Abstract
Human respiratory syncytial virus (RSV) is a major cause of respiratory tract infections worldwide. Several novel small-molecule inhibitors of RSV have been identified, but they are still in preclinical or early clinical evaluation. One such inhibitor is a recently discovered triphenol-based molecule, VP-14637 (ViroPharma). Initial experiments suggested that VP-14637 acted early and might be an RSV fusion inhibitor. Here we present studies demonstrating that VP-14637 does not block RSV adsorption but inhibits RSV-induced cell-cell fusion and binds specifically to RSV-infected cells with an affinity corresponding to its inhibitory potency. VP-14637 is capable of specifically interacting with the RSV fusion protein expressed by a T7 vaccinia virus system. RSV variants resistant to VP-14637 were selected; they had mutations localized to two distinct regions of the RSV F protein, heptad repeat 2 (HR2) and the intervening domain between heptad repeat 1 (HR1) and HR2. No mutations arose in HR1, suggesting a mechanism other than direct disruption of the heptad repeat interaction. The F proteins containing the resistance mutations exhibited greatly reduced binding of VP-14637. Despite segregating with the membrane fraction following incubation with intact RSV-infected cells, the compound did not bind to membranes isolated from RSV-infected cells. In addition, binding of VP-14637 was substantially compromised at temperatures of ≤22°C. Therefore, we propose that VP-14637 inhibits RSV through a novel mechanism involving an interaction between the compound and a transient conformation of the RSV F protein.
Human respiratory syncytial virus (RSV) is the major cause of upper and lower respiratory tract infections in the pediatric population. These infections are particularly problematic in infants that are born prematurely or with congenital heart disease or chronic lung disease or are otherwise immune compromised (reference 9 and references therein). Elderly and immunocompromised adults are also at increased risk for developing complications or even death associated with RSV infection (for a review, see reference 13).
The only antiviral agent approved for the treatment of RSV infection is ribavirin, but due to efficacy and toxicity issues, it has only limited utility (1). After more than 40 years of research, there is no approved vaccine, and the only prophylactic therapies available are RSV-IVIG, a polyclonal RSV immunoglobulin (19), and Synagis (palivizumab), a human monoclonal antibody targeting the RSV fusion protein (26). While effective, this treatment is only administered to high-risk pediatric patients. There is a clear need for new anti-RSV therapeutics, with improved efficacy and safety for broader applications (for a review, see reference 37).
RSV belongs to the Pneumovirinae subfamily of the Paramyxoviridae, which are enveloped viruses containing single-stranded, nonsegmented minus-strand RNA genomes (reviewed in reference 9). RSV encodes three surface glycoproteins, the fusion protein F, the attachment glycoprotein G, and the small hydrophobic protein SH. A distinguishing feature of the Pneumovirinae subfamily is the absence of a hemagglutinin-neuraminidase (HN) protein homolog. Both the F and G glycoproteins are required for efficient infectivity in vivo, but the F protein alone is sufficient for virus binding and entry into cells in vitro (30). The G protein does not seem to be directly involved in fusion but enhances viral entry by facilitating virus attachment to the surface of host cells (49). This is interesting considering the importance of the HN proteins for fusion in other paramyxoviruses (32). The function of the SH protein is unclear. One report indicates that fusion is enhanced in its presence (23), while others observed no effects (4, 28) or even an inhibitory effect (49). In the mouse model, SH-deleted viruses replicated as efficiently as wild-type RSV in the lungs but were attenuated in the upper respiratory tract (4).
No definitive cellular receptor for RSV has been identified. However, both the F and G glycoproteins have been shown to bind heparin in vitro, and cells treated with heparinase are not easily infected with RSV, suggesting that heparin may have at least an accessory role for RSV entry (3, 14, 20, 29, 35). Similar to F proteins of other paramyxoviruses, RSV F is expressed as a precursor, F0, which is cleaved by a cellular enzyme(s), probably furin, into two disulfide-linked subunits, F1 and F2 (10). This cleavage takes place in the Golgi or trans-Golgi and is essential for fusion activity and thus infectivity (17, 58). F1 contains the hydrophobic fusion peptide at the amino terminus, followed by two heptad repeats (HR1 and HR2) separated by almost 300 amino acids of intervening sequence, ending with a hydrophobic carboxy-terminal transmembrane domain, which anchors the protein to the viral membrane.
Various biochemical and structural analyses suggest that the F protein forms a homotrimer on the viral cell surface by interactions between the HR1 regions (5, 34, 36, 57). The fusion peptide is thought to be buried in the interior of the F protein trimer until attachment to the cell surface triggers a conformational change, which exposes the peptide and allows its insertion into the host cell membrane. In the second stage of the fusion process, another conformational change is thought to occur, presumably within the intervening domains, that results in the actual fusion of viral and cell membranes. This process appears to be driven by the formation of a stable six-helix bundle composed of the HR1 and HR2 regions from the three F protein monomers (38, 43). This model for the RSV fusion process has emerged from crystallographic data (57) and the characterization of fusion proteins from analogous systems such as human immunodeficiency virus type 1 (HIV-1) (gp41) (7, 27, 50), influenza virus (HA) (46), and other paramyxoviruses (F), including simian virus 5 (2, 24) and Newcastle disease virus (45).
Peptides comprised of sequences from either heptad repeat region of gp41 from HIV-1 (51, 52), the F protein from Sendai virus (40), human parainfluenza virus 3 (55), and Newcastle disease virus (56) are able to inhibit fusion of their respective viruses, presumably by disrupting the formation of the six-helix complex. In fact, for HIV-1, a peptide homologous to a portion of HR2, T-20, has been shown in clinical trials to significantly reduce plasma viral loads in a broad range of HIV-infected patients (31). In an analogous situation, a series of peptides derived from the HR2 sequence of the RSV F protein have been designed and shown to inhibit RSV fusion and infectivity in vitro (33). While peptide-based therapy might not be an optimal treatment approach, the discovery of these peptides has been indispensable for elucidating the mechanism of fusion and for validating fusion as a viable therapeutic target.
In recent years, several structurally distinct small molecules that appear to interfere with the RSV entry process have emerged from various corporate antiviral screens. These include the disulfonated stilbenes CL387626 and RFI-641 (Wyeth-Ayerst) (41, 54), the benzimidazole derivative R-170591 (Janssen Pharmaceutica) (K. Andries et al., Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., 2000, abstr. 1160), and the triphenol VP-14637 (ViroPharma) (D. C. Pevear et al., Abstr. 13th Int. Conf. Antiviral Res., 2000, abstr. 57). All of these compounds are potent inhibitors of RSV replication in vitro and can significantly reduce viral lung titers in the cotton rat model for RSV infection (25; D. C. Pevear et al., Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., 2000, abstr. 1854; K. Andries et al., Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., 2000, abstr. 1160). Time-of-addition experiments indicated that all of these compounds act early in the RSV replication cycle and that mutations conferring resistance to CL387626 and R-170591 map to various regions of the F protein (A. Gazumyan et al., Abstr. 12th Int. Conf. Antiviral Res., 1999, abstr. LB-9; K. Andries et al., Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., 2000, abstr. 1160). In addition, RFI-641 has been shown to inhibit RSV fusion, presumably via binding to the RSV F protein (41). However, no detailed mechanistic studies have been reported for either R-170591 or VP-14637. In the present study, we have undertaken an extensive analysis of VP-14637 as a model for understanding inhibitory mechanisms of RSV fusion.
MATERIALS AND METHODS
Cells and viruses.
Hep-2, Vero, and BHK-21 cell lines were cultured in minimal essential medium plus 2 mM l-glutamine, 0.1 mM nonessential amino acids, and 10% fetal bovine serum. The BHK-21 cells were also supplemented with 10% tryptose phosphate broth. Primary chicken embryo fibroblasts (CEF) and the CEF cell line were cultured in Dulbecco's modified Eagle's medium with 4.5 g of glucose per liter, 4 mM glutamine, and 10% fetal bovine serum at 39°C. All cell lines were obtained from the American Type Culture Collection. The RSV strain A2 (American Type Culture Collection) was grown and titered in Hep-2 cells as previously described (18). The attenuated vaccinia virus expressing T7 polymerase (MVA-T7), provided by Bernard Moss (National Institutes of Health, Bethesda, Md,), was grown in primary CEF cells and titered by a standard plaque assay (53).
Plasmids.
To generate the pCDNA-F construct, the F gene from RSV A2 was obtained by reverse transcription-PCR amplification of RNA isolated from RSV-infected Hep-2 cells and cloned into the pCDNA 3.1 expression vector (Invitrogen, Carlsbad, Calif.). To construct the F protein mutants F488Y and T400A, site-directed mutagenesis (QuikChange protocol from Stratagene) was performed on a region containing HR2 (nucleotides 916 to 1725), which had been subcloned into pBluescript II SK+ (Stratagene, La Jolla, Calif.). To verify the mutations, the inserts were sequenced with the ABI Prism BigDye terminator kit on the ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.). The fragments containing mutations were cloned back into the pCDNA-F plasmid. Plasmid pT7-Luc, which contains the luciferase gene under the control of a T7 polymerase promoter, was constructed by replacing the cat gene in pBH82 (22) with the luciferase gene from the pFR-Luc plasmid (Stratagene, La Jolla, Calif.). Plasmid pBH437 contains the T7 RNA polymerase gene driven by the cytomegalovirus promoter. Both the pBH82 and pBH437 constructs were kindly provided by Robert Lamb (Northwestern University, Evanston, Ill.).
Compounds.
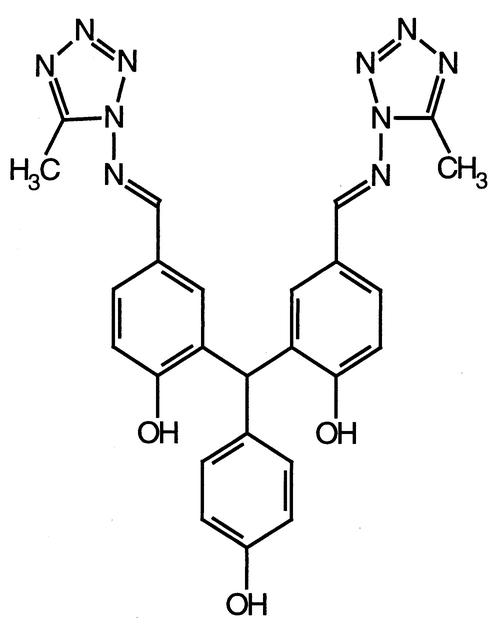
VP-14637 (5,5′-bis[1-(((5-amino-1H-tetrazolyl)imino)methyl)] 2,2′,4"-methylidynetrisphenol; Fig. 1) was synthesized essentially as described elsewhere (T. J. Nitz et al., Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., 2000, abstr. 1853). Briefly, 3-bromo-4-methoxybenzaldehyde was protected with neopentyl glycol to form the 1,3-dioxane derivative, which was treated with n-butyl lithium at −78°C, followed by the addition of methyl 4-methoxybenzoate in tetrahydrofuran to give 5,5′-bis(5,5-dimethyl-1,3-dioxan-2-yl)-2,2′,4′-trimethoxytriphenylmethanol. The triphenylmethanol derivative was dissolved in formic acid and heated at 100°C for 13 h to form 4,4′-dimethoxy-3,3′-(4-methoxyphenyl)methylenebisbenzaldehyde, which was then treated with boron tribromide solution to give unprotected methylenebisbenzaldehyde. The N,N-dimethylformamide solution of bisbenzaldehyde, 1-amino-5-methyl tetrazole, and p-toluenesulfonic acid was heated at 100°C overnight to produce the final compound. The tritiated compound, [3H]VP-14637 (specific activity, 4 Ci/mmol), was prepared by Moravek Biochemicals (Brea, Calif.). Ribavirin was obtained from Sigma. The HR2-derived peptide T-118 (33) was synthesized by Genemed Synthesis (South San Francisco, Calif.).
FIG. 1.
Structure of VP-14637.
Antiviral activity.
To determine the antiviral activity of the expected RSV inhibitors, a previously described cytopathic effect-inhibitory assay was performed (39, 44). Briefly, Hep-2 cells were seeded into 96-well plates at 1,000 cells per well. After 24 h, the cells were infected with RSV at a multiplicity of infection (MOI) of 0.05 to 0.1 and incubated at 37°C for 2 h. The inoculum was replaced with fresh medium containing serial dilutions of the tested inhibitors. Following a 4-day incubation, the medium was removed and the cells were stained with crystal violet (0.1% in 20% methanol). The RSV-induced cytopathic effect was quantitated spectrophotometrically by reading the absorbance at 630 nm, and the concentration of inhibitor that reduced cytopathic effect by 50% relative to the untreated control (EC50) was calculated by nonlinear regression. A difference of more than 10-fold was detected under the conditions described between the signals from uninfected and infected, untreated cells.
Temperature shift assay.
The antiviral assay was modified in the following manner to determine if VP-14637 could block RSV adsorption to cells. Hep-2 cells were plated as described above, but the RSV incubation was performed at 4°C for 1 h in the presence or absence of various amounts of compound. These conditions allow virus adsorption but not fusion. Heparin was used as a control that blocks RSV adsorption. After incubation with RSV, the cells were washed three times with phosphate-buffered saline and then shifted to 37°C. After 5 days at 37°C, the cells were stained, and cytopathic effect was quantitated as described in the previous section.
Viral fusion assay.
The fusion assay described previously for simian virus 5 (21) was adapted for RSV and modified in the following manner. BHK-21 cells in 96-well plates at approximately 50% confluence were infected at various MOIs with RSV for 22 to 24 h and then transfected with a pT7-Luc plasmid with Lipofectamine Plus according to the manufacturer's protocol for 3 to 5 h (Invitrogen, Carlsbad, Calif.). During the infection, another population of BHK-21 cells, plated in six-well plates, was transfected with plasmid pBH437. This second population of cells was trypsinized and overlaid onto the virus-infected, T7-Luc-transfected cells in a 1:1 ratio. After 6 to 7 h, the mixed population of cells was lysed and analyzed for luciferase activity with the luciferase assay system (Promega, Madison, Wis.). Luciferase activity was measured in a TopCount NXT (Packard Bioscience Co.) and is reported as relative light units.
Compound binding assays.
For time course and saturation binding experiments, Hep-2 cells were seeded into 12-well plates at 105 cells/well and infected with RSV at an MOI of ≈0.1 for 24 h. Various concentrations of [3H]VP-14637 in 2 ml of medium were added to the cells for various times at 37°C. After incubation, the medium was removed, and the cells were washed three times in phosphate-buffered saline, lysed in 1% Igepal, added to 5 ml of EcoLite scintillation fluid (ICN), and read in a LS6500 scintillation counter (Beckman Coulter, Fullerton, Calif.). For all other binding experiments, the infections were performed at an MOI of ≈0.01 to 0.05 for 48 h, followed by a 2-h incubation with 10 nM [3H]VP-14637. Where indicated, an excess of unlabeled VP-14637 (1 μM) was added together with [3H]VP-14637 to confirm specific binding. For the temperature-dependent binding experiment, the RSV-infected cells and compound were equilibrated on ice (0°C), at room temperature (22°C), and at 37°C before incubation together for 2 h at the indicated temperatures.
Cellular fractionation.
Cells were scraped into an isoosmotic buffer (10 mM triethanolamine, 10 mM acetic acid, 0.25 M sucrose, and 1 mM EDTA, pH 7.8) and disrupted by 30 strokes in a Dounce homogenizer. The nuclear fraction was obtained by pelleting at 1,000 × g for 10 min. The supernatant was then recentrifuged at 30,000 × g for 30 min to obtain the membrane fraction (pellet) and the cytoplasmic fraction (supernatant).
MVA-T7 vaccinia virus expression system.
CEF cells were plated at 4 × 105 cells/well in 12-well plates and incubated for 24 h at 39°C. After incubation, the cells were infected with MVA-T7 at an MOI of ≈1 for 1.5 to 2 h, washed in serum-free medium, and then transfected with F expression plasmid pCDNA-F or empty control plasmid pCDNA-3.1 with Lipofectamine Plus according to the manufacturer's directions (Invitrogen). After 24 h, the cells were incubated with [3H]VP-14637 for 6 h and analyzed as described above.
Immunoblotting, immunoprecipitation, and FACS analysis.
For total RSV F protein immunoblots, Hep-2 cells infected with wild-type or mutant RSV were trypsinized, pelleted, washed with phosphate-buffered saline, and lysed in 1% Igepal. A standard protein assay was performed (Bio-Rad, Hercules, Calif.), and 50 μg of total protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% gradient gel under nonreducing conditions. For the membrane fraction immunoblot, the entire sample was loaded. The proteins were transferred to nitrocellulose membranes and blocked with 5% powdered milk in Tris-buffered saline with Tween 20 (TBST). Detection of the F protein was performed with a 1:500 dilution of monoclonal antibody 0681 (ViroStat, Portland, Maine), followed by a 1:2,000 dilution of horseradish peroxidase-conjugated anti-mouse immunoglobulin antibody (Amersham Biosciences, Piscataway, N.J.). Proteins were visualized with enhanced chemiluminescence (Amersham Biosciences) and autoradiography.
For surface-expressed RSV F analysis, immunoprecipitations or fluorescence-activated cell sorting (FACS) analysis was performed. For immunoprecipitations, infected Vero cells were first treated with 20 μg of biotin-X-NHS (Calbiochem, San Diego, Calif.) following the cellular labeling protocol (Roche Applied Science, Indianapolis, Ind.). After surface biotinylation, the cells were lysed in 1% Igepal, and immunoprecipitations were performed. The lysates were precleared with normal rabbit serum (ICN, Costa Mesa, Calif.) for 1 h, followed by a 30-min incubation with fixed Staphylococcus aureus protein A, Zysorbin (Zymed Laboratories, South San Francisco, Calif.). The 0681 F monoclonal antibody (1 μg) was added for 3 to 24 h, followed by 1 μg of rabbit anti-mouse immunoglobulin (ICN) for 30 min and 100 μl of a 20% protein A-Sepharose slurry (Zymed Laboratories). All incubations were carried out at 4°C with rocking. The immune complexes were washed three times in lysis buffer, resuspended in protein sample buffer (Invitrogen), and then subjected to SDS-PAGE. The proteins were transferred to nitrocellulose, blocked in 5% powdered milk in TBST, and detected with a 1:1,000 dilution of streptavidin-horseradish peroxidase conjugate (Amersham).
For FACS analysis, CEF cells infected as previously described were washed with phosphate-buffered saline, trypsinized, washed with FACS buffer (1% fetal bovine serum in phosphate-buffered saline), resuspended in 100 μl of FACS buffer containing 0.1 μg of the 0681 monoclonal antibody, and incubated on ice for 30 min. After primary antibody incubation, the cells were washed twice in FACS buffer, resuspended in 100 μl of FACS buffer containing 0.1 μg of fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin antibody (Pharmingen, San Diego, Calif.), and incubated on ice for 30 min. After secondary-antibody incubation, the samples were washed twice, resuspended in 500 μl of FACS buffer, and analyzed on a FACSort (Becton Dickinson, San Jose, Calif.).
Selection and characterization of drug-resistant viruses.
RSV variants resistant to VP-14637 were selected by passaging the virus in Hep-2 cells in the presence of increasing concentrations of the inhibitor. The starting drug concentration was 5 nM, and after six passages, the viruses were able to grow in 50 nM VP-14637. Control viruses were grown in parallel to the same passage without drug. Susceptibility of the resultant viruses to VP-14637 was determined in the cytopathic effect assay as described above, and the F genes were sequenced. RNA was isolated from the infected cells by lysis in RNA STAT-60 (Tel-Test, Friendswood, Tex.), chloroform extraction, and isopropanol precipitation as per the manufacturer's protocol. cDNA for sequencing was then obtained by reverse transcription-PCR with the Titan One Tube reverse transcription-PCR kit with the following primer sequences that flank the F gene, 5′-AGTCTCTACAACATCCGAGT-3′ (nucleotides 5521 to 5540) and 5′-GAGAGTTTCGATGAAGTTCA-3′ (nucleotides 7471 to 7490). Sequence reactions were performed with the ABI Prism BigDye terminator kit with 60 ng of cDNA and a series of nine primers to completely span both strands of the entire F gene. The reactions were analyzed in the ABI Prism 3100 genetic analyzer.
RESULTS
Activity of VP-14637 in antiviral and cell fusion assays.
In various in vitro assays, VP-14637 was shown to effectively inhibit RSV replication at nanomolar concentrations (D. C. Pevear et al., Abstr. 13th Int. Conf. Antiviral Res., 2000. abstr. 57). To confirm these results, we performed a standard quantitative cytopathic effect-based RSV antiviral assay with VP-14637 and two other known inhibitors of RSV, ribavirin and the HR2 peptide T-118 (33). The results, depicted in Table 1, are consistent with the reported data and indicate that VP-14637 is approximately 1,000 times more potent than T-118 and 10,000 times more potent than ribavirin at inhibiting RSV.
TABLE 1.
Comparison of antiviral activity and fusion inhibition by several RSV inhibitorsa
| Compound | EC50 (nM)
|
|
|---|---|---|
| Cytopathic effect assay | Cell fusion assay | |
| VP-14637 | 1.4 | 5.4 |
| T-118 | 1,500 | 3,500 |
| Ribavirin | 33,000 | NA |
The cell-cell fusion assay was performed with wild-type RSV at an MOI of 0.6. NA, not applicable.
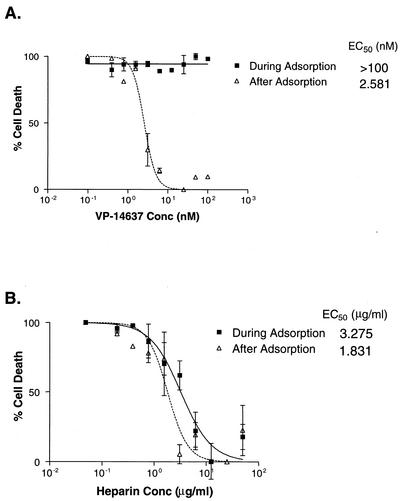
It has been suggested that VP-14637 acts as a fusion inhibitor. This is based on time-of-addition studies, which showed the greatest reduction in virus production when the compound was added 1 h before RSV infection (D. C. Pevear et al., Abstr. 13th Int. Conf. Antiviral Res., 2000, abstr. 57). While indicative, these data do not establish VP-14637 as a fusion inhibitor. To exclude inhibition of RSV attachment as a possible mode of action for VP-14637, the antiviral assay was performed with a temperature modification. RSV can bind to cells at 4°C, but fusion requires temperatures above 18°C (48). Therefore, a compound that can block virus attachment should have antiviral activity if added during a 4°C virus incubation. As shown in Fig. 2, VP-14637 had an EC50 of >100 nM when added only during the 4°C virus adsorption period, compared to 2.6 nM when present after adsorption. Conversely, a known inhibitor of RSV attachment, heparin, had similar EC50s whether added only during the virus adsorption period or throughout the entire assay, 3.3 and 1.8 μg/ml, respectively. These data indicate that VP-14637 acts at a stage subsequent to RSV attachment.
FIG. 2.
VP-14637 does not block RSV attachment. Hep-2 cells were incubated with RSV or mock infected at 4°C for 1 h in the presence (during adsorption) of serial dilutions of either VP-14637 (A) or heparin (B), washed to remove unbound virus and inhibitor, and then shifted to 37°C. The cytopathic effect was quantitated after 5 days and plotted as cell death with virus-only and cell-only controls. For comparison, a set of samples were treated with compounds after the RSV adsorption and washes (after adsorption).
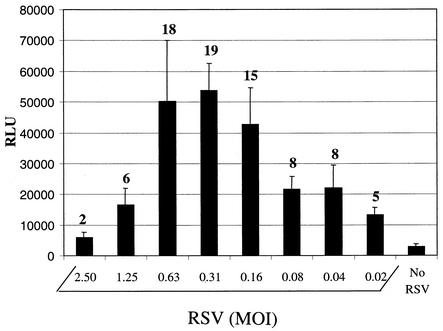
To further investigate the mechanism of RSV inhibition, we developed a cell-cell fusion assay similar to that used for simian virus 5 (21). One population of BHK-21 cells was infected with RSV and then transfected with a luciferase gene controlled by a T7 polymerase promoter. These cells were overlaid with a second population of BHK-21 cells that had been transfected with a T7 polymerase-expressing plasmid, and the fusion between the two cell populations was measured by quantitating the induced luciferase activity. The level of marker gene activation was dependent on viral input, with maximal expression reaching approximately 20-fold above background within an MOI range of 0.16 to 0.6 (Fig. 3). Optimal expression was also dependent on when the overlay was added relative to the time of infection and the duration of the coculture (data not shown). The assay function was verified by using T-118, a known RSV fusion inhibitor. Fusion, as measured by luciferase activity, was completely abolished when 20 μM T-118 was added to the RSV-infected population prior to the overlay step. The calculated EC50 for T-118 was 3.5 μM, a value similar to the EC50 obtained in the antiviral assay (Table 1). VP-14637 had an EC50 of approximately 5 nM in this assay, which is comparable to its activity in the antiviral assay. These results substantiate the view that VP-14637 acts by directly inhibiting RSV fusion.
FIG. 3.
RSV-dependent fusion assay. One population of BHK-21 cells was infected with the indicated amounts of RSV for 24 h, followed by transfection with pLuc-T7 plasmid containing a T7 promoter-driven luciferase gene. A second population of BHK-21 cells was transfected with T7 polymerase-expressing pBH437, trypsinized, and overlaid on top of the RSV-infected cells. After 6 h, the mixed cells were lysed and assayed for luciferase activity as a measure of cell-cell fusion. The luciferase activity is reported in relative light units (RLU). The numbers above the bars represent the increase (fold) over that in the control with no RSV.
Binding of VP-14637 to RSV-infected cells.
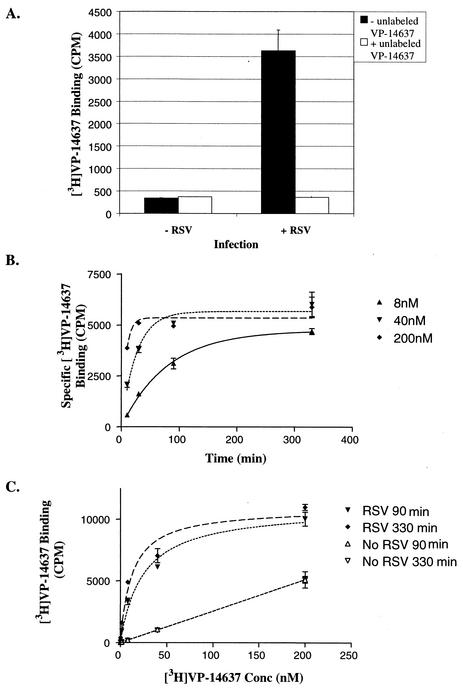
To further characterize the interactions important for the antiviral activity of VP-14637, its binding to RSV-infected cells was studied with radioactively labeled compound. As shown in Fig. 4A, [3H]VP-14637 bound the RSV-infected cells with a much greater efficiency than the control mock-infected cells. This binding was specific, as indicated by its inhibition by excess unlabeled compound. At a 10 nM concentration of the compound, the binding to infected cells reached levels about 15-fold higher than the nonspecific binding to uninfected cells. Interestingly, the presence of 20 μM T-118 had no effect on VP-14637 binding, suggesting that the compound may be acting at a different stage than the heptad repeat interaction (data not shown).
FIG. 4.
Binding of VP-14637 to RSV-infected cells. Hep-2 cells were infected with RSV or mock infected for 48 h (A) or 28 h (B and C) and incubated with various concentrations of [3H]VP-14637 for various lengths of time. After incubation, the cells were washed, lysed, and counted in a scintillation counter. (A) Specific binding of VP-14637 to RSV-infected cells was determined following the incubation of cells with 10 nM 3H-lableled compound for 150 min. Where indicated, a 1 μM excess of unlabeled compound was added. (B) Specific equilibrium binding was determined by subtracting the signal for mock-infected cells from that for RSV-infected cells. (C) The same data plotted as total binding versus drug concentration illustrate saturation binding for RSV-infected cells. Specific binding was used to calculate the reported Kd values for VP-14637.
Equilibrium binding was determined by incubating various concentrations of [3H]VP-14637 with RSV-infected cells for different time periods (Fig. 4B). As expected, equilibrium was reached sooner for higher concentrations of compound. The equilibrium t1/2 was 55, 18, and 5.5 min for 8, 50, and 200 nM compound concentrations, respectively. The compound exhibited saturable binding to the infected cells and exhibited a pattern typical of nonspecific binding to the uninfected cells (Fig. 4C). The calculated Kd for VP-14637 was 6 and 4 nM for 90-min and 6-h incubations, respectively, which correlated with the activity in both the antiviral and cell fusion assays.
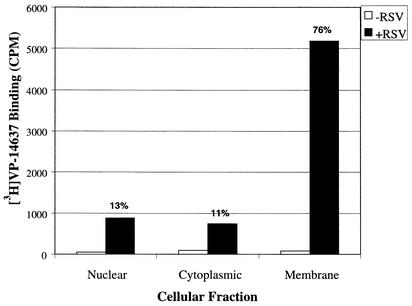
To investigate which components of the infected cells were interacting with VP-14637, cellular fractionation was performed on cells incubated with the labeled compound (Fig. 5). The majority (76%) of the compound remained bound to the membrane fraction, with only 13% segregating with the nuclear fraction and 11% with the cytoplasmic fraction. To verify that there was no cross-contamination between the fractions, a control fractionation was performed on RSV-infected cells and analyzed for F protein surface expression. As expected, the majority of F protein was in the membrane fraction, correlating with VP-14637 binding (data not shown). These data suggest that VP-14637 interacts with a component of RSV-infected cell membranes that could be the F protein.
FIG. 5.
Cellular fractionation following the binding of VP-14637. Hep-2 cells were infected with RSV at an MOI of 0.1 or mock infected for 48 h, followed by incubation with 10 nM [3H]VP-14637 for 2 h. The cells were subjected to Dounce homogenization and centrifugation to yield the indicated fractions. The samples were read in a scintillation counter to determine the level of bound [3H]VP-14637 in each fraction. The numbers above the bars indicate the percentage of compound in each fraction.
Binding of VP-14637 to F protein.
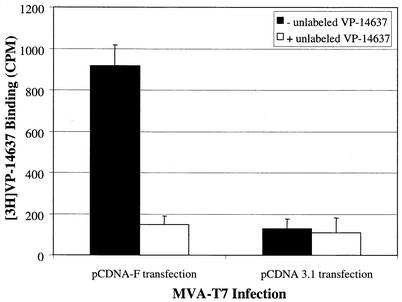
In order to test the potential interaction of VP-14637 with the RSV F protein, we used an expression system based on the attenuated recombinant vaccinia virus containing a T7 polymerase gene (MVA-T7) that allowed expression of only the F protein to levels equivalent to those produced by wild-type RSV. CEF cells were first infected with the vaccinia virus and then transfected with pCDNA-F. Twenty-four hours later, the cells were assayed for [3H]VP-14637 binding and F expression. Significant binding of VP-14637 occurred only in the vaccinia virus-infected cells, which also expressed the RSV F protein (Fig. 6). No significant binding was observed in cells infected with vaccinia virus and transfected with a control plasmid. Also, the binding of [3H]VP-14637 was completely and specifically inhibited by unlabeled VP-14637. Altogether, these data indicate that the RSV F protein is necessary and sufficient for VP-14637 interaction.
FIG. 6.
Binding of VP-14637 to cells expressing the RSV F protein. CEF cells were infected with a modified vaccinia virus expressing T7 polymerase, followed by transfection with pCDNA-F containing a T7 promoter-driven RSV F gene or a control plasmid, pCDNA-3.1. After 24 h, the cells were incubated with 10 nM [3H]VP-14637 (solid bars) or 10 nM [3H]VP-14637 plus 1 μM unlabeled VP-14637 (open bars). The cells were lysed, and measurements were obtained after 6 h.
Selection and characterization of VP-14637-resistant RSV variants.
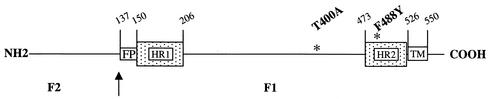
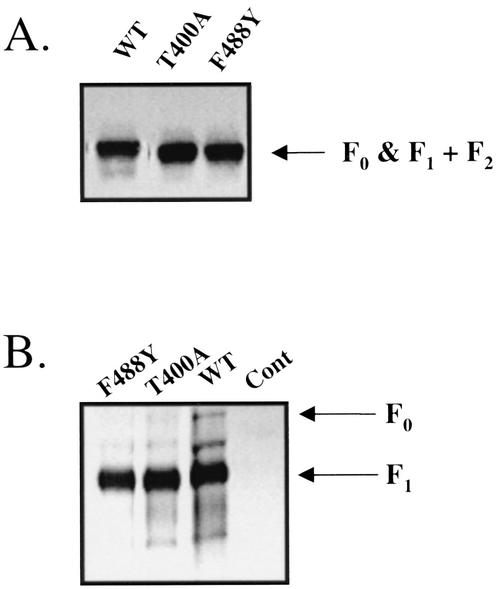
In order to verify the specific target for the inhibition of RSV fusion by VP-14637, wild-type RSV was passaged at least seven times in gradually increasing concentrations of the drug up to 50 nM. Untreated controls were similarly passaged to rule out any genotypic drift. Susceptibility to VP-14637 was determined for the resultant viruses, and their F genes were sequenced. The data from these analyses are presented in Table 2. Two independent selections resulted in viruses that each contained a unique point mutation, F488Y and T400A, that both conferred >1,000-fold-reduced susceptibility to VP-14637. These mutations were located in two separate regions of the F1 subunit. One was in the carboxy-terminal HR2 heptad repeat, and one was in the intervening region between the heptad repeats HR1 and HR2 (Fig. 7). Interestingly, no mutations were found in the amino-terminal heptad repeat HR1, which would be the expected site for resistance mutations generated against HR2 peptides. Neither of these two mutations appeared to have any deleterious effects on RSV replication, as measured by the standard cytopathic effect and fusion assays (data not shown). Additionally, the resistant viruses did not have any apparent defects in F protein expression, as shown by the immunoblot in Fig. 8A, or transport of F protein to the cell surface, as shown by surface labeling followed by immunoprecipitation (Fig. 8B). These data further suggest that F is involved in the mechanism of inhibition by VP-14637 and more closely define the regions of F protein that potentially participate in the interactions with the inhibitor.
TABLE 2.
Characterization of RSV variants resistant to VP-14637
| Virus | VP-14637 EC50 (nM)a | Resistance to VP-14637b (fold) | F genotype |
|---|---|---|---|
| RSV A2 | 0.90 | 1.0 | Wild type |
| Untreated Ac | 0.85 | 0.94 | Wild type |
| Untreated Bc | 0.72 | 0.80 | Wild type |
| VP-14637 A | >1,000 | >1,000 | F488Y |
| VP-14637 B | >1,000 | >1,000 | T400A |
EC50 values were determined in the standard antiviral assay described in Materials and Methods.
Resistance was calculated by dividing the EC50 of VP-14637 for the selected viral isolate by the EC50 for wild-type RSV A2.
Viruses passaged in the absence of compound. A and B represent two independent selections.
FIG. 7.
Location of VP-14637 resistance mutations. Schematic of the RSV F polyprotein indicating salient features: F1 and F2 subunits with an arrow representing the cleavage site. FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain. The numbers represent amino acid designations, and the asterisks indicate the resistance mutations that were selected by VP-14637.
FIG. 8.
Wild-type (WT) and resistant RSV-infected cells express equivalent levels of the F protein. Hep-2 or Vero cells were infected with the indicated viruses, and samples were processed for either total F protein expression via immunoblot analysis (A) or surface-expressed F protein via labeling with NHS-X-biotin followed by immunoprecipitation (B). The immunoblot and immunoprecipitation were performed with the same monoclonal anti-F antibody. In order to obtain optimal detection of the F protein for the immunoblot, electrophoresis was performed under nonreducing conditions. Cont, control.
VP-14637 binding to mutant F correlates with the resistance profile.
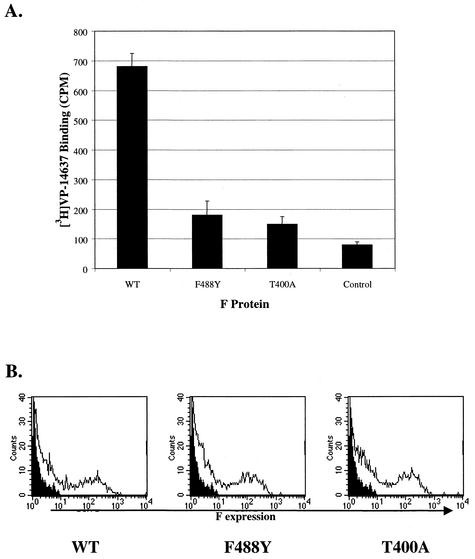
To determine whether the mechanism of resistance to VP-14637 might be a reduced ability of the compound to interact with the F protein, the binding assay was performed on individual F proteins containing the selected resistance mutations. The mutant F proteins were expressed in CEF cells with the MVA-T7 system. As shown in Fig. 9A, only the wild-type F protein allowed efficient binding of [3H]VP-14637. Binding of the compound to either the F488Y or T400A mutant was greatly reduced. To verify that this lack of binding was not due to decreased protein expression or inability to translocate to the cell surface, FACS analysis was performed on transfected CEF cells. Figure 9B indicates that neither F expression levels nor surface translocation was compromised for the mutant forms of F compared to wild-type F. Thus, the reduced susceptibility of RSV mutants to VP-14637 was due to the inability of the compound to interact with the mutant forms of the F protein.
FIG. 9.
VP-14637 does not bind to cells expressing F proteins that contain resistance mutations. (A) CEF cells were infected, transfected, and analyzed as described in the legend to Fig. 6. Genes encoding wild-type F (WT), one of the two point mutations (F488Y or T400A), or an empty control plasmid were used for transfection. (B) FACS analysis was performed on the cells to determine surface expression of the F proteins. The histograms are plotted as cell count versus F expression level. The solid histograms represent the control cells, and the open histograms correspond to cells expressing the indicated F proteins.
VP-14367 may bind F protein in a transient conformation.
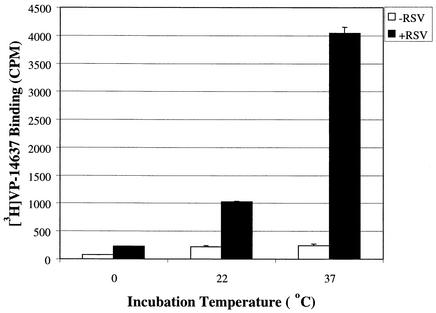
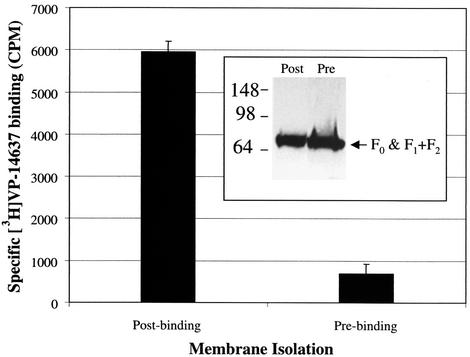
To further understand the interaction of VP-14637 with the F protein, the binding assay with RSV-infected cells was carried out at different temperatures (Fig. 10). Notably, 37°C was the only temperature that allowed efficient binding of [3H]VP-14637. At 22°C and 0°C, the specific binding was reduced >5-fold and >30-fold, respectively, suggesting that a dynamic temperature-sensitive process, such as a protein conformation change, may be required for efficient compound interaction. The ability of VP-14637 to bind RSV-infected cells and segregate with the membrane fraction led us to ask whether binding to isolated membranes would occur. The RSV-infected and mock-infected cells were first fractionated, and the isolated membranes were incubated with [3H]VP-14637 at 37°C. As shown in Fig. 11, there was no significant binding of VP-14637 to the isolated membrane fraction (prebinding sample). To rule out any appreciable protein loss or degradation due to the 37°C incubation and subsequent membrane washes, an immunoblot analysis of F protein expression was performed. The analysis confirmed that the levels of F in the samples were equivalent (Fig. 11, inset). Potentially, a transient conformation of F protein that may occur only during the process of active fusion is required for its interaction with VP-14637. The necessary conformational change in F protein may occur efficiently only at higher temperatures and may be present only in intact cells and not in isolated membranes.
FIG. 10.
VP-14637 binding is temperature dependent. Hep-2 cells were infected with wild-type RSV at an MOI of 0.01 (+RSV) or mock-infected (−RSV). After 48 h, the cells were incubated with 10 nM [3H]VP-14637 at the indicated temperatures for 2 h. After incubation, the cells were lysed, and compound binding was evaluated as described for Fig. 6.
FIG. 11.
VP-14637 does not bind to isolated membranes from RSV-infected cells. Hep-2 cells were infected with wild-type RSV at an MOI of 0.05. After 48 h, one set of cells (postbinding) were incubated with 10 nM [3H]VP-14637 or 10 nM [3H]VP-14637 plus 1 μM unlabeled VP-14637 for 2 h at 37°C, followed by membrane isolation, extensive washing, and either scintillation counting or anti-RSV F immunoblot analysis following SDS-PAGE under nonreducing conditions (inset). The other set of cells (prebinding) were first fractionated, and the isolated membranes were incubated with the same combination of compounds for 2 h at 37°C, followed by extensive washing, and then analyzed as for the previous set. Specific binding was determined by subtracting the background signal obtained from samples treated with excess unlabeled compound.
DISCUSSION
The majority of Food and Drug Administration-approved antiviral inhibitors to date target viral enzymes. Most of these are nucleoside and nucleotide analogs that inhibit viral DNA polymerases or HIV reverse transcriptase. The nucleoside analog ribavirin, licensed for the treatment of RSV infection, is the only clinically available drug that may act as a viral RNA polymerase inhibitor. Alternatively, its antiviral effect may be due more to its ability to inhibit IMP dehydrogenase (16) and/or its activity as a viral mutagen (11). Due to the insidious nature of HIV, extensive research into other viable viral targets has proceeded, and HIV protease inhibitors have become the first new class of clinically available antiviral therapeutics that target an enzyme other than a polymerase. Viral entry represents another attractive target for therapeutic intervention, particularly for acute viral infections such as RSV, because blocking virus replication at the earliest possible stage would be desirable. Although the various mechanisms of entry used by the most significant viral pathogens are not fully elucidated, much progress has been made. Again, HIV has become the model system for discovering a novel class of inhibitors, peptides that block a protein-protein interaction critical for the viral fusion process. Since this discovery, several groups have identified analogous peptides and novel small-molecule RSV inhibitors that also appear to block viral entry, presumably by interfering with the RSV fusion process. Here we have chosen one such molecule, VP-14637, as a model system to further understand RSV fusion and methods for its inhibition.
We have confirmed that VP-14637 is a potent inhibitor of RSV by a standard cytopathic effect assay, eliminated inhibition of virus attachment as a mechanism of action, and employed an RSV-dependent cell-cell fusion assay to demonstrate that the compound acts by inhibiting fusion. To investigate more directly the interactions of VP-14637 that are important for virus inhibition, a series of binding experiments with 3H-labeled compound were performed. The compound bound specifically to RSV-infected cells with an affinity comparable to the nanomolar EC50 values obtained from the antiviral and fusion assays. The majority of binding was associated with the membrane fraction, suggesting that an RSV-encoded membrane protein, presumably F, is a target for VP-14637 interaction. In order to confirm this hypothesis, recombinant F protein was expressed on the surface of CEF cells with a transient vaccinia virus-based system. Binding studies confirmed that F protein indeed interacts with VP-14637 in a specific manner. While our data indicate that the only RSV protein required for VP-14637 interaction is F, they do not completely rule out potential enhancing effects by other RSV proteins, such as the attachment protein G.
The interaction of VP-14367 with F protein was further confirmed by selecting RSV strains with reduced susceptibility to the inhibitor. Two viruses that both showed greater than 1,000-fold resistance to VP-14637 were identified. Each virus differed from wild-type RSV by a single point mutation in the F gene. The mutations occurred in two distinct regions of the F1 subunit, F488Y in HR2 and T400A in the intervening domain between HR1 and HR2. VP-14637 was not able to significantly bind to cells expressing either mutant F protein, indicating that the mutations have a direct role in reducing the virus's susceptibility to the inhibitor, presumably by preventing compound binding to the F protein.
One potential mechanism for the activity of VP-14637 is inhibition of the heptad repeat interactions, which would prevent the formation of a stabilized six-helix coiled coil that is believed to occur during the final stage of the viral fusion process. This is thought to be the inhibitory mechanism of the T-118 peptide derived from RSV HR2 (33) and the analogous carboxy-terminal heptad repeat peptide T-20 from HIV-1 gp41 (52). However, these peptides selected for resistant viruses that contained mutations only in their amino-terminal heptad repeat (42; S. J. Fischer-Mamalakis et al., Abstr. 20th Annu. Meet. Am. Soc. Virol., 2001, abstr. P2-4). In our study, the lack of resistance mutations in HR1 suggests that VP-14637 might be acting in an alternative manner.
Analysis of the crystal structure of the RSV fusion protein core formed by HR1 and HR2 peptides revealed the presence of a hydrophobic cavity formed by six amino acids from two adjacent HR1 repeats. This pocket can presumably accommodate two hydrophobic residues from HR2, F483 and F488, during the process of six-helix formation (57). Interestingly, F488 mutated to tyrosine in one of the VP-14637-resistant viruses. Potentially, this hydrophobic pocket is a site for VP-14637 binding, and the tyrosine substitution may prevent this interaction while still maintaining the proper function of the F protein. On the other hand, the HR2-derived peptide T-118, which contains F488 at its N terminus, does not interfere with the binding of VP-14637, suggesting that F488 might be involved in the interaction with VP-14637 at a different stage from the formation of the six-helix fusion core. Alternatively, the resistance mutations may be imparting a conformational change in a distal part of the protein that prevents the compound from interacting and may not be the direct site of compound binding.
An explanation for the involvement of T400 in the interaction with VP-14637 is not so readily available. Not much is known about the intervening domain located between the two heptad repeats of the F protein. The crystal structures of most viral fusion proteins contain only the helical core and therefore do not shed light on the potential role of the intervening sequences in fusion. Recently, a model structure of the RSV F protein was derived by homology modeling from the crystal structure of the Newcastle disease virus fusion protein (8, 47). The structures indicate that T400 is localized to an immunoglobulin-like β sandwich domain on the surface of the protein. It has been proposed that the RSV F protein as well as other viral fusion proteins go through multiple metastable conformations before reaching highly stabilized, fusion-competent conformations (6, 12, 32, 43). Perhaps the intervening sequences as well as HR2 are involved in these early transient conformations, and VP-14637 may be acting here instead of directly preventing the formation of the fusion core. This hypothesis is consistent with the lack of inhibition of VP-14637 binding in the presence of T-118. It also suggests that the fully formed six-helix core is not necessary for the interaction between VP-14637 and the F protein. Clearly, more extensive structural and mutational analyses need to be performed to elucidate the functions of these different regions in the viral fusion process and its inhibition.
The temperature dependence of VP-14637 binding further suggests that VP-14637 may interact with a transient conformation of the F protein. Significant binding occurred only at 37°C. The binding was considerably reduced at ≤22°C. Interestingly, active fusion also requires temperatures above 18°C (48). Perhaps the conformation of F protein necessary for VP-14637 interaction is only present during the active fusion process, which requires physiological temperatures. If this is the case, isolating the protein in a form capable of binding VP-14637 might be quite a challenge. To test this hypothesis, we decided to first determine if the compound would interact with isolated membranes from RSV-infected cells incubated at 37°C. VP-14637 did not specifically interact with the isolated membranes, although they contained the same amount of F protein as the intact RSV-infected cells. Apparently, the quality of the F protein was different between the isolated membranes and the intact cells. However, we cannot rule out the possibility that the membrane vesicles formed during fractionation were in the wrong orientation to allow efficient compound binding.
Further analysis of the VP-14637 interaction would benefit from the availability of purified F protein, but if the transient fusion-active conformation cannot be readily isolated, alternatives will have to be explored. It is not known what triggers the activation of F for competent fusion. Presumably, it is the interaction with a specific cellular receptor, a process analogous to the activation of the HIV-1 gp120/gp41 complex upon its interaction with CD4 and a specific chemokine coreceptor (15, 38). It is appealing to speculate that if the mechanism of activation were identified, perhaps the inactive conformation could be converted into the fusion-competent conformation in vitro. Another possible approach would be to isolate the F protein already bound to the inhibitor, provided that the complex is stable enough. This might allow mapping of the inhibitor binding site and the potential to obtain a crystal structure of the prefusion conformation of the F protein.
VP-14637 is one of several promising new anti-RSV compounds recently discovered through antiviral screens. These compounds are the first validation that small molecules can efficiently inhibit RSV fusion, presumably by blocking key protein-protein interactions or conformational changes within the F protein complex. Here we have presented data on the unique mode of action of VP-14637. This compound and others like it promise to be invaluable tools for dissecting the viral fusion machinery of RSV, which may finally lead to the design of treatment options that are safer and more effective than those currently available.
Acknowledgments
We thank Bernard Moss and Robert Lamb for providing necessary reagents. We also acknowledge Craig Gibbs, Bill Lee, and Mick Hitchcock for critical comments on the manuscript.
REFERENCES
- 1.Anderson, L. J., R. A. Parker, and R. L. Strikas. 1990. Association between respiratory syncytial virus outbreaks and lower respiratory tract deaths of infants and youg children. J. Infect. Dis. 161:640-646. [DOI] [PubMed] [Google Scholar]
- 2.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 3.Bourgeois, C., J. B. Bour, K. Lidholt, C. Gauthray, and P. Pothier. 1998. Heparin-like structures on respiratory syncytial virus are involved in its infectivity in vitro. J. Virol. 72:7221-7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukreyev, A., S. S. Whitehead, B. R. Murphy, and P. L. Collins. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J. Virol. 71:8973-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calder, L. J., L. Gonzalez-Reyes, B. Garcia-Barreno, S. A. Wharton, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2000. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology 271:122-131. [DOI] [PubMed] [Google Scholar]
- 6.Carr, C. M., C. Chaudhry, and P. S. Kim. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. USA 94:14306-14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9:255-266. [DOI] [PubMed] [Google Scholar]
- 9.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1443-1485. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 10.Collins, P. L., and G. Mottet. 1991. Post-translational processing and oligomerization of the fusion glycoprotein of human respiratory syncytial virus. J. Gen. Virol. 72:3095-3101. [DOI] [PubMed] [Google Scholar]
- 11.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. N. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 12.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion. Targets of opportunity. J. Cell Biol. 151:F9-F14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falsey, A. R., and E. E. Walsh. 2000. Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev. 13:371-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldman, S. A., S. Audet, and J. A. Beeler. 2000. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J. Virol. 74:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert, B. E., and V. Knight. 1986. Biochemistry and clinical applications of ribavirin. Antimicrob. Agents Chemother. 30:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Reyes, L., M. B. Ruiz-Arguello, B. Garcia-Barreno, L. Calder, J. A. Lopez, J. P. Albar, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. USA 98:9859-9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham, B. S., M. D. Perkins, P. F. Wright, and D. T. Karzon. 1988. Primary respiratory syncytial virus infection in mice. J. Med. Virol. 26:153-162. [DOI] [PubMed] [Google Scholar]
- 19.Groothuis, J. R., E. A. Simoes, M. J. Levin, C. B. Hall, C. E. Long, W. J. Rodriguez, J. Arrobio, H. C. Meissner, D. R. Fulton, R. C. Welliver, D. A. Tristram, G. R. Siber, G. A. Prince, M. Van Raden, and V. G. Hemming. 1993. Prophylactic administration of respiratory syncytial virus immune globulin to high-risk infants and young children. N. Engl. J. Med. 329:1524-1530. [DOI] [PubMed] [Google Scholar]
- 20.Hallak, L. K., D. Spillmann, P. L. Collins, and M. E. Peeples. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J. Virol. 74:10508-10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, B., G. P. Leser, R. G. Paterson, and R. A. Lamb. 1998. The paramyxovirus simian virus 5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology 250:30-40. [DOI] [PubMed] [Google Scholar]
- 22.He, B., W. T. McAllister, and R. K. Durbin. 1995. Phage RNA polymerase vectors that allow efficient gene expression in both prokaryotic and eukaryotic cells. Gene 164:75-79. [DOI] [PubMed] [Google Scholar]
- 23.Heminway, B. R., Y. Yu, Y. Tanaka, K. G. Perrine, E. Gustafson, J. M. Bernstein, and M. S. Galinski. 1994. Analysis of respiratory syncytial virus F, G, and SH proteins in cell fusion. Virology 200:801-805. [DOI] [PubMed] [Google Scholar]
- 24.Horvath, C. M., and R. A. Lamb. 1992. Studies on the fusion peptide of paramyxovirus fusion glycoprotein: roles of conserved residues in cell fusion. J. Virol. 66:2443-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huntley, C. C., W. J. Weiss, A. Gazumyan, A. Buklan, B. Feld, W. Hu, T. R. Jones, T. Murphy, A. A. Nikitenko, B. O'Hara, G. Prince, S. Quartuccio, Y. E. Raifeld, P. Wyde, and J. F. O'Connell. 2002. RFI-641, a potent respiratory syncytial virus inhibitor. Antimicrob. Agents Chemother. 46:841-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, S., C. Oliver, G. A. Prince, V. G. Hemming, D. S. Pfarr, S. C. Wang, M. Dormitzer, J. O'Grady, S. Koenig, J. K. Tamura, R. Woods, G. Bansal, D. Couchenour, E. Tsao, W. C. Hall, and J. F. Young. 1997. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 176:1215-1224. [DOI] [PubMed] [Google Scholar]
- 27.Judice, J. K., J. Y. K. Tom, W. Huang, T. Wrins, J. Vennari, C. J. Petropoulos, and R. S. McDowell. 1997. Inhibition of HIV type 1 infectivity by constrained a-helical peptides: Implications for the viral fusion mechanism. Proc. Natl. Acad. Sci. USA 94:13426-13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahn, J. S., M. J. Schnell, L. Buonocore, and J. K. Rose. 1999. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (RSV) glycoproteins: RSV fusion protein can mediate infection and cell fusion. Virology 254:81-91. [DOI] [PubMed] [Google Scholar]
- 29.Karger, A., U. Schmidt, and U. J. Buchholz. 2001. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. J. Gen. Virol. 82:631-640. [DOI] [PubMed] [Google Scholar]
- 30.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, b. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 94:13961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 32.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 33.Lambert, D. M., S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erickson, G. Merutka, and S. R. Petteway, Jr. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. USA 93:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawless-Delmedico, M. K., P. Sista, R. Sen, N. C. Moore, J. B. Antczak, J. M. White, R. J. Greene, K. C. Leanza, T. J. Matthews, and D. M. Lambert. 2000. Heptad-repeat regions of respiratory syncytial virus F1 protein form a six-membered coiled-coil complex. Biochemistry 39:11684-11695. [DOI] [PubMed] [Google Scholar]
- 35.Martinez, I., and J. A. Melero. 2000. Binding of human respiratory syncytial virus to cells: implication of sulfated cell surface proteoglycans. J. Gen. Virol. 81:2715-2722. [DOI] [PubMed] [Google Scholar]
- 36.Matthews, J. M., T. F. Young, S. P. Tucker, and J. P. Mackay. 2000. The core of the respiratory syncytial virus fusion protein is a trimeric coiled coil. J. Virol. 74:5911-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meanwell, N. A., and M. Krystal. 2000. Respiratory syncytial virus: recent progress towards the discovery of effective prophylactic and therapeutic agents. Drug Discov. Today 5:241-252. [DOI] [PubMed] [Google Scholar]
- 38.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moran, D. M., E. R. Kern, and J. C. J. Overall. 1985. Synergism between recombinant human interferon and nucleoside antiviral agents against herpes simplex virus: examination with an automated microtiter plate assay. J. Infect. Dis. 151:1116-1122. [DOI] [PubMed] [Google Scholar]
- 40.Rapaport, D., M. Ovadia, and Y. Shai. 1995. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 14:5524-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Razinkov, V., A. Gazumyan, A. Nikitenko, G. Ellestad, and G. Krishnamurthy. 2001. RFI-641 inhibits entry of respiratory syncytial virus via interactions with fusion protein. Chem. Biol. 8:645-659. [DOI] [PubMed] [Google Scholar]
- 42.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidtke, M., U. Schnittler, B. Jahn, H.-M. Dahse, and A. Stelzner. 2001. A rapid assay for evaluation of antiviral activity against coxsackie virus B3, influenza virus A, and herpes simplex virus type 1. J. Virol. Methods 95:133-143. [DOI] [PubMed] [Google Scholar]
- 45.Sergel-Germano, T., C. McQuain, and T. Morrison. 1994. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J. Virol. 68:7654-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 47.Smith, B. J., M. C. Lawrence, and P. M. Colman. 2002. Modelling the structure of the fusion protein from human respiratory syncytial virus. Protein Eng. 15:365-371. [DOI] [PubMed] [Google Scholar]
- 48.Srinivasakumar, N., P. L. Ogra, and T. D. Flanagan. 1991. Characteristics of fusion of respiratory syncytial virus with HEp-2 cells as measured by R18 fluorescence dequenching assay. J. Virol. 65:4063-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Techaarpornkul, S., N. Barretto, and M. E. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75:6825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wild, C., J. W. Dubay, T. Greenwell, T. Baird, T. G. Oas, C. McDanal, E. Hunter, and T. Matthews. 1994. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc. Natl. Acad. Sci. USA 91:12676-12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Mathews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: Correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Mathews. 1994. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyatt, L. S., B. Moss, and S. Rozenblatt. 1995. Replication-deficient vaccinia virus virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202-205. [DOI] [PubMed] [Google Scholar]
- 54.Wyde, P. R., D. K. Moore-Poveda, B. O'Hara, W. D. Ding, B. Mitsner, and B. E. Gilbert. 1998. CL387626 exhibits marked and unusual antiviral activity against respiratory virus in tissue culture and in cotton rats. Antivir. Res. 38:31-42. [DOI] [PubMed] [Google Scholar]
- 55.Yao, Q., and R. W. Compans. 1996. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology 223:103-112. [DOI] [PubMed] [Google Scholar]
- 56.Young, J. K., R. P. Hicks, G. E. Wright, and T. G. Morrison. 1997. Analysis of a peptide inhibitor of paramyxovirus (Newcastle disease virus) fusion with biological assays, NMR, and molecular modeling. Virology 238:291-304. [DOI] [PubMed] [Google Scholar]
- 57.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimmer, G., L. Budz, and G. Herrler. 2001. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J. Biol. Chem. 276:31642-31650. [DOI] [PubMed] [Google Scholar]