Abstract
Proteins of the mitochondrial carrier family (MCF) are located mainly in the inner mitochondrial membrane and mediate the transport of a large range of metabolic intermediates. The genome of Trypanosoma brucei harbors 29 genes encoding different MCF proteins. We describe here the characterization of MCP6, a novel T. brucei MCF protein. Sequence comparison and phylogenetic reconstruction revealed that MCP6 is closely related to different mitochondrial ADP/ATP and calcium-dependent solute carriers, including the ATP-Mg/Pi carrier of Homo sapiens. However, MCP6 lacks essential amino acids and sequence motifs conserved in these metabolite transporters, and functional reconstitution and transport assays with E. coli suggested that this protein indeed does not function as an ADP/ATP or ATP-Mg/Pi carrier. The subcellular localization of MCP6 is developmentally regulated: in bloodstream-form trypanosomes, the protein is predominantly glycosomal, whereas in the procyclic form, it is found mainly in the mitochondria. Depletion of MCP6 in procyclic trypanosomes resulted in growth inhibition, an increased cell size, aberrant numbers of nuclei and kinetoplasts, and abnormal kinetoplast morphology, suggesting that depletion of MCP6 inhibits division of the kinetoplast.
The order Kinetoplastida includes among its members a number of important human and veterinary pathogens, such as the African trypanosome Trypanosoma brucei (83). This parasite has a complex life cycle that alternates between the bloodstream form found in the blood and tissue fluids of mammals and the procyclic (insect) form in the midgut of the tsetse fly (46, 83). T. brucei undergoes a series of metabolic and morphological changes in order to adapt to these distinct host environments (45, 46). Bloodstream-form T. brucei metabolizes glucose to pyruvate and glycerol, obtaining ATP by substrate-level phosphorylation during glycolysis (48, 55). The mitochondria of this life form are highly reduced and lack key enzymes and components of the tricarboxylic acid cycle; their role in energy metabolism is probably restricted to the reoxidation of glycerol-3 phosphate by the mitochondrial alternative oxidase (20, 48). This is in contrast to the procyclic form, which metabolizes amino acids and has a well-developed mitochondrion in which ATP is generated by a combination of substrate-level and oxidative phosphorylation (16, 48, 73, 82).
In both life forms of T. brucei, most of the glycolytic enzymes are found within a peroxisome-like organelle called the glycosome (14, 48, 60, 61). This unique compartmentation of glycolytic enzymes has been shown to be essential for trypanosome survival (5, 11, 22, 25, 51). It has been proposed that the glycosomal membrane forms a barrier to ATP and ADP, insulating the enzymes of the glycosomal matrix from the ADP/ATP ratio in the cytosol (5).
So far, no glycosomal or mitochondrial metabolite carriers have been identified for T. brucei, although they most probably play a key role in the regulation of energy metabolism. The characterization of these carriers should make it possible to build more-accurate biochemical and mathematical models of trypanosome energy metabolism (5).
The mitochondrial carrier family (MCF) was initially defined as a group of proteins that are located in the inner mitochondrial membrane and mediate the transport of a large range of metabolic intermediates (3, 32, 35, 57-59, 86). Recently, several structurally and functionally related carrier proteins were also found in the membranes of peroxisomes (52, 81, 88, 90). We therefore wondered whether such proteins might also be found in kinetoplastid glycosomes. The recently completed genome sequence of T. brucei (8) contains about 29 genes encoding different proteins of the mitochondrial carrier family. In this paper, we describe the characterization of MCP6, a novel mitochondrial carrier protein homologue from T. brucei.
MATERIALS AND METHODS
Culture and transfection.
Trypanosoma brucei cell lines were cultured in either HMI-9 medium for bloodstream cell lines (29) or MEM-PROS medium for procyclic cell lines (56), supplemented with 10% (vol/vol) fetal calf serum (Sigma-Aldrich). Cells were transfected as described previously (9). In all experiments, “wild type” refers to bloodstream or procyclic T. brucei strain 449 cell lines, stably expressing the tetracycline (tet) repressor from the plasmid pHD449 (9).
Cloning and sequencing.
The open reading frame of MCP6 (1,155 bp) was amplified by PCR from genomic DNA of T. brucei strain Lister 427, using the sense primer 5′-GGACGGAAGCTTACCATGGGGGTTGAAGTAGCAGACTGCAGTAGTG-3′ and antisense primer 5′-GCTTGCAGTTAACGTCATCACCGACCAAAATGCTCTGAACCTTTTC-3′. The restriction enzyme sites HindIII and HpaI, which were used for subsequent cloning in different expression plasmids, are underlined. The resulting PCR product was cloned into the pGEM-T Easy TA cloning vector (Invitrogen) and sequenced (Medigenomix, Martinsried, Germany). Comparison of the MCP6 sequence from T. brucei Lister 427 with the sequence of the corresponding locus Tb927.4.1660 in the T. brucei strain 927 genome sequence database (available at http://www.genedb.org) revealed a few sequence differences at the DNA level but none in the predicted amino acid sequence.
Sequence alignment and phylogenetic reconstruction of MCP6.
Database searches of the deduced MCP6 sequence were done using the program BLASTP version 2.2.9 (http://www.ncbi.nlm.nih.gov/BLAST) and the sequence databases available at http://www.genedb.org (for T. brucei, Trypanosoma cruzi, and Leishmania major genome databases) and http://www.ncbi.nlm.nih.gov. Multiple alignments of amino acid sequences were obtained using Clustal X v1.83 (ftp.embl-heidelberg.de) (30) and were optimized manually using the SE-AL (Sequence Alignment Editor) program v2.0a11 (http://evolve.zoo.ox.ac.uk). Putative trans-membrane α-helices were predicted by the TMHMM (CBS, Denmark) and TMpred (EMBnet-CH) programs available at http://www.expasy.ch. The phylogenetic tree was constructed using the PHYLIP program package version 3.6a of J. Felsenstein available at http://evolution.genetics.washington.edu. Pairwise sequence distance matrices were calculated using Prodist (19). Unrooted phylogenetic trees were constructed from the distance matrices using the neighbor-joining method (69). The final phylogenetic tree was drawn with TreeView version 1.6.6. The statistical robustness of the resulting phylogenetic tree was assessed with the SEQBOOT program of the PHYLIP package by bootstrap resampling analysis generating 1,000 reiterated data sets. The resulting bootstrap values, expressed as percentages, were added manually at each node. Only bootstrap values above 60% are shown.
The GenBank (gb), EMBL (emb) and Swissprotein (sp) accession numbers are as follows: HsapGDC, Homo sapiens sp_P16260; BtauGDC, Bos taurus sp_Q01888; ScerLEU5, Saccharomyces cerevisiae NP_011865; CeleGDC, Caenorhabditis elegans NP_492333; ScerADT1, S. cerevisiae emb_CAA89766; TbruAAC, Trypanosoma brucei gb_AAC23561; HsapADT1, H. sapiens sp_P12235; HsapADT2, H. sapiens sp_P05141; ScerMRS4, S. cerevisiae sp_P23500; ScerMRS3, S. cerevisiae sp_P10566; ScerSAM5, S. cerevisiae NP_014395; BtauMPCP, B. taurus sp_P12234; HsapMPCP, H. sapiens sp_Q00325; ScerPic2, S. cerevisiae NP_010973; ScerMir1, S. cerevisiae NP_012611; HsapCMC2, H. sapiens sp_Q9UJS0; HsapCMC1, H. sapiens sp_O075746; HsapGHCl, H. sapiens sp_Q9H936; ScerTXTP, S. cerevisiae NP_014914; SpomTXTP, Schizosaccharomyces pombe NP_594262; RnorTXTP, Rattus norvegicus sp_P32089; HsapTXTP, H. sapiens sp_P53007; ScerACR1, S. cerevisiae sp_P33303; ScerODC1, S. cerevisiae NP_015191; HsapODC, H. sapiens sp_Q9BQT8; HsapORN1, H. sapiens NP_055067; HsapORT2, H. sapiens sp_Q9BXI2; RnorMCAT, R. norvegicus sp_P97521; HsapMCAT, H. sapiens sp_O43772; CeleDIF1, C. elegans sp_Q27257; NcraspARG1, Neurospora crassa sp_Q01356; HsapUCP1, H. sapiens sp_P25874; BtauUCP1, B. taurus sp_P10861; RnorDIC, R. norvegicus NP_596909; HsapDIC, H. sapiens sp_Q9UBX; HsapM2OM, H. sapiens NP_003553; BtauM2OM, B. taurus sp_P22292; NtabDIC-TX, Nicotiana tabacum emb_CAC84545; ScerPMT, S. cerevisiae NP_012802; ScerGGT1, S. cerevisiae sp_P38988; SpomFLX, S. cerevisiae NP_595541; HsapMFT, H. sapiens gb_AAG37834; ScerFLX, S. cerevisiae sp_P40464; MmusPMP34, Mus musculus sp_O70579; HsapPMP34, H. sapiens sp_O43808; XlaePMP34, Xenopus laevis emb_CAC21237; CboiPMP47A, Candida boidinii sp_P21245; HsapAPC, H. sapiens NP_077008; OcunperCSC, Oryctolagus cuniculus gb_AAB69156; HsapMCSC, H. sapiens NP_443133; AthaPCSC, Arabidopsis thaliana emb_CAB8792; ZmayBT1, Zea mays sp_P29518; TvagHMP31, Trichomonas vaginalis gb_AAF27626;t MCP6, T. brucei Tb927.4.1660.
RNA isolation and Northern blotting.
Total RNA was isolated from bloodstream or procyclic T. brucei cell lines using Trifast (PeqLab Biotechnology GmbH). mRNA was isolated from total RNA using a QuickPrep mRNA purification kit (Amersham Pharmacia Biotech). Total RNA (20 μg) and 4 μg mRNA were separated on a denaturing formaldehyde agarose gel and blotted onto a Hybond N nylon membrane (Amersham Pharmacia). The MCP6 DNA probe was labeled with [α32P]dCTP by random priming (Stratagene). After overnight hybridization, blots were subsequently washed in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate (SDS) for 30 min at room temperature, in prewarmed 1× SSC and 0.5% SDS for 45 min at 42°C, and in 0.1× SSC and 0.2% SDS for 30 min at 42°C before exposure to film.
Conditional double knockout of MCP6.
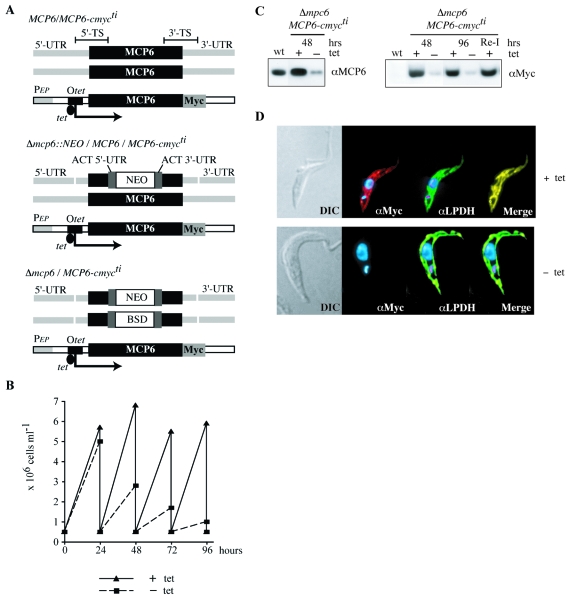
A schematic representation of the strategy used for the construction of the conditional double-knockout cell line Δmcp6/MCP6-cmycti is shown in Fig. 5A. The MCP6-encoding gene was cloned into the tetracycline-inducible expression vector pHD1484. This vector contains a hygromycin resistance gene (HYG), and the tetracycline-inducible expression from this vector will result in the addition of a double-myc tag (cmyc) to the carboxy terminus of the expressed protein. The resulting construct was used for the transfection of bloodstream- and procyclic-form T. brucei 449 cell lines, constitutively expressing the tet repressor (TETR BLE). Trypanosome clones resistant to hygromycin (HYG) and bearing a tetracycline inducible (ti) copy of MCP6-cmyc (MCP6-cmycti HYG) were isolated and used as the starting point for the double knockout of MCP6; their genotype is TETR BLE MCP6-cmycti HYG MCP6/MCP6. Tetracycline (0.5 μg ml−1) was added daily to the cultures to maintain the presence of functional MCP6, in this case a carboxy-terminal myc-tagged version of MCP6 (MCP6-cmyc).
FIG. 5.
Depletion (conditional double knockout) of MCP6 in procyclic-form trypanosomes. (A) Schematic representation of the strategy used for the construction of the procyclic conditional double-knockout cell line Δmcp6/MCP6-cmycti. Abbreviations: NEO, neomycin resistance cassette; BSD, blasticidin resistance cassette; ACT, actin; Otet, tet operator; PEP, procycline promoter; Myc, myc tag; TS, target sequence. (B) Growth curves of MCP6-expressing and MCP6-depleted procyclic trypanosomes. The procyclic conditional double-knockout cell line Δmcp6/MCP6-cmycti was grown for 96 h in medium with (triangles, solid line) or without (squares, dashed line) tetracycline. Every 24 h, samples were taken for the determination of cell density (cells ml−1), and grown cultures were diluted down to 5 × 105 cells/ml with fresh medium. (C) Western blot analysis of Δmcp6/MCP6-cmycti cells grown with (+) or without (−) tetracycline. Samples for Western blotting were taken at 48 and 96 h. (Lane Re-I) First, Δmcp6/MCP6-cmycti cells were grown for 96 h without tetracycline, after which tetracycline was added. After reinduction (Re-I) for 24 h, cells were harvested for Western blot analysis. Wild-type cells were used as a control. (D) Immunofluorescence microscopy of procyclic Δmcp6/MCP6-cmycti trypanosomes grown with (+tet) or without (−tet) tetracycline for 96 h. The antisera used were αMyc for the staining (red) of MCP6-cmyc and αLPDH for the staining (green) of mitochondrial dihydroxylipoamide dehydrogenase. DAPI staining of nucleus and kinetoplast DNA is shown in blue.
Subsequently, both MCP6 alleles were replaced by homologous recombination, using two different antibiotic resistance cassettes. Sense primer 5′-AGGGTGAGCTCTGGGCCACTTTCACACATTAGTTACCG-3′ and antisense primer 5′-ACCTGCAACTAGTAGCTGCTGTTGTGTACGAGTCTC-3′ were used to PCR amplify a 466-bp genomic fragment located upstream of the MCP6 open reading frame (ORF) (5′ untranslated region [UTR]), and sense primer 5′-CTCACCAGGATCCGCGCATACTTCTAAACTTCCTCATTG-3′ and antisense primer 5′-CCTTGGGCCCTAACCATCTTCACGAAACAGTCAGTCATG-3′ were used to amplify a 473-bp fragment downstream of the MCP6 ORF (3′ UTR). The restriction enzyme sites SacI, SpeI, BamHI, and ApaI, used for subsequent cloning, are underlined. As described previously for the construction of other T. brucei double-knockout cell lines (33, 41, 85), using the targeted gene replacement method, these targeting DNA sequences were inserted on either side of NEO and BSD genes bearing actin 5′ splice sites and actin 3′ untranslated regions, resulting in a NEO- and BSD-bearing double-knockout (dKO) construct, respectively (see Fig. 5A). After transfection with the NEO-dKO construct and subsequent selection with 15 μg ml−1 G418, different procyclic T. brucei cell lines were isolated bearing a single-allele knockout of MCP6 (Δmcp6::NEO/MCP6/MCP6-cmycti). The different double-knockout cell lines (Δmcp6::NEO/Δmcp6::BSD/MCP6-cmycti), in this paper further referred to as Δmcp6/MCP6-cmycti, were obtained after transfection of the single-allele knockout cell line Δmcp6::NEO/MCP6/MCP6-cmycti with the BSD-dKO construct and subsequent selection with both 15 μg ml−1 G418 and 10 μg ml−1 blasticidin. The correct integration of NEO- and BSD-dKO was monitored by Southern blotting.
Construction of RNAi cell lines.
For RNA interference (RNAi), primers were designed with the help of the RNAit program available at the TrypanoFan website (http://trypanofan.path.cam.ac.uk). A 181-bp DNA fragment of the coding sequence of MCP6 (nucleotides 7 to 187 of the ORF) was PCR amplified using the sense primer 5′-ACCGGATCCGTTGAAGTAGCAGACTGCAGTAGTG-3′ and antisense primer 5′-GGTCTCGAGCTGTCAGTGTGCGACTACATG-3′, with the BamHI and XhoI sites underlined. The PCR fragment was cloned into the vector p2T7TA-177 (13, 37), which contains two inducible and opposite T7 promoters for the generation of double-stranded RNA and is targeted to the transcriptionally silent 177-bp minichromosomal region. The resulting construct was linearized with NotI before transfection as described previously (9, 13). Tetracycline-dependent transcription, resulting in double-stranded RNA, was induced by the addition of 250 ng ml−1 tetracycline.
MCP6 polyclonal antibody.
The peptide SEAMTVGHEKAKEQHMHVKR (amino acids 19 to 39 of MCP6) was coupled to keyhole limpet hemocyanin and used for the immunization of guinea pigs (http://peptid.de, Heidelberg, Germany). Western blot analysis, using the generated antiserum, revealed the presence of several cross-reacting protein bands in both bloodstream and procyclic T. brucei cells, with molecular masses of 32, 43, 48, and 50 kDa (not shown), so the antiserum was further affinity purified using Affi-Gel 10 (Bio-Rad) and the MCP6 N-terminal peptide as ligand according to the manufacturer's protocol.
Western blotting.
Unless specified, for each lane, 2 ×106 cells of the different bloodstream or procyclic T. brucei cell lines were used and heated in Laemmli SDS-polyacrylamide gel electrophoresis (PAGE) buffer at 95°C for 5 min, and the proteins were subsequently separated on a denaturing 11% SDS-PAGE gel. Proteins were transferred to a nylon Hybond P membrane (Amersham Pharmacia) in transfer buffer (39 mM glycine, 48 mM Tris base, 20% methanol, pH 8.3) for 1 h at 100 V. The membranes were blocked by incubation in Tris-buffered saline (TBS) containing 7.5% (wt/vol) nonfat dry milk for 30 min at room temperature with gentle shaking and incubated with the appropriate primary antibodies for 1 h. The membranes were then washed once for 15 min and twice for 5 min in TBS supplemented with Tween 20 (0.2%, vol/vol) and incubated for 1 h at room temperature with the corresponding anti-rabbit or anti-guinea pig horseradish peroxidase-conjugated secondary antibody. Finally, the membranes were washed in TBS-Tween 20 once for 15 min and four times for 5 min and processed according to the manual of the ECL detection kit of Amersham Biosciences before exposure to film. The different antibodies were αMCP6 (this paper), αGIM5, antibody directed against the glycosomal membrane protein GIM5 of T. brucei (41), αLPDH, antibody directed against the mitochondrial dihydroxylipoamide dehydrogenase of T. brucei (70), and αMyc (Sigma-Aldrich).
Immunofluorescence microscopy.
For immunofluorescence microscopy, bloodstream and procyclic trypanosomes were centrifuged from culture medium at 2,000 × g and resuspended immediately in phosphate-buffered saline (PBS) containing 4% formaldehyde. Fixed cells were allowed to settle and attach onto poly-l-lysine-coated microscope slides. Labeling with 4′,6′-diamidino-2-phenylindole (DAPI) and different antibodies was performed as described previously (85). Cells were examined with a Leica DM RXA digital deconvolution microscope, and images were recorded using a digital charge-coupled-device camera (Hamamatsu).
Fluorescence-activated cell sorter (FACS) analysis.
2 × 107 cells each of wild-type, MCP6-cmyc-expressing (with tet), and MCP6-depleted (without tet) procyclic-form trypanosomes were collected by centrifugation, washed once with PBS, and resuspended in 70% (vol/vol) ethanol-PBS. After fixation overnight at 4°C, the cells were pelleted by centrifugation and resuspended in 1 ml of staining solution (RNase A [0.2 mg/ml], propidium iodide [50 μg/ml]). The cells were incubated at 37°C for 30 min and analyzed using a FACScan flow cytometer (Becton Dickinson). FL2-A fluorescence was recorded on 50,000 events gated according to forward and side scatter and analyzed using CellQuest Pro software.
Subcellular fractionation.
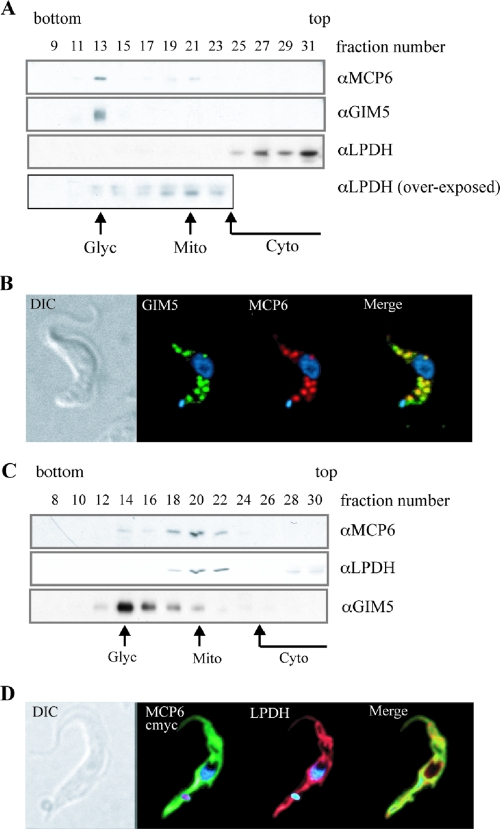
Bloodstream and procyclic T. brucei cells were grown to a maximum density of 2 × 106 and 1 × 107 cells ml−1, respectively, to a total of 2 × 1010 cells each. The cells were harvested by centrifugation at 2,000 × g and washed once in 100 ml of TEDS (25 mM Tris, 1 mM EDTA, 1 mM dithiothreitol, 250 mM sucrose, pH 7.8). The cells were resuspended in 2 ml of TEDS supplemented with complete (EDTA-free) protease inhibitor mix from Roche and ground with 1 volume of silicon carbide (<400 mesh; Crysalon, Norton Company) in a prechilled mortar. The cells were checked for at least 90% disruption using a light microscope. The lysate was then centrifuged at 1,000 × g for 10 min to remove the abrasive. The supernatant was layered onto a 30-ml semilinear 30 to 60% (wt/vol) sucrose gradient in 50 mM Tris-EDTA, pH 7.5, layered on top of a 5-ml 70% (wt/vol) sucrose cushion. Centrifugation was performed at 45,000 rpm for 1 h, with minimal acceleration and deceleration, at 4°C using 39-ml QuickSeal (Beckman) centrifuge tubes and a vertical Sorvall VTi 50 rotor. Aliquots of 1 ml were harvested from the bottom of the tube by puncture. An equal volume of each fraction was tricarboxylic acid precipitated, resuspended in denaturing SDS-PAGE buffer, and separated on an 11% SDS-PAGE gel. The fractions were blotted onto a Hybond P nylon membrane (Amersham Pharmacia) and probed with different antibodies directed against specific marker proteins for glycosomes and mitochondria (see legend for Fig. 4A and C).
FIG. 4.
Subcellular localization of MCP6 in bloodstream- and procyclic-form T. brucei cells. (A and C) Western blotting of sucrose gradient fractions obtained for bloodstream (A) and procyclic (C) trypanosomes. The bottom (densest) and top (lightest) fractions of the sucrose gradient are indicated. For both forms, MCP6 was detected using the affinity-purified αMCP6 antibody discussed in this paper. Mitochondrion-containing fractions (Mito indicates the mitochondrial peak fraction) were identified by using αLPDH (70), and the glycosome-containing fractions (Glyc indicates the glycosomal peak fraction) were identified by using αGIM5 (41). Cytosolic (Cyto) proteins were usually found in fractions 25 to 31. (B and D) Immunofluorescence microscopy of bloodstream (B) and procyclic (D) trypanosomes. For the bloodstream form (B), MCP6 was detected using the affinity-purified αMCP6 antibody discussed in this paper. For the procyclic T. brucei cell line MCP6/MCP6-cmycti (D), the myc-tagged version of MCP6 was detected by using a commercial αMyc antibody (Sigma-Aldrich) after 48 h of induction with tetracycline. Mitochondria were stained with αLPDH (70), whereas glycosomes were stained with αGIM5 (41). DAPI staining of nucleus and kinetoplast DNA is shown in blue.
Expression in Escherichia coli.
The complete MCP6 ORF was cloned into the expression vector pET16b (Novagen, Heidelberg, Germany) and used for the transformation of E. coli strain BL21 (DE3). Expression from this vector will result in the addition of a 10-residue-long histidine tag to the N terminus of the cloned gene product. The pET16b construct containing the complete ADP/ATP carrier AAC2 of Arabidopsis thaliana (26) was used as a positive control. The E. coli strains harboring the different constructs were grown in TB medium (2.5 g liter−1 KH2PO4, 12.5 g liter−1 K2HPO4, 12 g liter−1 peptone, 24 g liter−1 yeast extract, 0.4% [vol/vol] glycerin, pH 7.0) supplemented with 10 mM malate and 10 mM pyruvate. A larger volume of TB medium was inoculated with a fresh overnight culture and grown to an optical density at 600 nm (OD600) of about 0.5. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.012% (wt/vol) to initiate the T7 RNA polymerase-driven expression of MCP6 or A. thaliana AAC2.
Expression of recombinant full-length MCP6 in E. coli was confirmed by radiolabeling and enrichment of the histidine-tagged protein. E. coli cells (15 ml) harboring the MCP6 expression plasmid were grown to exponential phase, collected by centrifugation at an OD600 of 0.5 and resuspended in 3 ml of a methionine assay medium containing 42 mM Na2HPO4, 20 mM KH2PO4, 18 mM NH4Cl, 8.5 mM NaCl, 1 mM MgSO4, 0.1 mM CaCl2, 20 mM glucose, and 0.1 mg/ml thiamine (Difco Laboratories, Heidelberg, Germany). T7 RNA polymerase synthesis was induced by the addition of 1 mM IPTG. After shaking of the culture for 20 min at 37°C, 60 μl rifampin (20 mg/ml stock, dissolved in methanol) was added to inhibit the E. coli RNA polymerase. E. coli cells were shaken for an additional 15 min, after which 5 μl [35S]methionine (50 μCi) was added to label newly synthesized proteins for 30 min at 30°C. Cells were sedimented by centrifugation and transferred to liquid nitrogen to break the cells. After resuspension in a medium consisting of 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.1 mM Pefabloc, and 15% (vol/vol) glycerol, cells were further disrupted by ultrasonication (250 W, three cycles for 30 s, 4°C) and the suspension was centrifuged (10 min, 15,800 × g, 4°C) to remove unbroken cells and inclusion bodies. The membranes extracted in the supernatant were sedimented for 45 min at 100,000 × g (TFT 80 rotor; Kontron Instruments, Munich, Germany). The pellet was resuspended in 1 ml binding buffer A consisting of 10 mM imidazole, 300 mM NaCl, 100 mM Na2HPO4 (pH 8.0, HCl), and 0.1% Triton X-100. After incubation at 4°C for 2 h, the suspension was centrifuged for 2 min (15,800 × g, 4°C). The solubilized histidine-tagged MCP6 protein in the supernatant was further purified by Ni-chelating chromatography according to the supplier's instructions (Novagen, Heidelberg, Germany). Histidine-tagged protein was eluted with 1 M immidazol and desalted by Sephadex G 50 centrifugation. For SDS-PAGE, a protein aliquot was added to concentrated SDS-PAGE sample buffer medium and incubated on ice for 60 min. Finally, the preparation was applied to a polyacrylamide gel (3% stacking gel, 15% running gel) for electrophoresis in the presence of 0.1% sodium dodecyl sulfate (SDS-PAGE). After being dried, the gel was autoradiographed for 2 days.
Transport assays with E. coli.
For transport assays, IPTG-induced cells were grown for 1 h at 37°C and collected by centrifugation for 5 min at 5, 000 × g. The cell pellets were resuspended in 50 mM potassium phosphate buffer (pH 7.0) to an OD600 of about 5 and stored at 8°C until use. IPTG-induced E. coli cells (100 μl) expressing MCP6 or AAC2 of A. thaliana were added to 100 μl potassium phosphate buffer (50 mM, pH 7) containing 100 μM of [α-32P]ATP and [α-32P]ADP and up to 1,000 μM of Mg2+-[α-32P]ATP. Uptake experiments were carried out at 30°C in an Eppendorf reaction vessel incubator and terminated after the indicated time periods by transferring the cells to a 0.45-μm filter (25-mm diameter; Orange Scientific, Waterloo, Belgium) under vacuum and previously moistened with potassium phosphate buffer (50 mM, pH 7.0). The cells were further washed to remove nonimported label by the addition of three times 4 ml of potassium phosphate buffer (50 mM, pH 7.0). The filter was subsequently transferred into a 20-ml scintillation counter vessel filled with 10 ml of water, and the radioactivity was quantified using a Canberra-Packard Tricarb 2500 scintillation counter (Canberra-Packard, Frankfurt, Germany).
RESULTS
Predictions from the MCP6 sequence.
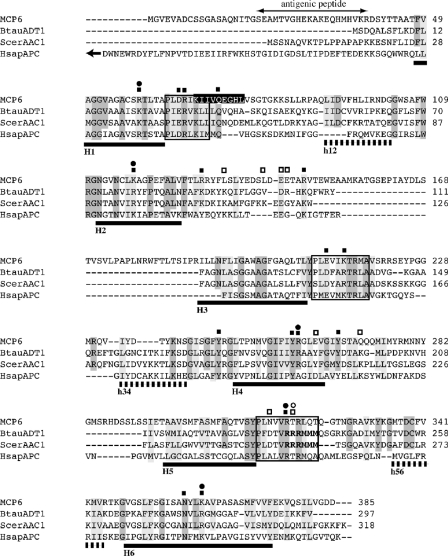
The T. brucei MCP6 gene (GeneDB accession no. Tb927.4.1660) was identified as a putative mitochondrial carrier protein gene by reciprocal BLAST analysis as a relative of the mitochondrial carrier family, showing the highest homology to mitochondrial ADP/ATP carriers (AACs) and mitochondrial calcium-dependent solute carriers (MCSC) (Table 1). The predicted protein, MCP6, has a length of 385 amino acid residues with a molecular mass of 42.7 kDa. The amino acid sequence (Fig. 1) exhibits the tripartite sequence structure typical of proteins belonging to the mitochondrial carrier family (31, 32, 35, 58, 86), with three repeats of about 100 amino acid residues separated by short linkers. Each 100-residue domain consists of two predicted membrane-spanning α-helices (H1 plus H2, H3 plus H4, and H5 plus H6) connected by large hydrophilic regions, each of which contains a short hydrophobic region (h12, h34, and h56). Downstream of, and adjacent to, the α-helices H1, H3, and H5, each repetitive domain also contains the sequence signature motif P-X-(DE)-X-(LIVAT)-(KR)-X-(LRH)-(LIVMFY)-(QGAIVM), which is found in all mitochondrial carriers characterized so far.
TABLE 1.
MCP6 is closely related to mitochondrial carrier proteins
| Organism | Gene name/accession no. | Amino acid identity (%) | Amino acid similarity (%) | Proposed function (reference) |
|---|---|---|---|---|
| B. taurus | ADT1/P02722 | 29 | 47 | Mitochondrial ATP/ADP carrier (64) |
| S. cerevisiae | AAC1/CAA89766 | 28 | 45 | Mitochondrial ATP/ADP carrier (23) |
| T. brucei | AAC/AAC23561 | 27 | 44 | Mitochondrial ATP/ADP carrier (annotated) |
| R. norvegicus | MCSC/AAL05592 | 29 | 47 | Mitochondrial Ca-dependent solute carrier (43) |
| H. sapiens | APC/CAF04060 | 29 | 47 | Mitochondrial ATP-Mg/Pi carrier (21) |
| T. cruzi | Tc00.1047053504057.140 | 66 | 81 | Unknown |
| L. major | LmjF34.3060 | 60 | 74 | Unknown |
FIG. 1.
Sequence alignment of MCP6 and different mitochondrial ATP/ADP and ATP-Mg/Pi carriers. Amino acid sequences were aligned using Clustal X (30) and SE-AL. The sequences used are MCP6 (this paper), BtauADT1, mitochondrial ATP/ADP carrier ADT1 of B. taurus, ScerAAC1, mitochondrial ATP/ADP carrier AAC1 of S. cerevisiae, and HsapAPC, mitochondrial ATP-Mg/Pi carrier of H. sapiens. For database accession numbers, see Materials and Methods. Putative trans-membrane α-helices, predicted by the TMHMM (CBS, Denmark) and TMpred (EMBnet-CH) programs available at http://www.expasy.ch, are underlined by solid black bars (numbered H1 through H6), whereas the short hydrophobic stretches between the trans-membrane α-helices are marked by dashed bars (numbered h12, h34, and h56). The putative mitochondrial carrier sequence signatures are boxed. The conserved mitochondrial ATP/ADP sequence motif RRRMMM is printed in boldface. Identical or similar amino acids, using MCP6 as the lead sequence, are shaded dark gray or light gray, respectively. Gaps are indicated by dashes. A double-headed arrow marks the peptide sequence used for immunization. A putative peroxisomal “PTS2” targeting signal is printed white on a black background but is at the wrong place in the sequence to function. The arrow at the start of the HsapAPC (21) sequence represents the N-terminal EF-hand calcium-binding domain, which is not shown in this alignment. Circles and squares mark functionally important amino acid residues identified in S. cerevisiae AAC1 (36, 53) and B. taurus ADT1 (62), respectively; when filled, they represent amino acid residues also conserved in MCP6, whereas open symbols stand for residues not conserved in MCP6.
Blasting of the MCP6 sequence against the genome databases of other kinetoplastids, such as Trypanosoma cruzi and Leishmania major, showed that the genomes of these parasites also harbor genes with high amino acid sequence similarities to MCP6 (Table 1).
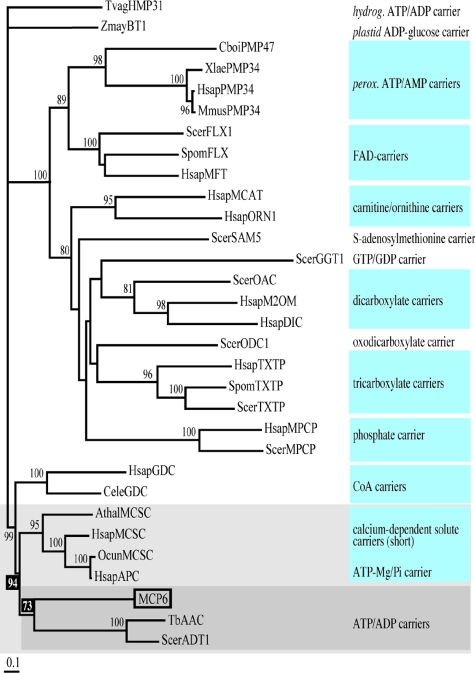
Analysis of selected MCF sequences by the neighbor-joining method (69) resulted in a phylogenetic tree (Fig. 2) with a topology similar to those previously described for proteins of the mitochondrial carrier family (34, 40, 84). For most of the groups, each containing sequences of MCF proteins with similar or related transport functions, the branching order is supported by high bootstrap values (Fig. 2). MCP6 clusters with high bootstrap support (94%) within a group of sequences encompassing two different groups of MCF proteins, e.g., the mitochondrial AACs and the MCSC, including the calcium-dependent ATP-Mg/Pi carrier (APC) of H. sapiens. The nodes for both groups are supported by high bootstrap values: 94% and 73%, respectively (Fig. 2). These results suggested that MCP6 is either an ADP/ATP carrier or a mitochondrial calcium-dependent solute carrier like the APC.
FIG. 2.
Phylogenetic reconstruction of MCP6. MCP6 clusters within a group (shaded light gray) of MCF protein sequences encompassing the mitochondrial ATP/ADP carriers (shaded dark gray) and MCSC, including the H. sapiens APC. The bootstrap values indicated at the nodes are expressed as percentages and are calculated after resampling analysis generating 1,000 reiterated data sets. Only significant bootstrap values, equal to or higher than 60%, are shown. The different groups of functionally related sequences, each supported by significant bootstrap values, are shaded blue. For database accession numbers of the sequences, see Materials and Methods. Abbreviations: hydrog, hydrogenosomal; perox, peroxisomal.
More-detailed analysis of the MCP6 sequence, however, cast doubt on the idea that MCP6 is an ATP/ADP carrier. In S. cerevisiae ADP/ATP carrier AAC1, the basic amino acid residues R96, R204, R252, R253, R294, and K38 were previously shown to be essential for ADP/ATP transport activity (36, 53). In MCP6, most of these basic amino acids are conserved (see Fig. 1 for details), with the exception of R253 of S. cerevisiae AAC1, which has been replaced by the hydroxylated amino acid T319 in MCP6. Almost all mitochondrial ADP/ATP carriers characterized so far contain a conserved RRRMMM amino acid sequence motif (31, 32, 34, 35, 58) in which only the first two arginines were shown to be essential for the ADP/ATP transport function of the carrier (53). In MCP6, however, the RRRMMM motif is not conserved at all; instead, the sequence RTRLQT is found.
Recently, the structure of B. taurus ADT1 was solved at a resolution of 2.2 Å by X-ray crystallography (62). Various polar amino acids were postulated to be important for the function of the translocation channel of ADT1. In MCP6, nearly all of the relevant amino acids are either identical or replaced by structurally similar residues, except for the above-described replacement of bovine R235 by T319 in MCP6 and the replacement of bovine D231 by N315 in MCP6 (Fig. 1). A cationic cluster near the bottom of the B. taurus ADT1 translocation channel and a “tyrosine ladder” motif within the channel (53) are only partially conserved in MCP6.
Sequence comparison and phylogenetic reconstruction also suggested a possible relationship of MCP6 with mitochondrial calcium-dependent solute carriers, such as the APC (Table 1 and Fig. 2.). In addition to the carboxy-terminal protein carrier domain, however, calcium-dependent solute carriers contain a calcium-binding amino-terminal domain (6, 21, 43, 88). This calcium-binding amino-terminal domain is apparently absent in MCP6.
Transport assays.
To test whether MCP6 functions as an ADP/ATP or ATP-Mg/Pi carrier, it was expressed in E. coli, using the mitochondrial ADP/ATP carrier AAC2 from Arabidopsis thaliana as a positive control as described by Haferkamp et al. (26). We first confirmed the reconstitution of sufficient amounts of MCP6 protein in the E. coli membrane. IPTG induction was carried out in the presence of [35S]methionine in order to detect even small amounts of newly synthesized protein in the membrane fraction. After induction, a significant fraction of MCP6 was incorporated into the cellular membrane of E. coli (Fig. 3A, lane 4). Since previous reports (24, 26, 27, 39, 49, 50, 74-76, 84) showed that virtually all heterologously expressed prokaryotic or eukaryotic membrane proteins, after integration into the Escherichia coli membrane, were properly folded and functional (including the A. thaliana AAC2 control; see below), we assumed that this is also the case for MCP6. The His-tagged MCP6 migrates according to the calculated molecular mass of about 45 kDa (Fig. 3A, lanes 2 and 4). In noninduced E. coli control cells, newly synthesized MCP6 protein could not be detected (Fig. 3A, lanes 1 and 3).
FIG. 3.
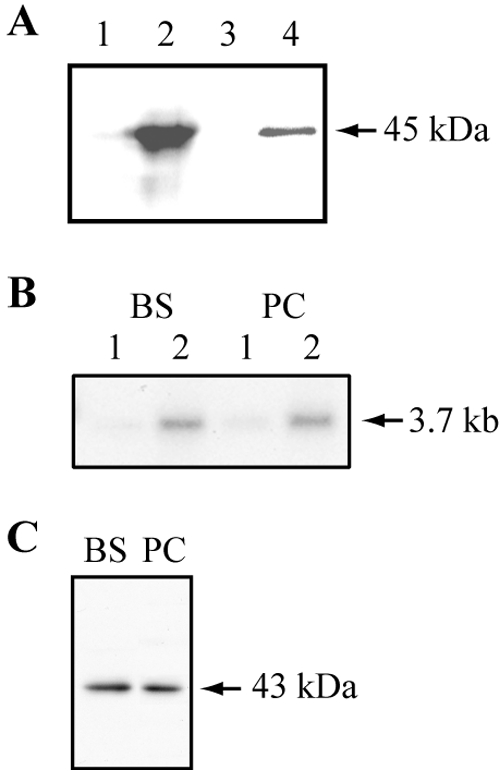
Expression of MCP6 in E. coli (A) and T. brucei (B and C). (A) Autoradiography after heterologous expression and membrane purification of [35S]methionine-labeled His-tagged MCP6 protein. E. coli cells harboring the MCP6 expression plasmid were IPTG induced (or noninduced for the control) for protein synthesis in the presence of [35S]methionine. Details of induction, purification, and autoradiography are given in Materials and Methods. Lane 1 (not induced, control) and lane 2 (induced) each contain 30 μg of total E. coli protein after heterologous expression; lane 3 (not induced, control) and lane 4 (induced) each contain 1 μg of purified E. coli membrane protein. (B) Northern blot analysis of RNA from bloodstream (BS)- and procyclic (PC)-form trypanosomes. Twenty micrograms of total RNA (lanes 1) and 4 μg of poly(A)+ RNA (lanes 2) were hybridized with [α32P]dCTP-labeled MCP6 DNA. (C) Western blotting analysis of bloodstream (BS) and procyclic (PC) trypanosomes, using the affinity-purified αMCP6 antibody (diluted 1:1,000) and 2 × 106 trypanosomes each.
Using E. coli cells expressing AAC2 of A. thaliana (control), a significant increase of [α32P]ATP uptake could be measured after induction with IPTG (Table 2). In contrast, in E. coli cells containing the MCP6 construct, no difference in uptake of [α32P]ATP, [α32P]ADP, or Mg2+-[α32P]ATP could be detected after induction with IPTG. These results suggested that MCP6 probably does not function as either an ADP/ATP or ATP-Mg/Pi carrier.
TABLE 2.
Results of transport assays with E. colia
| Substrate (concentration) | Uptake of indicated substrate for:
|
|||
|---|---|---|---|---|
| MCP6
|
AthaAAC2
|
|||
| +IPTG | −IPTG | +IPTG | −IPTG | |
| [α32P]ATP (100 μM) | 0.034 ± 0.002 | 0.037 ± 0.008 | 0.397 ± 0.014 | 0.036 ± 0.011 |
| Mg2+-[α32P]ATP (100 μM) | 0.016 ± 0.004 | 0.013 ± 0.003 | ND | ND |
| [α32P]ADP (100 μM) | 0.034 ± 0.005 | 0.025 ± 0.07 | ND | ND |
Uptake of [α32P]ATP, [α32P]ADP, or Mg2+-[α32P]ATP, under noninduced (−IPTG) or induced (+IPTG) conditions, is expressed as nmol per mg protein transported in 15 min. Data are the means ± standard deviations from four independent experiments. ND, not done; AthaAAC2, Arabidopsis thaliana AAC2.
MCP6 is not developmentally regulated at the mRNA level.
We next analyzed the expression of MCP6 in trypanosomes. Northern blotting showed that the MCP6 mRNA is found in comparable amounts in bloodstream- and procyclic-form trypanosomes (Fig. 3B). We examined the genomic environment of the MCP6 gene according to known rules for prediction of mRNA processing sites (7) and found that trans-splicing was likely to take place between positions −73 and −54 relative to ATG and that polyadenylation was likely to take place about 2.2 kb downstream of the stop codon, resulting in a predicted polyadenylated mRNA size of about 3.6 kb. The length of the detected transcript, approximately 3.7 kb, was consistent with this finding.
The affinity-purified polyclonal antiserum, raised against an N-terminal peptide (Fig. 1) of MCP6, detected a single 43-kDa protein band in both bloodstream- and procyclic-form T. brucei cells (Fig. 3C), corresponding well with the calculated molecular mass of MCP6, which is 42.7 kDa. The detected amounts were similar for both bloodstream- and procyclic-form T. brucei cells (Fig. 3C), which suggested that the expression of MCP6 is not developmentally regulated.
MCP6 is glycosomal in bloodstream-form trypanosomes.
The subcellular localization of MCP6 in bloodstream and procyclic T. brucei cells was examined by subcellular fractionation, followed by Western blot analysis, and by immunofluorescence microscopy. Western blot analysis (Fig. 4A) revealed that in bloodstream T. brucei, the majority of MCP6 cofractionated with the glycosomal marker protein GIM5 (41), suggesting that MCP6 is predominantly glycosomal. A small proportion of MCP6 was also detected in fractions 19 to 21 (Fig. 4A), which were identified as the mitochondrion-bearing fractions by the presence of small amounts of the mitochondrial marker protein lipoamide dehydrogenase (LPDH). Bloodstream-form T. brucei harbors highly reduced mitochondria with most of the LPDH located in the cytosol (70). Only a minor portion of LPDH is mitochondrial (70), as was shown here after prolonged exposure of the LPDH Western blot (Fig. 4A, bottom panel).
Immunofluorescence microscopy confirmed the predominantly glycosomal localization of MCP6 in bloodstream T. brucei. The staining patterns for αMCP6 and αGIM5 were identical, and the overlay of both patterns resulted in a perfect merge (Fig. 4B). The minor mitochondrial portion of MCP6 was not visible.
MCP6 is mitochondrial in procyclic-form trypanosomes.
In procyclic-form T. brucei, the mitochondrial marker protein LPDH is found predominantly in the well-developed mitochondria (70). Western blot analysis (Fig. 4C), using the affinity-purified MCP6 antibody, revealed that the majority of MCP6 cofractionated with LPDH (Fig. 4C, fraction 20), suggesting that MCP6 is mitochondrial in procyclic-form trypanosomes. Additionally, a minor portion of MCP6 was detected in the glycosomal peak fraction (Fig. 4C, fraction 14).
To confirm the mitochondrial localization of MCP6 in procyclic-form T. brucei, immunofluorescence microscopy was performed using the same affinity-purified MCP6 peptide antibody. However, in procyclics, no significant fluorescent signal could be detected; perhaps the peptide epitope is accessible to the antibody when MCP6 is in the glycosomal membrane (Fig. 4A and B) but is not accessible when MCP6 is located in the mitochondrial membrane. To overcome this problem, we decided to use the procyclic cell line MCP6-cmycti. These cells express a carboxy-terminal myc-tagged version of MCP6 in the presence of tetracycline. After 48 h of induction with tetracycline, a single cross-reacting protein could be detected by Western blotting with the expected molecular mass of about 45 kDa (Fig. 5C). The staining patterns for αMyc (MCP6-cmyc) and αLPDH obtained after immunofluorescence microscopy were identical, and the overlay of both patterns resulted in a perfect merge (Fig. 4D). These results provided additional evidence for a mitochondrial localization of MCP6 in procyclic-form T. brucei.
MCP6 is essential for the survival of T. brucei.
To find out whether MCP6 is essential for the survival of bloodstream- and procyclic-form T. brucei cells, we first attempted to replace both MCP6 alleles by conventional double knockout. After many attempts, however, only half-knockout clones (Δmcp6::BSD/MCP6 or Δmcp6::NEO/MCP6) could be obtained, suggesting that MCP6 is essential for survival. Attempts to deplete MCP6 by RNAi also failed: we were not able to retrieve any viable clones at all, even under “noninduced” conditions, perhaps because a low level of “leaky” expression of double-stranded RNA was sufficient to impair trypanosome growth (1, 13, 71, 80).
We next generated a procyclic-form knockout cell line containing a single, tetracycline-inducible copy of MCP6 with a carboxy-terminal myc tag (Fig. 5A) as described previously (33, 41, 85). The resulting Δmcp6/MCP6-cmycti cell line had to be grown in the constant presence of tetracycline (0.5 μg ml−1) in order to maintain a steady level of recombinant MCP6-cmyc expression. Western blot analysis, using the affinity-purified MCP6 antibody, revealed the presence of a threefold excess of MCP6 protein in induced (with tet) Δmcp6/MCP6-cmycti trypanosomes compared to that in wild-type trypanosomes (Fig. 5C, left panel). As expected, in induced (with tet) Δmcp6/MCP6-cmycti trypanosomes, the recombinant MCP6-myc protein could also be detected by the myc antibody (Fig. 5C, right panel).
To deplete the MCP6-cmyc protein in the Δmcp6/MCP6-cmycti cell line, tetracycline was withdrawn from the culture medium. After 48 h of tetracycline withdrawal, the amount of MCP6-cmyc protein was reduced, and the level stayed constant up to 96 h (Fig. 5C, right panel). The Δmcp6/MCP6-cmycti trypanosomes grew normally in the presence of tetracycline (Fig. 5B), but after tetracycline withdrawal, they grew poorly, and after 96 h, their growth nearly ceased (Fig. 5B). Growth of the MCP6-depleted trypanosomes could be restored to wild-type levels by the addition of tetracycline (not shown), resulting in normal MCP6-cmyc protein levels 24 h later (Fig. 5C, right panel).
Depletion of MCP6 affected cellular morphology and the numbers of kinetoplasts and nuclei.
MCP6-expressing and MCP6-depleted (96 h) procyclic-form trypanosomes were examined in more detail by immunofluorescence microscopy. As expected, no MCP6-cmyc protein could be detected in the cells grown without tetracycline (Fig. 5D), whereas myc-tagged protein was found in the mitochondria of MCP6-expressing trypanosomes (Fig. 4D and 5D). Staining of the mitochondria with antibodies to LPDH did not reveal any visible deviations in mitochondrial morphology in either the MCP6-(over)expressing or MCP6-depleted trypanosomes (Fig. 5D).
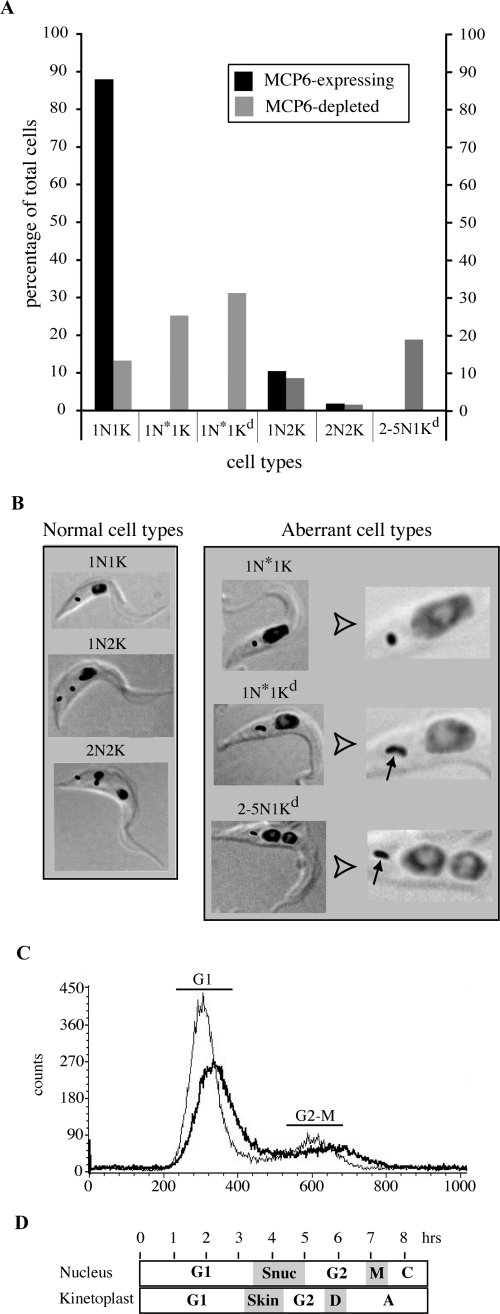
To determine the cell cycle status of the MCP6-depleted cells, we stained both the mitochondrial DNA (kinetoplast) and the nucleus with DAPI. Cells in G1 and S phases have one nucleus and one kinetoplast (1N1K), cells in G2/M phase have one nucleus and two kinetoplasts (1N2K), and cells undergoing cytokinesis have two nuclei and two kinetoplasts (2N2K) (Fig. 6D) (46). In the MCP6-myc-expressing cell line (Fig. 6A), the majority of the cells (88%) were found to have a normal-sized single nucleus and a single kinetoplast; the rest of the cells were either 1N2K (10.5%) or 2N2K (1.5%). These numbers are similar to those reported previously for wild-type trypanosomes (68).
FIG. 6.
Immunofluorescence microscopy and FACS analysis of MCP6-depleted procyclic Δmcp6/MCP6-cmycti trypanosomes. (A) Tabulation of the different cell types found for MCP6-expressing and MCP6-depleted procyclic Δmcp6/MCP6-cmycti trypanosomes. Two hundred MCP6-expressing (with tet, 96 h) and 200 MCP6-depleted (without tet, 96 h) cells were stained with DAPI and examined for the sizes and numbers of nuclei and kinetoplasts in individual cells. Abbreviations: N, normal nucleus; N*, enlarged nucleus; K, kinetoplast; Kd, dumbbell-shaped kinetoplast. (B) Phase/DAPI photographs of the different normal (left) and aberrant (right) cell types found. DAPI staining of nuclear and kinetoplast DNA is shown in black. Arrows point to dumbbell-shaped kinetoplasts. (Right panel) On the right side of this panel, magnifications are shown of the phase/DAPI photographs from the left for a better view of aberrant nuclei and kinetoplasts. (C) FACS analysis of the DNA content of procyclic-form Δmcp6/MCP6-cmycti trypanosomes. MCP6-expressing (with tet, 96 h, thin line) and MCP6-depleted (without tet, 96 h, thick line) Δmcp6/MCP6-cmycti trypanosomes were stained with propidium iodide and analyzed (50,000 cells each) with a FACScan flow cytometer for their DNA content. The x axis shows the channel number, which is proportional to the fluorescence signal and indicates DNA content. The y axis shows the number of events (counts) in each channel, which is proportional to cell numbers. G1, wild-type cells in the G1 cell cycle phase with a 2N (normal diploid) DNA content; G2-M, wild-type cells after DNA replication in the G2-M cell cycle phase. (D) Schematic representation of the T. brucei cell cycle, illustrating the temporally coordinated kinetoplast and nuclear phases. Abbreviations: Sn, nuclear S phase; M, mitosis; C, cytokinesis; Sk, kinetoplast S phase; D, kinetoplast division; A, kinetoplast segregation.
Examination of MCP6-depleted trypanosomes revealed a reduction in the proportion of cells in G1/S (1N1K) phase and an increase in cells with an aberrant nuclear and kinetoplast phenotype (Fig. 6A). The majority (57.5%) of the MCP6-depleted cells contained an enlarged nucleus (1N*). Within this 1N* population, two different cell types could be discriminated: (i) cells containing a single normal kinetoplast (1N*1K), representing 26% of the total population, and (ii) cells containing a single but dumbbell-shaped kinetoplast (1N*1Kd), representing 31% of the total population (Fig. 6). About 20% of the cells contained a single dumbbell-shaped kinetoplast and two or more nuclei (2-5N1Kd). MCP6-depleted trypanosomes of the 1N*1K, 1N*1Kd, and 2-5N1Kd cell types appeared to be thicker than normal 1N1K-type cells (Fig. 5D and 6B). Trypanosomes containing a kinetoplast but no nucleus (zoids) were not observed at all.
The observed aberrant nuclear phenotype was supported by FACS analysis of the cellular DNA content by staining of MCP6-expressing and MCP6-depleted Δmcp6/MCP6-cmycti trypanosomes with propidium iodide. The patterns for wild-type (not shown) and MCP6-myc expressing (Fig. 6C) trypanosomes were identical and showed, as expected, that the majority of the cells were in the G1 cell cycle phase (1N1K), with a minor portion of the cells in G2 phase (2N2K). The pattern for the MCP6-depleted trypanosomes, however, was significantly different: both major peaks were clearly shifted to the right and were less high and wider than those in the MCP6-expressing cell lines. The overall cell population appeared to be more fluorescent, indicating an increased DNA content per cell. The pattern is consistent with the presence of cell types other than 1N1K and 2N2K and correlates with the observation that the majority (about 78%) of the MCP6-depleted cells contained an enlarged nucleus or even multiple nuclei.
Overall, these results suggested that the cells lacking MCP6 were unable to complete mitosis or cytokinesis because of a defect in the division of the kinetoplast.
DISCUSSION
Implications of the MCP6 sequence for substrate specificity.
Sequence analysis of MCP6 revealed conserved sequence motifs and a structural topology typical for MCF proteins. Phylogenetic reconstruction showed further that MCP6 is most closely related to mitochondrial ADP/ATP carriers and MCSC. Detailed sequence comparison revealed, however, that MCP6 lacked some highly conserved amino acid residues that were previously proposed to be important for its function as an ADP/ATP carrier. The logical conclusion that MCP6 probably does not transport ATP, ADP, or ATP-Mg/Pi was confirmed by transport assays using E. coli expressing MCP6. The absence of a calcium-binding domain also precludes a function as a calcium-regulated carrier. It remains possible, however, that MCP6 has an (non-calcium-dependent) APC-like activity, such as the transport of nucleotides other than ATP and ADP.
Subcellular fractionation and immunofluorescence microscopy revealed that MCP6 is localized in two different compartments in bloodstream and procyclic trypanosomes. In the bloodstream form, MCP6 was found predominantly in the glycosomes, with a minor amount in the mitochondria, whereas in procyclic forms, MCP6 was predominantly mitochondrial. This is the first time such a life form-dependent dual glycosomal (peroxisomal) versus mitochondrial localization has been described for a membrane protein from a kinetoplastid parasite. So far, there are only a few reports of mammalian proteins with a dual mitochondrial versus peroxisomal localization, e.g., alanine:glyoxylate aminotransferase (AGT) (17), 2-methylacyl-coenzyme A racemase (2), and the membrane-associated calcium-independent phospholipase A2 gamma (42). The best-studied example is the mammalian AGT, which in humans is predominantly peroxisomal, whereas in other animals, its mitochondrial versus peroxisomal localization was reported to be dependent on diet (10). This variable localization of AGT is the result of a differential expression of N-terminal mitochondrial and C-terminal peroxisomal targeting sequences by the use of alternative transcription and translation initiation sites (17, 54).
It is currently not possible to predict the localization of MCF proteins—or indeed, of other mitochondrial and peroxisomal membrane proteins—from sequence alone. Targeting signals of peroxisomal membrane proteins are mostly ill defined but are thought to include multiple segments of the proteins (reference 4 and references therein). The dual regulated localization of MCP6 may be a consequence of the differing degree of mitochondrial elaboration in bloodstream and procyclic trypanosomes. In bloodstream trypanosomes, the mitochondrion is poorly developed (20, 48), and the import capacity of the organelle in this life form may be rather limited in correlation with the small available mitochondrial membrane area. This is illustrated by the fact that most LPDH is cytosolic in bloodstream forms (Fig. 4A). In addition, there is evidence that the developmental regulation of expression of cytochrome c and cytochrome c reductase is due to degradation of the proteins in the bloodstream form, perhaps because of failure of mitochondrial import (65, 66, 77, 78). Experiments with trypanosomes with a soluble protein containing an N-terminal targeting signal for the mitochondrial matrix, and a C-terminal glycosomal targeting signal, have also shown that such signals can compete (28). We speculate, therefore, that MCP6 contains targeting signals for both the mitochondrial and the glycosomal membranes. When a large area of mitochondrial membrane is present, the mitochondrial targeting is dominant; when the mitochondrial membrane capacity is limiting, the protein is instead incorporated into the glycosomes.
Depletion of MCP6 in procyclic trypanosomes resulted in a severe growth defect, indicating that it is essential for the survival of T. brucei. The phenotype of the cells suggested that they were defective in kinetoplast division. The kinetoplast is made up of concatenated circular DNA molecules, which have to be disentangled during kinetoplast division. During the division process, elongated dumbbell-shaped kinetoplasts are seen (63, 67, 68, 89). After segregation of the daughter kinetoplasts, mitosis of the nucleus occurs. The replication and division of the kinetoplast DNA (kDNA) and the nucleus are temporarily coordinated during the T. brucei cell cycle, as shown in Fig. 6D (63, 67, 68, 89). Segregation of kDNA occurs in the middle of the nuclear G2 period. The trypanosome cell cycle is remarkable because it appears to lack the strict checkpoints present in multicellular eukaryotes and yeasts (44, 46, 47), thus allowing the appearance of aberrant karyotypes after disruption of cell cycle control (reference 79 and references therein).
Examination of the MCP6-depleted trypanosomes by immunofluorescence microscopy revealed that most of the cells were thicker than wild-type cells and contained a single enlarged nucleus with either a single normal (1N*1K) or dumbbell-shaped (1N*1Kd) kinetoplast and that other cells had a single dumbbell-shaped kinetoplast and multiple nuclei. These results suggested that the MCP6-depleted cells are arrested in their cell cycle, which could be explained by defects in either mitosis or cytokinesis. However, no anucleated trypanosomes (zoids) were observed, which are frequently seen as a result of blocking nuclear DNA synthesis or mitosis with the nuclear DNA synthesis inhibitor aphidicolin or the antimicrotubule agent rhizoxin (68, 79, 82). Also, the number of cells with multiple well-separated nuclei remained rather low in comparison to the previously reported karyotype for procyclics in which cytokinesis was inhibited by okadaic acid (18).
A more likely explanation for the observed aberrant nuclear and kinetoplast phenotype is a defect in the division of the kinetoplast. The cell cycle arrest phenotype cannot, however, be solely a consequence of a defect in kDNA segregation, since down-regulation of proteins required for kDNA replication was previously shown to cause accumulation of 1N0K (dyskinetoplastic) cells with consequent cell death (87). Our results therefore suggest that MCP6 depletion inhibits not only kDNA replication/division but also other processes.
Kinetoplastid protists are incapable of de novo purine synthesis (8). They obtain their purines by taking up nucleobases and nucleosides from their environment (12, 38) and interconvert them using various purine phosphoribosyltransferases. Several of the purine salvage enzymes have been shown to be localized in the glycosome (15, 72, 91), implying that the glycosomal membrane must contain appropriate transporters. Also, the mitochondria must contain such transporters, since nucleotides required for DNA replication and RNA synthesis need to be imported from the cytosol. It is, therefore, attractive to speculate that MCP6 functions as a nucleotide transporter. Given our inability to predict the specificity of MCP6 based on sequence alone, however, a wide variety of possible substrates will need to be tested in order to determine its function.
Acknowledgments
This research was financially supported through a grant (VO965/1-1) from the “Deutsche Forschungsgemeinschaft” (DFG) to F.V. V.P.A. was supported by the UNDP/WHO Program on Research and Training in Tropical Diseases (TDR).
REFERENCES
- 1.Alibu, V. P., L. Storm, S. Haile, C. Clayton, and D. Horn. 2005. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol. Biochem. Parasitol. 139:75-82. [DOI] [PubMed] [Google Scholar]
- 2.Amery, L., M. Fransen, K. De Nys, G. P. Mannaerts, and P. P. Van Veldhoven. 2000. Mitochondrial and peroxisomal targeting of 2-methylacyl-CoA racemase in humans. J. Lipid Res. 41:1752-1759. [PubMed] [Google Scholar]
- 3.Aquila, H., T. A. Link, and M. Klingenberg. 1987. Solute carriers involved in energy transfer of mitochondria form a homologous protein family. FEBS Lett. 212:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Baerends, R. J., K. N. Faber, J. A. Kiel, I. J. van der Klei, W. Harder, and M. Veenhuis. 2000. Sorting and function of peroxisomal membrane proteins. FEMS Microbiol. Rev. 24:291-301. [DOI] [PubMed] [Google Scholar]
- 5.Bakker, B. M., F. I. C. Mensonides, B. Teusink, P. van Hoek, P. A. M. Michels, and H. V. Westerhoff. 2000. Compartmentation protects trypanosomes from the dangerous design of glycolysis. Proc. Natl. Acad. Sci. USA 97:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassi, M. T., M. Manzoni, R. Bresciani, M. T. Pizzo, A. Della Monica, S. Barlati, E. Monti, and G. Borsani. 2005. Cellular expression and alternative splicing of SLC25A23, a member of the mitochondrial Ca2+-dependent solute carrier gene family. Gene 345:173-182. [DOI] [PubMed] [Google Scholar]
- 7.Benz, C., D. Nilsson, B. Andersson, C. Clayton, and D. L. Guilbride. 2005. Messenger RNA processing sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 143:125-134. [DOI] [PubMed] [Google Scholar]
- 8.Berriman, M., et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416-422. [DOI] [PubMed] [Google Scholar]
- 9.Biebinger, S., L. E. Wirtz, and C. E. Clayton. 1997. Vectors for inducible over-expression of potentially toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 85:99-112. [DOI] [PubMed] [Google Scholar]
- 10.Birdsey, G. M., J. Lewin, A. A. Cunningham, M. W. Bruford, and C. J. Danpure. 2004. Differential enzyme targeting as an evolutionary adaptation to herbivory in carnivora. Mol. Biol. Evol. 21:632-646. [DOI] [PubMed] [Google Scholar]
- 11.Blattner, J., S. Helfert, P. Michels, and C. E. Clayton. 1998. Compartmentation of phosphoglycerate kinase in Trypanosoma brucei plays a critical role in parasite energy metabolism. Proc. Natl. Acad. Sci. USA 95:11596-11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter, N. S., N. Rager, and B. Ullman. 2003. Purine and pyrimidine transport and metabolism, p. 179-223. In J. J. Marr, T. Nilsen, and R. Komuniecki (ed.), Molecular and medical parasitology. Academic Press, London, United Kingdom.
- 13.Clayton, C. E., A. M. Esteacutevez, C. Hartmann, V. P. Alibu, M. Field, and D. Horn. 2005. Down-regulating gene expression by RNA interference in Trypanosoma brucei. Methods Mol. Biol 309:39-60. [DOI] [PubMed] [Google Scholar]
- 14.Clayton, C. E., and P. Michels. 1996. Metabolic compartmentation in trypanosomes. Parasitol. Today 12:465-471. [DOI] [PubMed] [Google Scholar]
- 15.Colasante, C., M. Ellis, T. Ruppert, and F. Voncken. 2006. Comparative proteomics of glycosomes from bloodstream form and procyclic culture form Trypanosoma brucei brucei. Proteomics 6:3275-3293. [DOI] [PubMed] [Google Scholar]
- 16.Coustou, V., S. Besteiro, M. Biran, P. Diolez, V. Bouchaud, P. Voisin, P. A. Michels, P. Canioni, T. Baltz, and F. Bringaud. 2003. ATP generation in the Trypanosoma brucei procyclic form: cytosolic substrate level is essential, but not oxidative phosphorylation. J. Biol. Chem. 278:49625-49635. [DOI] [PubMed] [Google Scholar]
- 17.Danpure, C. J. 1997. Variable peroxisomal and mitochondrial targeting of alanine: glyoxylate aminotransferase in mammalian evolution and disease. Bioessays 19:317-326. [DOI] [PubMed] [Google Scholar]
- 18.Das, A., M. Gale, Jr., V. Carter, and M. Parsons. 1994. The protein phosphatase inhibitor okadaic acid induces defects in cytokinesis and organellar genome segregation in Trypanosoma brucei. J. Cell Sci. 107:3477-3483. [DOI] [PubMed] [Google Scholar]
- 19.Dayhoff, M. O., and B. C. Orcutt. 1979. Methods for identifying proteins by using partial sequences. Proc. Natl. Acad. Sci. USA 76:2170-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fairlamb, A. H., and F. R. Opperdoes. 1986. Carbohydrate metabolism in African trypanosomes, with special reference to the glycosome, p. 183-224. In M. J. Morgan (ed.), Carbohydrate metabolism in cultured cells. Plenum Publishing Corporation, New York, N.Y.
- 21.Fiermonte, G., F. De Leonardis, S. Todisco, L. Palmieri, F. M. Lasorsa, and F. Palmieri. 2004. Identification of the mitochondrial ATP-Mg/Pi transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 279:30722-30730. [DOI] [PubMed] [Google Scholar]
- 22.Furuya, T., P. Kessler, A. Jardim, A. Schnaufer, C. Crudder, and M. Parsons. 2002. Glucose is toxic to glycosome-deficient trypanosomes. Proc. Natl. Acad. Sci. USA 99:14177-14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gawaz, M., M. G. Douglas, and M. Klingenberg. 1990. Structure-function studies of adenine nucleotide transport in mitochondria. II. Biochemical analysis of distinct AAC1 and AAC2 proteins in yeast. J. Biol. Chem. 265:14202-14208. [PubMed] [Google Scholar]
- 24.Geigenberger, P., C. Stamme, J. Tjaden, A. Schulz, P. W. Quick, T. Betsche, H. J. Kersting, and H. E. Neuhaus. 2001. Tuber physiology and properties of starch from tubers of transgenic potato plants with altered plastidic adenylate transporter activity. Plant Physiol. 125:1667-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerra-Giraldez, C., L. Quijada, and C. E. Clayton. 2002. Compartmentation of enzymes in a microbody, the glycosome, is essential in Trypanosoma brucei. J. Cell Sci. 115:2651. [DOI] [PubMed] [Google Scholar]
- 26.Haferkamp, I., J. H. Hackstein, F. G. Voncken, G. Schmit, and J. Tjaden. 2002. Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur. J. Biochem. 269:3172-3181. [DOI] [PubMed] [Google Scholar]
- 27.Haferkamp, I., S. Schmitz-Esser, N. Linka, C. Urbany, A. Collingro, M. Wagner, M. Horn, and H. E. Neuhaus. 2004. A candidate NAD+ transporter in an intracellular bacterial symbiont related to Chlamydiae. Nature 432:622-625. [DOI] [PubMed] [Google Scholar]
- 28.Häusler, T., Y. D. Stierhof, E. Wirtz, and C. E. Clayton. 1996. Import of a DHFR-hybrid protein into glycosomes in vivo is not inhibited by the folate-analogue aminopterin. J. Cell Biol. 132:311-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirumi, H., and K. Hirumi. 1989. Continuous cultivation of Trypanosoma brucei bloodstream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985-989. [PubMed] [Google Scholar]
- 30.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 31.Jezek, P., and J. Jezek. 2003. Sequence anatomy of mitochondrial anion carriers. FEBS Lett. 534:15-25. [DOI] [PubMed] [Google Scholar]
- 32.Kaplan, R. S. 2001. Structure and function of mitochondrial anion transport proteins. J. Membr. Biol. 179:165-183. [DOI] [PubMed] [Google Scholar]
- 33.Krieger, S., W. Schwarz, M. R. Ariyanagam, A. Fairlamb, L. Krauth-Siegel, and C. E. Clayton. 2000. Trypanosomes lacking trypanothione reductase are avirulent and show increased sensitivity to oxidative stress. Mol. Microbiol. 35:542-552. [DOI] [PubMed] [Google Scholar]
- 34.Kuan, J., and M. H. Saier, Jr. 1993. The mitochondrial carrier family of transport proteins: structural, functional, and evolutionary relationships. Crit. Rev. Biochem. Mol. Biol 28:209-233. [DOI] [PubMed] [Google Scholar]
- 35.Kunji, E. R. 2004. The role and structure of mitochondrial carriers. FEBS Lett. 564:239-244. [DOI] [PubMed] [Google Scholar]
- 36.Kunji, E. R., and M. Harding. 2003. Projection structure of the atractyloside-inhibited mitochondrial ADP/ATP carrier of Saccharomyces cerevisiae. J. Biol. Chem. 278:36985-36988. [DOI] [PubMed] [Google Scholar]
- 37.LaCount, D. J., S. Bruse, K. L. Hill, and J. E. Donelson. 2000. Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol. Biochem. Parasitol. 111:67-76. [DOI] [PubMed] [Google Scholar]
- 38.Landfear, S. M., B. Ullman, N. S. Carter, and M. A. Sanchez. 2004. Nucleoside and nucleobase transporters in parasitic protozoa. Eukaryot. Cell 3:245-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leroch, M., S. Kirchberger, I. Haferkamp, M. Wahl, H. E. Neuhaus, and J. Tjaden. 2005. Identification and characterization of a novel plastidic adenine nucleotide uniporter from Solanum tuberosum. J. Biol. Chem. 280:17992-18000. [DOI] [PubMed] [Google Scholar]
- 40.Loytynoja, A., and M. C. Milinkovitch. 2001. Molecular phylogenetic analyses of the mitochondrial ADP-ATP carriers: the Plantae/Fungi/Metazoa trichotomy revisited. Proc. Natl. Acad. Sci. USA 98:10202-10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maier, A., P. Lorenz, F. Voncken, and C. E. Clayton. 2001. An essential dimeric membrane protein of trypanosome glycosomes. Mol. Microbiol. 39:1443-1451. [DOI] [PubMed] [Google Scholar]
- 42.Mancuso, D. J., C. M. Jenkins, H. F. Sims, J. M. Cohen, J. Yang, and R. W. Gross. 2004. Complex transcriptional and translational regulation of iPLAgamma resulting in multiple gene products containing dual competing sites for mitochondrial or peroxisomal localization. Eur. J. Biochem. 271:4709-4724. [DOI] [PubMed] [Google Scholar]
- 43.Mashima, H., N. Ueda, H. Ohno, J. Suzuki, H. Ohnishi, H. Yasuda, T. Tsuchida, C. Kanamaru, N. Makita, T. Iiri, M. Omata, and I. Kojima. 2003. A novel mitochondrial Ca2+-dependent solute carrier in the liver identified by mRNA differential display. J. Biol. Chem. 278:9520-9527. [DOI] [PubMed] [Google Scholar]
- 44.Matthews, K. R. 2005. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 118:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthews, K. R. 1999. Developments in the differentiation of Trypanosoma brucei. Parasitol. Today 15:76-80. [DOI] [PubMed] [Google Scholar]
- 46.Matthews, K. R., J. R. Ellis, and A. Paterou. 2004. Molecular regulation of the life cycle of African trypanosomes. Trends Parasitol. 20:40-47. [DOI] [PubMed] [Google Scholar]
- 47.McKean, P. G. 2003. Coordination of cell cycle and cytokinesis in Trypanosoma brucei. Curr. Opin. Microbiol. 6:600-607. [DOI] [PubMed] [Google Scholar]
- 48.Michels, P. A. M., V. Hannaert, and F. Bringaud. 2000. Metabolic aspects of glycosomes in trypanosomatidae—new data and views. Parasitol. Today 16:482-489. [DOI] [PubMed] [Google Scholar]
- 49.Mohlmann, T., J. Tjaden, G. Henrichs, W. P. Quick, R. Hausler, and H. E. Neuhaus. 1997. ADP-glucose drives starch synthesis in isolated maize endosperm amyloplasts: characterization of starch synthesis and transport properties across the amyloplast envelope. Biochem. J. 324:503-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohlmann, T., J. Tjaden, C. Schwoppe, H. H. Winkler, K. Kampfenkel, and H. E. Neuhaus. 1998. Occurrence of two plastidic ATP/ADP transporters in Arabidopsis thaliana L.—molecular characterisation and comparative structural analysis of similar ATP/ADP translocators from plastids and Rickettsia prowazekii. Eur. J. Biochem. 252:353-359. [DOI] [PubMed] [Google Scholar]
- 51.Moyersoen, J., J. Choe, A. Kumar, F. G. Voncken, W. G. Hol, and P. A. Michels. 2003. Characterization of Trypanosoma brucei PEX14 and its role in the import of glycosomal matrix proteins. Eur. J. Biochem. 270:2059-2067. [DOI] [PubMed] [Google Scholar]
- 52.Nakagawa, T., T. Imanaka, M. Morita, K. Ishiguro, H. Yurimoto, A. Yamashita, N. Kato, and Y. Sakai. 2000. Peroxisomal membrane protein Pmp47 is essential in the metabolism of middle-chain fatty acid in yeast peroxisomes and is associated with peroxisome proliferation. J. Biol. Chem. 275:3455-3461. [DOI] [PubMed] [Google Scholar]
- 53.Nelson, D. R. 1996. The yeast ADP/ATP carrier. Mutagenesis and second-site revertants. Biochim. Biophys. Acta 1275:133-137. [DOI] [PubMed] [Google Scholar]
- 54.Oatey, P. B., M. J. Lumb, and C. J. Danpure. 1996. Molecular basis of the variable mitochondrial and peroxisomal localisation of alanine-glyoxylate aminotransferase. Eur. J. Biochem. 241:374-385. [DOI] [PubMed] [Google Scholar]
- 55.Opperdoes, F. R. 1987. Compartmentation of carbohydrate metabolism in trypanosomes. Annu. Rev. Microbiol. 41:127-151. [DOI] [PubMed] [Google Scholar]
- 56.Overath, P., J. Czichos, and C. Haas. 1986. The effect of citrate/cis-aconitate on oxidative metabolism during transformation of Trypanosoma brucei. Eur. J. Biochem. 160:175-182. [DOI] [PubMed] [Google Scholar]
- 57.Palmieri, F. 1994. Mitochondrial carrier proteins. FEBS Lett. 346:48-54. [DOI] [PubMed] [Google Scholar]
- 58.Palmieri, F. 2004. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. Eur. J. Physiol. 447:689-709. [DOI] [PubMed] [Google Scholar]
- 59.Palmieri, F., C. Indiveri, F. Bisaccia, and V. Iacobazzi. 1995. Mitochondrial metabolite carrier proteins: purification, reconstitution, and transport studies. Methods Enzymol. 260:349-369. [DOI] [PubMed] [Google Scholar]
- 60.Parsons, M. 2004. Glycosomes: parasites and the divergence of peroxisomal purpose. Mol. Microbiol. 53:717-724. [DOI] [PubMed] [Google Scholar]
- 61.Parsons, M., T. Furuya, S. Pal, and P. Kessler. 2001. Biogenesis and function of peroxisomes and glycosomes. Mol. Biochem. Parasitol. 115:19-28. [DOI] [PubMed] [Google Scholar]
- 62.Pebay-Peyroula, E., C. Dahout-Gonzalez, R. Kahn, V. Trezeguet, G. J. Lauquin, and G. Brandolin. 2003. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426:39-44. [DOI] [PubMed] [Google Scholar]
- 63.Ploubidou, A., D. R. Robinson, R. C. Docherty, E. O. Ogbadoyi, and K. Gull. 1999. Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J. Cell Sci. 112:4641-4650. [DOI] [PubMed] [Google Scholar]
- 64.Powell, S. J., S. M. Medd, M. J. Runswick, and J. E. Walker. 1989. Two bovine genes for mitochondrial ADP/ATP translocase expressed differences in various tissues. Biochemistry 28:866-873. [DOI] [PubMed] [Google Scholar]
- 65.Priest, J. W., and S. L. Hajduk. 1994. Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J. Bioenerg. Biomembr. 26:179-191. [DOI] [PubMed] [Google Scholar]
- 66.Priest, J. W., and S. L. Hajduk. 1994. Developmental regulation of Trypanosoma brucei cytochrome c reductase during bloodstream to procyclic differentiation. Mol. Biochem. Parasitol. 65:291-304. [DOI] [PubMed] [Google Scholar]
- 67.Robinson, D. R., and K. Gull. 1991. Basal body movement as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352:731-734. [DOI] [PubMed] [Google Scholar]
- 68.Robinson, D. R., T. Sherwin, A. Ploubidou, E. H. Byard, and K. Gull. 1995. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J. Cell Biol. 128:1163-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 70.Schoneck, R., O. Billaut-Mulot, P. Numrich, M. A. Ouaissi, and R. L. Krauth-Siegel. 1997. Cloning, sequencing and functional expression of dihydrolipoamide dehydrogenase from the human pathogen Trypanosoma cruzi. Eur. J. Biochem. 243:739-747. [DOI] [PubMed] [Google Scholar]
- 71.Shi, H., A. Djikeng, T. Mark, E. Wirtz, C. Tschudi, and E. Ullu. 2000. Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA 6:1069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shih, S., P. Stenberg, and B. Ullman. 1998. Immunolocalization of Trypanosoma brucei hypoxanthine-guanine phosphoribosyltransferase to the glycosome. Mol. Biochem. Parasitol. 92:367-371. [DOI] [PubMed] [Google Scholar]
- 73.Tielens, A. G. M., and J. J. van Hellemond. 1998. Differences in energy metabolism between Trypanosomatidae. Parasitol. Today 14:265-272. [DOI] [PubMed] [Google Scholar]
- 74.Tjaden, J., I. Haferkamp, B. Boxma, A. G. Tielens, M. Huynen, and J. H. Hackstein. 2004. A divergent ADP/ATP carrier in the hydrogenosomes of Trichomonas gallinae argues for an independent origin of these organelles. Mol. Microbiol. 51:1439-1446. [DOI] [PubMed] [Google Scholar]
- 75.Tjaden, J., C. Schwoppe, T. Mohlmann, P. W. Quick, and H. E. Neuhaus. 1998. Expression of a plastidic ATP/ADP transporter gene in Escherichia coli leads to a functional adenine nucleotide transport system in the bacterial cytoplasmic membrane. J. Biol. Chem. 273:9630-9636. [DOI] [PubMed] [Google Scholar]
- 76.Tjaden, J., H. H. Winkler, C. Schwoppe, M. Van Der Laan, T. Mohlmann, and H. E. Neuhaus. 1999. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J. Bacteriol. 181:1196-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Torri, A. F., K. I. Bertrand, and S. L. Hajduk. 1993. Protein stability regulates the expression of cytochrome c during the developmental cycle of Trypanosoma brucei. Mol. Biochem. Parasitol. 57:305-316. [DOI] [PubMed] [Google Scholar]
- 78.Torri, A. F., and S. L. Hajduk. 1988. Posttranscriptional regulation of cytochrome c expression during the developmental cycle of Trypanosoma brucei. Mol. Cell. Biol. 8:4625-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tu, X., and C. C. Wang. 2004. The involvement of two cdc2-related kinases (CRKs) in Trypanosoma brucei cell cycle regulation and the distinctive stage-specific phenotypes caused by CRK3 depletion. J. Biol. Chem. 279:20519-20528. [DOI] [PubMed] [Google Scholar]
- 80.Ullu, E., A. Djikeng, H. Shi, and C. Tschudi. 2002. RNA interference: advances and questions. Philos. Trans. R. Soc. Lond. B 357:65-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Roermund, C. W. T., R. Drissen, M. van den Berg, L. Ijlst, E. H. Hettema, H. F. Tabak, H. R. Waterham, and R. J. A. Wanders. 2001. Identification of a peroxisomal ATP carrier required for medium-chain fatty acid β-oxidation and normal peroxisome proliferation in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:4321-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Weelden, S. W., J. J. van Hellemond, F. R. Opperdoes, and A. G. Tielens. 2005. New functions for parts of the Krebs cycle in procyclic Trypanosoma brucei, a cycle not operating as a cycle. J. Biol. Chem. 280:12451-12460. [DOI] [PubMed] [Google Scholar]
- 83.Vickerman, K. 1985. Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull. 41:105-114. [DOI] [PubMed] [Google Scholar]
- 84.Voncken, F., B. Boxma, J. Tjaden, A. Akhmanova, M. Huynen, F. Verbeek, A. G. Tielens, I. Haferkamp, H. E. Neuhaus, G. Vogels, M. Veenhuis, and J. H. Hackstein. 2002. Multiple origins of hydrogenosomes: functional and phylogenetic evidence from the ADP/ATP carrier of the anaerobic chytrid Neocallimastix sp. Mol. Microbiol. 44:1441-1454. [DOI] [PubMed] [Google Scholar]
- 85.Voncken, F., J. J. van Hellemond, I. Pfisterer, A. Maier, S. Hillmer, and C. Clayton. 2003. Depletion of GIM5 causes cellular fragility, a decreased glycosome number and reduced levels of ether-linked phospholipids in trypanosomes. J. Biol. Chem. 278:35299-35310. [DOI] [PubMed] [Google Scholar]
- 86.Walker, J. E., and M. J. Runswick. 1993. The mitochondrial transport protein superfamily. J. Bioenerg. Biomembr. 25:435-446. [DOI] [PubMed] [Google Scholar]
- 87.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
- 88.Weber, F. E., G. Minestrini, J. H. Dyer, M. Werder, D. Boffelli, S. Compassi, E. Wehrli, R. M. Thomas, G. Schulthess, and H. Hauser. 1997. Molecular cloning of a peroxisomal Ca2+-dependent member of the mitochondrial carrier superfamily. Proc. Natl. Acad. Sci. USA 94:8509-8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Woodward, R., and K. Gull. 1990. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 95:49-57. [DOI] [PubMed] [Google Scholar]
- 90.Wylin, T., M. Baes, C. Brees, G. P. Mannaerts, M. Fransen, and P. P. Van Veldhoven. 1998. Identification and characterization of human PMP34, a protein closely related to the peroxisomal integral membrane protein PMP47 of Candida boidinii. Eur. J. Biochem. 258:332-338. [DOI] [PubMed] [Google Scholar]
- 91.Zarella-Boitz, J. M., N. Rager, A. Jardim, and B. Ullman. 2004. Subcellular localization of adenine and xanthine phosphoribosyltransferases in Leishmania donovani. Mol. Biochem. Parasitol. 134:43-51. [DOI] [PubMed] [Google Scholar]