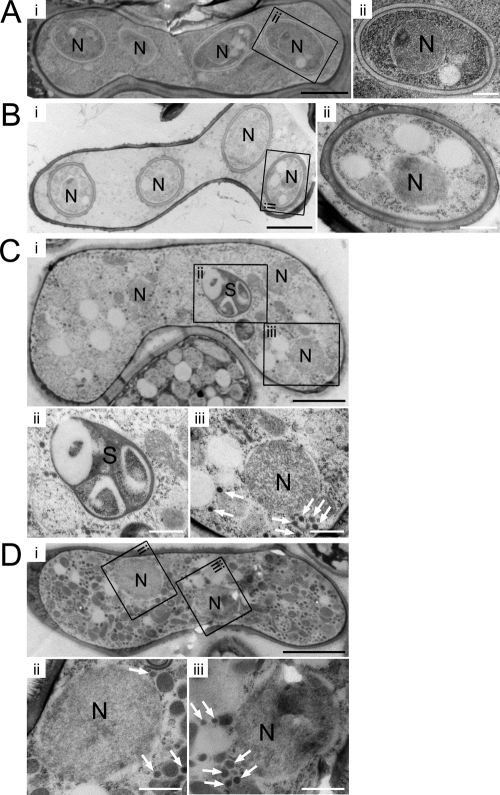
Spore morphology as observed by electron microscopy.
To investigate the structure of the forespore membrane and the spore wall of spo5Δ cells in more detail, we examined their morphologies by thin-section EM. The EM images confirmed that the spores of spo5 cells, which are spo5Δ cells that express native Spo5 (Fig. 3B), resemble the spores of WT cells (Fig. 3A). In contrast, the EM images revealed that spo5Δ cells did not form a spore wall at all (Fig. 3C). Enlarged pictures show many small vesicles and the spore-like body without a nucleus (Fig. 3C, panels ii and iii) in most of the spo5Δ cells. Since these small vesicles are thought to be the precursors of the forespore membrane (30), this abnormal accumulation of small vesicles appears to be the causative phenotype that leads to the failure of spo5Δ cells to form a forespore membrane. The spore-like body may represent a structural body in which the spore wall materials are abnormally organized. Similar abnormal EM images were obtained from Spo5FAFA cells (Fig. 3D), which further indicates that these two amino acids in the RRM motifs are important for Spo5 function.
FIG. 3.
Thin-section EM of WT and spo5 mutant cells after the commencement of spore wall formation. N denotes the nucleus. (A) EM images of the WT (CRL266) strain. (ii) An enlarged view of the indicated region shows normal spores. (B) EM images of the Spo5 (TK155) strain. (ii) An enlarged view shows that this strain formed normal spore walls. (C) EM images of a spo5Δ (AS18) cell that carries three nuclei, indicating the meiotic process has ceased prematurely at meiosis II. Enlarged views of the spore-like body (S) (ii) and a nucleus that lacks forespore membrane and spore wall (iii) are shown. Cells contain small vesicles in the cytoplasm (white arrows). (D) EM images of a Spo5FAFA (TK163-FAFA) cell that carries two nuclei, probably because the cell had stopped in the meiotic process before meiosis II was initiated. Enlarged views of the indicated region show the nuclei that lack a spore wall (ii and iii). The white arrows indicate small vesicles which accumulate abnormally throughout most of the cytoplasm in this strain. White scale bar, 0.5 μm. Black scale bar, 2 μm.
Spo5 is essential for forespore membrane formation and structural modification of SPB.
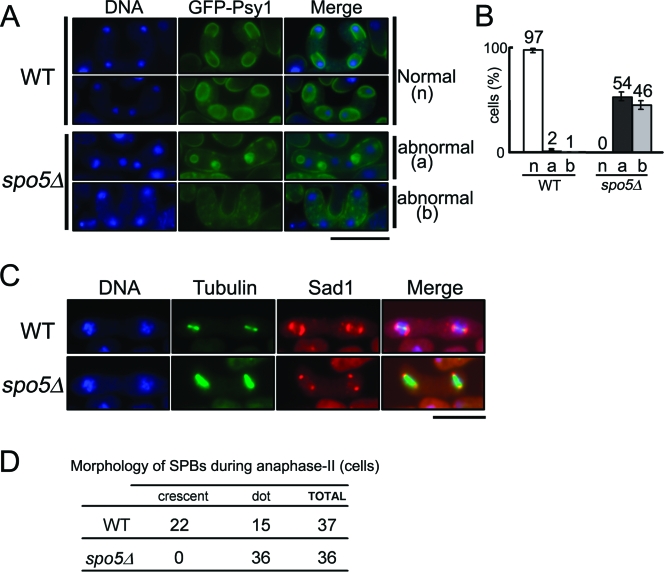
For the spore wall to be correctly constructed, it is essential for the postmeiotic nuclei to be properly encapsulated by the forespore membrane (21, 29). To examine whether the forespore membrane is formed normally in spo5Δ cells, we observed by microscopy the subcellular localization of GFP-tagged Psy1, which is a component of the forespore membrane (19). As shown in Fig. 4A, first and second rows, the GFP-Psy1 signal was detected as four strong rings (open or closed) in the cells at metaphase II in almost all WT cells (Fig. 4B). However, in all spo5Δ cells (Fig. 4B), no forespore membrane rings were observed. Moreover, GFP-Psy1 appears to either accumulate outside the nuclei, where it forms aggregated cores (Fig. 4A, third row), or remain in the plasma membrane (Fig. 4A, fourth row). These results indicate that the defective sporulation of spo5Δ cells is due to their failure to form proper forespore membranes.
FIG. 4.
The spo5Δ cells fail to form proper forespore membranes and display aberrant structural transformation of SPB. (A) Psy1, which is a component of the forespore membrane, localizes abnormally in spo5Δ cells. YN68 (GFP-psy1+) and AS44 (GFP-psy1+ spo5Δ) cells were induced to enter meiosis, after which they were stained by Hoechst 33342 (blue) and GFP-Psy1 (green) was observed by fluorescence microscopy. The localizations of Psy1-GFP in WT cells were classified as normal (n), while the localizations of Psy1-GFP in spo5Δ cells were classified as abnormal (a and b). The bar represents 10 μm. (B) Bar graphs showing the frequency of a, b, and n in WT and spo5Δ cells. The data show the averages of three independent experiments with standard errors (error bars). (C) spo5Δ cells are defective in the structural transformation of SPBs that occurs during meiosis II. WT cells (AS6) and spo5Δ cells (AS17) were induced to enter meiosis and then fixed and immunostained with Hoechst 33342 (blue), TAT-1 (green), or anti-Sad1 antibody (red). Merged images are also shown. The bar represents 10 μm. (D) The frequency of crescent SPBs in anaphase II differs between WT and spo5Δ cells.
At meiosis II, the SPBs of WT cells are known to serve as the starting site for the formation of the forespore membrane (prospore membrane in budding yeast) and prior to forespore membrane formation to transform from a dot into a crescent shape (15). This transformation is not observed in spo15Δ cells, which are defective in forespore membrane formation (7). Thus, we examined whether the structural modification of SPBs occurs in spo5Δ cells by immunostaining them with anti-Sad1 antibody, which detects SPBs. As shown in Fig. 4C and D, the spo5Δ cells did not exhibit any crescent forms during meiosis II. These results suggest that the defective formation of the forespore membrane in spo5Δ cells is induced by the defective structural modification of SPB.
Spo5 plays a role in prophase I.
To accurately determine the timing of Spo5 function, we examined the meiotic progression of h90 spo5Δ diploid cells after they were induced to enter zygotic meiosis. We found that spo5Δ cells are normal in the initiation of the horsetail phase and meiosis I. After that, however, spo5Δ cells harboring two nuclei did not disappear even at 16 h after meiotic induction, by which time most WT cells had completed sporulation (see Fig. S4A in the supplemental material).
We next examined the synchronous meiotic progression of spo5Δ pat1-114 diploid cells after they were induced to enter meiosis by temperature shift. We found that these cells also showed abnormal meiotic progression, as they displayed a delayed peak of cells with two nuclei, and only 50% of the cells were found to carry three or four nuclei 8 h after the temperature shift, at which point almost all spo5+ pat1-114 diploid cells carried three or four nuclei (Fig. 5C, upper panels). After 8 h, many of the spo5Δ cells with four nuclei lysed and died. Fluorescence-activated cell sorter analysis indicated that premeiotic DNA synthesis in spo5Δ pat1-114 diploid cells looked normal (see Fig. S3 in the supplemental material). The examination of the timing of meiotic DSB formation by PFGE analysis revealed that the formation and/or repair of DSB were retarded in spo5+ pat1-114 diploid cells (Fig. 5A). We also found that the frequency of intragenic recombination of the Spo5FAFA mutant was largely reduced compared with that of the WT, indicating that Spo5 plays a role in meiotic recombination at prophase I (Fig. 5B).
To compare Spo5 expression with that of other meiotic regulators, we used HB buffer to prepare whole-cell extracts from pat1-114 diploid cells every 30 min after inducing meiosis, and the extracts were subjected to Western blot analysis (see Materials and Methods). As shown in Fig. 5C (lower panels), during the synchronized meiosis of pat1-114 diploid cells, Spo5 appeared early in prophase I (2 h), peaked at the middle of prophase I (3 to 4 h), and then abruptly disappeared during meiosis I and meiosis II. Notably, Spo5 displayed a band shift during prophase I (Fig. 5C, uppermost panel), with the intensity of the upper band gradually increasing after 3 h, while the lower band almost disappeared at 4 h. This suggests that Spo5 is somehow modified during prophase I. It is not yet clear whether this band shift is due to phosphorylation because Spo5 protein is extracted in an insoluble fraction and resists biochemical analysis. Spo5 appeared slightly later than the tyrosine 15 phosphorylation of Cdc2 and Cdc13 (cyclin B), which appears around premeiotic S phase (2 to 3 h). Spo5 disappeared at about the same time as the tyrosine 15 phosphorylation of Cdc2 and slightly earlier than that of Cdc13. Cdc2 and Cdc13 are lost on MII entry. Taken together, Spo5 is expressed in the interval between premeiotic S phase and MII.
When we compared this pattern to that in spo5Δ pat1-114 diploid cells, we found that, while the band showing the tyrosine 15 phosphorylation of Cdc2 became weaker, it did not disappear even 8 h after meiosis had been induced (Fig. 5C, right panels). The band for Cdc13 also weakened but continued to remain until 7 to 8 h after the induction of meiosis. In contrast, in WT cells, these bands disappeared sharply 4 to 5 h after meiotic induction (Fig. 5C, left panels). We also obtained similar data using the cell extracts dissolved in TCA (see Fig. S4B in the supplemental material). These results indicate that the tyrosine 15 dephosphorylation of Cdc2 and the degradation of Cdc13 in meiosis I are incomplete in spo5Δ cells, which suggests that Spo5 plays a pivotal role in the early events of meiosis I (see Discussion).
Spo5 appears predominantly during meiosis I.
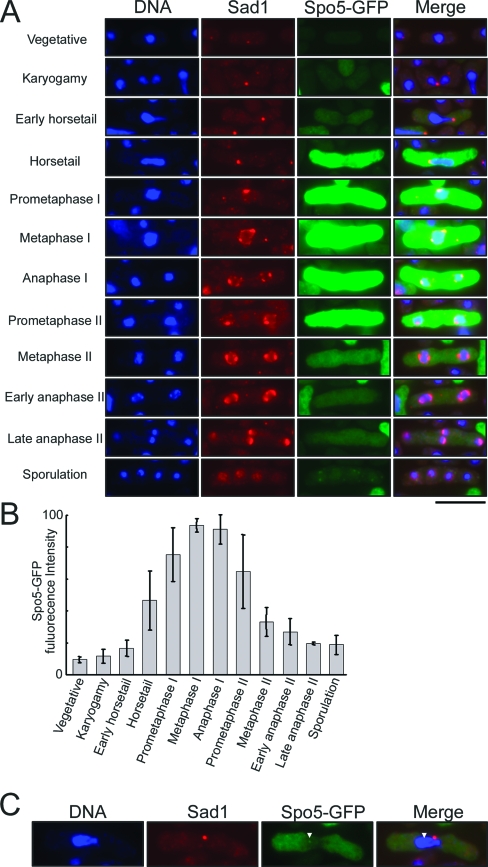
To accurately determine when Spo5 appears and disappears in meiosis, we examined the subcellular localization of Spo5 under a microscope. To do so, we prepared a Spo5-GFP-expressing strain in the h90 genetic background. The cells were induced to enter zygotic meiosis, fixed by glutaraldehyde and paraformaldehyde, and stained with Hoechst 33342 to detect the nucleus and with anti-Sad1 antibody to mark the SPBs for the purpose of monitoring the timing of meiotic progression. Successful expression of intact Spo5-GFP fusion protein of the expected size was confirmed by Western blot analysis (see Fig. S2B in the supplemental material). As shown in Fig. 6A (third row), no GFP signal was detected during vegetative growth phase (uppermost panels). Upon mating, however, the Spo5-GFP fusion protein appeared in the cytoplasm at the horsetail phase and peaked in the middle of meiosis I, probably around the metaphase I to anaphase I transition. Thereafter, the intensity of the GFP signal weakened slightly at prometaphase II and suddenly decreased at metaphase II. By anaphase II, the GFP signal had almost disappeared. The bar graph showing the intensity of the GFP signal clearly reveals that the most intense peaks occurred around metaphase I and anaphase I (Fig. 6B). This subcellular behavior of Spo5-GFP protein is consistent with the results obtained by Western blot analysis (Fig. 5C).
FIG. 6.
Subcellular localization of Spo5-GFP in chemically fixed AS17 (h90 spo5+-GFP) cells. (A) Fluorescence from Spo5-GFP was observed at various stages of meiosis after meiosis was induced by nitrogen starvation. To correctly monitor the stages of meiotic progression, the cells were immunostained with the anti-Sad1 antibody to mark the SPB. Images analyzed by fluorescence microscopy for DNA (blue), Spo5-GFP (green), and SPB (red) were merged and are shown in the rightmost panels. Bar, 10 μm. (B) Bar graphs showing the frequency of cells bearing strong Spo5-GFP signals at each stage of meiosis. The fluorescence intensity from each cell was measured under a microscope and averaged. The number of counted cells is denoted in each bar. Error bars indicate standard deviations. (C) The Spo5-GFP signal occurs as a nuclear dot in some AS17 cells that are undergoing meiosis.
Interestingly, in some of the cells at the horsetail phase, the GFP signals appeared as one or two dots in the nucleus that resemble a Mei2 dot (Fig. 6C). It was difficult to determine whether these Spo5 dots also occur in the cells at prometaphase I and anaphase I since the GFP signals were too strong. On the other hand, no Spo5 dots were detected in the cells at prometaphase II.
Spo5 forms a Mei2 dot-like focus in the nucleus.
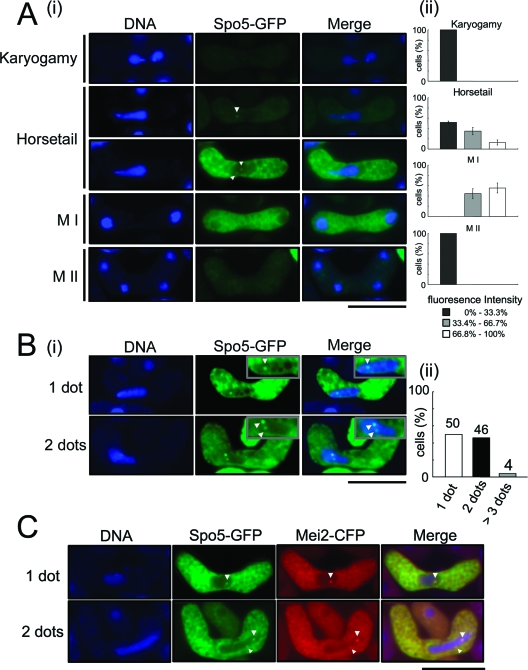
To examine the Spo5 dot in more detail in live cells, we obtained images of meiotic Spo5-GFP-expressing cells under a microscope. As shown in Fig. 7A, this analysis revealed that, during meiosis (panel i, second and third rows), Spo5-GFP has a punctuate distribution in the cytoplasm that peaks around prophase I (horsetail phase) and meiosis I (panel ii). Moreover, we could observe the GFP signal as dots in the nucleus during the horsetail phase more frequently than we could in fixed cells (Fig. 7A, panel i, second and third rows). The Spo5 dot appears at the horsetail phase and later disappears at late meiosis I and meiosis II. The number of Spo5 dots in the nucleus varies; about half of the cells had one or two dots, while 4% of all cells carried aberrant number of dots, namely, more than three dots (Fig. 7B, lower panel).
FIG. 7.
Live observation of Spo5-GFP cells. (A) Spo5-GFP fluorescence during meiosis. (i) AS17 (h90 spo5+-GFP) cells induced to enter meiosis were analyzed by fluorescence microscopy for DNA (blue) and Spo5-GFP (green). The merged images of the two signals are shown. Spo5 dots, which are indicated by the white arrowheads, appear in the nucleus at meiotic prophase I. Bar, 10 μm. (ii) A bar graph showing the Spo5-GFP fluorescence intensity in cells at each stage. The strongest and weakest GFP signal intensities that were observed were set to 100% and 0%, respectively. On the basis of this, the fluorescent intensity of each cell was classified into one of three groups. The data show the averages of three independent experiments with standard errors (error bars). (B) Examples of cells showing more than one dot in the prophase I nucleus. (i) The localization of Spo5-GFP in cells at meiotic prophase was observed, and some typical images are shown. The frequency of cells with one, two, or three Spo5-GFP dots was assessed by counting 120 cells in prophase I. The insets show enlarged images of the Spo5 dots in the nucleus, as indicated by the white arrowheads. Bar, 10 μm. Error bars indicate standard deviations. (ii) The bar graph shows the frequency of cells with one, two, or three Spo5 dots at prophase I. (C) Colocalization of Spo5 dots with Mei2 dots during meiotic prophase I. AS24 (h90 spo5+-GFP mei2+-CFP) cells induced to enter meiosis were analyzed by fluorescence microscopy for DNA (blue), Spo5-GFP (green), and Mei2-CFP (red) expression. The merged images of these signals are shown in the rightmost panels. The dots are indicated by white arrowheads. Bar, 10 μm.
To examine whether the Spo5 dot colocalizes with the Mei2 dot, we constructed the h90 spo5-GFP mei2-CFP strain and observed the Mei2-cyan fluorescent protein (CFP) signal in these cells under a microscope 6 h after meiosis was induced by nitrogen starvation. We found that when a single Spo5 dot was present in the cell, it colocalized with a single Mei2 dot in the nucleus (Fig. 7C, upper panels). Moreover, all cells carrying two Spo5 dots carry two Mei2 dots, and both of these dots colocalized with each other (Fig. 7C, lower panels). In these experiments, we confirmed that the Spo5-GFP signal was not detected by the CFP filter and that the Mei2-CFP signal was not detected by the GFP filter (data not shown); this excludes the possibility that the colocalizations are due to artifacts. These results suggest that Spo5 and Mei2 behave similarly in the nucleus during meiosis.
It is known that the Mei2 dot localizes at the sme2 locus on chromosome II (28) and that Mei2 dot formation is dependent on meiRNA expressed from the sme2 gene. To determine whether Spo5 dot formation is also dependent on the sme2 gene, we constructed the h90 spo5-GFP sme2Δ strain. As shown in Fig. S6A in the supplemental material, these cells contain Spo5 dots, which indicates that the formation of the Spo5 dot is independent of the sme2 gene. Similarly, Mei2 dots can be observed in spo5Δ cells, which indicate that the Mei2 dot forms independently of the spo5+ gene (see Fig. S6B in the supplemental material). To examine whether the Spo5 and Mei2 proteins interact, we performed a two-hybrid assay and found they interact only weakly, if at all (see Fig. S5 in the supplemental material). We also performed pull-down assays with affinity-purified GST-Spo5 fusion protein and the cell lysate of the mei2-9myc+ strain; this technique also failed to detect a significant interaction between the two proteins (data not shown). These results suggest the existence of a putative ncRNA gene that is required for Spo5 dot formation in the vicinity of the sme2 gene. Its gene product may form a complex with Spo5 and play an important role in meiotic progression in a manner similar to, but independent of, the Mei2/meiRNA complex.
Subcellular localization of mutated Spo5.
To determine which part of the Spo5 molecule is important for its subcellular localization, we induced the h90 spo5 mutant strains described previously (Fig. 2C) to enter meiosis by nitrogen starvation (see Fig. S7 in the supplemental material). We found that, at the horsetail phase, both Spo5N and Spo5C display a ubiquitous distribution in the cytoplasm and nucleus, whereas WT Spo5 is present as one or two dots in the nucleus (see Fig. S7A in the supplemental material). Since Spo5FAFA shows a punctuate distribution in both the cytoplasm and nucleus at the horsetail phase, it is difficult to determine whether it forms a dot in the nucleus.
In cells at meiosis I, similar abnormal subcellular localizations were observed (see Fig. S7B in the supplemental material). Notably, Spo5N showed intense nuclear signals compared to those shown by the WT and other Spo5 mutants, which suggests that the C-terminal portion of Spo5, containing both RRM motifs (F341 and F427), plays an important role in directing the correct subcellular localization of Spo5.
In sporulated mutant cells that failed to form spore walls, the mutated Spo5N and Spo5C proteins appeared to remain in the nucleus of the spores (see Fig. S5C in the supplemental material). Spo5FAFA protein was also visible throughout the asci. In contrast, no WT Spo5 was visible in the asci, which is consistent with the results shown above indicating that Spo5 is absent at the late stages of meiosis (Fig. 5C and 6A).
DISCUSSION
In the present study, we report that the putative RNA-binding protein Spo5 displays meiosis-specific expression that peaks around prophase I (Fig. 1 and 5). An examination of its subcellular localization indicated that Spo5 appears at the horsetail phase (prophase I) and disappears at the beginning of meiosis II (Fig. 5 and 6). These kinetics of expression were confirmed by Western blot analysis (Fig. 5). The disruption of Spo5 induced abnormal sporulation due to failed forespore membrane formation (Fig. 4) and the absence of a spore wall (Fig. 2 and 3). Indeed, these phenotypes caused spo5+ to be described originally as the sporulation mutant spo5 (6). We also found that the SPB modification that normally occurs around the end of meiosis II was aberrant in spo5Δ cells (Fig. 4). Since, in spo5Δ cells, the premeiotic DNA synthesis looks normal, as determined by fluorescence-activated cell sorter analysis (see Fig. S3 in the supplemental material), and meiosis I starts in a timely fashion, as judged by Western blot analysis of the tyrosine 15 phosphorylation of Cdc2 (Fig. 5), we conclude that Spo5 functions somewhere between prophase I and meiosis II. Given that, in spo5Δ cells, the peaking of cells bearing two nuclei was delayed by more than 1 h (Fig. 5C) and the Spo5-GFP signal in spo5+-GFP-expressing cells was maximal between metaphase I and anaphase I (Fig. 6), we speculate that Spo5 primarily regulates the progression from prophase I to anaphase I and its putative function around meiosis I and meiosis II (see below) may be its secondary function. Retarded formation and/or repair of DSBs (Fig. 5A) and the reduced frequency of intragenic recombination (Fig. 5B) in spo5 mutant cells also indicate that Spo5 mainly functions at prophase I.
In the fission yeast, meiosis-specific alternative splicing plays an essential role in controlling gene expression during meiosis (1). We examined whether Spo5 regulates the splicing of these meiotically regulated genes by RT-PCR using the strategy described in Fig. 1B. However, spo5Δ cells did not show any abnormalities in mRNA splicing (see Fig. S8 in the supplemental material). Thus, Spo5 is not involved in the alternative splicing of these genes.
Of the various phenotypes of spo5Δ cells, one was that their abnormal progression of meiosis I caused the incomplete degradation of Cdc13 (cyclin B) and the insufficient tyrosine 15 dephosphorylation of Cdc2 (Fig. 5C and see Fig. S4B in the supplemental material). During meiosis in S. pombe, the incomplete degradation of Cdc13 occurs as the cells exit from meiosis I and initiate meiosis II; this degradation event is regulated by the meiosis-specific small protein Mes1 (8). Cdc13 and Mes1 can interact with the same domain in Slp1 (Cdc20 in Saccharomyces cerevisiae), which functions as a substrate adaptor that recruits substrates to APC/C, thereby activating this ubiquitin ligase (31). Consequently, Mes1 may be a competitive inhibitor of the binding of Cdc13 to APC/CSlp1 (22). Thus, Mes1 regulates ubiquitination activity to a level that permits sufficient separase activation but is not enough to result in complete Cdc13 inactivation.
Since the degradation of Cdc13 is partially inhibited in spo5Δ cells and Mes1 plays an essential role in the progression of meiosis from meiosis I to meiosis II, we surmised that Spo5 may suppress the role Mes1 plays in regulating Cdc13 stability. To test this, we examined whether the absence of Mes1 expression in spo5Δ cells would cause them to recover their abilities to progress through meiosis II normally. However, when we prepared the spo5Δ mes1Δ double mutant strain and examined its meiotic progression, we found that it ceased meiosis before entering meiosis II, which is a phenotype of the mes1Δ single mutant cells (data not shown). Thus, Spo5 acts earlier than Mes1 in meiotic progression, and Spo5 does not seem to regulate the function of Mes1 in determining Cdc13 stability.
We also report here that Spo5 forms a dot structure in the horsetail nucleus during meiotic prophase I (Fig. 7C), and that this dot colocalizes with the Mei2 dot, which is formed off the sme2 gene locus on chromosome II (28). Mei2 dot formation requires the meiosis-specific RNA species meiRNA, which is expressed from the sme2 gene (35). Consequently, we expected that the Spo5 dot would, like the Mei2 dot, also disappear in sme2Δ cells. However, it appears that the Spo5 dot is formed independently of the sme2 gene (see Fig. S6A in the supplemental material). Moreover, while Mei2 is essential for inducing both premeiotic DNA synthesis and meiosis I (34), premeiotic DNA synthesis was normal in spo5Δ cells (see Fig. S3 in the supplemental material). This indicates that Spo5 functions later in meiotic progression than Mei2 does. Furthermore, the structure of Spo5 is essentially distinct from that of Mei2 since it carries two RRMs while Mei2 carries three RRMs. In addition, Mei2 dot formation is independent of Spo5 since Mei2 dots can be observed in spo5Δ cells (see Fig. S6B in the supplemental material). Finally, Spo5 and Mei2 were not found to interact directly (see Fig. S5 in the supplemental material). These results indicate that the Spo5 dot is essentially distinct from the Mei2 dot and that Spo5 and Mei2 behave independently during meiosis. Nonetheless, we speculate that these two molecules may still interact, albeit indirectly, by associating with an as-yet-uncharacterized large complex that localizes in the vicinity of the sme2 gene locus on chromosome II.
Like Mei2, which intrinsically undergoes nucleocytoplasmic shuttling (26), the primary part of Spo5 is excluded from the nucleus and is localized in the cytoplasm (Fig. 7). However, in Spo5 mutant cells that are defective in sporulation, the Spo5-GFP signal is observed homogeneously in both the cytoplasm and nucleus throughout meiosis (see Fig. S7 in the supplemental material). This indicates that the exclusion of Spo5 from the nucleus is defective in these mutants, even though no nuclear export signal was found in Spo5. Thus, it is not clear whether the Spo5 dot is formed in the nucleus in these mutant cells. Since both Spo5N and Spo5C mutants are unable to be efficiently exported, it appears that the proper export of Spo5 requires its full size. The functions of Spo5 in both the cytoplasm and nucleus remain to be determined.
Supplementary Material
Acknowledgments
We are obliged to Mitsuhiro Yanagida, Chikashi Shimoda, and Masayuki Yamamoto for S. pombe strains and antibodies. We thank the Yeast Genetics Resource Center Japan (http://bio3.tokyo.jst.go.jp/jst/) for S. pombe strains. We also thank Zhao Hanjun and Jun Sato for technical advice in preparing the anti-Spo5 antibody, Takashi Morishita for PFGE, and Patrick Hughes for critically reading the manuscript.
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas and Scientific Research (S) from the Ministry of Education, Science, Sports and Culture of Japan.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Averbeck, N., S. Sunder, N. Sample, J. A. Wise, and J. Leatherwood. 2005. Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol. Cell 18:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Carninci, P., T. Kasukawa, S. Katayama, J. Gough, M. C. Frith, N. Maeda, R. Oyama, T. Ravasi, B. Lenhard, C. Wells, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309:1559-1563. [DOI] [PubMed] [Google Scholar]
- 3.Gregan, J., P. K. Rabitsch, B. Sakem, O. Csutak, V. Latypov, E. Lehmann, J. Kohli, and K. Nasmyth. 2005. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr. Biol. 15:1663-1669. [DOI] [PubMed] [Google Scholar]
- 4.Grimm, C., J. Kohli, J. Murray, and K. Maundrell. 1988. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable maker. Mol. Gen. Genet. 215:81-86. [DOI] [PubMed] [Google Scholar]
- 5.Hagan, I., and S. J. Hyams. 1988. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 89:45-52. [DOI] [PubMed] [Google Scholar]
- 6.Hirata, A., and C. Shimoda. 1994. Structural modification of spindle pole bodies during meiosis II is essential for the normal formation of ascospores in Schizosaccharomyces pombe: ultrastructural analysis of spo mutants. Yeast 10:173-183. [DOI] [PubMed] [Google Scholar]
- 7.Ikemoto, S., T. Nakamura, M. Kubo, and C. Shimoda. 2000. S. pombe sporulation-specific coiled-coil protein Spo15p is localized to the spindle pole body and essential for its modification. J. Cell Sci. 113:545-554. [DOI] [PubMed] [Google Scholar]
- 8.Izawa, D., M. Goto., A. Yamashita, H. Yamano, and M. Yamamoto. 2005. Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature 434:529-533. [DOI] [PubMed] [Google Scholar]
- 9.Jeffares, D. C., M. J. Phillips, S. Moore, and B. Veit. 2004. A description of the Mei2-like protein family; structure, phylogenetic distribution and biological context. Dev. Genes Evol. 214:149-158. [DOI] [PubMed] [Google Scholar]
- 10.Kakihara, Y., K. Nabeshima, A. Hirata, and H. Nojima. 2003. Overlapping omt1+ and omt2+ genes are required for spore wall maturation in Schizosaccharomyces pombe. Genes Cells 8:547-558. [DOI] [PubMed] [Google Scholar]
- 11.Kanaoka, Y., and H. Nojima. 1994. SCR: novel human suppressors of cdc2/cdc13 mutants of Schizosaccharomyces pombe harbour motifs for RNA binding proteins. Nucleic Acids Res. 22:2687-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin-Castellanos, C., M. Blanco, A. Rozalen, L. Perez-Hidalgo, A. I. Garcia, F. Conde, J. Mata, C. Ellermeier, L. Davis, P. San-Segundo, G. R. Smith, and S. Moreno. 2005. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr. Biol. 15:2056-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mata, J., R. Lyne, G. Burns, and J. Bahler. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32:143-147. [DOI] [PubMed] [Google Scholar]
- 14.Molnar, M., S. Parisi, Y. Kakihara, H. Nojima, A. Yamamoto, Y. Hiraoka, A. Bozsik, M. Sipiczki, and J. Kohli. 2001. Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics 157:519-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno-Borchart, A. C., K. Strasser, M. G. Finkbeiner, A. Shevchenko, A. Shevchenko, and M. Knop. 2001. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 20:6946-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morishita, T., Y. Tsutsui, H. Iwasaki, and H. Shinagawa. 2002. The Schizosaccharomyces pombe rad60 gene is essential for repairing double-strand DNA breaks spontaneously occurring during replication and induced by DNA-damaging agents. Mol. Cell. Biol. 22:3537-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nabeshima, K., Y. Kakihara, Y. Hiraoka, and H. Nojima. 2001. A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J. 20:3871-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura, T., M. Kishida, and C. Shimoda. 2000. The Schizosaccharomyces pombe spo6+ gene encoding a nuclear protein with sequence similarity to budding yeast Dbf4 is required for meiotic second division and sporulation. Genes Cells 5:463-479. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura, T., M. Nakamura-Kubo, A. Hirata, and C. Shimoda. 2001. The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1+-encoding syntaxin-like protein. Mol. Biol. Cell 12:3955-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura, T., M. Nakamura-Kubo, T. Nakamura, and C. Shimoda. 2002. Novel fission yeast Cdc7-Dbf4-like kinase complex required for the initiation and progression of meiotic second division. Mol. Cell. Biol. 22:309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okuzaki, D., W. Satake, A. Hirata, and H. Nojima. 2003. Fission yeast mue14+ is required for proper nuclear division and accurate forespore membrane formation during meiosis II. J. Cell Sci. 116:2721-2735. [DOI] [PubMed] [Google Scholar]
- 22.Peters, J. M. 2005. Cyclin degradation: don't Mes(s) with meiosis. Curr. Biol. 15:R461-R463. [DOI] [PubMed] [Google Scholar]
- 23.Saito, T. T., T. Tougan, T. Kasama, D. Okuzaki, and H. Nojima. 2004. Mcp7, a meiosis-specific coiled-coil protein of fission yeast, associates with Meu13 and is required for meiotic recombination. Nucleic Acids Res. 32:3325-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito, T. T., T. Tougan, D. Okuzaki, T. Kasama, and H. Nojima. 2005. Mcp6, a meiosis-specific coiled-coil protein of Schizosaccharomyces pombe, localizes to the spindle pole body and is required for horsetail movement and recombination. J. Cell Sci. 118:447-459. [DOI] [PubMed] [Google Scholar]
- 25.Saito, T. T., D. Okuzaki, and H. Nojima. 2006. Mcp5, a meiotic cell cortex protein, is required for nuclear movement mediated by dynein and microtubules in fission yeast. J. Cell Biol. 173:27-33. [DOI] [PMC free article] [PubMed]
- 26.Sato, M., S. Shinozaki-Yabana, A. Yamashita, Y. Watanabe, and M. Yamamoto. 2001. The fission yeast meiotic regulator Mei2p undergoes nucleocytoplasmic shuttling. FEBS Lett. 499:251-255. [DOI] [PubMed] [Google Scholar]
- 27.Shimada, M., K. Nabeshima, T. Tougan, and H. Nojima. 2002. The meiotic recombination checkpoint is regulated by checkpoint rad+ genes in fission yeast. EMBO J. 11:2807-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimada, T., A. Yamashita, and M. Yamamoto. 2003. The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol. Biol. Cell 14:2461-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimoda, C. 2004. Forespore membrane assembly in yeast: coordinating SPBs and membrane trafficking. J. Cell Sci. 117:389-396. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka, K., and A. Hirata. 1982. Ascospore development in the fission yeasts Schizosaccharomyces pombe and S. japonicus. J. Cell Sci. 56:263-279. [DOI] [PubMed] [Google Scholar]
- 31.Vodermaier, H. C. 2001. Cell cycle: waiters serving the destruction machinery. Curr. Biol. 11:R834-R837. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe, T., K. Miyashita, T. T. Saito, T. Yoneki, Y. Kakihara, K. Nabeshima, Y. A. Kishi, C. Shimoda, and H. Nojima. 2001. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 29:2327-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe, T., K. Miyashita, T. T. Saito, K. Nabeshima, and H. Nojima. 2002. Abundant poly(A)-bearing RNAs that lack open reading frames in Schizosaccharomyces pombe. DNA Res. 9:209-215. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe, Y., and M. Yamamoto. 1994. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78:487-498. [DOI] [PubMed] [Google Scholar]
- 35.Yamashita, A., Y. Watanabe, N. Nukina, and M. Yamamoto. 1998. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell 95:115-123. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto, M. 1996. The molecular control mechanisms of meiosis in fission yeast. Trends Biochem. Sci. 21:18-22. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.