Abstract
Human immunodeficiency virus type 1 (HIV-1) entry is mediated by the consecutive interaction of the envelope glycoprotein gp120 with CD4 and a coreceptor such as CCR5 or CXCR4. The CCR5 coreceptor is used by the most commonly transmitted HIV-1 strains that often persist throughout the course of infection. Compounds targeting CCR5-mediated entry are a novel class of drugs being developed to treat HIV-1 infection. In this study, we have identified the mechanism of action of two inhibitors of CCR5 function, SCH-350581 (AD101) and SCH-351125 (SCH-C). AD101 is more potent than SCH-C at inhibiting HIV-1 replication in primary lymphocytes, as well as viral entry and gp120 binding to cell lines. Both molecules also block the binding of several anti-CCR5 monoclonal antibodies that recognize epitopes in the second extracellular loop of CCR5. Alanine mutagenesis of the transmembrane domain of CCR5 suggests that AD101 and SCH-C bind to overlapping but nonidentical sites within a putative ligand-binding cavity formed by transmembrane helices 1, 2, 3, and 7. We propose that the binding of small molecules to the transmembrane domain of CCR5 may disrupt the conformation of its extracellular domain, thereby inhibiting ligand binding to CCR5.
A new generation of antiviral drugs intended to counter human immunodeficiency virus type 1 (HIV-1) entry into susceptible cells is now under development. These compounds, generally referred to as fusion or entry inhibitors, are expected to have different toxicity and resistance profiles than the existing reverse transcriptase and protease inhibitors (7, 8, 20, 28). HIV-1 entry inhibitors that target CD4-gp120 interactions, coreceptor function, and gp41-mediated membrane fusion already are in different phases of preclinical or clinical development (7, 8, 20, 28). The HIV-1 coreceptors are particularly attractive from the perspective of identifying new antiviral compounds, since they are seven-transmembrane-domain G protein-coupled receptors, a family of proteins that is a well-validated target for drug development (31).
Among the many chemokine receptors that can mediate HIV-1 entry in vitro, only CCR5 and CXCR4 are of frontline pharmacological importance, since they are the coreceptors used by HIV-1 to enter primary CD4+ T cells, dendritic cells, and macrophages (2, 8, 20, 42). In particular, CCR5 is essential for viral transmission and replication during the early, clinically latent phase of disease (2, 14, 21). Moreover, in more than half of HIV-1-infected people, CCR5-using viruses are found exclusively even during late-stage disease, whereas in the remaining cases viruses that use the CXCR4 coreceptor are also present (2, 8). In vitro experiments indicate that a lower level of CCR5 expression can reduce cellular infection by HIV-1 (26, 39). This observation might have clinical relevance, because individuals carrying a mutant CCR5 allele that codes for a nonfunctional protein have a reduced rate of disease progression, presumably because of the lower CCR5 levels on their cells (6, 14, 21). Furthermore, blocking the natural function of CCR5 may not significantly impact human health, since individuals entirely lacking CCR5 do not exhibit any overt immune dysfunctions (6, 18, 29).
For the reasons outlined above, the identification of inhibitors of CCR5-mediated HIV-1 fusion and entry has been a focus of antiviral drug development in recent years. The first such inhibitors to be studied were the CC-chemokines macrophage inflammatory proteins 1α and 1β and RANTES (3). Variants of chemokines with increased potency in vitro have since been developed (5, 19, 32, 40, 41; F. Arenzana-Seisdedos, J. L. Virelizier, D. Rousset, I. Clark-Lewis, P. Loetscher, B. Moser, and M. Baggiolini, Letter, Nature 383:400, 1996). CCR5-specific monoclonal antibodies (MAbs), in particular those that recognize epitopes in the second extracellular loop (ECL2), efficiently inhibit HIV-1 fusion and entry (16, 17, 22, 38). Chemokines and MAbs, however, would not be orally available drugs because they are proteins, so an alternative strategy has been to identify small-molecule inhibitors of CCR5 coreceptor function based on their ability to block chemokine binding and/or signaling (20, 31). The first such small-molecule CCR5 antagonist to be described was TAK-779 (1). This particular compound is no longer being pursued as a drug candidate, but other small molecules that specifically target coreceptor function have now entered phase I clinical trials, specifically SCH-351125 (SCH-C) against CCR5 and AMD-3100 against CXCR4 (9, 15, 24, 30, 33). Both SCH-C and AMD-3100 have shown an ability to reduce plasma viremia in HIV-1-infected people, validating coreceptor function as a clinical drug target (31; J. Reynes, R. Rouzier, T. Kanouni, V. Baillat, B. Baroudy, A. Keung, C. Hogan, M. Markowitz, and M. Laughlin, 9th Conf. Retrovir. Opportun. Infect., p. 53, 2002; D. Schols, S. Claes, E. De Clercq, C. Hendrix, G. Bridger, G. Calandra, G. Henson, S. Fransen, W. Huang, J. Whitcomb, and C. Petropoulos, 9th Conf. Retrovir. Opportun. Infect., p. 53, 2002). SCH-C is a receptor antagonist that potently inhibits RANTES binding as well as HIV-1 entry and replication and has excellent oral bioavailability in rats, dogs, monkeys, and humans (33; Reynes et al., 9th Conf. Retrovir. Opportun. Infect.).
Small-molecule CCR5 antagonists are thought to inhibit HIV-1 entry by blocking the gp120-CCR5 interaction. For example, TAK-779 prevents gp120 and CC-chemokines, but not anti-CCR5 MAbs, from binding to CCR5 (1, 12). Inhibition of ligand binding by TAK-779 probably occurs through an allosteric mechanism, because this compound binds within a putative ligand-binding cavity formed by transmembrane (TM) helices 1, 2, 3, and 7 of CCR5 (12). Here we show that SCH-350581 (AD101) and SCH-C, two structurally related compounds, inhibit CCR5-mediated viral replication and entry, as well as gp120-CCR5 binding. Both inhibitors are active in the nanomolar range, but AD101 is about an order of magnitude more potent than SCH-C. Unlike TAK-779, AD101 and SCH-C also block the binding to CCR5 of several MAbs directed principally against the ECL2 of CCR5. Alanine scanning mutagenesis of the CCR5 TM domain shows that the activities of both AD101 and SCH-C depend on residues in helices 1, 2, 3, 5, and 7. AD101 relies on additional residues in helices 2 and 3, compared to SCH-C. According to a new CCR5 TM domain model developed by Seibert et al. (C. Seibert, W. Ying, S. O. Smith, B. Baroudy, S. McCombie, J. P. Moore, T. Dragic, and T. P. Sakmar, unpublished data), some residues are predicted to directly interact with the inhibitors, whereas others indirectly affect their activity by an allosteric mechanism. The residues involved in CCR5-inhibitor interactions fall within a putative ligand-binding cavity that we previously showed to be the binding site for TAK-779 (12). We propose that binding of small molecules to the TM domain of CCR5 disrupts the conformation of the extracellular domain, thus inhibiting the binding of chemokines, MAbs, and gp120 to CCR5.
MATERIALS AND METHODS
Reagents.
AD101 is a piperidino-piperazine amide, and SCH-C is an oximino-piperidino-piperidine amide. The compounds have molecular weights of 502.6 and 557.5, respectively (23, 24, 34, 35). Anti-CCR5 MAbs (PA8, PA10, PA11, PA12, and PA14), gp120JR-FL, and CD4-immunoglobulin G2 (IgG2) were generously provided by William C. Olson (Progenics Pharmaceuticals, Tarrytown, N.Y.). MAbs 23, 29, 02, 49, and 31 were purchased from R&D Systems (Minneapolis, Minn.).
Inhibition of HIV-1 replication and entry.
Mitogen-stimulated peripheral blood mononuclear cells (PBMC) (2 × 105) were incubated for 1 h with AD101 or SCH-C at 1.3 times the final concentration before the addition of HIV-1JR-FL (100 50% tissue culture infective doses), as previously described (37). The inhibitors were present throughout the duration of the culture. The production of p24 antigen was determined after 4 to 6 days as a measure of HIV-1 replication. NLluc+env− viruses complemented in trans by envelope glycoproteins (Env) from the HIV-1JR-FL isolate were generated as previously described (11). U87-CD4+ cells were lipofected with a plasmid expressing wild-type CCR5 and infected 24 h later with Env-pseudotyped reporter viruses in the presence of various concentrations of AD101 or SCH-C. The viral inoculum was equivalent to ∼106 relative light units (RLU) of luciferase activity, and inhibitors were present throughout the first 16 h of infection. Lysis buffer (Promega, Madison, Wis.) was added to the cells 48 h postinfection, and luciferase activity (in RLU) was measured as described elsewhere (11). In both the replication and entry assays, inhibitors were diluted 100-fold in dimethyl sulfoxide prior to being added to the cells.
Inhibition of gp120 and MAb binding to CCR5.
A soluble gp120-CD4 complex formed from monomeric gp120JR-FL (100 nM) and biotinylated CD4-IgG2 (50 nM) was added to L1.2-CCR5 cells (106) in the presence of different concentrations of AD101 or SCH-C for 1 h at 37°C. Inhibitors were diluted 100-fold in dimethyl sulfoxide prior to being added to the cells. The mean fluorescence intensity (MFI) was measured by flow cytometry after addition of phycoerythrin-labeled streptavidin (Pharmingen, San Diego, Calif.). Alternatively, L1.2-CCR5 cells (106) were incubated with 10 μg (50 nM) of anti-CCR5 MAb/ml with or without 100 nM AD101 or SCH-C. MAb binding was detected using a phycoerythrin-labeled goat anti-mouse antibody (Caltag Laboratories, Burlingame, Calif.).
HIV-1 entry mediated by CCR5 mutants.
U87-CD4+ cells were lipofected with a plasmid expressing wild-type or mutant CCR5 as described previously (11). After 24 h, the cells were infected with NLluc+env− virus pseudotyped with HIV-1JR-FL Env, with or without 100 nM AD101 or SCH-C, a concentration that causes >95% entry inhibition in this assay. The viral inoculum was equivalent to ∼106 RLU of luciferase activity, and inhibitors were present throughout the first 16 h of infection. The cells were lysed 48 h postinfection, and luciferase activity (in RLU) was measured. A typical RLU value (± standard deviation [SD]) for wild-type CCR5 without inhibitor was 75,000 ± 15,000. The uninhibited value, in each experiment, was defined as 100%. With inhibitor present, the extent of HIV-1 entry was reduced to 1,000 ± 200 RLU for wild-type CCR5. This residual entry was defined as 0%. For each CCR5 mutant, the extent of HIV-1 entry with and without inhibitor was normalized in the range of 0 to 100%. A few mutants, apparently more sensitive than the wild-type receptor to AD101 and SCH-C, yielded entry levels <0%. These effects were small and probably have no functional implication. For clarity, values of <0% are represented as 0%.
Identification of CCR5 mutants that are significantly insensitive to AD101 and SCH-C.
We estimate that a variation of ±20% from the mean is not significant in luciferase-based assays of HIV-1 entry, so all mutants yielding entry levels <20% with the inhibitors were considered to be not significantly different from wild-type CCR5. The normalized mean entry level (± SD) for this subset of mutant receptors was 2% ± 3%. Since a deviation from the mean of >2 SD is considered significant, a level of ≥16% entry in the presence of 100 nM AD101 or SCH-C was used as a cutoff for significance. Mutants with <1% wild-type CCR5 coreceptor activity are unreliable for studies of entry inhibitors. The following CCR5 mutants were therefore excluded from the study based on their poor function as HIV-1 coreceptors: Y15A, Y68A, F85A, C101A, S169A, Y251A, and N293A.
RESULTS
Inhibition of HIV-1 replication and entry.
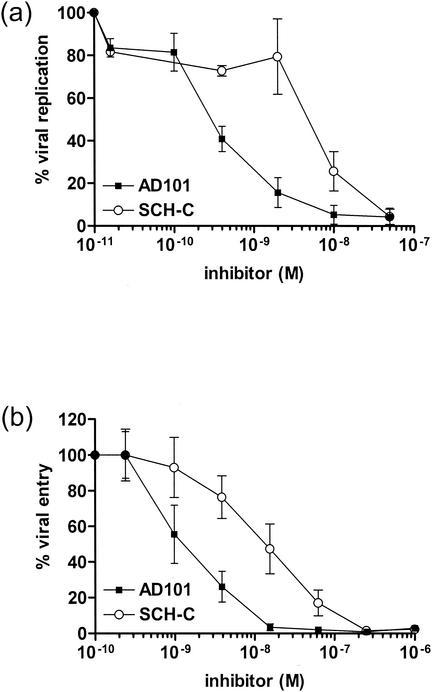
AD101 and SCH-C have been shown to potently and specifically inhibit the replication of a selection of CCR5-using (R5) primary HIV-1 isolates in human PBMC. The mean half-maximal inhibitory concentrations (IC50) determined using panels of test isolates were 1.5 nM for AD101 and 2.3 nM for SCH-C (33, 38). However, when the two inhibitors were compared directly, i.e., in the same PBMC-based replication assay, AD101 was generally found to be at least an order of magnitude more potent than SCH-C for a number of R5 isolates (J. P. Moore, A. Trkola, and T. Ketas, unpublished observations). For example, the IC50 for HIV-1JR-FL replication in human PBMC is 0.3 nM for AD101 and 6.1 nM for SCH-C (Fig. 1a). We also found AD101 to be about 10-fold more potent than SCH-C in blocking a single cycle of entry mediated by HIV-1JR-FL envelope glycoproteins into CCR5-expressing U87-CD4 cells. Half-maximal inhibition in this assay was observed at 1.0 nM for AD101 and 13.4 nM for SCH-C (Fig. 1b). The discrepancies in IC50 values between the replication and entry assays are probably due to differences in cell types, CCR5 levels, and input inocula.
FIG. 1.
Inhibition of viral replication and entry by AD101 and SCH-C. (a) Replication of HIV-1JR-FL was measured in mitogen-stimulated human PBMC in the presence of different concentrations of AD101 (black squares) or SCH-C (white circles). The perccent HIV-1JR-FL replication is defined as (nanograms of p24 with inhibitor/nanograms of p24 without inhibitor) × 100% and is plotted as a function of AD101 or SCH-C concentration. (b) Entry of HIV-1JR-FL Env-pseudotyped reporter viruses into U87-CD4 cells transiently expressing the CCR5 coreceptor was measured in the presence of different concentrations of AD101 (black squares) or SCH-C (white circles). The percent viral entry is defined as (RLU with inhibitor/RLU without inhibitor) × 100% and is plotted as a function of AD101 or SCH-C concentration. All values are means ± SD of three independent experiments.
Inhibition of gp120 and MAb binding to CCR5.
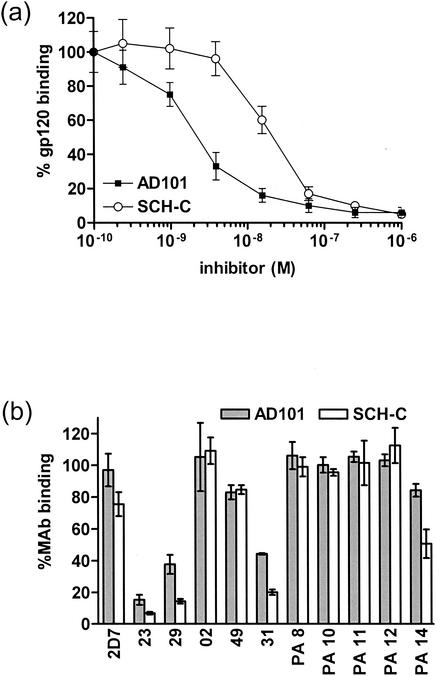
To investigate the mechanisms by which AD101 and SCH-C inhibit CCR5-mediated HIV-1 entry, we tested their abilities to block binding of a soluble gp120-CD4 complex to CCR5. The IC50 for binding of gp120JR-FL-CD4-IgG2 to CCR5-expressing L1.2 cells was 1.8 nM for AD101 and 19.3 nM for SCH-C (Fig. 2a). The differences between inhibitor concentrations necessary for half-maximal inhibition of viral entry and gp120-CCR5 binding are probably due to the use of monomeric gp120 and/or variation in the expression of CCR5 on different cell types.
FIG. 2.
Inhibition of gp120 and MAb binding by AD101 and SCH-C. (a) Binding of gp120JR-FL-CD4-IgG2 to L1.2 cells expressing the CCR5 coreceptor was measured in the presence of different concentrations of AD101 (black squares) or SCH-C (white circles). The percent inhibition of gp120-CCR5 binding is defined as (MFI with inhibitor/MFI without inhibitor) × 100% and is plotted as a function of AD101 or SCH-C concentration. (b) Alternatively, binding of anti-CCR5 MAbs to L1.2 cells was measured in the presence of 100 nM AD101 (gray bars) or SCH-C (white bars). The percent inhibition of MAb binding is defined as (MFI with inhibitor/MFI without inhibitor) × 100%. All values are means ± SD of three independent experiments.
We also tested the abilities of AD101 and SCH-C to inhibit the binding of a panel of anti-CCR5 MAbs to the CCR5-expressing L1.2 cells. The binding of MAbs 23, 29, 31, and PA14 was inhibited by 30 to 90%, depending on the combination of MAb and inhibitor (Fig. 2b). Similar binding inhibition was observed when using 10- and 100-fold-lower MAb concentrations (data not shown). All of the MAbs for which binding was inhibited by AD101 and SCH-C recognize epitopes that lie in ECL2, with the exception of PA14, for which the epitope straddles the Nt and ECL2 (16, 22).
Effects of AD101 and SCH-C on HIV-1 entry mediated by extracellular CCR5 mutants.
To examine the binding sites for AD101 and SCH-C on CCR5, we tested their inhibition of HIV-1 entry mediated by an extensive selection of CCR5 alanine mutants. The aim was to identify mutants that still supported viral entry but were less sensitive or insensitive to AD101 and SCH-C inhibition than was wild-type CCR5. We initially evaluated a panel of well-characterized alanine-substituted mutants with changes in the extracellular domain of CCR5, because residues in the Nt and ECL2 are important for coreceptor function (11, 13, 27). Alanine substitutions in the extracellular domains of CCR5 had little or no effect on the inhibitory activities of AD101 and SCH-C (Table 1).
TABLE 1.
Effects of AD101 and SCH-C on HIV-1 entry mediated by CCR5 extracellular domain mutantsa
| Domain | Mutant | Relative % entry in presence of:
|
|
Domain | Mutant | Relative % entry in presence of:
|
||
|---|---|---|---|---|---|---|---|---|
| AD101 | SCH-C | AD101 | SCH-C | |||||
| Nt | D2 | 0.0 ± 0.0 | 0.0 ± 0.0 | ECL2 | R168 | 0.3 ± 0.8 | 0.0 ± 0.0 | |
| Y3 | 0.0 ± 0.0 | 0.0 ± 0.0 | S169 | NA | NA | |||
| Q4 | 0.0 ± 0.0 | 0.0 ± 0.0 | Q170 | 0.5 ± 0.8 | 2.5 ± 0.6 | |||
| S6 | 0.3 ± 2.7 | 0.0 ± 0.0 | K171A/E172A | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| S7 | 0.3 ± 4.2 | 0.2 ± 1.4 | H175 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| P8 | 0.8 ± 0.4 | 1.3 ± 2.1 | Y176 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| Y10 | 0.0 ± 0.0 | 0.0 ± 0.0 | T177 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| D11 | 0.0 ± 0.0 | 0.0 ± 0.0 | C178 | 2.7 ± 2.3 | 2.6 ± 2.2 | |||
| N13 | 0.0 ± 0.0 | 0.0 ± 0.0 | S179 | 1.5 ± 1.8 | 0.0 ± 0.0 | |||
| Y14 | 0.4 ± 1.5 | 0.0 ± 0.0 | S180 | 0.7 ± 2.3 | 1.8 ± 0.8 | |||
| Y15 | NA | NA | H181 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| T16 | 0.0 ± 0.0 | 0.0 ± 0.0 | F182 | 4.0 ± 1.0 | 3.4 ± 0.9 | |||
| S17 | 0.0 ± 0.0 | 0.0 ± 0.0 | P183 | 1.0 ± 1.9 | 4.2 ± 1.1 | |||
| E18 | 0.0 ± 0.0 | 0.0 ± 0.0 | Y184 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| P19 | 0.0 ± 0.0 | 0.0 ± 0.0 | S185 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| C20 | 0.9 ± 1.0 | 0.0 ± 0.0 | Q186 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| Q21 | 0.0 ± 0.0 | 0.0 ± 0.0 | Y187 | 1.2 ± 0.1 | 0.0 ± 0.0 | |||
| K22 | 0.0 ± 0.0 | 0.0 ± 0.0 | Q188 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| N24 | 0.0 ± 0.0 | 0.0 ± 0.0 | F189 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| K26 | 0.0 ± 0.0 | 0.2 ± 1.5 | W190 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| Q27 | 0.0 ± 0.0 | 0.0 ± 0.0 | K191A/N192A | 0.9 ± 1.3 | 3.8 ± 0.8 | |||
| R31 | 2.9 ± 1.2 | 0.0 ± 0.0 | F193 | 0.1 ± 0.7 | 0.0 ± 0.0 | |||
| ECL1 | H88 | 0.4 ± 1.6 | 0.9 ± 0.2 | Q194 | 0.3 ± 0.5 | 0.0 ± 0.0 | ||
| Y89 | 0.0 ± 0.0 | 0.0 ± 0.0 | ECL3 | E262 | 0.0 ± 0.0 | 0.0 ± 0.0 | ||
| Q93 | 1.7 ± 1.5 | 1.5 ± 0.2 | F263 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| W94 | 0.0 ± 0.0 | 0.0 ± 0.0 | F264 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| D95 | 0.0 ± 0.0 | 0.0 ± 0.0 | N267 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| F96 | 0.0 ± 0.0 | 4.2 ± 2.8 | N268 | 0.0 ± 1.8 | 0.0 ± 0.0 | |||
| N98 | 1.3 ± 1.6 | 3.7 ± 1.4 | C269 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| T99 | 0.2 ± 2.2 | 0.0 ± 0.0 | S270 | 0.1 ± 2.8 | 0.0 ± 0.0 | |||
| M100 | 0.5 ± 1.7 | 0.0 ± 0.0 | S271 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| C101 | NA | NA | S272 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| Q102 | 0.0 ± 0.0 | 0.0 ± 0.0 | N273 | 0.0 ± 0.0 | 0.0 ± 0.0 | |||
| R274A/D276A | 2.2 ± 1.6 | 6.3 ± 1.0 | ||||||
| Q277 | 0.6 ± 2.1 | 0.0 ± 0.0 | ||||||
Entry of HIV-1JR-FL Env-pseudotyped reporter viruses into U87-CD4 cells transiently expressing CCR5 alanine mutants was measured in the presence of 100 nM AD101 or SCH-C. Relative HIV-1 entry in the presence of inhibitors was calculated as (RLU with inhibitor/RLU without inhibitor) × 100%. All values are means ± SD of three independent experiments. NA, not applicable; alanine substitutions of residues Y15 in the Nt, C101 in ECL1, and S169 in ECL2 were excluded from the study because they are substantially defective at mediating HIV-1 entry (<1% of wild-type CCR5 coreceptor function) (12, 13).
Effect of AD101 and SCH-C on HIV-1 entry mediated by TM CCR5 mutants.
We also evaluated a panel of alanine substitutions in the CCR5 TM domain for their effects on the inhibitory activities of AD101 and SCH-C (12). Our initial mutagenesis effort was guided by a molecular model of the CCR5 TM domain based on the corresponding region of rhodopsin (12). The CCR5 structure predicted by this model suggested that a cavity formed by the TM helices might be a binding site for small-molecule antagonists of CCR5 function. We therefore studied residues with side chains that protrude into this cavity. Residues found to be important for the inhibitory activity of both AD101 and SCH-C include L33 and Y37 in TM1, D76 and W86 in TM2, F113 in TM3, I198 in TM5, and E283 in TM7 (Table 2). Residues F79 in TM2 and Y108 in TM3 were essential for AD101 but not SCH-C activity (Table 2). All of these mutants exhibited between 50 and 150% of wild-type CCR5 activity, except for D76A and F113A, which exhibited approximately 5% of wild-type CCR5 activity. In all cases, coreceptor activity correlated with expression levels of the mutants (data not shown).
TABLE 2.
Effects of AD101 and SCH-C on HIV-1 entry mediated by CCR5 TM mutantsa
| TM | Mutant | Relative % entry in presence ofb:
|
|
|---|---|---|---|
| AD101 | SCH-C | ||
| TM1 | L33 | 34.2 ± 2.2 | 19.9 ± 2.7 |
| Y37 | 71.5 ± 2.2 | 32.1 ± 5.5 | |
| F41 | 2.6 ± 1.9 | 4.3 ± 0.8 | |
| N48 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| L55 | 1.8 ± 2.7 | 0.0 ± 0.0 | |
| I56 | 0.7 ± 3.0 | 0.0 ± 0.0 | |
| TM2 | Y68 | NA | NA |
| L69 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| N71 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| D76 | 44.7 ± 5.1 | 20.6 ± 3.3 | |
| F79 | 30.1 ± 6.2 | 0.0 ± 0.0 | |
| T82 | 0.8 ± 2.0 | 0.0 ± 0.0 | |
| F85 | NA | NA | |
| W86 | 93.8 ± 12.4 | 62.0 ± 0.8 | |
| TM3 | L104 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| T105 | 2.3 ± 1.3 | 6.6 ± 1.0 | |
| Y108 | 42.6 ± 1.0 | 9.6 ± 0.2 | |
| F112 | 0.9 ± 3.0 | 9.1 ± 0.5 | |
| F113 | 58.2 ± 8.7 | 18.1 ± 2.3 | |
| F117 | 11.8 ± 2.0 | 5.4 ± 0.7 | |
| F118 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| L121 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| L122 | 0.3 ± 0.6 | 0.0 ± 0.0 | |
| T123 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| TM4 | F144 | 1.5 ± 2.6 | 0.0 ± 0.0 |
| TM5 | T195 | 0.5 ± 1.4 | 0.0 ± 0.0 |
| I198 | 79.6 ± 6.7 | 72.2 ± 3.3 | |
| TM6 | Y251 | NA | NA |
| N252 | 6.1 ± 1.4 | 11.1 ± 2.0 | |
| L255 | 3.7 ± 1.5 | 2.8 ± 1.1 | |
| N258 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| T259 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| TM7 | M279 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| E283 | 118.7 ± 18.4 | 94.6 ± 13.3 | |
| H289 | 0.5 ± 2.2 | 0.0 ± 0.0 | |
| N293 | NA | NA | |
| Y297 | 2.9 ± 2.8 | 0.0 ± 0.0 | |
Entry of HIV-1JR-FL Env-pseudotyped reporter viruses into U87-CD4 cells transiently expressing CCR5 alanine mutants was measured in the presence of 100 nM AD101 or SCH-C. Relative HIV-1 entry in the presence of inhibitors was calculated as (RLU with inhibitor/RLU without inhibitor) × 100%. All values are means ± SD of three independent experiments. NA = not applicable; alanine substitutions of residues Y68 and T82 in TM2, Y251 in TM6, and N293 in TM7 were excluded from the study because they are substantially defective at mediating HIV-1 entry (<1% of wild-type CCR5 coreceptor function) (13).
Values shown in bold are significantly higher than the cutoff level of inhibition (16%).
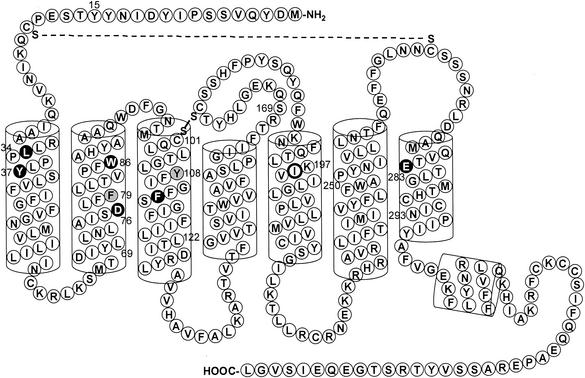
A new CCR5 TM domain model developed by Seibert et al. (unpublished data), based on the crystal structure of rhodopsin (25), predicts that residues L33, Y37, D76, W86, F113, and E283 are common to the binding sites of AD101 and SCH-C, whereas residues F79 and Y108 are only involved in AD101 binding (Fig. 3). The I198 residue, found to affect the activity of AD101 and SCH-C, is probably involved in stabilizing the conformation of the ligand-binding site (Fig. 3). Analyses of novel alanine and nonalanine substitutions based on the new CCR5 model have also been described by Seibert et al. and further refine our understanding of the ligand-binding cavity formed by the TM helices of CCR5 (Seibert et al., unpublished observations).
FIG. 3.
Residues important for AD101 and SCH-C activity. A two-dimensional model of CCR5 depicts the extracellular loops, the seven TM helices, and the intracellular loops of the coreceptor. Residues important for binding of AD101 and SCH-C are in black; residues important only for AD101 binding are in gray. Residues thought to exert indirect effects on the activities of the inhibitors are circled in bold.
DISCUSSION
In this study we have analyzed the mechanisms of action of two structurally related small-molecule inhibitors of CCR5 function, AD101 and SCH-C. These compounds were initially characterized by their ability to inhibit RANTES binding and signaling, and then they were shown to be potent and specific inhibitors of CCR5-mediated HIV-1 entry into target cells (33, 36). Both AD101 and SCH-C efficiently block the replication of multiple primary and molecularly cloned isolates in PBMC (33, 36). Moreover, SCH-C has now shown antiviral efficacy in phase I clinical trials (Reynes et al., 9th Conf. Retrovir. Opportun. Infect.). Small-molecule CCR5 antagonists are therefore credible candidates for effective antiviral drugs, so it is necessary that we understand as much as possible about their interactions with CCR5.
AD101 and SCH-C inhibit HIV-1 entry into target cells by directly blocking the interaction between gp120 and CCR5. This is how TAK-779, another small-molecule CCR5 inhibitor, and anti-CCR5 MAbs are also thought to act (12, 22). Unlike TAK-779, AD101 and SCH-C also inhibit the binding of several anti-CCR5 MAbs (12). The specific MAbs that are blocked by AD101 or SCH-C all recognize epitopes in ECL2, with the exception of the PA14 MAb, for which the epitope straddles the Nt and ECL2 domains (16, 22). In contrast, MAbs that continue to bind to CCR5 in the presence of AD101 or SCH-C recognize epitopes in the Nt, with the exception of the 2D7 MAb, which has an epitope in ECL2 (16, 22). All the anti-CCR5 MAbs described to date recognize elements of the Nt and/or ECL2, suggesting that these are the two most exposed or immunogenic regions of the extracellular domain of CCR5. Furthermore, both the Nt and ECL2 have been shown to be crucial for gp120-CCR5 interactions and viral entry (reviewed in reference 10). MAbs directed against either one of these regions inhibit gp120-CCR5 binding and viral entry.
Mapping studies with a panel of CCR5 alanine mutants, coupled with molecular modeling, show that the activities of both AD101 and SCH-C are dependent on residues in TM helices 1, 2, 3, 5, and 7. Some of these residues are involved in direct interactions with the inhibitors, whereas others probably influence inhibitor activity indirectly by affecting the conformation and stability of the ligand-binding pocket (Fig. 3). Additional residues are involved in AD101 binding to CCR5 that do not appear to influence SCH-C binding. Other TM domain residues that we have not yet studied by mutagenesis may also exclusively interact with one or the other of the compounds. Some, but not all, of the residues found to be important for AD101 and SCH-C function were also previously shown to be important for the inhibitory activity of TAK-779 (12). In particular, residues R31 (TM1) and T82 (TM2) affect TAK-779 activity, whereas D76, F79 (TM2), and F113 (TM3) do not. TAK-779 has significant antagonist activity against CCR2 as well as CCR5, whereas AD101 and SCH-C are pure CCR5 antagonists. The subtle differences between the binding sites of TAK-779, AD101, and SCH-C may account for the greater specificity of the latter compounds for CCR5. They may also explain why TAK-779 does not inhibit MAb binding to the CCR5 surface, whereas AD101 and SCH-C do.
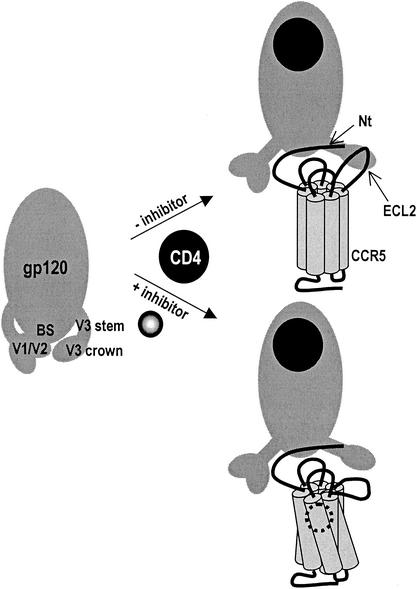
This study and general knowledge about gp120-CCR5 interactions enabled us to propose a model explaining the inhibitory effect on viral entry of small molecules that bind to the putative ligand-binding pocket of CCR5. Docking of gp120 to CCR5 involves interactions between conserved residues in the V3 stem-C4 region and the coreceptor Nt, as well as binding of the V3 crown to a second region of CCR5, which a number of studies suggest to be primarily ECL2 (reviewed in references 5 and 10). It is also possible that the V3 crown interacts with TM domain residues, although none of the TM domain mutants that we have studied were significantly impaired for viral entry. We previously showed that TAK-779 does not disrupt the gp120-Nt interaction (12). Taking all the data together, we propose that the binding of small-molecule antagonists to the TM domain of CCR5 induces a conformational change in the region of CCR5 that interacts with the V3 crown, which we suggest is likely to be ECL2 (Fig. 4). Disruption of the binding site on CCR5 for the V3 crown would inhibit the gp120-CCR5 interaction and viral entry. In general, this may be the most effective way to inhibit HIV-1 entry, since MAbs that recognize ECL2 are 10-fold more efficient inhibitors of viral entry than MAbs that recognize epitopes in the Nt (17, 22). If the gp120-CCR5 interaction occurs in two steps, wherein V3 crown binding to ECL2 (or another region of CCR5) is followed by the binding of the V3 stem-C4 domain to the Nt, then inhibitors of the latter interaction might be less potent simply because gp120 and CCR5 are already in contact by this time. Clearly, however, other explanations for the actions of CCR5 inhibitors can be envisioned, so additional studies will be required to refine our understanding of how these compounds prevent HIV-1 entry.
FIG. 4.
Model of the mechanism of action of small-molecule inhibitors of CCR5 coreceptor function. gp120 is initially in a closed state, wherein the V1/V2 and V3 loops conceal the coreceptor binding site. Upon CD4 binding to gp120, conformational changes create and/or expose the coreceptor binding site. In the absence of inhibitor, the CCR5 Nt interacts with residues in the bridging sheet (BS) and the V3 stem, whereas ECL2 interacts with the V3 crown. In the presence of inhibitor, the conformation of ECL2 is modified such that it can no longer interact with the V3 crown, thus inhibiting viral entry.
Our study suggests that the binding of antagonists such as AD101 and SCH-C profoundly alters the structure and function of CCR5, both as a chemokine receptor and as an HIV-1 coreceptor. The creation of new generations of CCR5 antagonists with more potency, greater specificity, and improved pharmacological properties will no doubt be accomplished by medicinal chemists over the coming years for applications both within and perhaps also outside the HIV-1 therapeutic arena. As more and more information on the binding sites for these antagonists within the CCR5 TM domain becomes available from studies such as the present one and our previous work on TAK-779, some aspects of the design of these new compounds might be facilitated. Perhaps the next challenge for developers of CCR5-binding compounds with anti-HIV-1 activity will be to produce “discriminatory” molecules that only disrupt gp120 binding and viral entry without affecting chemokine binding and signaling. For this to be achieved by a rational approach, much more knowledge of the precise conformational changes induced by these small molecules in the CCR5 extracellular domains will be required. This is likely to involve a combination of further molecular modeling and biochemical studies of the receptor and its various ligands.
Acknowledgments
We thank William Olson of Progenics Pharmaceuticals for his generous gift of key reagents.
This work was funded by the National Institutes of Health grants R01 AI43847 to T.D., R01 AI41420 to J.P.M., and R01 DK54718 to T.P.S. and by the Schering Plough Research Institute. The Department of Microbiology and Immunology at the Weill Medical College gratefully acknowledges the support of the William Randolph Hearst Foundation. J.P.M. is a Stavros S. Niarchos Scholar.
REFERENCES
- 1.Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 96:5698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 3.Cocchi, F., A. L. DeVico, A. Garzino-Demo, S. K. Arya, R. C. Gallo, and P. Lusso. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811-1815. [DOI] [PubMed] [Google Scholar]
- 4.Cormier, E. G., D. N. Tran, L. Yukhayeva, W. C. Olson, and T. Dragic. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75:5541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dealwis, C., E. J. Fernandez, D. A. Thompson, R. J. Simon, M. A. Siani, and E. Lolis. 1998. Crystal structure of chemically synthesized [N33A] stromal cell-derived factor 1α, a potent ligand for the HIV-1 “fusin” coreceptor. Proc. Natl. Acad. Sci. USA 95:6941-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, and S. J. O'Brien. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273:1856-1862. (Erratum, 274:1069.) [DOI] [PubMed] [Google Scholar]
- 7.De Clercq, E. 2001. New developments in anti-HIV chemotherapy. Curr. Med. Chem. 8:1543-1572. [DOI] [PubMed] [Google Scholar]
- 8.Doms, R. W. 2001. Chemokine receptors and HIV entry. AIDS 15(Suppl. 1):S34-S35. [DOI] [PubMed] [Google Scholar]
- 9.Donzella, G. A., D. Schols, S. W. Lin, J. A. Este, K. A. Nagashima, P. J. Maddon, G. P. Allaway, T. P. Sakmar, G. Henson, E. De Clercq, and J. P. Moore. 1998. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 4:72-77. [DOI] [PubMed] [Google Scholar]
- 10.Dragic, T. 2001. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J. Gen. Virol. 82:1807-1814. [DOI] [PubMed] [Google Scholar]
- 11.Dragic, T., A. Trkola, S. W. Lin, K. A. Nagashima, F. Kajumo, L. Zhao, W. C. Olson, L. Wu, C. R. Mackay, G. P. Allaway, T. P. Sakmar, J. P. Moore, and P. J. Maddon. 1998. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J. Virol. 72:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dragic, T., A. Trkola, D. A. Thompson, E. G. Cormier, F. A. Kajumo, E. Maxwell, S. W. Lin, W. Ying, S. O. Smith, T. P. Sakmar, and J. P. Moore. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. USA 97:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genoud, S., F. Kajumo, Y. Guo, D. Thompson, and T. Dragic. 1999. CCR5-mediated human immunodeficiency virus entry depends on an amino-terminal gp120-binding site and on the conformational integrity of all four extracellular domains. J. Virol. 73:1645-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez, E., R. Dhanda, M. Bamshad, S. Mummidi, R. Geevarghese, G. Catano, S. A. Anderson, E. A. Walter, K. T. Stephan, M. F. Hammer, A. Mangano, L. Sen, R. A. Clark, S. S. Ahuja, M. J. Dolan, and S. K. Ahuja. 2001. Global survey of genetic variation in CCR5, RANTES, and MIP-1α: impact on the epidemiology of the HIV-1 pandemic. Proc. Natl. Acad. Sci. USA 98:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrix, C. W., C. Flexner, R. T. MacFarland, C. Giandomenico, E. J. Fuchs, E. Redpath, G. Bridger, and G. W. Henson. 2000. Pharmacokinetics and safety of AMD-3100, a novel antagonist of the CXCR-4 chemokine receptor, in human volunteers. Antimicrob. Agents Chemother. 44:1667-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill, C. M., D. Kwon, M. Jones, C. B. Davis, S. Marmon, B. L. Daugherty, J. A. DeMartino, M. S. Springer, D. Unutmaz, and D. R. Littman. 1998. The amino terminus of human CCR5 is required for its function as a receptor for diverse human and simian immunodeficiency virus envelope glycoproteins. Virology 248:357-371. [DOI] [PubMed] [Google Scholar]
- 17.Lee, B., M. Sharron, C. Blanpain, B. J. Doranz, J. Vakili, P. Setoh, E. Berg, G. Liu, H. R. Guy, S. R. Durell, M. Parmentier, C. N. Chang, K. Price, M. Tsang, and R. W. Doms. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274:9617-9626. [DOI] [PubMed] [Google Scholar]
- 18.Liu, R., W. Paxton, S. Choe, D. Ceradini, S. Martin, R. Horuk, M. MacDonald, H. Stuhlmann, R. Koup, and N. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-378. [DOI] [PubMed] [Google Scholar]
- 19.Loetscher, P., J. H. Gong, B. Dewald, M. Baggiolini, and I. Clark-Lewis. 1998. N-terminal peptides of stromal cell-derived factor-1 with CXC chemokine receptor 4 agonist and antagonist activities. J. Biol. Chem. 273:22279-22283. [DOI] [PubMed] [Google Scholar]
- 20.Moore, J. P., and M. Stevenson. 2000. New targets for inhibitors of HIV-1 replication. Nat. Rev. Mol. Cell. Biol. 1:40-49. [DOI] [PubMed] [Google Scholar]
- 21.O'Brien, S. J., and J. P. Moore. 2000. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol. Rev. 177:99-111. [DOI] [PubMed] [Google Scholar]
- 22.Olson, W. C., G. E. Rabut, K. A. Nagashima, D. N. Tran, D. J. Anselma, S. P. Monard, J. P. Segal, D. A. Thompson, F. Kajumo, Y. Guo, J. P. Moore, P. J. Maddon, and T. Dragic. 1999. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J. Virol. 73:4145-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palani, A., S. Shapiro, J. W. Clader, W. J. Greenlee, K. Cox, J. Strizki, M. Endres, and B. M. Baroudy. 2001. Discovery of 4-[(Z)-(4-bromophenyl)-(ethoxyimino)methyl]-1′-[(2,4-dimethyl-3-pyridinyl)carbonyl]-4′-methyl-1,4′-bi-piperidine N-oxide (SCH 351125): an orally bioavailable human CCR5 antagonist for the treatment of HIV infection. J. Med. Chem. 44:3339-3342. [DOI] [PubMed] [Google Scholar]
- 24.Palani, A., S. Shapiro, H. Josien, T. Bara, J. W. Clader, W. J. Greenlee, K. Cox, J. M. Strizki, and B. M. Baroudy. 2002. Synthesis, SAR, and biological evaluation of oximino-piperidino-piperidine amides. 1. Orally bioavailable CCR5 receptor antagonists with potent anti-HIV activity. J. Med. Chem. 45:3143-3160. [DOI] [PubMed] [Google Scholar]
- 25.Palczewski, K., T. Kumasaka, T. Hori, C. A. Behnke, H. Motoshima, B. A. Fox, I. Le Trong, D. C. Teller, T. Okada, R. E. Stenkamp, M. Yamamoto, and M. Miyano. 2000. Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289:739-745. [DOI] [PubMed] [Google Scholar]
- 26.Paxton, W. A., R. Liu, S. Kang, L. Wu, T. R. Gingeras, N. R. Landau, C. R. Mackay, and R. A. Koup. 1998. Reduced HIV-1 infectability of CD4+ lymphocytes from exposed-uninfected individuals: association with low expression of CCR5 and high production of beta-chemokines. Virology 244:66-73. [DOI] [PubMed] [Google Scholar]
- 27.Rabut, G. E., J. A. Konner, F. Kajumo, J. P. Moore, and T. Dragic. 1998. Alanine substitutions of polar and nonpolar residues in the amino-terminal domain of CCR5 differently impair entry of macrophage- and dual-tropic isolates of human immunodeficiency virus type 1. J. Virol. 72:3464-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 29.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 30.Schols, D., S. Struyf, J. Van Damme, J. A. Este, G. Henson, and E. De Clercq. 1997. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J. Exp. Med. 186:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz, M. K., and T. N. Wells. 2002. New therapeutics that modulate chemokine networks. Nat. Rev. Drug Disc. 1:347-358. [DOI] [PubMed] [Google Scholar]
- 32.Simmons, G., P. R. Clapham, L. Picard, R. E. Offord, M. M. Rosenkilde, T. W. Schwartz, R. Buser, T. N. C. Wells, and A. E. Proudfoot. 1997. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science 276:276-279. [DOI] [PubMed] [Google Scholar]
- 33.Strizki, J. M., S. Xu, N. E. Wagner, L. Wojcik, J. Liu, Y. Hou, M. Endres, A. Palani, S. Shapiro, J. W. Clader, W. J. Greenlee, J. R. Tagat, S. McCombie, K. Cox, A. B. Fawzi, C. C. Chou, C. Pugliese-Sivo, L. Davies, M. E. Moreno, D. D. Ho, A. Trkola, C. A. Stoddart, J. P. Moore, G. R. Reyes, and B. M. Baroudy. 2001. SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. USA 98:12718-12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tagat, J. R., S. W. McCombie, R. W. Steensma, S. Lin, D. V. Nazareno, B. Baroudy, N. Vantuno, S. Xu, and J. Liu. 2001. Piperazine-based CCR5 antagonists as HIV-1 inhibitors. I: 2(S)-methyl piperazine as a key pharmacophore element. Bioorg. Med. Chem. Lett. 11:2143-2146. [DOI] [PubMed] [Google Scholar]
- 35.Tagat, J. R., R. W. Steensma, S. W. McCombie, D. V. Nazareno, S. I. Lin, B. R. Neustadt, K. Cox, S. Xu, L. Wojcik, M. G. Murray, N. Vantuno, B. M. Baroudy, and J. M. Strizki. 2001. Piperazine-based CCR5 antagonists as HIV-1 inhibitors. II. Discovery of 1-[(2,4-dimethyl-3-pyridinyl)carbonyl]-4-methyl-4-[3(S)-methyl-4-[1(S)-[4-(trifluoromethyl)phenyl]ethyl]-1-piperazinyl]-piperidine N1-oxide (Sch-350634), an orally bioavailable, potent CCR5 antagonist. J. Med. Chem. 44:3343-3346. [DOI] [PubMed] [Google Scholar]
- 36.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trkola, A., W. A. Paxton, S. P. Monard, J. A. Hoxie, M. A. Siani, D. A. Thompson, L. Wu, C. R. Mackay, R. Horuk, and J. P. Moore. 1998. Genetic subtype-independent inhibition of human immunodeficiency virus type 1 replication by CC and CXC chemokines. J. Virol. 72:396-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, L., G. LaRosa, N. Kassam, C. J. Gordon, H. Heath, N. Ruffing, H. Chen, J. Humblias, M. Samson, M. Parmentier, J. P. Moore, and C. R. Mackay. 1997. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, L., W. Paxton, N. Kassam, N. Ruffing, J. Rottman, N. Sullivan, H. Choe, J. Sodroski, W. Newman, R. Koup, and C. Mackay. 1997. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1 in vitro. J. Exp. Med. 185:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, O. O., S. L. Swanberg, Z. Lu, M. Dziejman, J. McCoy, A. D. Luster, B. D. Walker, and S. H. Herrmann. 1999. Enhanced inhibition of human immunodeficiency virus type 1 by Met-stromal-derived factor 1β correlates with down-modulation of CXCR4. J. Virol. 73:4582-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ylisastigui, L., J. Vizzavona, E. Drakopoulou, P. Paindavoine, C. F. Calvo, M. Parmentier, J. C. Gluckman, C. Vita, and A. Benjouad. 1998. Synthetic full-length and truncated RANTES inhibit HIV-1 infection of primary macrophages. AIDS 12:977-984. [PubMed] [Google Scholar]
- 42.Zhang, Y. J., and J. P. Moore. 1999. Will multiple coreceptors need to be targeted by inhibitors of human immunodeficiency virus type 1 entry? J. Virol. 73:3443-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]