Abstract
A Vibrio cholerae deletion mutant lacking VS2773, a parA partitioning gene homolog located in a parAB operon on the large chromosome, displays altered positioning of the large chromosome origin. Deletion of a second parA homolog on the large chromosome (VC2061) does not affect its origin positioning. The origin position of the small chromosome is unchanged by either or both of these deletions, suggesting that VC2773 function is specific to the replicon on which it is carried. VC2773 and VC2772 form a parABS system with inverted repeats found near the large chromosome origin.
In recent years, it has become possible to study the internal organization of the bacterial cell in great detail. DNA positioning and movement have been studied in various bacteria, using both live- and fixed-cell models. While it is clear that the chromosome is highly organized within the cell, the mechanisms underlying these processes are still not fully understood (18). Studies of bacteria with single chromosomes, such as Escherichia coli, Bacillus subtilis, and Caulobacter crescentus, have revealed variations in origin positioning and the timing of this placement (13, 24, 29). In the multireplicon-containing alpha-proteobacteria Agrobacterium tumefaciens and Sinorhizobium meliloti, origins localize predominantly to the cell poles, although they remain nonoverlapping (15). Fogel and Waldor have studied origin positioning in the bipartite genome of Vibrio cholerae, using a live-cell model and a fluorescent repressor-operator system (6). Their findings of distinct positioning patterns and cell cycle timing of movement for the two origins were independently confirmed by Fiebig et al. (5a) and suggest that a separate segregation mechanism may exist for each chromosome.
Early studies of bacterial DNA partitioning focused on the maintenance of low-copy-number plasmids, such as F factor, P1, and R1. The genetic loci responsible for plasmid partitioning encode members of the ParA and ParB protein families. Homologs of these genes have been discovered on many bacterial chromosomes, with the exception of E. coli and related enteric organisms, and work in model systems such as B. subtilis, C. crescentus, Streptomyces coelicolor, and Pseudomonas putida has demonstrated a role for ParA and ParB in chromosome segregation. However, the segregation defects in strains with mutants of these genes have often been mild, with stronger phenotypes manifested under specific growth conditions or during developmental processes such as sporulation (9, 12, 16, 19, 23).
The Vibrio species studied to date all have a divided genome consisting of two chromosomes; each chromosome carries a parAB locus (1, 10, 21, 26). The locus on the large chromosome is related more closely to other bacterial chromosomal par loci than that on the small chromosome, which bears homology to plasmid-carried loci (10). Given the distinct localization patterns of the replication origins in V. cholerae, the parAB loci are factors that may confer specificity in positioning. Vibrio species also have a second parA homolog carried separately on the large chromosome, but its function is unclear. In this study, we provide evidence that the parA homolog VC2773, found in the parAB operon on the large chromosome of V. cholerae, can specifically alter positioning of the large chromosome origin if it is deleted or present in multiple copies. In contrast, the parA homolog VC2061 does not appear to affect the origin positioning of either chromosome. We also demonstrate that the large chromosome parAB locus can function in DNA segregation and stabilizes an unstable mini-F plasmid carrying putative parS sequences unique to the large chromosome of V. cholerae.
A fixed-cell model of origin positioning recapitulates a live-cell model in V. cholerae.
We investigated the role of the parA homologs carried on the large chromosome of V. cholerae, visualizing positioning of the V. cholerae replication origins by using fluorescence in situ hybridization (FISH) in fixed cells. An ∼10-kb region of each V. cholerae chromosome near the origin was synthesized by PCR and verified using restriction analysis and sequencing. The sequences of oligonucleotides used in this study are available upon request. FISH analysis was performed as described by Jensen and Shapiro (13), and probes were labeled with either Cy3-dCTP (Molecular Probes) or digoxigenin-11-dUTP (Roche) (15). Bacteria were grown with shaking in LB medium at 37°C to an optical density at 600 nm (OD600) of 0.6 to 0.8 and fixed immediately prior to processing. Fluorescence and phase-contrast images were acquired using a Leica DM4000B microscope with a 100× objective and a cooled charge-coupled device camera (Qimaging). Image processing and analysis were performed using ImageJ (NIH) and ImagePro software (MediaCybernetics, Silver Spring, MD). Hybridizations were performed with multiple independent cultures and slides. The strains and plasmids used in this study are listed in Table 1. As needed, bacteria were grown in Luria-Bertani (LB) medium, AB minimal medium with 0.2% glycerol as the carbon source (2), or M9-CSA medium (M9 medium with 0.2% Casamino Acids and 0.4% glucose) (5) with appropriate antibiotics.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| DH5αλpir | F−endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 φ80dlacZΔM15 λpir | Laboratory collection |
| DH10B | F−mcrA Δ(mrr hsdRMS mcrBC) φ80dlacZΔM15 ΔlacX74 endA1 recA1 deoR Δ(ara leu)7697 araD139 galU galK nupG rpsL | Laboratory collection |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpir pro endA hsdA hsdR supF | Laboratory collection |
| V. cholerae strains | ||
| N16961 | Wild-type El Tor strain, Smr | 10 |
| LK626 | N16961 ΔVC2773 | This study |
| LK681 | N16961/pJK4 | This study |
| LK705 | N16961 ΔVC2061 | This study |
| LK706 | N16961 ΔVC2773 ΔVC2061 | This study |
| LK712 | LK626 with reconstituted VC2773 | This study |
| KFV10 | N16961 ΔlacZ | 7 |
| Plasmids | ||
| pBluescript SKII+ | Cloning vector, Ampr | Stratagene |
| pCVD442 | oriR6K mobRP4 bla sacB | 4 |
| pWM91 | oriR6K oriTRP4bla sacB | 22 |
| pRKlac290 | Broad-host-range vector with promoterless lacZ | 28 |
| pWKS30-Tc | Low-copy-number broad-host-range vector, Tcr | 14 |
| pJK4 | pWKS30-Tc with parAB upstream region and parA | This study |
| pSD15 | VC2773 in-frame deletion construct in pCVD442 | This study |
| pJK11 | VC2061 in-frame deletion construct in pWM91 | This study |
| pSD19 | pRKlac290 with 450-bp parAB upstream region | This study |
| pDAG114 | Stable mini-F, Cmr | 25 |
| pDAG203 | Unstable mini-F, Δ(sopOPABC) Cmr | 5 |
| pSD38 | pDAG203 with IR1 cloned into AflIII-Bsu36I sites | This study |
| pSD39 | pDAG203 with IR2 cloned into AflIII-Bsu36I sites | This study |
| pSD44 | pDAG203 with IR3 cloned into AflIII-Bsu36I sites | This study |
| pSD25 | pBluescript II SK(+) with parAB and upstream region | This study |
| pSD9 | pBluescript II SK(+) with parB and in-frame deletion of parA | This study |
| pSD21 | pBluescript II SK(+) with parA and in-frame deletion of parB | This study |
Ampr, ampicillin resistance; Tcr, tetracycline resistance; Cmr, chloramphenicol resistance; Smr, streptomycin resistance.
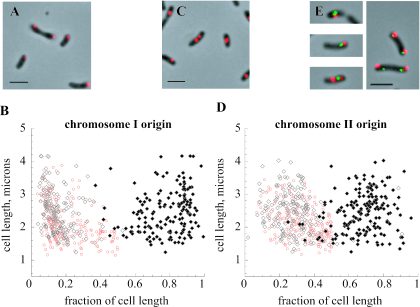
The positions of fluorescent foci in wild-type cells were measured from the nearest cell pole and expressed as fractional proportions of the length of the cell body as visualized by phase-contrast microscopy (Fig. 1). In cells containing a single fluorescent focus, the large chromosome origin (oriCI) was often positioned centrally in the smallest cells but could be found near the cell pole in larger cells (median position, 19% of cell length). The origin of the second chromosome (oriCII) was also centrally located in the smallest cells but was found closer to the quarter-cell position in longer cells (median, 34% of cell length). In cells with two foci, the large chromosome origin was asymmetrically localized at both polar and subpolar positions (medians, 13% and 80%, respectively), while the small chromosome origin was found, on average, near the quarter-cell positions (medians, 28% and 68%, respectively). These results were confirmed using reciprocal labeling, and no signal was detected with a probe made from empty vector.
FIG. 1.
Positions of V. cholerae chromosome origins in wild-type strain N16961. (A and B) Positioning of the large chromosome origin in cells. (C and D) Positioning of the small chromosome origin. Graphs show positions of signals in cells containing one (red) or two (black) fluorescent foci; they depict the lengths of the cells on the y axis and the positions of the fluorescent foci along the length of the cell on the x axis. Photomicrographs depict the Cy3 probe signals superimposed on phase-contrast images of cell bodies. Bars, 2 μm. (E) Examples of dually labeled cells, where the oriCI probe signal is red (Cy3) and the oriCII probe signal is green (fluorescein).
The distribution of origin positions in our study is consistent with that found by Fogel and Waldor (6). The median position for cells with single foci was closer to the cell pole in our work, but this may reflect incomplete detection of second foci in larger cells (typically, 90 to 95% of cells contained foci). The distributions of large chromosome and small chromosome origin focus positions overlapped to a greater extent in this work; however, an evaluation of 400 sets of closely paired foci showed that the majority (72%) were noncoincident (Fig. 1E) (11). We observed dual oriCII foci more readily in small cells by comparison with the live-cell technique. While integrated arrays may have some effect on the timing of segregation in the live-cell model, we also found cells in which two oriCI foci were present with a single oriCII focus, consistent with a model where duplicated oriCII foci segregate later than oriCI. Our findings may also be related to faster growth conditions, as the cultures had a doubling time of approximately 25 min. Indeed, while the majority of cells contained one or two fluorescent foci for each probe, we observed that a small number contained three or four foci.
A deletion mutant lacking the parA homolog VC2773 is viable and has altered oriCI positioning.
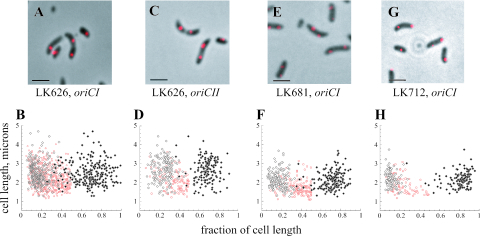
The VC2773 and VC2772 genes constitute a parAB locus near the origin of the large V. cholerae chromosome. To examine the role of VC2773, the gene was replaced with an in-frame deletion construct lacking the sequence for amino acids 11 to 244 of the predicted protein. All constructs described in this study were confirmed by restriction digestion and sequencing. As needed, constructs were cloned into the suicide vector pCVD442 (4) or pWM91 (22), transferred into V. cholerae by conjugation with the E. coli donor strain SM10λpir, and used for allelic exchange with sucrose counterselection (7). As shown in Fig. 2A and B, positioning of oriCI in the VC2773 deletion strain (LK626) was less polar than that in the wild-type parent strain; the median position in cells with single foci was 26% of the cell length. In cells with two foci, we often observed that neither focus was at the pole of the cell. However, oriCI still appeared able to attain a position closer to the cell pole than that of oriCII. The localization pattern of oriCII was unchanged from that of the wild-type strain (Fig. 2C and D). In cells with two foci, we assessed the absolute distances from the nearest pole of the cell of all foci. oriCI foci were found at median distances of 15% of cell length in strain N16961 and 23% of cell length in strain LK626, as opposed to 28.8% for oriCII foci in both strains; these differences were statistically significant. For cells with two foci, the median distances between the foci were 65% of cell length for N16961 and 51% of cell length for LK626.
FIG. 2.
Positions of V. cholerae chromosome origins in strains LK626, LK681, and LK712. Graphs show positions of signals in cells containing one (red) or two (black) fluorescent foci. The labeling of axes and scale are the same as those in Fig. 1. (A and B) Positioning of the large chromosome origin in LK626. (C and D) Positioning of the small chromosome origin in LK626. (E and F) LK681 (N16961 with pJK4) labeled with the oriCI probe. (G and H) LK712 (LK626 with “knocked-in” VC2773) labeled with the oriCI probe.
While the doubling times in LB medium were indistinguishable between the wild type and LK626, there was a small but reproducible slowing of the doubling time in minimal medium (AB minimal medium with 0.2% glycerol) (2), with doubling times of 97.4 min for strain N16961 and 106.8 min for LK626. The cells were normally motile, and we did not observe a cell division defect. Studies of chromosomal par genes in other bacteria have suggested that they have various roles in development and at growth-phase transitions. Specifically, mutations in one or both genes of the parAB locus on the single chromosome of P. putida give rise to a low frequency of anucleate cells during exponential growth and a significantly increased number at the entrance to stationary phase in minimal medium (9, 19). There was no significant difference in average cell size between N16961 and LK626; the culture density and CFU correlated well with each other and were comparable to those of the wild-type parent strain in both rich and minimal media. Because the loss of the large chromosome would yield cells which still stained with 4′,6′-diamidino-2-phenylindole (DAPI), we also utilized propidium iodide staining, which did not reveal any increase in inviable cells for LK626 over a range of growth, including the transition to stationary phase and late stationary phase (data not shown). We also did not note any increase in cells that did not give a signal for oriCI.
The parA homolog VC2061 does not appear to play a role in origin positioning.
We assessed the possibility of functional redundancy with another parA homolog, VC2061, carried on the large chromosome of V. cholerae. We created an in-frame deletion of VC2061, eliminating the sequence encoding amino acids 9 to 166. A VC2061 deletion strain was recently independently reported; although the gene is located in an operon with flagellar and chemotaxis genes, it does not appear to play a significant role in motility or flagellar biogenesis (3). Our ΔVC2061 strain (LK705) had normal growth rates in both rich and minimal media, with wild-type oriCI and oriCII positioning. A ΔVC2061 ΔVC2773 double mutant (LK706) had no additive positioning phenotype (data not shown). In minimal and rich media, the growth rate of LK705 was similar to that of the N16961 parent strain, and the growth rate of LK706 was similar to that of LK626. Thus, the VC2061 parA homolog does not appear to influence origin positioning.
Multiple copies of VC2773 alter oriCI positioning in wild-type cells.
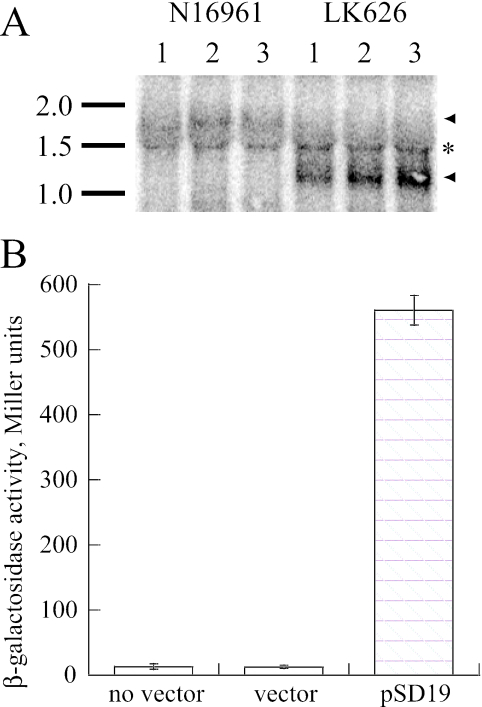
Our aim was to complement the VC2773 mutation by using its endogenous promoter. The VC2773-VC2772 locus on the large V. cholerae chromosome is carried immediately downstream of gidAB, suggesting that these four genes may form an operon. We performed Northern blotting, using a 374-bp internal fragment of VC2772 as a probe, and detected a transcript of approximately 1.8 kb in the wild-type strain that was appropriately shortened in LK626 (Fig. 3A); thus, VC2773-VC2772 is an independently transcribed operon. We cloned a 450-bp segment of DNA upstream of the VC2773 coding region into the plasmid pRKlac290 (28), which contains a promoterless lacZ gene. This construct (pSD19) was tested in KFV10, a ΔlacZ derivative of N16961 (7), and promoter activity was detected in beta-galactosidase assays (27) (Fig. 3B).
FIG. 3.
Isolation of the promoter for the V. cholerae chromosome 1 parAB locus. (A) Northern blot showing transcripts detected using a parB probe in three separate samples of each strain. The positions of RNA markers and their sizes in kb are depicted to the side of the blot. Arrowheads, transcripts; asterisk, background band from rRNA. (B) Beta-galactosidase activities (in Miller units) assayed in bacteria grown in LB at 37°C to an OD600 of 0.4 to 0.6, using the following strains: wild-type parent strain KFV10, KFV10 with a plasmid containing a promoterless lacZ gene (pRKlac290), and KFV10 with pSD19 (pRKlac290 containing the upstream region of parA) (averages of three experiments are shown).
To complement the VC2773 mutation, we cloned the promoter and VC2773 coding region into the low-copy-number vector pWKS30-Tc (14). The position of oriCI was not restored to the wild-type position in the LK626 background; in fact, the construct caused aberrant, less polar positioning of the large chromosome origin in the N16961 wild-type strain (LK681) (Fig. 2E and F) that was not observed in a strain containing the empty vector. In cells with single foci, oriCI was found at a mean distance of 34% of cell length, and in cells with two foci, oriCI was found at 18% and 71% of cell length. The positioning of oriCII was not affected. When we regenerated a wild-type copy of VC2773 in the deletion strain through allelic exchange (strain LK712), oriCI was restored to its wild-type position (Fig. 2G and H). Thus, both excess copies and the absence of VC2773 appear to lead to isolated oriCI positioning abnormalities. LK681 had normal morphology and growth; in contrast, in P. putida, parAB overexpression causes anucleate cells and abnormal morphology during deceleration and stationary phase (9). In C. crescentus, constitutive overexpression of either ParA or ParB causes cell division defects that are less severe when both proteins are overexpressed together, suggesting that proper stoichiometry between these two components is also important (23).
VC2773 and VC2772 encode gene products that exhibit activity with IRs partition located on the large chromosome of V. cholerae.
We attempted to investigate the role of parB (VC2772) as well as that of parA; however, despite extensive efforts, we were unable to generate a parB or parAB deletion mutant. Thus, to study the partitioning activity of the parAB locus, we assayed its function using the unstable mini-F plasmid pDAG203, which has been employed in characterizing parABS systems from other bacteria (5, 9). Plasmid stability assays were performed according to the method of Dubarry et al. (5). We tested pDAG203 and its derivatives in E. coli DH10B in the presence of coresident plasmids carrying parAB genes (Fig. 4). To find candidate parS sequences, we searched the genome of V. cholerae for inverted repeats (IRs) with similarity to chromosomal parS sequences characterized in other bacteria (5, 9, 20). Three highly similar IRs were located only on the large chromosome, approximately 8° from the origin: they are IR1 (5′ CGTTTCACGTGAAACA [bp 62423 to 62438]), IR2 (5′ TGTTTCACGTGAAACC [bp 66117 to 66132]), and IR3 (5′ CGTTTCACGTGGAACA [bp 69417 to 69432]). We used complementary oligonucleotides with appropriate overhangs to clone these repeats individually into pDAG203, generating pSD38, pSD39, and pSD44, respectively.
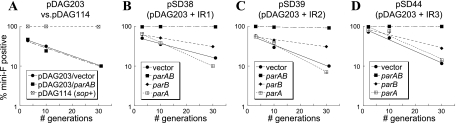
FIG. 4.
Stabilization of mini-F plasmids containing putative parS sequences by coresident plasmids carrying the VC2773-VC2772 parAB locus. Experiments were performed multiply, and the graphs are from a representative experiment. Bacteria were grown in M9-CSA with chloramphenicol to maintain the mini-F derivative; at an OD of ∼0.3, they were back diluted into medium lacking chloramphenicol (time zero) and plated at the specified generation times. Colonies were patched onto chloramphenicol to assay for the presence of the mini-F plasmid. The y axes in all panels show the percentages of colonies still retaining the mini-F derivative. (A) Stability of the sop+ mini-F plasmid pDAG114, as well as its unstable derivative pDAG203, in the presence of a coresident pBluescript vector or pSD25 (carrying both parA and parB). (B to D) Stabilities of pDS38, pSD39, and pSD44 (pDAG203 carrying IR1, IR2, and IR3, respectively). Each plasmid was assayed in the presence of empty pBluescript vector, pSD9 (with an in-frame deletion of VC2773, thus carrying parB), pSD21 (with an in-frame deletion of VC2772, thus carrying parA), or pSD25 (carrying both parA and parB). The legend for each graph indicates the par genes supplied in trans.
Consistent with previously published results, pDAG203 was unstable in host strain DH10B, while its parent sop+ plasmid, pDAG114, was completely stable (5, 25). pDAG203 remained unstable in the presence of a pBluescript vector and its derivative, pSD25, containing the VC2773-VC2772 parAB locus and its upstream region (Fig. 4A). As shown in Fig. 4B to D, pSD38, pSD39, and pSD44 were all stable in the presence of pSD25. However, they were unstable in the presence of empty vector, pSD9 (with an in-frame deletion of VC2773), or pSD21 (with an in-frame deletion of VC2772, eliminating the sequence encoding amino acids 19 to 280). Thus, stabilization of an unstable mini-F plasmid requires the presence of a parS-like inverted repeat sequence and both VC2773 and VC2772. A recent study of the four parABS systems in the multipartite genome of Burkholderia cenocepacia showed that each parAB locus displays partitioning activity with its cognate parS gene; our results suggest that this may also be the case for V. cholerae, although, as in B. cenocepacia, interactions between components of par systems in the same genome cannot be ruled out (5).
Concluding remarks.
In conclusion, we have used a fixed-cell FISH model to study origin positioning in V. cholerae and observed that deletion or extra copies of VC2773, a parA gene homolog found in a parAB operon on the large V. cholerae chromosome, can selectively affect large chromosome origin positioning while leaving the position of the small chromosome origin unaffected. Thus, the function of VC2773 appears to be specific to the replicon on which it is located, and its operon forms a parABS system with three inverted repeats unique to the large V. cholerae chromosome. The subtlety of the mutant phenotype is not attributable to the presence of a second parA homolog carried on the large chromosome. However, Lee and Grossman have recently demonstrated that in B. subtilis, a soj mutation in an smc mutant background reveals a significant partitioning defect not seen in a Δsoj single mutant (17). Thus, as is the case for a growing number of bacteria, it will be of great interest to identify the additional factors that influence the critical process of proper origin placement in V. cholerae (8).
Acknowledgments
We are grateful to Karla Fullner Satchell and David Lane for strains, plasmids, and advice and to V. Viswanathan, L. Kenney, our lab members, and the reviewers for helpful comments.
This work was supported by grants from the Glaxo Institute for Digestive Health and the Department of Veterans Affairs (Merit Review) and by University of Illinois funds.
REFERENCES
- 1.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark, D., and O. Maaloe. 1967. DNA replication and the cell cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 3.Correa, N. E., F. Peng, and K. E. Klose. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J. Bacteriol. 187:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubarry, N., F. Pasta, and D. Lane. 2006. ParABS systems of the four replicons of Burkholderia cenocepacia: new chromosome centromeres confer partition specificity. J. Bacteriol. 188:1489-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Fiebig, A., K. Keren, and J. A. Theriot. 2006. Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes. Mol. Microbiol. 60:1164-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fogel, M. A., and M. K. Waldor. 2005. Distinct segregation dynamics of the two Vibrio cholerae chromosomes. Mol. Microbiol. 55:125-136. [DOI] [PubMed] [Google Scholar]
- 7.Fullner, K. J., and J. J. Mekalanos. 1999. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect. Immun. 67:1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 9.Godfrin-Estevenon, A. M., F. Pasta, and D. Lane. 2002. The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Mol. Microbiol. 43:39-49. [DOI] [PubMed] [Google Scholar]
- 10.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, T. Q., Z. Zhong, S. Aung, and J. Pogliano. 2002. Compatible bacterial plasmids are targeted to independent cellular locations in Escherichia coli. EMBO J. 21:1864-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ireton, K., N. W. Gunther IV, and A. D. Grossman. 1994. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176:5320-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen, R. B., and L. Shapiro. 1999. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl. Acad. Sci. USA 96:10661-10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julio, S. M., D. M. Heithoff, D. Provenzano, K. E. Klose, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2001. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect. Immun. 69:7610-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahng, L. S., and L. Shapiro. 2003. Polar localization of replicon origins in the multipartite genomes of Agrobacterium tumefaciens and Sinorhizobium meliloti. J. Bacteriol. 185:3384-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, H. J., M. J. Calcutt, F. J. Schmidt, and K. F. Chater. 2000. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J. Bacteriol. 182:1313-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, P. S., and A. D. Grossman. 2006. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol. Microbiol. 60:853-869. [DOI] [PubMed] [Google Scholar]
- 18.Leonard, T. A., J. Moller-Jensen, and J. Lowe. 2005. Towards understanding the molecular basis of bacterial DNA segregation. Philos. Trans. R. Soc. Lond. B 360:523-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis, R. A., C. R. Bignell, W. Zeng, A. C. Jones, and C. M. Thomas. 2002. Chromosome loss from par mutants of Pseudomonas putida depends on growth medium and phase of growth. Microbiology 148:537-548. [DOI] [PubMed] [Google Scholar]
- 20.Lin, D. C., and A. D. Grossman. 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92:675-685. [DOI] [PubMed] [Google Scholar]
- 21.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 22.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 23.Mohl, D. A., and J. W. Gober. 1997. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88:675-684. [DOI] [PubMed] [Google Scholar]
- 24.Niki, H., and S. Hiraga. 1998. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev. 12:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravin, N., and D. Lane. 1999. Partition of the linear plasmid N15: interactions of N15 partition functions with the sop locus of the F plasmid. J. Bacteriol. 181:6898-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Stephens, C. M., G. Zweiger, and L. Shapiro. 1995. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J. Bacteriol. 177:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webb, C. D., A. Teleman, S. Gordon, A. Straight, A. Belmont, D. C. Lin, A. D. Grossman, A. Wright, and R. Losick. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88:667-674. [DOI] [PubMed] [Google Scholar]