Abstract
We previously identified retroperitoneal fibromatosis-associated herpesvirus (RFHV) as a simian homolog of Kaposi's sarcoma-associated herpesvirus (KSHV) in a fibroproliferative malignancy of macaques that has similarities to Kaposi's sarcoma. In this report, we cloned 4.3 kb of divergent locus B (DL-B) flanking the DNA polymerase gene from two variants of RFHV from different species of macaque with a consensus degenerate hybrid oligonucleotide primer approach. Within the DL-B region of RFHV, viral homologs of the cellular interleukin-6, dihydrofolate reductase, and thymidylate synthase genes were identified, along with a homolog of the gammaherpesvirus open reading frame (ORF) 10. In addition, a homolog of the KSHV ORF K3, the modulator of immune recognition-1, was identified. Our data show a close similarity in sequence conservation, gene content, and genomic structure between RFHV and KSHV which strongly supports the grouping of these viral species within the same RV-1 rhadinovirus lineage and the hypothesis that RFHV is the macaque homolog of KSHV.
The most recently discovered human tumor virus, Kaposi's sarcoma-associated herpesvirus (KSHV), has been implicated in the pathogenesis of Kaposi's sarcoma, primary effusion lymphoma, and Castleman's disease (for a review, see reference 41). KSHV was assigned to the Rhadinovirus genus of the gammaherpesviruses based on similarities at the levels of nucleotide sequence, gene content, and genomic structure with the Rhadinovirus prototype, Herpesvirus saimiri (HVS) of the South American squirrel monkey (30). Like other gammaherpesviruses, KSHV has numerous genes with homology to cellular host genes which have been captured during virus evolution (37). Viral homologs of cellular genes contribute to the unique biological properties and pathogenic effects of different viral species and, in the case of KSHV, function in the disruption of antiviral responses, cytokine-regulated cell growth, apoptosis, and cell cycle control (29). As in other gammaherpesviruses, these cellular homologs cluster in a restricted number of divergent genomic loci, and the presence and orientation of these genes are uniquely characteristic of KSHV.
We have identified a simian homolog of KSHV in macaque retroperitoneal fibromatosis (36), a vascular fibroproliferative malignancy with morphological and histological similarities to Kaposi's sarcoma (12, 45). A consensus degenerate hybrid oligonucleotide primer (CODEHOP) technique (35) utilizing PCR primers derived from highly conserved amino acid motifs within the herpesvirus DNA polymerase genes was used to amplify DNA sequences from retroperitoneal fibromatosis lesions of two macaque species. These sequences were identified as portions of the DNA polymerase genes of two closely related novel viruses, retroperitoneal fibromatosis-associated herpesvirus of Macaca mulatta (RFHVMm) and of Macaca nemestrina (RFHVMn) (36). Subsequently, it was shown that these macaque species are also host to another closely related herpesvirus with similarity to KSHV, rhesus rhadinovirus (RRV) (11) and Macaca nemestrina rhadinovirus 2 (RV-2) (40). This virus has been alternatively named pig-tail rhadinovirus (26) and pig-tailed monkey rhadinovirus (4).
Phylogenetic analysis of the DNA polymerase sequences demonstrated that RFHVMm and RFHVMn clustered together with KSHV within a lineage designated rhadinovirus 1 (RV-1), while RRV and M. nemestrina RV-2 cluster into a more distantly related lineage designated rhadinovirus 2 (RV-2) (40). Additional studies have identified viral species belonging to both the RV-1 and RV-2 lineages of KSHV-related viruses in a number of Old World primate host species, including African green monkeys, drills, mandrills, gorillas, and chimpanzees (13, 14, 20-22).
Classification of viral species within the RV-1 and RV-2 rhadinovirus lineages has been based mainly on comparisons of partial sequences of DNA polymerase genes. In this report, we cloned 4.3 kb of divergent locus B (DL-B) flanking the DNA polymerase gene of RFHVMm and RFHVMn with the CODEHOP strategy. As in the corresponding DL-B locus in KSHV, viral homologs of the cellular interleukin-6 (vIL-6), dihydrofolate reductase (vDHFR), and thymidylate synthase (vTS) genes were identified, along with a homolog of the conserved gammaherpesvirus open reading frame (ORF) 10. In addition, a homolog of the KSHV ORF K3, the modulator of immune recognition-1 (MIR-1) (10), was identified. Our data show a close similarity between the nucleotide sequences, gene content, and genomic structure of RFHV and KSHV, supporting the hypothesis that these viral species belong to the same RV-1 viral lineage and that RFHV is the macaque homolog of KSHV. Genetic differences were identified between the RV-1 and RV-2 macaque viruses, suggesting important biological differences between the two rhadinovirus lineages.
MATERIALS AND METHODS
Tissue samples.
Retroperitoneal fibromatosis tumor samples were obtained from a simian retrovirus 2-infected Maccaca mulatta (MmuYN-91) provided by Harold McClure, Yerkes National Primate Research Center, Atlanta, Ga. Retroperitoneal fibromatosis tumor and spleen samples from a simian retrovirus 2-infected M. nemestrina (Mne442N) diagnosed with retroperitoneal fibromatosis were obtained from Riri Shibata, National Institutes of Health, Bethesda, Md.
DNA isolation.
Frozen tissue samples were quickly thawed in the presence of a standard proteinase K extraction buffer containing 0.1% sodium dodecyl sulfate by homogenization in a disposable homogenizer. Samples were digested at 55°C for several hours, and the DNA was purified by standard phenol-chloroform extraction and ethanol precipitation.
Partial inverse PCR.
In order to clone the region flanking the DNA polymerase gene of RFHVMm, 5 μg of high-molecular-weight DNA extracted from a retroperitoneal fibromatosis tumor sample obtained from MmuYN-91 was partially digested with MboI (New England Biolabs) at enzyme dilutions ranging from 2 U/μg of DNA to 0.031 U/μg of DNA. Appropriately sized fragments were circularized with 4 U of T4 DNA ligase/μl (Promega) for 6 h at 22°C, essentially as described (33). PCR amplification was then performed with the DRIPA (5′-TTCACGACAGGATACCCTACG-3′) sense and QIRQB (5′-CAGCTCCTCTTGTCTGATTTG-3′) antisense primers derived from the 5′ end of the RFHVMm DNA polymerase sequence (Fig. 1), and the resulting PCR product was isolated with a QIAEX gel extraction kit (Qiagen), cloned (Perfectly blunt cloning kit; Novagen), and sequenced.
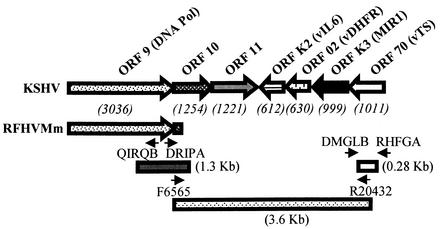
FIG. 1.
PCR strategy for cloning a region of divergent locus B (DL-B) of RFHVMm. The order and orientation of the genes within the DL-B region of KSHV flanking the DNA polymerase genes are shown (the size of each ORF is indicated in base pairs). The positions of the DNA polymerase-specific PCR primers QIRQB and DRIPA and the PCR product obtained by partial inverse amplification of the ORF 10 flanking region of RFHVMm are indicated. The positions of the DMGLB and RHFGA primer pools used for CODEHOP amplification of a region of the RFHVMm TS gene are also shown. Finally, the 3.6-kb PCR fragment of RFHVMm obtained by long-range PCR amplification between the ORF 10-specific primer F6565 and the TS-specific primer R20432 is indicated.
Thermal gradient CODEHOP PCR amplification of thymidylate synthase homolog.
CODEHOPs were designed with an alignment of the vTS genes from KSHV, herpesvirus saimiri (HVS), and equine herpesvirus 2 (EHV2) (see Table 1 for accession numbers) with the method previously described (35). Two conserved amino acid sequence motifs, RHFG and DMGL, were chosen as targets. The CODEHOPs RHFGA (5′-CCTGTTTACGGTTTCCARTGGAGRCAYTTYGG-3′) and DMGLB (5′-GGCAATGTTAAAAGGAACTCCNARNCCCATRTC-3′) were derived from these motifs and designated A (sense orientation) or B (antisense orientation), where Y = C or T; R = A or G; and N = A, C, G, or T. Approximately 1 μg of DNA extracted from the retroperitoneal fibromatosis tumors of MmuYN-91 and Mne442N was used as the template in a PCR in 0.067 M Tris-HCl (pH 8.8)-4 mM MgCl2-0.016 M (NH4)2SO4-0.01 M β-mercaptoethanol-100 μg of bovine serum albumin per ml (24)-100 nM deoxynucleoside triphosphates-1 μM each oligonucleotide-2.5 U of Platinum Taq polymerase (Life Technologies) in a 50-μl reaction volume. Amplification was preceded by a 1-min incubation at 95°C to activate the polymerase. Cycling conditions consisted of a 30-s 94°C denaturing step, a 30-s 55 to 70°C annealing step, and a 30-s 72°C extension step for 40 cycles in a thermal gradient iCycler thermocycler (Bio-Rad).
TABLE 1.
Herpesviruses and sequence accession numbers
| Virus | Host | Gene(s)a | Accession no. |
|---|---|---|---|
| Rhadinoviruses | |||
| RFHV Mn | Pig-tailed macaque | DNA polymerase and partial flanks | AF204166 |
| RFHV Mm | Rhesus macaque | DNA polymerase and partial flanks | AF005479 |
| RRV | Rhesus macaque | CG | AF210726 |
| KSHV | Human | CG | NC_003409 |
| HVS | South American squirrel monkey | CG | NC_001350 |
| AtHV3 | South American spider monkey | CG | NC_001987 |
| AHV1 | Wildebeest | CG | NC_002531 |
| BHV4 | Cow | CG | NC_002665 |
| EHV2 | Horse | CG | NC_001650 |
| MHV68 | Mouse | CG | NC_001826 |
| Lymphocrypto- virus | |||
| EBV | Human | CG | NC_001345 |
CG, complete genome (long unique region).
Long-range PCR amplification.
High-molecular-weight DNA isolated from retroperitoneal fibromatosis tissue of MmuYN-91 and spleen tissue from Mne442N was used as the template in long-range PCR amplification of the region between the ORF 10 and TS genes of RFHVMm and between the DNA polymerase and TS genes of RFHVMn. The gene-specific PCR primers F6565 (5′-TGAACTATTTTGCTACCCCAACACCGCTAT-3′; ORF 10, RFHVMm), R20432 (5′-CTTTGCCCCCCTGTCACGTTCTCTGTCAGT-3′; vTS, RFHVMm), PolF1LR 5′-CCACCGTCCCAGACCAACGAAAGCGCCAGA-3′; DNA polymerase, RFHVMn), and TSR1LR (5′-GTCTGCCTGGAATCCCGTGGATATACCAAA-3′; vTS, RFHVMn) were designed with Oligonucleotide v5 software (NBI) with a standard length of 30 nucleotides to ensure high specificity and annealing temperature (see Fig. 1). Long-range PCR amplification was performed with the Expand Long Template PCR system (Roche) with the recommended protocol. PCR products were analyzed on a 1% agarose gel and purified with a QIAquick PCR purification kit (Qiagen).
DNA sequencing and sequence assembly.
PCR products were either cloned and sequenced with vector-specific primers or sequenced directly with CODEHOP or gene-specific primers. Additional internal gene-specific primers were used to obtain overlapping sequences within the larger PCR products. Multiple PCR products and clones were sequenced in both orientations to avoid artifacts and Taq polymerase errors. Sequencing was performed on an ABI model 310 automated sequencer with Prism Big Dye terminator cycle sequencing ready reaction kit with Amplitaq DNA polymerase FS (Applied Biosystems). DNA sequences were assembled with Sequencher 4.0.5b10 (GeneCodes). Sequence redundancy was three- to fourfold.
Sequence and phylogenetic analysis.
ORFs were identified in Sequencher and compared to existing sequences with Blast analysis of the NCBI nonredundant database. RFHV ORFs were named according to the nomenclature proposed for KSHV (37), with ORFs not found in HVS designated with an RF preface, for example, RF3. Pairwise nucleotide and encoded amino acid alignments were performed with GenePro software (Riverside Scientific, Bainbridge Island, Wash.). Multiple sequence alignment was done with ClustalW (EMBL, Heidelberg, Germany). Phylogenetic analysis of amino acid sequences was done with parsimony, neighbor-joining analysis, and protein maximum likelihood (ProML) from the Phylip package version 3.6 of phylogenetic analysis programs (University of Washington, Seattle). Bootstrap analysis was performed with the programs Seqboot and Consense from the Phylip package. The Consense treefile was displayed with TreeView (32). Transmembrane helix predictions were performed with TMHMM version 2.0 at the Centralbureau voor Schimmelcultures (http://www.cbs.dtu.dk/services/TMHMM/) (19) and TMpred at EMBNET http://www.ch.embnet.org/software/TMPRED_form.html). Table 1 indicates the herpesvirus sequences used in the various analyses and their NCBI accession numbers.
RESULTS
Partial inverse PCR amplification of ORF 10 sequences from RFHVMm.
We have previously used the CODEHOP technique, described in reference 35, to clone the entire DNA polymerase genes and flanking sequences of the macaque rhadinoviruses RFHVMn and RFHVMm (40). At present, this is the only information available regarding the genetic make-up of these viruses. We have had to rely on PCR amplification for the characterization of these viruses because of the minimal copy number of the viral genomes within the available tissue samples and the inability to culture the viruses in vitro.
In an attempt to extend the sequences of these viral genomes into the region flanking the DNA polymerase gene, we initially employed a partial inverse PCR technique for cloning flanking sequences. Specific oligonucleotide primers, DRIPA and QIRQB, were derived from opposite strands of the DNA sequence at the 3′ end of the RFHVMm DNA polymerase gene, as shown in Fig. 1. These primers were used to amplify circularized DNA templates obtained from partial restriction digests of DNA isolated from retroperitoneal fibromatosis tumor samples of MmuYN-91, essentially as described before (33). A 1.3-kb PCR fragment was obtained, cloned, and sequenced. Sequence analysis revealed that half of the fragment was identical to the previously determined DNA polymerase gene, and the other half contained new sequence derived from the downstream ORF 10 gene (see Fig. 1).
CODEHOP-based PCR amplification of sequences within the vTS genes of RFHVMm and RFHVMn.
Although the inverse PCR technique resulted in new sequences flanking our original sequence, the amount of sequence was limited. Therefore, we pursued long-range amplification to obtain larger DNA fragments. Previous comparison of the nucleotide sequences of the DNA polymerase genes of KSHV and the two RFHV species suggested that the genomes of the macaque viruses would be similar to the KSHV genome (36, 40). Flanking the KSHV DNA polymerase gene is divergent locus B (DL-B), which contains a number of viral homologs of cellular genes that have been captured during the evolution of the virus (Fig. 1) (31). Because the presence and orientation of the genes within this region are uniquely characteristic of KSHV, analysis of this region within the genomes of RFHVMn and RFHVMm would allow a more definitive characterization of these viruses to determine their relationship to KSHV. The vTS gene was identified as a suitably conserved candidate target for cloning with the CODEHOP technique. Successful amplification of a portion of the RFHV vTS gene would provide the sequence information to prepare a TS-specific primer allowing long-range amplification between the vTS gene and known DNA polymerase/ORF 10 sequences within the RFHV genomes (Fig. 1).
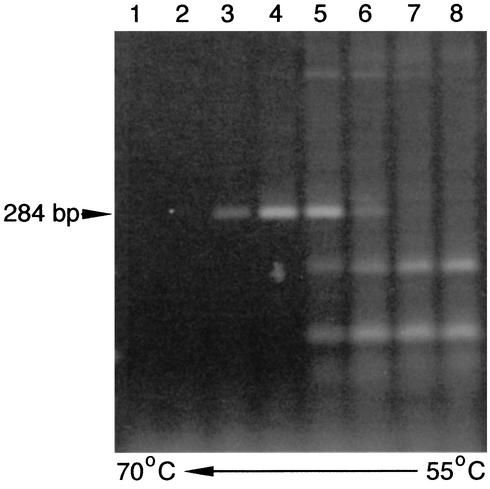
Alignment of the available herpesvirus vTS homologs revealed the presence of two conserved amino acid motifs, RHFG and DMGL, which were separated by approximately 200 bp (data not shown). These motifs were chosen as CODEHOP targets, and RHFGA (sense orientation) and DMGLB (antisense orientation) CODEHOPs were derived from these motifs. Thermal gradient PCR amplification was performed with these CODEHOP primer pools with DNA isolated from retroperitoneal fibromatosis tissues from MmuYN-91 and Mne442N. Thermal gradient amplification empirically determines the optimal annealing temperature of CODEHOPs for amplification of unknown DNA templates by allowing multiple amplification reactions to proceed at different annealing temperatures. The PCR products obtained from amplification at different temperatures were analyzed by gel electrophoresis. An annealing temperature of ≈64.3°C (Fig. 2, lane 4) yielded a single strong PCR product of the predicted size (284 bp) from both DNA templates. These DNA products were chosen for analysis and sequenced directly with both of the TS CODEHOPs used in the amplification. A comparison of the sequences revealed a close similarity with the sequence of the KSHV vTS homolog, indicating that these PCR products were derived from the vTS genes of the closely related RFHVMm and RFHVMn (see below).
FIG.2.
CODEHOP PCR amplification of a region of the RFHVMm vTS gene with a thermal gradient. PCR amplification of DNA from the retroperitoneal fibromatosis tumor from MmuYN91 was performed with CODEHOPs derived from two conserved TS motifs, RHFGA and DMGLB. A gradient of annealing temperatures ranging from 55 to 70oC was used. The expected 284-bp fragment is indicated. Lane temperatures: 1, 70°C; 2, 68.9°C; 3, 67.1°C; 4, 64.3°C; 5, 60.5°C; 6, 57.9°C; 7, 56.1°C; 8, 55.0°C.
Long-range PCR amplification of divergent locus B located between DNA polymerase and vTS genes of RFHVMm and RFHVMn.
To characterize the DL-B region of the RFHVMm genome, gene-specific primers were derived from the ORF 10 sequence obtained by partial inverse PCR (primer F6565) and the TS sequence obtained by CODEHOP PCR (primer R20432) (see Fig. 1). To characterize the RFHVMn genome, a DNA polymerase-specific primer was derived from our previous sequence of the DNA polymerase (PolF1LR), and a TS-specific primer (TSR1LR) was derived from the RFHVMn CODEHOP TS PCR product. Based on the relative positions of the DNA polymerase/ORF 10 and vTS genes within the KSHV genome, it was predicted that amplification of the RFHV region would yield a PCR product of approximately 5 kb (Fig. 1).
Long-range PCR amplification was performed on DNA from MmuYN-91 and Mne442N tissues containing RFHVMm and RFHVMn viral DNA, respectively. A single 3.6-kb PCR product was obtained from the MmuYN-91 retroperitoneal fibromatosis tissue with the ORF 10 and TS primers, while a 4.1-kb product was obtained from the Mne442N spleen tissue with the DNA polymerase and TS primers. The DNA sequences of these products were obtained and analyzed by Blast for similarities to known sequences. Five ORFs were identified downstream of the DNA polymerase gene in both the RFHVMm and RFHVMn PCR products (see Fig. 3), including ORF 10, ORF RF2, a viral homolog of cellular IL-6; ORF 02, a viral homolog of dihydrofolate reductase (vDHFR); ORF RF3, a homolog of the ORF K3 modulator of immune response-1 (MIR-1); and ORF 70, a viral homolog of the thymidylate synthase (vTS). These ORFs have been named according to the HVS designations and their similarities to counterparts in the KSHV genome. ORFs not found in HVS were assigned labels beginning with RF (for retroperitoneal fibromatosis), similar to the convention used for naming KSHV ORFs, as described in Materials and Methods.
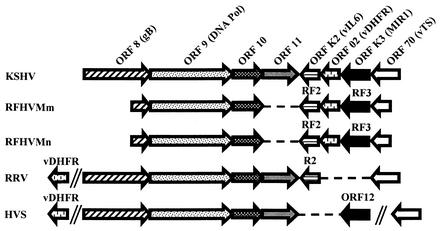
FIG. 3.
Relative organization of ≈7.7 kb of the RFHV genome between the glycoprotein B (gB) and vTS genes in comparison to the genomes of other primate rhadinoviruses, including human KSHV, macaque RRV, and South American squirrel monkey HVS. Ateline herpesvirus 3 from the South American spider monkey displays the same organization as HVS, a close relative, except it lacks a vDHFR homolog (2). The relative position and orientation of the ORFs identified within the respective 7,739- and 7,756-bp fragments of the RFHVMm and RFHVMn genomes assembled from overlapping sequences from the previously published partial gB and complete DNA polymerase (DNA Pol) (40) and the 3.6 to 4.1 kb of the DL-B region targeted in the present study are shown. Missing genes are indicated with a dashed line, and noncontiguous regions of the genome are indicated with a double slanted line.
Analysis of RFHV ORFs. (i) ORF 70 (vTS homolog).
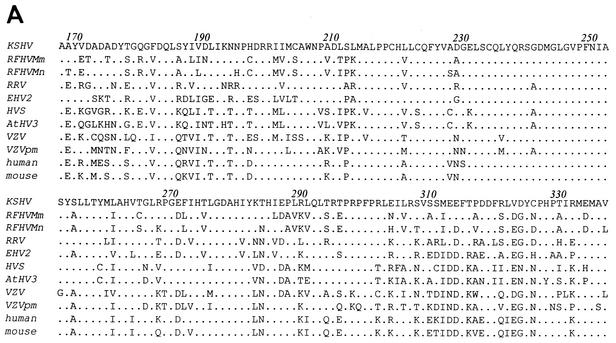
Blast analyses of the sequences obtained from the RFHVMn and RFHVMm long-range PCR products identified sequences homologous to the 3′ end of the KSHV vTS gene, which was designated ORF 70 because of homology with the HVS ORF 70 vTS gene (Fig. 3). These sequences overlapped those obtained with the TS-specific CODEHOP primers described above, and a consensus was derived for each of the viral species. An alignment of the RFHVMm and RFHVMn ORF 70 vTS sequences with the homologous regions from other viral and cellular TS homologs is shown in Fig. 4A. The amino acids encoded by the RFHVMn and RFHVMm vTS sequences were 86% identical to each other and 74% identical to the C-terminal 170 amino acids of the 337-amino-acid KSHV vTS (Table 2). The RFHV vTS sequences were 69% and 66% identical to the corresponding regions of the RRV and HVS vTS sequences, respectively. Comparison of the RRV and KSHV vTS sequences revealed 74% identical residues, equivalent to that seen between RFHV and KSHV vTS sequences (Table 3).
FIG. 4.
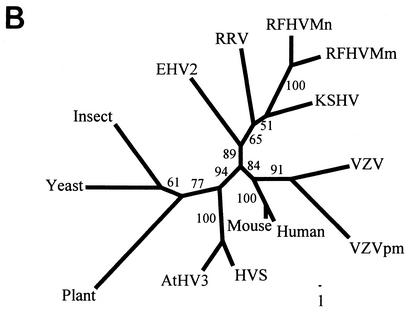
Comparison of viral ORF 70 and cellular TS homologs. (A) Amino acid sequence alignment of the partial vTS sequences obtained from RFHVMm and RFHVMn with the comparable regions of other viral and cellular TS homologs. The numbering is derived from the KSHV vTS sequence. (B) Phylogenetic analysis of the alignment in A with the protein maximum-likelihood method. Bootstrap values from 100 replica samplings and the scale for substitutions per site are provided. VZV, human varicella-zoster virus (NP_040136); VZVpm, patas monkey varicella-zoster virus (NP_077428); human TS (YXHUT); mouse TS (YXMSI); yeast TS, Saccharomyces cerevisiae (P06785); insect TS, Anopheles sp. (EAA07160); plant TS, Arabidopsis thaliana (AAA32788). See Table 1 for other accession numbers.
TABLE 2.
Putative RFHVMn and RFHVMm gene products and comparisons with homologs present in other gammaherpesviruses
| RFHV ORF designationa | Location RFHVMm/ RFHVMnb | Orien- tation | ORF size in amino acids (% identity with the RFHVMm ORF)
|
Possible function | |||||
|---|---|---|---|---|---|---|---|---|---|
| RFHVMm | RFHVMn | KSHV | RRV | HVS | EBV | ||||
| ORF 8 (3′ end) | 1-331/1-331 | + | 110c | 110c (90) | 847 (64) | 829 (53) | 808 (32) | 857 (36) | Glycoprotein B |
| ORF 9 | 373-3411/373-3411 | + | 1,013 | 1,013 (87) | 1,012 (74) | 1,014 (67) | 1,009 (60) | 1,015 (53) | DNA polymerase |
| ORF 10 | 3514-4746/3528-4760 | + | 411 | 411 (75) | 418 (46) | 416 (32) | 407 (23) | 422d (12) | Unknown |
| ORF 11 | NPe | NP | NP | 407 | 409 | 405 | 429f | Unknown | |
| RF2 | 5624-4998/5632-5006 | − | 209 | 209 (74) | 204 (35) | 208 (19) | NP | NP | IL-6 homolog |
| ORF02 | 6252-5629/6260-5637 | − | 208 | 208 (84) | 210 (52) | 188 (41) | 187 (45) | NP | Dihydrofolate reductase |
| RF3/ORF 12g | 7059-6360/7059-6363 | − | 233 | 232 (85) | 333 (40) | NP | 169 (18) | NP | Modulator of immune response |
| ORF 70 (3′ end) | 7739-7229/7756-7246 | − | 170c | 170c (86) | 337 (74) | 333 (69) | 294 (66) | NP | Thymidylate synthase |
Derived from the HVS and KSHV ORF nomenclature, with non-HVS ORFs labeled with the prefix RF.
Position within genomic DNA fragments of RFHVMm (7,739 bp) and RFHVMn (7,756 bp) extending from within ORF 8 to within the vTS gene assembled from the partial sequence of ORF 8 and complete sequence of ORF 9 (RFHVMm, AF005479; RFHVMn, AF204166) and the partial sequence of ORF 70 and the complete sequences of ORF 10, RF2, ORF 02, and RF3 (this study).
Partial sequence of the 3′ end.
LF1 gene present in the EBV Raji insertion.
NP, not present.
LF2 gene present in the EBV Raji insertion.
ORF K3 in KSHV was not recognized as homologous to ORF 12 in HVS and thus was given a K designation in the original KSHV nomenclature (37). For this reason, the RFHV ORF was given the RF designation even though it is homologous to HVS ORF 12.
TABLE 3.
Amino acid sequence comparison between the KSHV DL-B ORFs and homologous ORFs of RFHV and other gammaherpesviruses
| Gamma- herpes- virus | % Homologya
|
||||||
|---|---|---|---|---|---|---|---|
| ORF 9 (DNA poly- merase) | ORF 10 | ORF 11 | ORF K2 (vIL6) | ORF 02 (vDHFR) | ORF K3 | ORF 70 (vTS) | |
| RFHV | 76 | 45 | NP | 35 | 52 | 39 | 74 |
| RRV | 67 | 34 | 32 | 14 | 45* | NP | 74 |
| HVS | 61 | 21 | 30 | NP | 48* | 17 | 65* |
| EBV | 54 | 14 | 27 | NP | NP | NP | NP |
NP, not present. *, homologous genes located outside the DL-B region (see Fig. 1).
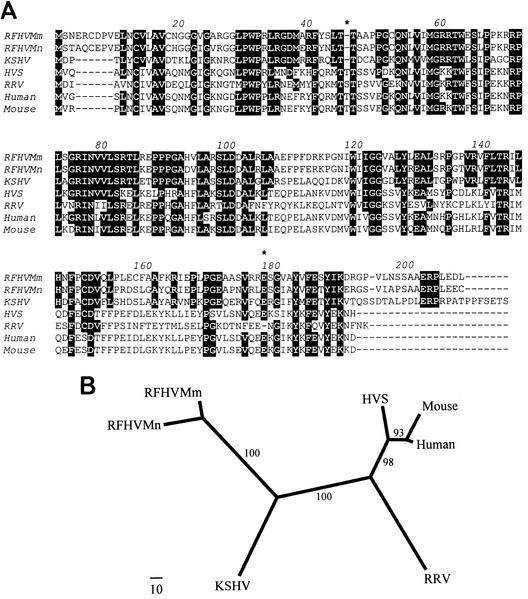
Phylogenetic analysis of the TS sequence alignment in Fig. 4A (supplemented with outgroup sequences from the TS homologs from an insect, a yeast, and a plant) by the protein maximum-likelihood method produced a tree with an ln likelihood of −2613.62538, with bootstrap support as indicated on the nodes (Fig. 4B). In this tree, the RFHVMn and RFHVMm vTS sequences grouped together, as expected for virus variants obtained from closely related macaque host species. The RFHV vTS sequences grouped with the KSHV vTS sequence in a branch separate from RRV vTS and the other TS sequences. Although the bootstrap value for this branch pattern was 51%, the likelihood was significantly positive, P < 0.01. Maximum-likelihood, neighbor-joining, and parsimony methods with both protein and DNA sequences confirmed this branch pattern (data not shown). This pattern mirrors that obtained previously with DNA polymerase sequences, supporting the grouping of RFHV and KSHV together within the RV-1 lineage of Old World primate rhadinoviruses distinct from RRV (40). Interestingly, the vTS homologs of the alphaherpesviruses and the New World primate rhadinoviruses grouped distinctly with the mammalian and eukaryotic cellular TS homologs (Fig. 4B).
(ii) ORF 10 homolog.
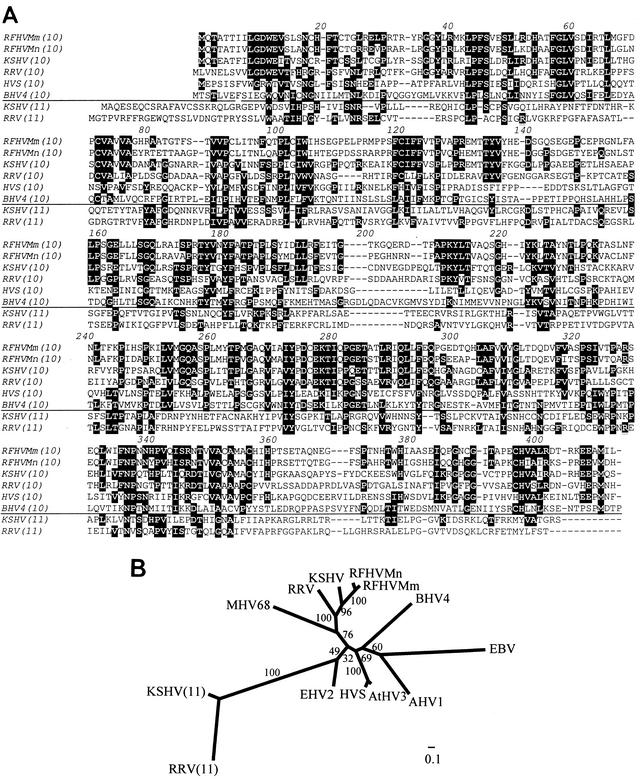
ORFs which encoded 411 amino acids that were 75% identical to each other and 46% identical to the KSHV ORF 10 were identified downstream of the DNA polymerase gene in the PCR fragments from RFHVMm and RFHVMn (Table 2). Blast (protein) analysis against the NCBI databases revealed similarities with the ORF 10 homologs of other gammaherpesviruses, including RRV (32% identity), HVS (23% identity), and Epstein-Barr virus (EBV) (12% identity, LF1 gene). Alignment of the amino acid sequences revealed the presence of numerous blocks of sequences conserved between the ORF 10 homologs of RFHVMn, RFHVMm, KSHV, and RRV (Fig. 5A). The alignment showed much less sequence homology within the other ORF 10 homologs analyzed. A much stronger sequence similarity was detected between the ORF 10 homologs of RFHV and KSHV (46% identical residues) than was detected between the ORF 10 homologs of RRV and KSHV (34% identical residues) (Table 3).
FIG. 5.
Comparison of ORF 10 homologs. (A) Alignment of the gammaherpesvirus ORF 10 homologs and the distantly related ORF 11 homologs of KSHV and RRV. Residues conserved in four of the six ORF 10 homologs are highlighted to show ORF 10 conservation. Residues in the ORF 11 homologs which were conserved in two other ORF 10 or ORF 11 homologs are also highlighted to indicate the similarity between the ORF 10 and ORF 11 homologs. (B) Phylogenetic analysis with the protein maximum-likelihood method. ORF 10 homolog sequences from the Old World primate rhadinoviruses RFHVMn, RFHVMm, KSHV, and RRV, the New World primate rhadinoviruses HVS and AtHV3, the ungulate rhadinoviruses BHV4, AHV1, and EHV2, the murine rhadinovirus MHV68, and the lymphocryptovirus EBV (LF1 gene) were aligned with ClustalW and analyzed with the PROML program. Bootstrap values from 100 replica samplings and the scale for substitutions per site are provided. The ORF 11 homologs of KSHV and RRV were included as an outgroup.
A phylogenetic analysis was performed with the protein maximum-likelihood procedure, and a tree with an ln likelihood of −11551.56201 was obtained (Fig. 5B). Like the vTS sequences, the RFHVMn and RFHVMm ORF 10 sequences clustered together with the KSHV ORF 10 sequence in a lineage distinct from that of the macaque RRV. This clustering was well supported by the bootstrap analysis and was confirmed by neighbor-joining and parsimony methods. The extended branch lengths observed for the ORF 10 homologs of the nonprimate rhadinoviruses murine herpesvirus 68 (MHV68), bovine herpesvirus 4 (BHV-4), alcelaphine herpesvirus 1 (AHV1), and equine herpesvirus 2 (EHV2) and the lymphocryptovirus EBV indicate the weak sequence conservation within the more distantly related members of this gene family; the amino acid identity between the ORF 10 homologs of the rhadinoviruses and lymphocryptoviruses was less than 20% (Table 2).
To search for possible functional motifs, iterated searching of the nonredundant NCBI protein database with PSI-Blast was performed. While no obvious sequence similarity was detected between the RFHV ORF 10 and genes of known function, a similarity was detected with the ORF 11 sequences of KSHV, RRV, and other gammaherpesviruses. Figure 5A shows an alignment of ORF 10 and 11 homologs in which few gaps were needed to align a number of conserved amino acid residues and motifs.
(iii) RFHV genome lacks an ORF 11 homolog.
Downstream of ORF 10 in the KSHV genome is the 407-amino-acid ORF 11, which is homologous to the HVS ORF 11 and the EBV LF2 gene (37). ORF 11 homologs have been identified in analogous positions in all other gammaherpesviruses characterized except BHV4, which lacks a homologous ORF 11 gene (49). In our initial long-range PCR amplification studies, the PCR fragment obtained from RFHVMm was approximately 2 kb shorter than the fragment size predicted from the KSHV genome (Fig. 1). Sequence analysis indicated that this difference in size was due to the absence of an ORF 11 homolog downstream of ORF 10. To determine whether this was a true characteristic of the RFHV genome, we obtained the sequence of the analogous fragment of the RFHVMn genome, as described above. As shown in Fig. 3, the DL-B regions of both the RFHVMn and RFHVMm genomes, like that of BHV4, lack an ORF 11 homolog.
(iv) ORF RF2 homolog (vIL-6).
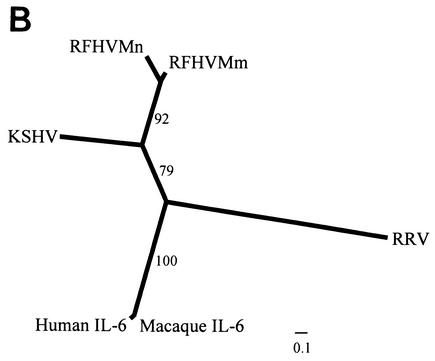
Blast analysis of the sequences obtained from the long-range PCR products from RFHVMn and RFHVMm revealed the presence of a gene homologous to the ORF K2 gene of KSHV and the cellular cytokine IL-6. The ORF K2 gene has been identified previously as a viral homolog of IL-6 (vIL-6) (37). While the sequence similarity between the KSHV vIL6 and the cellular IL-6 is low (24%), the KSHV vIL-6 has been shown to have many biological functions in common with cellular IL-6 (29). Consistent with the naming convention used for the KSHV genome, we have designated the vIL-6 homolog of RFHV ORF RF2. ORF RF2 is similar in genomic location and orientation to the KSHV ORF K2 gene (Fig. 3). A vIL-6 homolog, designated ORF R2, has been found in the macaque RRV genome (11).
The amino acid sequences of the RFHV vIL-6 homologs were aligned with the vIL-6 homologs from KSHV and RRV and the cellular human and macaque IL-6 sequences. Only 12 of more than 200 amino acids (<6%) were conserved between the cellular and viral IL-6 homologs. However, examination of the alignment revealed significant sequence similarity between the vIL-6 homologs of RFHV and KSHV which did not extend to either the vIL-6 homolog of RRV or the human and macaque cellular IL-6 homologs (Fig. 6A). Specific differences with RRV include the sequence length variations observed at the N and C termini, the cysteine at position 49, the potential N-linked glycosylation site at position 89, and four internal amino acid insertions present only in the RRV vIL-6. Overall, the vIL-6 homologs from RFHVMn and RFHVMm were 74% identical to each other and 35% identical to the KSHV vIL-6. In contrast, the RRV vIL6 was only 14% identical to the KSHV vIL-6 homolog and 19% identical to the RFHV vIL-6 homologs (Tables 2 and 3). These relationships are shown graphically in the phylogenetic analysis obtained with protein maximum likelihood (Fig. 6B). The RFHVMm and RFHVMn sequences clustered together with KSHV vIL-6 in a branch separate from the RRV and cellular IL-6 homologs. The long branch lengths between RRV and both the KSHV/RFHV vIL-6 and cellular macaque/human IL-6 clusters are indicative of the large sequence variation seen in the RRV sequence.
FIG. 6.
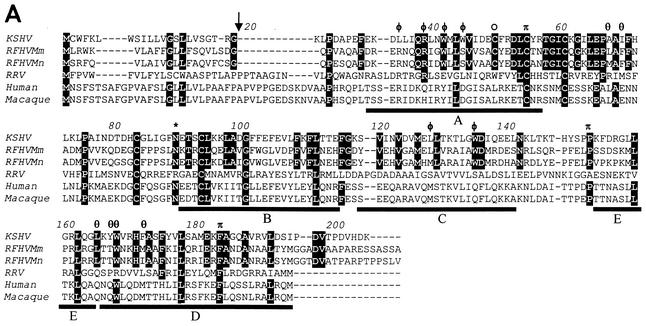
Comparison of viral and cellular IL-6 homologs. (A) Amino acid sequence alignment of the RFHVMm and RFHVMn vIL-6 homologs compared to the vIL-6 of KSHV and RRV and the human (NP_000591) and rhesus macaque (L26028) cellular IL-6. Amino acids conserved between the KSHV and RFHV sequences are highlighted; a potential N-linked glycosylation site and a cysteine residue conserved between KSHV and RFHV are indicated with an asterisk and an open circle, respectively. The predicted signal peptide cleavage site for the vIL-6 homologs is shown (arrow) and corresponds to the cleavage site determined for human IL-6. The four major alpha-helical regions (A, B, C, and D) and the minor helical region (E) determined for human IL-6 are indicated (47). The residues in sites II and III of KSHV vIL-6 which interact with the gp130 receptor subunit (see text) are indicated with φ and θ, respectively (7). The residues in site I which interact with the IL-6R subunit are indicated with π (23). The amino acid numbering is relative to the KSHV vIL-6 sequence. (B) Phylogenetic analysis of the alignment in A with the protein maximum-likelihood method. Bootstrap values from 100 replica samplings and the scale for substitutions per site are provided.
(v) ORF 02 homolog (vDHFR).
Dihydrofolate reductase (DHFR) is a ubiquitous protein in both prokaroytes and eukaryotes that is responsible for de novo synthesis of purines and deoxythymidine monophosphate for DNA synthesis. Viral homologs of DHFR have been identified in a restricted number of gammaherpesviruses within the genus Rhadinovirus, including KSHV (37), macaque RRV (42), and HVS from the New World squirrel monkey (44). As shown in Fig. 3, a homolog of DHFR was detected flanking the vIL-6 gene within the PCR products obtained from RFHVMm and RFHVMn, in a position identical to that seen in KSHV. This differs from the situation in RRV and HSV, in which the vDHFR genes are located near the left ends of their respective genomes (Fig. 3). The RFHVMm and RFHVMn vDHFR homologs were 208 amino acids in length and 84% identical to each other (Table 2). They were similar in size to the 210-amino-acid KSHV vDHFR, which was 52% identical to the RFHV homologs. This similarity is greater than that seen between the RRV and KSHV vDHFR sequences (45%; Table 3). Furthermore, the RFHV and KSHV homologs contained a 17- to 24-amino-acid C-terminal region that was not found in either the cellular DHFR genes or the vDHFR homologs of HVS and RRV, all of which were 187 to 188 amino acids in length (Fig. 7A). Additionally, the RFHV and RRV sequences differed at two insertion-deletion positions, as indicated in Fig. 7A. Although the C-terminal regions of the RFHVMn and RFHVMm vDHFR homologs were highly conserved, little homology was seen with the KSHV C-terminal region except for a conserved ERP motif (Fig. 7A). Phylogenetic analysis revealed a clustering of the RFHV and KSHV vDHFR genes that was distinct from the RRV, HVS, and cellular DHFR genes (Fig. 7B).
FIG. 7.
Comparison of viral and cellular DHFR homologs. (A) Amino acid sequence alignment of the vDHFR of KSHV, RFHVMn, RFHVMm, RRV, and HVS and comparison with the human (NP_000782) and mouse (NP_034179) cellular DHFR. Residues conserved between KSHV, RFHVMn, and RFHVMm are highlighted. An asterisk indicates the presence of an insertion or deletion difference with RRV. (B) Phylogenetic analysis of the alignment in A with the protein maximum-likelihood method. Bootstrap values from 100 replica samplings and the scale for substitutions per site are provided.
(vi) ORF RF3 (MIR-1) homolog.
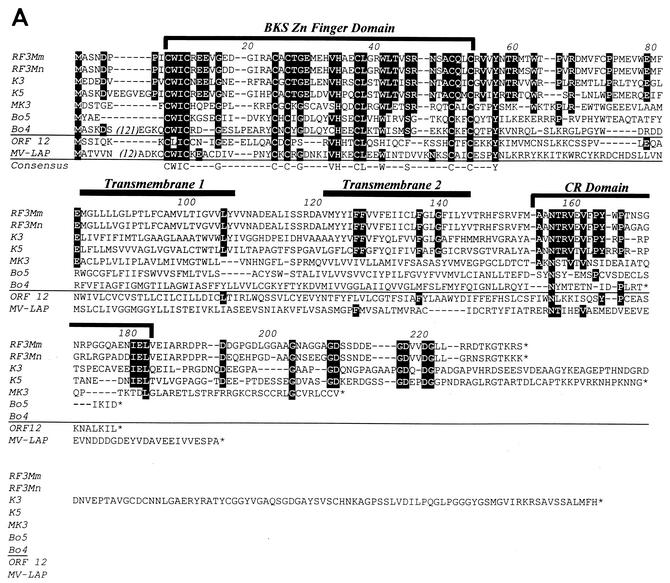
ORFs encoding 233 and 232 amino acids were identified between the vDHFR and vTS genes of the RFHVMn and RFHVMm PCR fragments, respectively. A Blast similarity search demonstrated that these ORFs were closely related to the ORF K3 and K5 genes of KSHV. Because the RFHV ORFs were in a location identical to that of the K3 gene within the KSHV genome, they were designated ORF RF3 (see Fig. 3). While the RF3 homologs from RFHVMn and RFHVMm were 85% identical to each other, they were only 40% and 38% identical to the K3 and K5 genes of KSHV, respectively (Table 2). Blast analysis identified similarities with other herpesvirus proteins, including two ORFs, Bo4 (nonspliced) and Bo5 (spliced), derived from the immediate-early-1 gene transcript of BHV4 and the ORF K3 homolog in MHV68, herein designated MK3. In addition, sequence similarity was identified with several poxvirus proteins, including the C7 protein of swinepox (swine host), the MV-LAP protein of myxoma virus (rabbit host), and the 5L protein of yaba-like disease virus (monkey host). A distinct but more distant sequence similarity was also noted with HVS ORF 12. An ORF RF3 homolog was not detected within the genome of the macaque RRV. Within the herpesviruses, all of the RF3-related ORFs were colocalized in analogous positions within their respective genomes.
Alignment of the RFHVMn and RFHVMm RF3 genes with the other related sequences revealed a strong sequence similarity centered around a conserved cysteine and histidine motif within the N-terminal domain of these proteins. This motif belongs to the BKS (BHV4/KSHV/swinepox) subset of the C4HC3 PHD/LAP zinc finger motifs, as described before (31). The PHD/LAP motif family consists of a very general grouping of a number of proteins containing a C4HC3 motif, which has been implicated in DNA binding and chromatin-mediated transcriptional regulation (1, 38). A consensus sequence of highly conserved residues was determined for the BKS motif of RF3, K3, K5, Bo4, Bo5, and MK3 which consisted of C1-W-I-C2-X(2-4)-G-X(5-8)C3-X-C4-X-G-X4-V-H5-X2-C6-L-X2-W-X3-S-X4-C7-X2-C8-X3-Y. Whereas the domains containing the zinc finger motif (amino acids 9 to 53, Fig. 8A) in RF3 from RFHVMn and RFHVMm were 98% identical, with only a single amino acid difference, Asp-Glu at position 19, they were only 70% identical to the domains found in K3 and K5 and approximately 40% identical to the domains present in the MK3, Bo5, and Bo4 homologs. Although HVS ORF 12 contained the conserved cysteine and histidine residues characteristic of the C4HC3 BKS motif, it lacked many of the additional residues which were completely conserved in the other herpesvirus proteins within this domain (Fig. 8A).
FIG. 8.
Comparison of ORF RF3 homologs. (A) Amino acid sequence alignment of the MIR homologs of KSHV (K3 and K5), RFHVMn (RF3Mn), RFHVMm (RF3Mm), BHV4 (Bo5 and Bo4), MHV68 (MK3), HVS (ORF 12), and myxoma virus (MV-LAP; AAK00734). The BKS zinc finger domain is indicated, with the positions of the hydrophobic transmembrane domains and the conserved region (CR) in the C-terminal domain shown. Residues identical within the K3, K5, RF3Mn, and RF3Mm sequences are highlighted. A consensus sequence for the BKS zinc finger domain is shown. (B) Phylogenetic analysis of the complete ORFs with the protein maximum-likelihood method with the addition of the 5L protein of yaba-like disease virus (NP_073390). Bootstrap values from 100 replica samplings and the scale for substitutions per site are provided.
Like the other viral homologs, the RF3 sequences contained two extended hydrophobic regions downstream of the zinc finger motif which were predicted to be membrane-spanning domains, as indicated in Fig. 8A. While the transmembrane domains and flanking regions (amino acids 55 to 156, Fig. 8A) of the RF3 homologs from RFHVMn and RFHVMm were 95% identical to each other, they were only 31% and 27% identical to the corresponding regions of KSHV K3 and K5, respectively. Only a few residues within this region were identical between the RF3, K3, and K5 sequences, as seen in Fig. 8A. The transmembrane domains and flanking regions of the other viral homologs were not well conserved and were aligned based on the positions of their hydrophobic regions. The C-terminal domains downstream of the second hydrophobic domain (amino acids 157 to 236) within the RF3 sequences from RFHVMn and RFHVMm were only 50% identical but contained a conserved region (CR) in which several sequence motifs were also conserved with the K3 and K5 sequences, for example, the NTRV motif (amino acids 156 to 159), the PY motif (amino acids 163 to 164), and the IEL motif (amino acids 180 to 182). The Asn and Pro at positions 156 and 163, respectively, were also highly conserved within the C-terminal domains of the other K3/K5-related homologs. The C-terminal domains of RF3, K3, and K5 also contained several G-N/D and N/D-G dipeptide motifs which were not found in the other homologous sequences. While the RF3 and K5 sequences terminated shortly after these dipeptide motifs, the C-terminal region of the K3 sequence extended for an additional 100 amino acids.
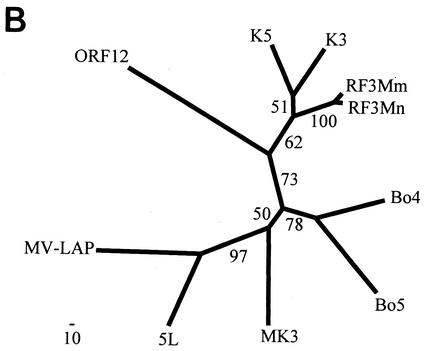
Phylogenetic analysis of the RF3 homologs with several different methods, including neighbor-joining and protein maximum likelihood, revealed an obvious clustering of RF3, K3, and K5 (see Fig. 8B) which was distinct from that of the other herpesvirus and poxvirus homologs. However, the K3 homolog did not cluster exclusively with the RF3 homologs, as expected from their homologous positions within their genomes. Instead, the branch pattern between K3, RF3, and K5 was inconsistent. In some analyses, K3 clustered with the RF3 homologs with the exclusion of K5, while in other trees K3 clustered with K5 with the exclusion of the RF3 homologs. This was also evident from the low bootstrap score of 51 for this branch point in the protein maximum-likelihood analysis in Fig. 8B. The clustering of the two proteins from the myxoma and yaba-like disease virus poxviruses and the clustering of the two BHV4 homologs, Bo4 and Bo5, was consistent in the different analyses.
DISCUSSION
We and others have previously reported that a number of Old World primate species are each host to two distinct RV-1 and RV-2 lineages of KSHV-like rhadinoviruses (13, 14, 20-22, 40). While the complete sequences have been determined for the genomes of human KSHV and macaque RRV, the classification of these and other KSHV-like rhadinoviruses into the RV-1 and RV-2 lineages of rhadinoviruses has been based solely on comparison of DNA polymerase sequences. Sequence analysis of the complete genomes of KSHV and RRV has revealed numerous similarities in gene structure and content which support a close evolutionary relationship (11). However, the genetic content of these viruses is not identical, with numerous examples of genes which are present in one virus and absent in the other (42). It has not been clear whether these differences are simply because of speciation of the human and macaque viruses or whether they reflect important structural and functional properties of the two distinct viral lineages.
Identification of genetic properties unique to the RV-1 and RV-2 rhadinovirus lineages has been hampered by the absence of an identified RV-2 rhadinovirus from a human host and the difficulty in obtaining further genomic sequence from the other identified RV-1 rhadinoviruses from nonhuman hosts. Among the identified nonhuman RV-1 rhadinoviruses, the greatest amount of sequence information has been obtained for RFHV, the putative macaque RV-1 rhadinovirus, with sequence covering the entire DNA polymerase gene and flanking sequences. However, there has still been some uncertainty concerning the exact nature of the relationship between RFHV and the other macaque virus, RRV, because of the relatively small amount of sequence data and information on gene content available for RFHV.
In the present study, we have added to the amount of available RFHV genetic information by determining 4.3 kb of contiguous sequence within the DL-B region of the RFHVMm and RFHVMn genomes. These sequences overlap the DNA polymerase sequences obtained previously (36, 40), extending the available sequence for both RFHV species to approximately 7.7 kb. Within this 7.7-kb genomic region, open reading frames encoding homologs of the KSHV glycoprotein B, DNA polymerase, ORF 10, vIL-6, vDHFR, ORF K3/MIR-1, and vTS genes were identified. These genes were present in the same order and relative positions as in the KSHV genome (37). However, unlike KSHV, the RFHVMn and RFHVMm genomes lacked an ORF 11 homolog. Blast comparisons and phylogenetic analysis of each RFHVMn and RFHVMm ORF demonstrated the closest similarity to the corresponding ORF within the KSHV genome. These results strongly support the grouping of RFHV with KSHV within the RV-1 rhadinovirus lineage and provide evidence that RFHVMm and RFHVMn are the rhesus and pig-tailed macaque homologs of KSHV.
Comparison of the macaque RFHV and RRV genomes within the region of the DL-B targeted in this study revealed important differences in the colinear organization and genetic content of the genomes. One such difference was the presence of the RF3 homolog of MIR-1/ORF K3 in RFHV which was absent from RRV. MIR-1 homologs have been identified in other herpesviruses, including BHV4, MHV68, and HVS, and in several poxviruses, including swinepox and myxoma virus (31, 46). MIR-1 and the closely related MIR-2 of KSHV, ORF MK3 of MHV68, and the MV-LAP protein of myxoma virus have been shown to function in immune evasion through downregulation of members of major histocompatibility complex class I (MHC-I) and other molecules involved in immune recognition (8, 9, 15, 16, 48). For this reason, the KSHV proteins have been termed modulators of immune response (MIR), while the MV-LAP protein have been described as endoplasmic reticulum-resident surface cellular receptor abductor proteins, called scrapins (15).
MIR-1 and MIR-2 function as membrane-bound E3 ubiquitin ligases which ubiquitinate the cytosolic tail of immune recognition proteins, targeting them for endolysosomal degradation (10, 25). The ability to impair host cytotoxic T-lymphocyte recognition of virus-infected cells is believed to be an important factor in the viral persistence characteristic of KSHV and other herpesviruses. The presence of the RF3 MIR homolog in RFHV and its absence in RRV demonstrates for the first time a clear difference between the genetic repertoire of the two distinct lineages of KSHV-like macaque rhadinoviruses and suggests either that the persistence of RRV in infected cells is different from that seen with RFHV and KSHV or that RRV employs another means of enhancing viral persistence. Further analysis of the genomes of the other RV-1 Old World primate rhadinoviruses will determine whether this genetic feature is characteristic of the RV-1 lineage in general.
In addition to the conservation within the BKS zinc finger domain of the MIR homologs, sequence similarity was detected within the C-terminal conserved region (CR) domains of RF3, MIR-1, and MIR-2 (Fig. 8A). Deletion-insertion studies have shown that the conserved region within MIR-2 is necessary but not sufficient for the downregulation of MHC proteins (39). Although this region is conserved between the MIR-1, MIR-2, and RF3 sequences, it is poorly conserved with the corresponding regions of the other herpesvirus and poxvirus proteins. The upstream flanking hydrophobic transmembrane domains are even less well conserved, with only isolated residues, including three phenylalanine residues, conserved between the transmembrane domains of RF3, MIR-1, and MIR-2. Even less conservation with other herpesvirus and poxvirus proteins was noted across the transmembrane domains.
Only the RF3 transmembrane regions from RFHVMn and RFHVMm were well conserved between each other, with 95% amino acid identity between amino acids 80 and 145. Replacement studies have shown that the transmembrane domains are responsible for the target specificity of MIR activity (39). Thus, the close similarity between the RF3 homologs of RFHVMn and RFHVMm might reflect target similarities within the two macaque species, while the differences between the RF3 and MIR transmembrane domains could reflect the different specificities required for interactions with macaque and human immune proteins. Experiments are under way to examine the ubiquitin ligase activity and target specificity of macaque RF3.
Other differences in the genome structures of RFHV and RRV were also evident from our analysis of the DL-B region. Whereas the genomes of both RFHV and KSHV contain a vDHFR gene immediately to the left and downstream of the RF3/K3 homolog, the RRV vDHFR is positioned at the far left end of the viral genome. The positioning of the RRV vDHFR gene is similar to that seen in the New World primate rhadinovirus HVS (see Fig. 3). Furthermore, the sequence of the RRV vDHFR gene is quite distinct from the that of RFHV and KSHV and groups phylogenetically more closely with the HVS and mammalian cellular DHFR homologs. In fact, the RFHV and KSHV vDHFR sequences are more similar to HVS vDHFR than to RRV vDHFR (Tables 2 and 3). Specific differences include the presence of an additional amino acid at position 45 in the sequences of human, mouse, HVS, and RRV DHFR homologs which is absent in the KSHV and RFHV vDHFR homologs and the unique C-terminal extensions found only in the KSHV and RFHV vDHFR homologs. These findings suggest that the vDHFR homologs of KSHV and RFHV were acquired from an ancestral host in a different acquisition event than the vDHFR homologs of HVS and RRV. The differences in the genomic location of the vDHFR genes further support independent acquisition events. While the KSHV and RFHV vDHFR genes are more closely related to each other than to the other DHFR genes, they have evolved to differ considerably from each other, especially when compared to the minimal differences seen between the human and murine DHFR genes, which are 90% identical.
Our studies show that RFHV, like KSHV and RRV, contains a viral homolog of the cellular IL-6 in a conserved orientation within the DL-B region. Of all the herpesviruses identified to date, only these KSHV-like rhadinoviruses contain a vIL6 homolog (31, 37). While our studies show that the IL-6 homologs of RFHV and KSHV have a conserved gene structure and sequence, the RRV vIL-6 has diverged considerably, with substantial changes in amino acid sequence and three internal insertions or deletions with respect to the other vIL-6 and cellular IL-6 homologs. These structural differences suggest the possibility of functional differences. Cellular IL-6 is one of a family of related cytokines which has been implicated in the pathogenesis of several B-cell neoplasias (18), and the presence of a vIL6 homolog in KSHV suggests a potential role for this protein in the induction and progression of Kaposi's sarcoma and the B-cell neoplasias associated with KSHV.
Whereas cellular IL-6 requires the presence of the nonsignaling receptor subunit (IL-6R) and the signaling receptor subunit gp130 for signal transduction, the KSHV vIL-6 can interact and signal directly through the ubiquitous gp130 receptor subunit in the absence of IL-6R (28). Recent results suggest that the gp130-specific signaling by KSHV vIL-6 plays an important role in bypassing the antiviral effects of interferon (6). Crystallographic studies of the KSHV vIL6-gp130 extracellular receptor complex have identified major sites of protein interaction which are primarily hydrophobic (7). The site II interface of KSHV vIL-6 is dominated by Trp41, Trp44, and Trp134, while the site III interface is dominated by Leu165, Tyr166, Trp167, and Phe171. These interactions are predicted to allow two molecules of vIL-6 to bind to two gp130 subunits in the absence of the IL-6R subunit. Most of these residues are conserved within the RFHVMn and RFHVMm vIL-6 homologs, suggesting similarities in receptor binding (Fig. 6A).
Interestingly, very few of these hydrophobic residues are conserved within the RRV vIL-6. In RRV, Trp41 is replaced by a negatively charged glutamic acid and Trp167 is replaced by a positively charged arginine. This is similar to the situation with the cellular IL-6 homologs, which lack the conserved hydrophobic residues in the site II and site III domains and instead have small polar residues in these positions. The lack of hydrophobic residues available for site II and III binding suggests that, like cellular IL-6, RRV vIL-6 may not bind and signal directly through interactions with gp130, but instead could require the presence of the IL-6R subunit. Studies with RRV vIL-6 support this hypothesis. Antibodies directed against the IL-6R subunit completely inhibit RRV vIL-6-induced proliferation of B9 mouse cells (17). This contrasts with the situation with KSHV vIL-6, where antibodies against gp130 inhibited vIL-6 activity while antibodies against IL-6R did not (5, 28). These results suggest that the vIL-6 homologs from members of the RV-1 lineage of rhadinoviruses could exert their biological effect on any cell which expresses a sufficient quantity of the gp130 signaling receptor, whereas the effects of vIL-6 homologs from members of the RV-2 lineage of rhadinoviruses would be limited to cells expressing both the IL-6R nonsignaling subunit and the gp130 signaling subunit. We are currently examining the receptor binding properties of the two macaque vIL6 homologs.
Our results demonstrate that RFHV contains a vTS homolog in a genomic position and orientation similar to those of the vTS homologs of the other Old World primate rhadinoviruses KSHV and RRV. This contrast with the position of the vTS homologs within the genomes of the New World primate rhadinoviruses HVS and ateline herpesvirus 3 and the ungulate rhadinovirus EHV2, in which the vTS genes are located much further downstream at the right end of the genome (2, 3, 43). While TS homologs are also present in the human and simian varicella-zoster virus homologs (as shown in Fig. 4A and B), the genome structures of these alphaherpesviruses are not analogous to the rhadinovirus genomes and cannot be easily compared. Three separate clusters of the herpesvirus vTS homologs were evident from our phylogenetic analysis: (i) the Old World primate and ungulate rhadinoviruses vTS homologs, (ii) the human and monkey varicella-zoster virus-related alphaherpesvirus and the mammalian TS homologs, and (iii) the New World primate rhadinovirus and lower eukaryote TS homologs. This suggests that at least three separate acquisitions of cellular TS genes into the herpesviridae may have occurred. A second possibility is that the sequence and genomic location differences between the TS homologs may derive from a combination of different mutational rates and translocations between the two genomic terminal regions (27).
In contrast to KSHV and RRV, RFHV lacks an ORF 11 homolog downstream of ORF 10. This was confirmed by sequencing this region in both RFHVMn and RFHVMm species. Although the presence of an ORF 11 homolog is a conserved feature within most gammaherpesviruses, the ungulate rhadinovirus BHV4 also lacks an ORF 11 homolog (49). Analysis of the ORF 11 homologs of other gammaherpesviruses with repetitive PSI-Blast searches of the NCBI databases revealed a distant similarity to the gammaherpesvirus ORF 10 homologs (see alignment in Fig. 5A). This similarity, though weak, suggests a possible functional overlap between ORFs 10 and 11. However, no function has yet been attributed to either ORF 10 or 11, nor have obvious similarities been detected with proteins of known function. ORFs 10 and 11 do not appear to be required for viral replication, as both gene homologs (LF1 and LF2) are deleted in the prototypical EBV variant which replicates within the B95-8 marmoset cell line (34). Our results indicate that sometime after host speciation and the evolutionary split between RFHV and KSHV, the RFHV lineage lost the ORF 11 homolog within the DL-B region. Future cloning studies will reveal whether or not an ORF 11 homolog is alternatively present within the remainder of the uncharacterized RFHV genome.
Our studies demonstrate a strong similarity in sequence conservation, genetic complement, and colinear organization of genes within the DL-B region of the genomes of macaque RFHV and human KSHV. These findings strongly support the hypothesis that RFHV is the macaque equivalent of KSHV and substantiate the grouping of these viruses together within the RV-1 lineage of Old World primate rhadinoviruses. Conversely, the differences detected between the sequence conservation, genetic complement, and colinear organization of the macaque RFHV and RRV genomes support the grouping of these two macaque viruses into distinct RV-1 and RV-2 rhadinovirus lineages, respectively. However, the ultimate comparison of these macaque rhadinoviruses awaits the complete sequencing of the RFHV genome. Even though the RV-1 and RV-2 lineages of rhadinoviruses are more closely related to each other than to other members of the Herpesviridae, our findings suggest that the two macaque rhadinovirus lineages have diverged considerably over time, yielding differences in gene content, organization, and structure that would provide the basis for biological differences between the members of these two viral lineages. No evidence of interlineage recombination between the macaque RV-1 and RV-2 rhadinoviruses was detected, even though mixed infections were identified in numerous individual macaques.
Acknowledgments
This work was supported by grants to T. M. Rose from the National Institutes of Health (R55-CA72237 and RO1-RR13154). T. M. Rose was the recipient of an Independent Scientist Award, K02-AI49275.
We acknowledge Riri Shibata and Harold McClure for generous gifts of tissue samples and Greg Bruce and Jacques Garrigues for helpful advice and discussion. We also thank John Nicolas for providing unpublished information concerning the structure of KSHV during the initiation of this research.
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht, J. C. 2000. Primary structure of the herpesvirus ateles genome. J. Virol. 74:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albrecht, J. C., J. Nicholas, D. Biller, K. R. Cameron, B. Biesinger, C. Newman, S. Wittmann, M. A. Craxton, H. Coleman, B. Fleckenstein, et al. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 66:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auerbach, M. R., S. C. Czajak, W. E. Johnson, R. C. Desrosiers, and L. Alexander. 2000. Species specificity of macaque rhadinovirus glycoprotein B sequences. J. Virol. 74:584-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burger, R., F. Neipel, B. Fleckenstein, R. Savino, G. Ciliberto, J. R. Kalden, and M. Gramatzki. 1998. Human herpesvirus type 8 interleukin-6 homologue is functionally active on human myeloma cells. Blood 91:1858-1863. [PubMed] [Google Scholar]
- 6.Chatterjee, M., J. Osborne, G. Bestetti, Y. Chang, and P. S. Moore. 2002. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science 298:1432-1435. [DOI] [PubMed] [Google Scholar]
- 7.Chow, D., X. He, A. L. Snow, S. Rose-John, and K. C. Garcia. 2001. Structure of an extracellular gp130 cytokine receptor signaling complex. Science 291:2150-2155. [DOI] [PubMed] [Google Scholar]
- 8.Coscoy, L., and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 97:8051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coscoy, L., and D. Ganem. 2001. A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T cell costimulation. J. Clin. Investig. 107:1599-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coscoy, L., D. J. Sanchez, and D. Ganem. 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desrosiers, R. C., V. G. Sasseville, S. C. Czajak, X. Zhang, K. G. Mansfield, A. Kaur, R. P. Johnson, A. A. Lackner, and J. U. Jung. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J. Virol. 71:9764-9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giddens, W. E., Jr., C. C. Tsai, W. R. Morton, H. D. Ochs, G. H. Knitter, and G. A. Blakley. 1985. Retroperitoneal fibromatosis and acquired immunodeficiency syndrome in macaques. Pathologic observations and transmission studies. Am. J. Pathol. 119:253-263. [PMC free article] [PubMed] [Google Scholar]
- 13.Greensill, J., and T. F. Schulz. 2000. Rhadinoviruses (gamma2-herpesviruses) of Old World primates: models for KSHV/HHV8-associated disease? AIDS 14:S11-S19. [PubMed] [Google Scholar]
- 14.Greensill, J., J. A. Sheldon, N. M. Renwick, B. E. Beer, S. Norley, J. Goudsmit, and T. F. Schulz. 2000. Two distinct gamma-2 herpesviruses in African green monkeys: a second gamma-2 herpesvirus lineage among Old World primates? J. Virol. 74:1572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerin, J. L., J. Gelfi, S. Boullier, M. Delverdier, F. A. Bellanger, S. Bertagnoli, I. Drexler, G. Sutter, and F. Messud-Petit. 2002. Myxoma virus leukemia-associated protein is responsible for major histocompatibility complex class I and Fas-CD95 down-regulation and defines scrapins, a new group of surface cellular receptor abductor proteins. J Virol. 76:2912-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishido, S., C. Wang, B. S. Lee, G. B. Cohen, and J. U. Jung. 2000. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaleeba, J. A., E. P. Bergquam, and S. W. Wong. 1999. A rhesus macaque rhadinovirus related to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 encodes a functional homologue of interleukin-6. J. Virol. 73:6177-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kishimoto, T., S. Akira, M. Narazaki, and T. Taga. 1995. Interleukin-6 family of cytokines and gp130. Blood 86:1243-1254. [PubMed] [Google Scholar]
- 19.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 20.Lacoste, V., P. Mauclere, G. Dubreuil, J. Lewis, M. C. Georges-Courbot, and A. Gessain. 2000. KSHV-like herpesviruses in chimps and gorillas. Nature 407:151-152. [DOI] [PubMed] [Google Scholar]
- 21.Lacoste, V., P. Mauclere, G. Dubreuil, J. Lewis, M. C. Georges-Courbot, and A. Gessain. 2001. A novel gamma 2-herpesvirus of the rhadinovirus 2 lineage in chimpanzees. Genome Res. 11:1511-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacoste, V., P. Mauclere, G. Dubreuil, J. Lewis, M. C. Georges-Courbot, J. Rigoulet, T. Petit, and A. Gessain. 2000. Simian homologues of human gamma-2 and betaherpesviruses in mandrill and drill monkeys. J. Virol. 74:11993-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, H., H. Wang, and J. Nicholas. 2001. Detection of direct binding of human herpesvirus 8-encoded interleukin-6 (vIL-6) to both gp130 and IL-6 receptor (IL-6R) and identification of amino acid residues of vIL-6 important for IL-6R-dependent and -independent signaling. J. Virol. 75:3325-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lisitsyn, N., and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 25.Lorenzo, M. E., J. U. Jung, and H. L. Ploegh. 2002. Kaposi's sarcoma-associated herpesvirus K3 utilizes the ubiquitin-proteasome system in routing class major histocompatibility complexes to late endocytic compartments. J. Virol. 76:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansfield, K. G., S. V. Westmoreland, C. D. DeBakker, S. Czajak, A. A. Lackner, and R. C. Desrosiers. 1999. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J. Virol. 73:10320-10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGeoch, D. J. 2001. Molecular evolution of the gamma-Herpesvirinae. Phil. Trans. R. Soc. Lond. B Biol. Sci. 356:421-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molden, J., Y. Chang, Y. You, P. S. Moore, and M. A. Goldsmith. 1997. A Kaposi's sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J. Biol. Chem. 272:19625-19631. [DOI] [PubMed] [Google Scholar]
- 29.Moore, P. S., and Y. Chang. 2001. Molecular virology of Kaposi's sarcoma-associated herpesvirus. Phil. Trans. R. Soc. Lond. B Biol. Sci. 356:499-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore, P. S., S. J. Gao, G. Dominguez, E. Cesarman, O. Lungu, D. M. Knowles, R. Garber, P. E. Pellett, D. J. McGeoch, and Y. Chang. 1996. Primary characterization of a herpesvirus agent associated with Kaposi's sarcoma. J. Virol. 70:549-558. (Erratum, J. Virol. 70:9083.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholas, J., V. Ruvolo, J. Zong, D. Ciufo, H. G. Guo, M. S. Reitz, and G. S. Hayward. 1997. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J. Virol. 71:1963-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 33.Pang, K. M., and D. A. Knecht. 1997. Partial inverse PCR: a technique for cloning flanking sequences. BioTechniques 22:1046-1048. [DOI] [PubMed] [Google Scholar]
- 34.Parker, B. D., A. Bankier, S. Satchwell, B. Barrell, and P. J. Farrell. 1990. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology 179:339-346. [DOI] [PubMed] [Google Scholar]
- 35.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose, T. M., K. B. Strand, E. R. Schultz, G. Schaefer, G. W. Rankin, Jr., M. E. Thouless, C. C. Tsai, and M. L. Bosch. 1997. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J. Virol. 71:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saha, V., T. Chaplin, A. Gregorini, P. Ayton, and B. D. Young. 1995. The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins. Proc. Natl. Acad. Sci. USA 92:9737-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez, D. J., L. Coscoy, and D. Ganem. 2002. Functional organization of MIR-2, a novel viral regulator of selective endocytosis. J. Biol. Chem. 277:6124-6130. [DOI] [PubMed] [Google Scholar]
- 40.Schultz, E. R., G. W. Rankin, Jr., M. P. Blanc, B. W. Raden, C. C. Tsai, and T. M. Rose. 2000. Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:4919-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulz, T. F., and P. S. Moore. 1999. Kaposi's sarcoma-associated herpesvirus: a new human tumor virus, but how? Trends Microbiol. 7:196-200. [DOI] [PubMed] [Google Scholar]
- 42.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telford, E. A., M. S. Watson, H. C. Aird, J. Perry, and A. J. Davison. 1995. The DNA sequence of equine herpesvirus 2. J. Mol. Biol. 249:520-528. [DOI] [PubMed] [Google Scholar]
- 44.Trimble, J. J., S. C. Murthy, A. Bakker, R. Grassmann, and R. C. Desrosiers. 1988. A gene for dihydrofolate reductase in a herpesvirus. Science 239:1145-1147. [DOI] [PubMed] [Google Scholar]
- 45.Tsai, C. C., W. E. Giddens, Jr., H. D. Ochs, W. R. Morton, G. H. Knitter, G. A. Blakley, and R. E. Benveniste. 1986. Retroperitoneal fibromatosis and acquired immunodeficiency syndrome in macaques: clinical and immunologic studies. Lab. Anim. Sci. 36:119-125. [PubMed] [Google Scholar]
- 46.Virgin, H. W. t., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, G. Y., H. A. Yu, J. Hong, M. Stahl, T. McDonagh, L. E. Kay, and D. A. Cumming. 1997. Solution structure of recombinant human interleukin-6. J. Mol. Biol. 268:468-481. [DOI] [PubMed] [Google Scholar]
- 48.Yu, Y. Y., M. R. Harris, L. Lybarger, L. A. Kimpler, N. B. Myers, H. W. t. Virgin, and T. H. Hansen. 2002. Physical association of the K3 protein of gamma-2 herpesvirus 68 with major histocompatibility complex class I molecules with impaired peptide and beta2-microglobulin assembly. J. Virol. 76:2796-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmermann, W., H. Broll, B. Ehlers, H. J. Buhk, A. Rosenthal, and M. Goltz. 2001. Genome sequence of bovine herpesvirus 4, a bovine rhadinovirus, and identification of an origin of DNA replication. J. Virol. 75:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]