Abstract
The immortalization of human B lymphocytes by Epstein-Barr virus (EBV) requires the virus-encoded transactivator EBNA2 and the products of both viral and cellular genes which serve as EBNA2 targets. In this study, we identified BATF as a cellular gene that is up-regulated dramatically within 24 h following the infection of established and primary human B cells with EBV. The transactivation of BATF is mediated by EBNA2 in a B-cell-specific manner and is duplicated in non-EBV-infected B cells by the expression of mammalian Notch proteins. In contrast to other target genes activated by EBNA2, the BATF gene encodes a member of the AP-1 family of transcription factors that functions as a negative regulator of AP-1 activity and as an antagonist of cell growth. A potential role for BATF in promoting EBV latency is supported by studies in which BATF was shown to negatively impact the expression of a BZLF1 reporter gene and to reduce the frequency of lytic replication in latently infected cells. The identification of BATF as a cellular target of EBV provides important new information on how programs of viral and cellular gene expression may be coordinated to promote viral latency and control lytic-cycle entry.
Epstein-Barr virus (EBV) is causally associated with Burkitt's lymphoma (BL), nasopharyngeal carcinoma, unusual T-cell lymphomas, and other lymphoproliferative diseases (reviewed in references 35 and 59). EBV has a restricted host range and primarily infects CD21+ B lymphocytes and epithelial cells (reviewed in references 37 and 59). EBV establishes a latent infection in B cells in which the viral episome persists and expresses proteins that promote indefinite B-cell proliferation (immortalization) (reviewed in reference 35). Latency in cultured B cells is associated with the expression of six nuclear proteins (EBNAs). Of these, only EBNA1, EBNA2, EBNA3A, and EBNA3C are essential for immortalization (5, 18, 73). In addition to the EBNAs, three integral membrane proteins are expressed during latent infection (latent membrane proteins [LMPs]). Of these, only LMP1 is required for B-cell immortalization, and it serves as the major transforming protein encoded by the EBV genome (3, 34, 75).
EBNA2 is one of the first viral gene products to be expressed after EBV infection and plays a crucial role in the process by transcriptionally activating viral genes encoding EBNA1, -3A, -3B, -3C, LMP1, and itself (reviewed in reference 35). In addition to viral targets, EBNA2 can enhance or repress the transcription of cellular genes whose encoded proteins contribute to the alterations in cellular growth that are associated with EBV infection. EBNA2 does not bind DNA directly but instead activates gene expression by interacting with the cellular CBF1 (RBP-Jκ) protein complex (17, 21, 42, 74, 83) to mask its transcriptional repressor function (25). The ability of cellular Notch proteins to activate gene transcription is also facilitated by its interaction with CBF1 (72; reviewed in references 2 and 51), implying that there may be overlap between the genes induced by Notch signaling and those up-regulated during EBV latency (26). In support of this, constitutively active Notch can substitute for EBNA2 in B-cell immortalization (16, 22). Interestingly, of the identified cellular genetic targets of EBNA2 in B cells (8, 30, 32, 38, 69), only CD21 is a common target for up-regulation by EBNA2 and activated Notch (71).
In this paper, we report the novel finding that the human gene encoding BATF is up-regulated in B lymphocytes following infection with EBV. BATF is a member of the AP-1/ATF superfamily of basic leucine zipper transcription factors and forms heterodimers with the Jun proteins to bind preferentially to AP-1 consensus sites (9, 10). In vitro and in vivo studies have demonstrated that BATF:Jun heterodimers have a reduced transcriptional activity relative to Fos:Jun heterodimers and thus inhibit the expression of AP-1 target genes and retard AP-1-mediated cell growth (10, 79). BATF is expressed predominantly in hematopoietic tissues, suggesting a tissue-specific function for BATF as a negative regulator of AP-1 activity.
The induction of BATF gene expression by EBV is mediated by EBNA2 and is duplicated in non-EBV-infected B cells by an activated form of the Notch protein. Interestingly, the induction of BATF by EBNA2 and Notch is B cell specific, suggesting that additional B-cell transcription factors (or signaling events) are required for BATF gene expression. The documented role of AP-1 in the transactivation of several EBV genes, including BZLF1 (reviewed in references 35 and 68), provides a context for investigating the biological significance of BATF induction in EBV-infected cells. Our studies demonstrate that BATF negatively impacts the expression of a BZLF1 reporter gene and reduces the efficiency of lytic-cycle induction. The identification of BATF as a target of EBNA2 activity early in infection provides important new insight into how coordinated programs of viral and cellular gene expression can positively impact viral latency by decreasing the number of cells entering the lytic cycle in response to lytic-cycle induction.
MATERIALS AND METHODS
Cell culture and EBV infections.
BJAB (EBV-negative B-cell lymphoma) (36) and 721 (in vitro immortalized lymphoblastoid cell line) (65) cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 100 U of penicillin/ml-100 μg of streptomycin/ml (P/S). DG75 (EBV-negative B-cell lymphoma) (4), Raji (EBV-positive BL) (ATCC CCL86), B95-8 (EBV-positive marmoset B-cell line) (62), and HH514 (EBV-positive BL clone derived from the P3HR1 clone of Jijoye BL) (56) cells were maintained in RPMI 1640 medium supplemented with 10% calf serum and P/S. RPMI 2650 cells (ATCC CCL30) were cultured in minimum essential medium-10% FBS-P/S, and HeLa cells (ATCC CCL2) were grown in basal modified Eagle medium-10% FBS-P/S. Primary human B lymphocytes were isolated from 200 ml of whole blood by Ficoll-Hypaque gradient, followed by positive selection with bovine serum albumin (63), and were cultured in RPMI 1640 medium-10% FBS-P/S. All medium components were obtained from Gibco-BRL Life Technologies.
Virus supernatants were prepared from B95-8 and HH514 cells as described previously (11). BJAB cells at a density of 2 × 105 cells/ml were incubated with virus supernatant for 3 h at 37°C with continuous rocking. Infected cells were harvested by centrifugation and cultured in complete RPMI 1640 medium. The efficiency of infection was monitored 72 h postinfection by EBNA staining (58) and/or by immunoblotting for LMP1 expression as described previously (47). Primary B lymphocytes were infected with B95-8 virus as described above and cultured for 5 days prior to analysis.
RNA analysis.
Total RNA was prepared from cells as described previously (53), and poly(A+) mRNA was isolated by using the FastTrack 2.0 kit (Invitrogen). RNA hybridizations were performed as described previously (53) by using 2 μg of poly(A+) mRNA and 32P-labeled cDNA probes for BATF (10) and human β-actin (Clontech) mRNA. Reverse transcription (RT) involved 1 μg of total RNA, 50 pmol of oligo(dT) primer, and RT reagents purchased from Gibco-BRL. The PCR was performed with 1/10 volume of each RT reaction and 20 pmol of 5′ (GGAGCAGTCCCTCTGCACC) and 3′ (CAGTTCCTCTGTGAGCTGC) BATF primers in 50 μl containing 1× buffer (Promega), 4 mM MgCl2, and 5 U of Taq polymerase (Promega). Thirty cycles of denaturation at 94°C for 1 min, annealing at 60°C for 30 s, and extending at 72°C for 30 s, with the final extension performed for 5 min, defined each amplification reaction. Amplification with 5′ (CGAGCACGGCATCGTCACC) and 3′ (GTCAGGCAGCTCGTAGCTC) β-actin primers served as the control. Semiquantitative PCR was performed as described previously (54) by using the PCR Mimic construction kit (Clontech) to generate a heterologous DNA standard amplified by the BATF primers described above. An equal amount of mimic DNA was added to each BATF reaction. Amplification was performed in the presence of radiolabeled nucleotide and quantified by densitometry. The results were normalized by using the signal from the internal mimic.
Plasmids.
Flag EBNA2, a simian virus 40-driven EBNA2 expression vector, and p288, an simian virus 40-driven LMP1 expression vector, have been described previously (43, 80). The CBF1-chloramphenicol acetyltransferase (CAT) reporter gene and the pPDL151 and pPDL152 plasmids encoding CBF1-interacting and noninteracting EBNA2 proteins, respectively, were obtained from P. Ling (42, 43). mNotch IC expresses a constitutively active mouse Notch1 protein and was a gift of J. Nye (50). Human Notch1 expression plasmids for activated Notch1 (NIC), an NIC variant appended with a strong nuclear localization signal (NLS), and an NIC NLS protein deleted for the CBF1 interaction domain (ΔR) were obtained from T. Capobianco (29). pDCRBATF expresses an amino-terminal, hemagglutinin antigen (HA)-tagged BATF protein (9). pCS2+MT-BATF was generated as part of this study to express a BATF protein tagged at the N terminus with seven tandem Myc epitopes (MT). pCS2+MT-BATF(1-49) encodes a nonfunctional derivative of BATF lacking the basic leucine zipper region. CMV-c-Jun-HA was a gift from S. Rhodes (Indiana University-Purdue University at Indianapolis). BMRF1-CAT and pCMV-Z were obtained from S. Kenney (81). Luciferase derivatives of the ZII and mutant ZII (mut ZII) CAT reporter genes described in Ruf and Rawlins (60) were generously provided by I. Ruf. The pCMV-lacZ and pRL normalization vectors used in the transient expression studies were obtained from S. Konieczny (Purdue University) and Promega, respectively.
Gene transfer and expression analysis.
A total of 2 × 107 BJAB cells in a volume of 400 μl of RPMI 1640 medium were electroporated at 200 V and 975 μF with 20 μg of the indicated plasmid DNA and transferred to complete medium for 48 h. RNA was prepared from 107 cells. Protein extracts were prepared from the remaining cells, normalized for protein content, and resolved by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE). Proteins were transferred to a nitrocellulose membrane and immunoblotted by using the transfer, blocking, hybridization, and wash conditions described in reference 57 and a 1:500 dilution of monoclonal EBNA2 antibody (Novocastra Laboratories) or a 1:5,000 dilution of LMP1 antiserum as primary antibody. Immune complexes were detected by using peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) or anti-rabbit IgG (Vector Laboratories), enhanced chemiluminescence (Amersham Pharmacia Biotech), and autoradiography. Gene transfer to the RPMI 2650 and HeLa cell lines utilized standard Ca2PO4-DNA transient transfection as described previously (53). Cell extracts were prepared, normalized to the β-galactosidase activity of pCMV-lacZ, and assayed for CAT activity as described previously (53). A total of 2 × 106 DG75 cells in 250 μl were electroporated at 310 V and 950 μF with the amounts of DNA indicated in the legend to Fig. 6. Forty-eight hours after transfer of extracts to complete medium, luciferase activities were measured and normalized by using a dual-activity luciferase kit (Promega). All reporter gene assays were performed a minimum of three times in duplicate.
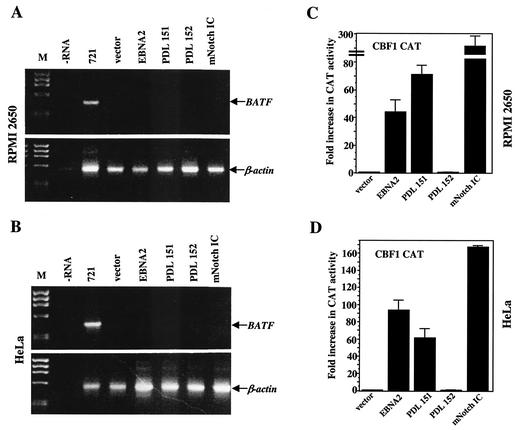
FIG. 6.
Induction of BATF mRNA expression is B cell specific. The adherent RPMI 2650 cell line (A and C) and the HeLa cell line (B and D) were transfected as described in Materials and Methods with 5 μg of the indicated activators, 5 μg of the CBF-1 CAT reporter, and 2 μg of a β-galactosidase expression plasmid. Forty-eight hours following transfection, the cells were harvested for the preparation of RNA and protein. (A and B) RNA was analyzed by RT-PCR for BATF. As a control, the same RT samples were analyzed for the expression of β-actin mRNA by PCR. RNA from 721 B cells provided a positive control. A no-RNA (−RNA) reaction and total RNA from cells transfected with empty vector DNA served as the negative controls. ØX174 HaeIII DNA (M) was the molecular weight marker. Results indicate no activation of BATF gene expression in either cell line. (C and D) Protein extracts were normalized to β-galactosidase activity and assayed for CAT as described in Materials and Methods. The transfection was performed a minimum of three independent times to arrive at the average activities presented. Error bars indicate the standard errors of the means.
BATF protein expression analysis.
Total protein was prepared from DG75 and 721 cells by lysis in standard radioimmunoprecipitation assay (RIPA) buffer plus protease inhibitors (Sigma) and normalized for protein content by a Bio-Rad protein assay. Total protein from BJAB cells mock infected or infected with B95-8 virus was harvested by lysis in 1.5× SDS-PAGE sample buffer, separated by SDS-15% PAGE, transferred to a polyvinylidene difluoride membrane, and blotted by following the procedure described in reference 57. A 1:250 dilution of affinity-purified rabbit anti-BATF antiserum or a 1:1,000 dilution of LMP1 antiserum was used as primary antibody. Immune complexes were detected by using a 1:5,000 dilution of peroxidase-conjugated goat anti-mouse IgG or anti-rabbit IgG (Vector Laboratories), SuperSignal West Dura extended-duration substrate (Pierce), and autoradiography. The BJAB blot was stripped by incubating it for 30 min at room temperature in 2% (wt/vol) SDS-62.5 mM Tris-HCl (pH 6.8)-100 mM β-mercaptoethanol and then reprobed as described above with a 1:5,000 dilution of clone C4 mouse monoclonal β-actin antibody (ICN) and goat anti-mouse IgG secondary antibody.
Electrophoretic mobility shift assays (EMSA).
Complementary double-stranded oligonucleotides containing a consensus AP-1 binding site (Promega) were labeled by using T4 DNA polynucleotide kinase and [γ-32P]ATP (6,000 Ci/mmol; NEN Life Science Products). Nuclear extracts were prepared from HH514 cells and 721 cells as described previously (39). A total of 10 μg of nuclear extract was incubated with 40 fmol (1.0 × 105 cpm) of probe DNA on ice for 15 min in 20 μl of buffer containing 10 mM HEPES (pH 7.9), 50 mM KCl, 0.1 mM EDTA, 5 mM dithiothreitol, 5 mM MgCl2, 10% glycerol, 1 μg of bovine serum albumin, and 2 μg of poly(dI-dC) (Sigma). Binding specificity was assayed by using a 100-fold molar excess of cold AP-1 or nonspecific E-box competitor DNA (positive strand, AGTGTTAATCCCCAGTTGTTTTTGAAGTTG; negative strand, CAACTTCAAAAACAACTGGGGATTACT). For the supershift experiments, the incubation with AP-1 DNA was followed by the addition of 4 μl of affinity-purified rabbit polyclonal anti-BATF antiserum (K. Williams and E. Taparowsky, unpublished data) or 4 μl of preimmune rabbit serum (PI) and a further incubation on ice for 15 min. Protein/DNA complexes were resolved on a 4% native polyacrylamide gel in Tris-glycine buffer (25 mM Tris, 190 mM glycine, 1 mM EDTA, pH 8.5) and visualized by autoradiography.
Coimmunoprecipitation assays.
DG75 cells were electroporated as described above with 10 μg of pCS2+MT-BATF, 10 μg of CMV-c-Jun-HA, or both plasmids; transferred to complete medium; and incubated for 36 h, after which the cells were lysed in standard RIPA buffer plus protease inhibitors (Sigma) and normalized for protein content. A fraction of each normalized extract was resolved by SDS-12.5% PAGE, transferred to a nitrocellulose membrane (Bio-Rad), and immunoblotted with a 1:1,000 dilution of anti-Myc monoclonal antibody 9E10 (Developmental Studies Hybridoma Bank) or anti-HA monoclonal antibody (12CA; Boehringer Mannheim) by using the transfer, blocking, hybridization, and wash conditions outlined in reference 57. Immune complexes were detected by incubating extracts for 60 min with a 1:5,000 dilution of peroxidase-conjugated goat anti-mouse IgG (Vector Laboratories) and visualized with SuperSignal chemiluminescence reagent (Pierce) and autoradiography. The remaining volume of each extract was incubated with anti-HA monoclonal antibody bound to protein A-Sepharose (Amersham Pharmacia Biotech) for 90 min at 4°C. Immunoprecipitates were washed extensively with RIPA buffer plus protease inhibitors, and proteins were eluted by boiling for 5 min in 1.5× SDS sample buffer. Proteins were resolved by SDS-12.5% PAGE, transferred to a nitrocellulose membrane, and immunoblotted with anti-HA antibody as described above. The blot was stripped by incubating it for 30 min at 60°C in 2% (wt/vol) SDS-62.5 mM Tris-HCl (pH 6.8)-100 mM β-mercaptoethanol and reprobed as described above by using the 9E10 anti-Myc antibody.
Immunofluorescence analysis.
HH514 cells were electroporated with 5 μg of plasmid DNA encoding MT-tagged wild-type BATF (MT-BATF) or a nonfunctional derivative of BATF [MT-BATF(1-49)] in which the bZIP dimerization domain had been deleted. After 24 h in complete medium, EBV's lytic cycle was induced with 20 ng of 12-O-tetradecanoylphorbol-13-acetate (TPA)/ml-3.5 mM sodium butyrate. Seventy-two hours after electroporation, cells were washed three times in phosphate-buffered saline and applied to microscope slides (104 cells/μl) and dried. Cells were fixed in acetone (−20°C) and were costained with a 1:10 dilution of anti-gp350/220 mouse monoclonal antibody (a gift from L. Hutt-Fletcher) and a 1:200 dilution of anti-mouse Alexa Fluor 594-conjugated secondary antibody (Molecular Probes) and with a 1:50 dilution of anti-Myc antibody (A14; Santa Cruz Biotechnology) and a 1:50 dilution of anti-rabbit fluorescein isothiocyanate-conjugated secondary antibody (Sigma). Slides were mounted by using VECTASHIELD with DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratories), viewed with a Nikon Eclipse E800 fluorescence microscope at a 40× lens objective, and photographed at 100× magnification with a Cooke SensiCam Digital camera and SlideBook software.
RESULTS
EBV infection induces BATF mRNA and protein in human B cells.
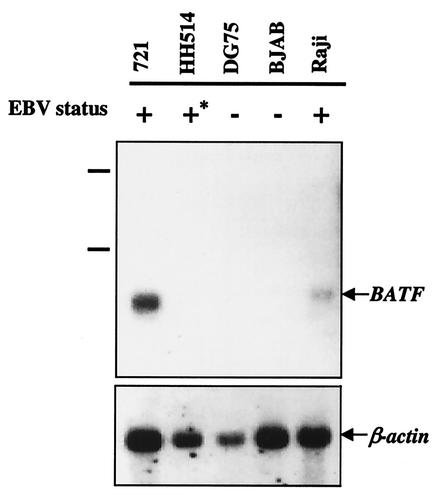
The BATF gene is expressed predominantly in human hematopoietic tissues and in the BL cell line Raji (9, 10). The elevated level of BATF expression in Raji, an EBV+ B-cell line, prompted us to investigate if BATF gene transcription is a general feature of EBV-infected cells. Poly(A+) mRNA prepared from a panel of human B-cell lines was analyzed by Northern blot hybridization. As shown in Fig. 1, the two cell lines harboring a fully functional EBV episome (Raji and 721) expressed BATF while the lines negative for EBV (BJAB and DG75) did not. Interestingly, BATF mRNA was not detected in HH514 cells, a human BL cell line harboring a transformation-defective strain (HH514) of EBV, indicating that events associated with transformation by EBV are necessary to induce endogenous BATF expression.
FIG. 1.
Expression of BATF mRNA in EBV-positive (+) and negative (−) human B cell lines. +*, presence of a transformation-defective EBV episome. Poly(A+) mRNA was hybridized with a radiolabeled probe for BATF mRNA (upper panel) as described in Materials and Methods. The filter was stripped and reprobed for β-actin mRNA as a control (lower panel). The migration of the 18S and 28S rRNA in a total RNA sample resolved in parallel is indicated to the left of the upper panel.
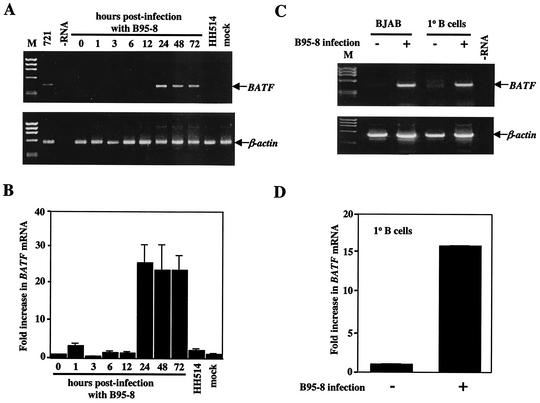
To test if BATF mRNA expression is induced in human B cells in response to EBV infection, BJAB cells were infected with viral supernatant from B95-8 cells (harboring a transforming strain of EBV). Controls included BJAB cells infected with virus supernatant prepared from HH514 cells or mock infected with medium only. The efficiency of viral infection was monitored by immunostaining for EBNAs or LMP1 and was estimated to be 50 to 80% for all groups (data not shown). At specific times following infection, total RNA was isolated and analyzed by RT-PCR for the presence of BATF mRNA. As early as 24 h postinfection, BATF mRNA was induced and remained at an elevated level for the duration of the experiment (Fig. 2A). In contrast, BATF mRNA was not detected in mock-infected cells or in cells infected with HH514 virus. Duplicate samples were analyzed by using semiquantitative RT-PCR, and results showed a 25-fold induction of BATF mRNA 24 h postinfection (Fig. 2B).
FIG. 2.
BATF mRNA is induced in human B cells following infection with EBV. (A) RT-PCR analysis of total RNA isolated at the indicated times (hours) from BJAB cells infected with B95-8 EBV or transformation-defective HH514 EBV. Amplification of BATF from the 721 B-cell line provided a positive control for expression. No RNA (−RNA) and RNA from mock-infected BJAB cells (mock) served as negative controls. ØX174 HaeIII DNA (M) served as a molecular weight marker. Each RT reaction was PCR amplified with primers for β-actin to control for sample integrity and amount. (B) Semiquantitative RT-PCR of the RT samples shown in panel A was performed as described in Material and Methods. The fold increase in BATF mRNA expression was quantified following the densitometry of ethidium bromide-stained gels. The experiment was performed three independent times with error bars indicating the standard error of the mean for each averaged set of values. (C) Primary (1° B) cells were isolated from human blood and mock infected (−) or infected with B95-8 EBV (+) as described in Materials and Methods. Total RNA from the primary B cells or control BJAB cells was isolated and examined for BATF mRNA expression by RT-PCR (upper panel). The amplification of β-actin served as the positive control. A no-RNA (−RNA) reaction served as the negative control. ØX174 HaeIII DNA (M) provided a marker for molecular weight. (D) Semiquantitative RT-PCR was performed on the 1° B-cell samples, and the fold increase in BATF mRNA was quantified as described for panel B.
In an effort to relate the EBV-induced expression of BATF observed in BJAB cells to the process of EBV infection in vivo, primary B cells were purified from human peripheral blood, treated with B95-8 virus supernatant, and cultured for 5 days prior to the preparation of RNA. Whereas noninfected primary B cells do not express detectable levels of BATF mRNA, cells infected with B95-8 virus rapidly induce BATF gene transcription (Fig. 2C). Semiquantitative RT-PCR was used to estimate the induction at 15 times the normal BATF level (Fig. 2D). These results provide evidence that transcription of the BATF gene is an early response of B lymphocytes to EBV infection in vivo.
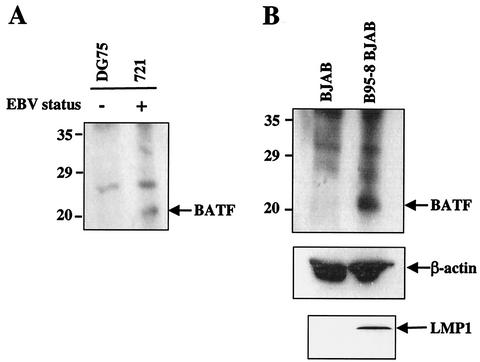
The induction of BATF transcripts observed in EBV-infected cells should reflect an increase in BATF protein. Extracts prepared from DG75 (BATF−) and 721 (BATF+) cells (Fig. 3A) or from BJAB cells mock infected and infected with B95-8 virus (Fig. 3B) were resolved by SDS-PAGE and immunoblotted with a polyclonal rabbit antiserum to BATF (see Materials and Methods for details). The results show expression of BATF protein in the 721 and B95-8 samples. Reprobing the BJAB blot with an antibody to β-actin demonstrated equal loading of the samples; probing a parallel blot with an antibody to LMP1 confirmed the infection of the BJAB cells with EBV.
FIG. 3.
BATF protein is detected in EBV-infected cells. (A) Total protein from 721 (EBV+) and DG75 (EBV−) cells was immunoblotted with anti-BATF antiserum as described in Materials and Methods to detect endogenous BATF protein expression. (B) Total protein from BJAB cells mock infected or infected with B95-8 virus was immunoblotted as described for panel A to detect endogenous BATF protein (top panel). To ensure equal loading of the samples, the membrane was stripped and reprobed with anti-β-actin antibody (middle panel). An immunoblot performed in parallel was probed for LMP1 expression to confirm viral infection of the B95-8 sample (lower panel). The migration of molecular weight markers is indicated to the left of BATF blots.
BATF is a cellular target of the EBNA2 transactivator.
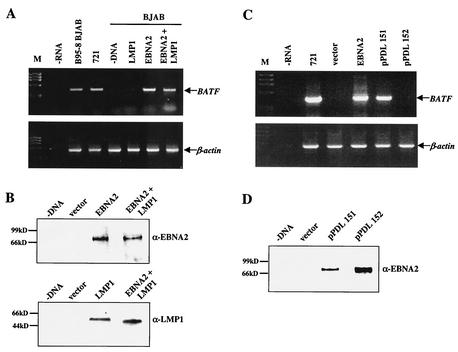
Infection with B95-8, the prototypical transforming strain of EBV, resulted in a dramatic up-regulation of BATF mRNA and protein. In contrast, infection with the nontransforming HH514 strain had no effect on BATF gene expression (Fig. 2). The HH514 virus is nontransforming due to a deletion of the latent genes EBNA2 and EBNA-LP (18). While EBNA2 is absolutely required for the immortalization of B cells by EBV (17), EBNA-LP plays an ancillary role in latency by enhancing EBNA2 function (19, 49, 52, 67). EBNA2 is a well characterized transactivator that regulates both viral and cellular genes (reviewed in references 35, 37, and 66). Among the viral genes activated by EBNA2 is LMP1 (1, 12, 76), which encodes the major transforming protein of the EBV genome (3, 34, 75). Thus, cells infected with HH514 virus do not express LMP1. To test the possibility that EBNA2 and/or LMP1 is responsible for BATF mRNA induction in B cells, expression vectors for the proteins were introduced into BJAB cells by electroporation. After 48 h, total RNA was prepared and analyzed by RT-PCR. As shown in Fig. 4A and B, BATF mRNA was induced in cells expressing EBNA2 but not in cells expressing LMP1 alone. These data identify the EBNA2 transactivator as both necessary and sufficient to induce BATF gene expression in human B cells.
FIG. 4.
The BATF gene is a target for transactivation by EBNA2. (A) BJAB cells were electroporated as described in Materials and Methods with 10 μg of each indicated plasmid DNA (20 μg of total DNA in each group). Total RNA was isolated after 48 h and analyzed for BATF expression and control β-actin gene expression by RT-PCR. RNA isolated from EBV-infected BJAB cells (B95-8 BJAB) and 721 B cells provided positive controls. A no-RNA reaction (−RNA) and a control BJAB reaction (−DNA) served as the negative controls. ØX174 HaeIII DNA (M) provided a marker for molecular weight. (B) Immunoblot analysis of EBNA2 and LMP1 protein expression in cell extracts prepared from BJAB cells electroporated as described for panel A with the indicated expression plasmids. The migration of molecular mass protein markers is indicated to the left of each autoradiography gel. (C) BJAB cells were electroporated as described for those in panel A with two EBNA2 expression plasmids (EBNA2, pPDL151) and a plasmid directing the production of a nontransactivating EBNA2 variant (pPDL152). Total RNA was isolated after 48 h and analyzed for BATF and β-actin mRNA expression by RT-PCR. RNA from 721 cells served as the positive control. A no-RNA (−RNA) reaction and RNA from BJAB cells electroporated with empty vector DNA served as negative controls. ØX174 HaeIII DNA (M) provided a molecular weight marker. (D) Immunoblot analysis of EBNA2 protein expression in the indicated groups analyzed for BATF gene expression as for that in panel C. The migration of molecular mass protein markers is indicated to the left of the autoradiography gel.
EBNA2 possesses a potent transcription activation domain but lacks a DNA binding domain (6, 7). EBNA2 exerts its transcriptional influence on viral and cellular target genes by interacting with a cellular sequence-specific DNA binding protein called CBF1 or RBP-Jκ (17, 21, 42, 74, 83). To examine if the induction of BATF gene expression relies on the interaction of EBNA2 with CBF1, an expression vector for wild-type EBNA2 (pDL151) and a matched vector for an EBNA2 protein containing a mutation that abolishes binding to CBF1 (pDL152) were expressed in BJAB cells. RT-PCR results show that BATF mRNA was induced only by the fully functional EBNA2 protein (Fig. 4C). Immunoblot analysis of cell extracts prepared in parallel demonstrate that both EBNA2 proteins were expressed at roughly equivalent levels in the cells (Fig. 4D). These data link the induction of BATF gene expression with the formation of an EBNA2/CBF1 complex and suggest that BATF may be a direct cellular target for regulation by EBNA2.
BATF is a putative target for regulation by the Notch signaling pathway.
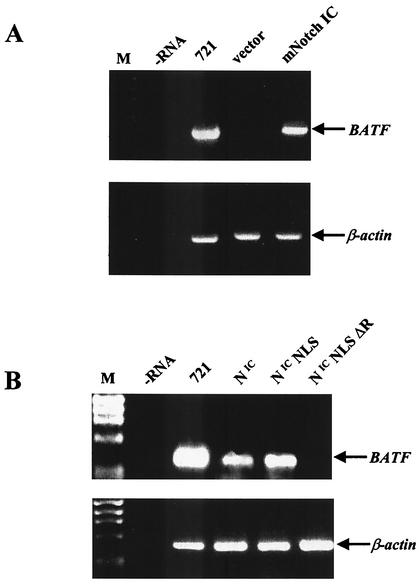
Having identified the viral EBNA2 transactivator as an inducer of BATF gene expression in B cells, we next sought to identify an endogenous regulator of BATF gene expression. The Notch family of proteins is comprised of transmembrane receptor proteins that are processed into nuclear-targeted transcriptional regulators following activation (reviewed in references 2 and 51). In recent years, a number of studies have shown that activated Notch functions in a way similar to that of EBNA2 to positively impact cellular gene expression via an interaction with CBF1 (26, 45, 70, 72) and can function as a substitute for EBNA2 in B-cell immortalization (16, 22). To examine if Notch activates BATF gene expression, a constitutively activated derivative of Notch1 (mNotch IC) was expressed in BJAB cells. Efficient transactivation occurred in cells expressing mNotch IC but not in control cells (Fig. 5A). An equivalent level of activation was observed when the experiment was repeated with a constitutively active form of mouse Notch2, the Notch family member that predominates in B cells (27, 78) (data not shown).
FIG. 5.
The BATF gene is a target for transactivation by Notch. (A) BJAB cells were electroporated as described in Fig. 3 with 10 μg of the indicated plasmid DNA. After 48 h, total RNA was isolated and analyzed for BATF and control β-actin gene expression by RT-PCR. RNA from 721 B cells served as a positive control. A no-RNA (−RNA) reaction and RNA from BJAB cells electroporated with empty vector DNA served as negative controls. ØX174 HaeIII DNA (M) was the molecular weight marker. (B) The experiment shown in panel A was repeated with activated human Notch 1 (NIC), NIC with a nuclear import signal (NIC NLS), and NIC NLS with a deletion (ΔR) of the CBF1 interaction domain. RNA for 721 B cells served as a positive control. A no-RNA reaction (−RNA) served as the negative control.
In order to test if the Notch-mediated activation of BATF relies on the interaction with CBF1, the experiment was repeated with Notch IC and variants were appended with a strong nuclear import signal (NLS) or appended with an NLS with a deletion (ΔR) of the CBF1 interaction motif. Although all three proteins were expressed equivalently (data not shown), BATF mRNA induction was observed with only Notch IC and Notch IC NLS and not with Notch IC NLS ΔR (Fig. 5B). These data strongly suggest that nuclear Notch activates BATF in B cells and that this activation depends upon an intact CBF1 interaction domain.
Induction of BATF gene expression by EBNA2 and Notch is B cell specific.
Our results implicate CBF1 as a DNA binding protein that is important to the transcriptional regulation of the BATF gene. CBF1 is a member of a highly conserved and ubiquitously expressed family of proteins that mediate the activation or repression of gene transcription depending on the cofactors available for interaction (25, 28, 30, 82; reviewed in reference 51). To examine the role of CBF1 in BATF gene regulation, we selected HeLa cells, a cell line that has been used previously to investigate CBF1 function (45), and RPMI 2650 cells, a human nasal epithelial cell line. Both cell types express CBF1, but not BATF (data not shown). DNA precipitates containing expression vectors for the indicated EBNA2 proteins or Notch IC were introduced into both cell types with the goal of activating endogenous BATF mRNA expression. To ensure that the introduced activators functioned as expected, a CAT reporter gene controlled by CBF1 binding sites was included in each precipitate. After 48 h, the cells were harvested and processed for the extraction of RNA and protein. As shown in Fig. 6A and B, BATF mRNA was not detected in any of the groups even though the CBF1 CAT reporter was efficiently activated by wild-type EBNA2 and constitutively active Notch1 (Fig. 6C and D). We concluded from these experiments that CBF1 and its interacting transactivator(s) play an important role in the up-regulation of BATF gene transcription in B cells but that transcription activation via CBF1 is not sufficient to activate BATF gene expression in non-B-cell types.
Protein complexes containing BATF bind to target AP-1 sites.
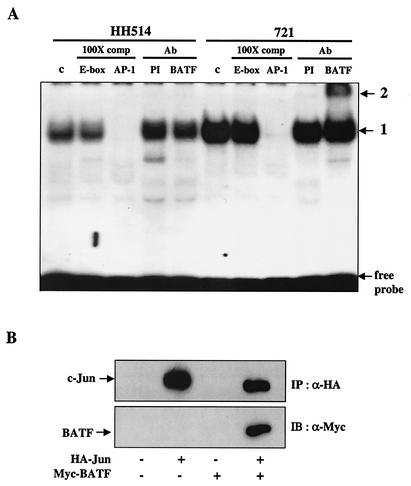
Previous studies have shown that BATF forms heterodimers with the Jun family of bZIP proteins to bind AP-1 DNA (9, 10). To demonstrate that BATF is part of the AP-1 DNA binding activity in EBV-infected B cells, nuclear extracts were prepared from 721, a cell line that carries a fully transforming EBV episome and expresses BATF, and HH514, a cell line that carries a nontransforming EBV variant in which BATF expression is undetectable. EMSA with 32P-labeled AP-1 DNA as a probe revealed a significantly higher level of AP-1 DNA binding activity in extracts prepared from the cells expressing BATF (Fig. 7A). The AP-1 binding observed was specific since it was competed by unlabeled AP-1 DNA and not by consensus E-box DNA to which basic helix-loop-helix transcription factors bind. The presence of BATF in the AP-1 DNA complex from 721 cells was established by adding anti-BATF antiserum or control PI to the binding reactions. As expected, anti-BATF antiserum supershifted the AP-1 complex derived from 721 cells but not from the non-BATF-expressing HH514 cells (Fig. 7A).
FIG. 7.
BATF is a component of AP-1 DNA binding activity in B cells. (A) Nuclear extracts were prepared from HH514 (BATF−) and 721 (BATF+) B cells as described in Materials and Methods and used for EMSA with a 32P-labeled AP-1 DNA probe. The major complex formed in the control (c) reactions (1) was competed by the addition of unlabeled AP-1 DNA but not by adding an unrelated DNA sequence (E-box). Addition of PI or anti-BATF antiserum (BATF) to the reaction resulted in a supershift (2) only in the extracts from 721 cells. Ab, antibody. (B) Coimmunoprecipitation of BATF:c-Jun complexes from B cell extracts. DG75 cells were electroporated as described in Materials and Methods with 10 μg of an expression plasmid for HA-tagged c-Jun, 10 μg of an expression plasmid for a Myc-tagged BATF, both plasmids, and empty vector DNA (where needed) to adjust the total DNA to 20 μg. After 36 h, the cells were lysed and protein complexes were immunoprecipitated (IP) with anti-HA antiserum (α-HA). The proteins were resolved by SDS-PAGE and immunoblotted (IB) initially with α-HA and then with anti-Myc antiserum (α-Myc) to detect BATF. Results indicate the presence of coimmunoprecipitated BATF only in extracts from DG75 cells expressing both proteins.
The ability to detect BATF-containing AP-1 DNA complexes in nuclear extracts prepared from EBV-positive B cells prompted us to examine if BATF forms heterodimers with the c-Jun protein in B cells. DG75 B cells were electroporated with vectors expressing Myc-tagged BATF and/or HA-tagged c-Jun. Cell extracts were prepared, and c-Jun protein complexes were immunoprecipitated with anti-HA antibody. Following resolution by SDS-PAGE, the proteins were immunoblotted with anti-HA antibody to detect c-Jun and then anti-Myc antibody to detect BATF protein. Control immunoblots in which whole-cell extracts were incubated with anti-HA and anti-Myc antibodies established protein mobilities and antibody specificity (data not shown). The results of this experiment revealed that BATF is present in c-Jun precipitates prepared from cells coexpressing both proteins (Fig. 7B). We concluded that the intracellular environment of human B cells supports the formation of BATF:c-Jun heterodimers that bind AP-1 target DNA.
BATF modulates EBV's lytic cycle.
The transcriptional activity of AP-1 is known to be essential for the transition from EBV latency to lytic replication in cultured B lymphocytes (reviewed in reference 35). A standard method of lytic-cycle induction in vitro employs treatment with the phorbol ester TPA to stimulate intracellular signaling events involving the activation of protein kinase C and AP-1 (reviewed in reference 68). Consensus AP-1 sites are found within the promoter regions of a number of immediate-early viral genes and must be occupied to achieve full transcriptional activation of the genes (reviewed in references 35 and 68). Thus, the accumulation of a molecular inhibitor of AP-1 activity is predicted to promote a delay in the onset of the lytic cycle.
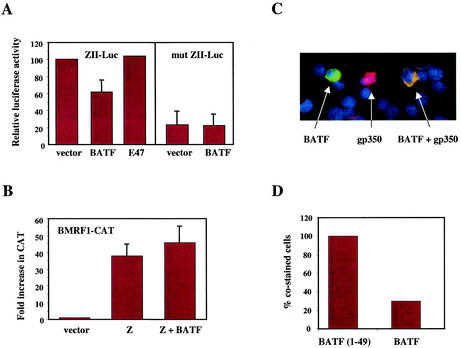
To investigate the potential of BATF to modulate immediate-early events in the EBV lytic cycle, we tested the impact of ectopic BATF expression on a reporter gene containing the ZII element from the promoter of BZLF1, a viral gene encoding a transactivator of early-EBV-lytic gene expression. Previous studies have shown that ZII binds an AP-1 complex of unknown composition (60) and that AP-1 binding is required for full activation of the BZLF1 gene (14; reviewed in reference 68). Electroporation of the EBV− BATF− B-cell line DG75 with the ZII-luciferase reporter resulted in efficient activation, which was reduced by 40% following the cointroduction of BATF (Fig. 8A). The repression was specific for BATF since it was not duplicated by E47, an E-box binding transcription factor. In addition, the effect of BATF depended upon the AP-1 site in ZII since the basal level of expression of a ZII construct containing a point mutation in the AP-1 site (mut ZII) was unaffected by coexpressed BATF. These results indicate that BATF exerts a negative regulatory impact on the AP-1 complexes involved in transcriptionally activating the BZLF1 gene.
FIG. 8.
Overexpression of BATF inhibits BZLF1 gene expression and reduces the frequency of lytic-cycle induction. (A) Five micrograms of a luciferase reporter gene driven by the ZII element of the BZLF1 promoter (ZII-Luc) or a matched reporter gene containing a mutated ZII element (mut ZII-Luc) were electroporated into DG75 cells along with 5 μg of expression plasmids for the indicated proteins and 2 μg of pRL DNA. After 48 h, cell extracts were prepared, normalized to the control luciferase activity from pRL, and assayed for ZII-directed luciferase expression. The experiment was repeated three times in duplicate, and the average luciferase values are expressed relative to the levels in the ZII-Luc/vector-alone group which were set at 100. Error bars indicate the standard errors of the means. (B) HeLa cells were transfected as described in the legend to Fig. 5 with a Z-responsive BMRF1-CAT reporter gene (5 μg) and 5 μg of the indicated expression plasmids. The experiment was performed three times, the CAT activities were averaged, and results were expressed relative to the basal level of BMRF1-CAT expression that was set to 1.0. Error bars indicate the standard errors of the means. (C) Immunofluorescence of HH514 cells electroporated with BATF or BATF(1-49) expression vectors and induced with TPA-butyrate (see Materials and Methods for details). Shown is an example of a field of cells stained for BATF (green), gp350/220 (red), or both BATF and gp350/220 (yellow). (D) Captured images were used to score the number of gp350/220-positive cells (red) for the lytic cycle, BATF- and BATF(1-49)-positive cells (green), total cells (DAPI), and gp350/220- and BATF-costained or gp350/220- and BATF(1-49)-costained cells (yellow). The number of costained cells in the BATF-transfected population was expressed as a percentage of the number of costained cells in the BATF(1-49)-transfected population. These results are representative of three experiments with similar results.
The BZLF1 gene encodes a dimerizing transactivator, referred to as ZEBRA, Zeta (Zta), or Z, that shows functional and structural similarity to AP-1 family members (13). Z binds AP-1-related DNA sites in the promoters of a number of viral genes, including the early-lytic BMRF1 gene (24). To test the possibility that BATF, in addition to inhibiting Z production, also interferes with the efficient activation of Z target genes, we examined the transactivation of a BMRF1 reporter gene in HeLa cells. BMRF1-CAT is not expressed in HeLa cells unless activated by Z (24, 55) (Fig. 8B). When assayed in the presence of Z and BATF, no reduction in CAT expression was observed, demonstrating that in this cell system, BATF does not impair the efficiency with which Z activates BMRF1 gene expression.
The studies described above suggest that the induction of BATF in latently infected B cells may influence the lytic switch by modulating the efficient production of Z. In order to examine if BATF expression inhibits the efficiency of lytic-cycle induction in B cells, HH514 cells (EBV+, BATF−) were electroporated with an expression plasmid for Myc-tagged BATF or for a Myc-tagged BATF variant rendered nonfunctional by a deletion of the leucine zipper dimerization domain. HH514 cells are permissive for viral replication, and the lytic cycle can be induced by TPA-butyrate treatment. Twenty-four hours following electroporation, the cells were treated with TPA-butyrate for 48 h, at which time the cultures were analyzed by immunofluorescence and individual cells were scored for BATF expression, for the expression of the viral late protein gp350, and for the coexpression of both proteins (Fig. 8C). The efficiency of lytic-cycle induction (gp350+ cells/total cells) was 11% for each group, and the efficiency of gene transfer (BATF+ cells/total cells) was between 3 and 4% on average and was independent of the input plasmid. Importantly, the number of BATF- and gp350-costained cells was 32% of the number of BATF(1-49)- and gp350-costained cells (Fig. 8D), indicating that BATF expression inhibits lytic-cycle entry in response to induction with TPA-butyrate. Thus, expression of BATF in EBV-positive lymphoblastoid cells may play a crucial role in the maintenance of latency by modulating lytic-cycle-promoting AP-1 activity.
DISCUSSION
EBV establishes a latent infection in B cells in which a precise program of viral gene expression maintains the viral episome and orchestrates cellular changes leading to cell immortalization and transformation (reviewed in reference 35). The role of the EBNA2 protein in transactivating the expression of EBV latency genes, including the LMP1 oncogene, has been well studied (reviewed in reference 66). Only recently, however, has the role of EBNA2 as an activator of cellular gene expression been fully appreciated and the efforts to identify EBNA2-regulated cellular genes been revitalized (32, 33, 69; reviewed in reference 35). In most cases, the genes activated by EBNA2 encode proteins that function directly, or indirectly, to activate B cells and promote cell cycle progression (8, 38). In this paper, we have identified the BATF gene as a cellular target of the EBNA2 transactivator. In contrast to other cellular genes up-regulated early following infection, the BATF gene encodes a protein that functions as a negative regulator of AP-1 activity and an antagonist of cell growth (10, 79). Thus, our results are the first to link infection of B cells with EBV to a transcriptional regulatory pathway that inhibits cell growth.
The BATF gene is expressed predominantly in mammalian hematopoietic tissues (10, 79). In addition to the dramatic increase in BATF gene expression observed in EBV-infected B cells, increased BATF expression has also been observed following receptor engagement on T helper cells (79) and the infection of T cells with human T-cell leukemia virus type 1 (20). Most recently, BATF mRNA was shown to be elevated following the activation of NF-κB in pre-B cells (41). Given the similarity between the molecular mechanism of transcription activation by EBNA2 and the Notch proteins (23, 26, 61, 70), we were able to establish that BATF mRNA is induced in B cells by nuclear-localized Notch proteins containing an intact CBF1 interaction domain. Interestingly, EBNA2 and Notch activation of the BATF gene is B cell specific even though CBF1 is expressed ubiquitously (reviewed in reference 51) and a simple CBF1 reporter gene is responsive to these transactivators in non-B-cell types (45). Cell specificity has been described for the EBNA2-mediated activation of LMP1 (12, 76), suggesting that a major factor in EBNA2 inducibility is the collaboration of EBNA2 with additional B-cell transcription factors and/or cellular signaling events. In this regard, CBF2/AUF and the Ets domain transcription factors PU.1/Spi1 and Spi-B have been identified as additional DNA binding proteins that interact with EBNA2 response elements (15, 31, 40). These factors, plus the molecules activated in response to other stimuli associated with the induction of BATF gene expression in T and B lymphocytes (41, 79), will provide the starting point for future studies to identify EBNA2 collaborators.
The human BATF gene has been cloned, and although the 5′ flanking region directs the B-cell-specific expression of a CAT reporter gene (48), the expression of this reporter is not restricted to EBV-infected B cells and is not induced to higher levels by EBNA2 or Notch (N. Meyer and E. Taparowsky, unpublished observations). There is a near-consensus CBF1 binding site within the first intron of the BATF gene, and future experiments will examine if this region (or additional extra- or intragenic regions) function together with the 5′ flanking region to confer inducibility. It remains a formal possibility that the induction of BATF gene expression by EBNA2 and Notch is indirect and requires the de novo synthesis of a second protein(s). Evidence to support this possibility was generated by using EBNA2-inducible B-cell lines where, in the presence of a protein synthesis inhibitor, BATF mRNA was induced, but not to the level observed in the absence of inhibitor (G. Laux, personal communication). A similar observation has been described for the induction of the C-MYC gene by EBNA2 (32).
Although the precise mechanism(s) through which EBNA2 and Notch activate BATF gene expression remains to be determined, the accumulation of this AP-1 inhibitor in EBV-infected B cells is intriguing. There have been numerous studies on the importance of AP-1 transcription complexes during the transition from EBV latency to lytic replication (reviewed in references 35 and 68). In particular, AP-1 activity is essential for the expression of BZLF1 (14), the lytic-cycle switch gene encoding Z, a transactivator that coordinates the expression of the immediate-early viral genes required for productive infection (reviewed in references 35 and 68). A cis-acting DNA element referred to as ZII mediates the AP-1 induction of BZLF1 expression and is occupied by proteins prior to and following the treatment of B cells with lytic-phase inducers (14). While the composition of the complexes bound to ZII is still under investigation, there is clear evidence pointing to occupancy by a c-Jun heterodimer during latency and either an ATF1 or ATF2 dimer in cells actively expressing Z (44, 46, 77). We found that the high basal activity of a ZII-luciferase reporter gene in DG75 cells (presumably the result of the constitutive expression of AP-1 family activators in these immortalized EBV− B cells) is reduced by 40% following expression of BATF. This suggests that BATF forms dimers with endogenous AP-1 activators, depleting the pool of AP-1 dimers with full transactivation potential. Additional experiments are required to identify the partners of BATF during viral latency and to test if these heterodimers bind the ZII element. Regardless of the outcome of these studies, the significant reduction in BZLF1 promoter activity observed in the presence of BATF suggests that, during lytic-cycle induction, BATF delays accumulation of the levels of Z necessary to activate immediate-early lytic-gene expression.
To obtain evidence that BATF can play a role in regulating the lytic-phase switch in vitro, expression vectors for wild-type and nonfunctional BATF were used to transfect HH514 cells and the lytic program was induced by treatment with TPA-butyrate. By counting the number of cells coexpressing the lytic-cycle protein gp350/220 and either BATF or BATF(1-49), we observed 60% fewer gp350/220-positive cells in the BATF-expressing population than in the BATF(1-49)-expressing population. Thus, BATF expression decreased the number of TPA-butyrate-induced HH514 cells in the lytic cycle. Based on our knowledge of BATF, we predicted that BATF would interfere with the AP-1 transcriptional activity triggered by the treatment of cells with phorbol ester tumor promoters. As we have shown, one target of BATF-mediated inhibition of AP-1 activity is the viral BZLF1 gene. However, the ability of BATF to change the composition and overall function of cellular AP-1 activity suggests that the expression of additional genes also will be affected.
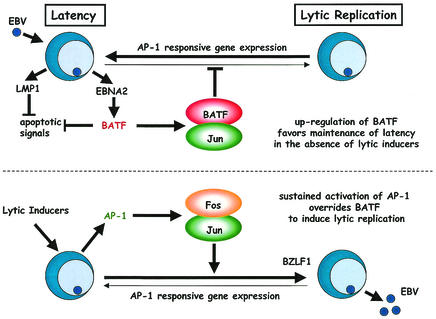
The findings presented here are the first to describe a cellular gene product induced immediately following EBV infection that has the potential to modulate viral latency by reducing the efficiency of lytic-cycle induction. The model we present in Fig. 9 provides an explanation as to why B cells expressing BATF resist entry into the lytic phase unless appropriately induced. It also considers that the subversion of cellular apoptosis that is associated with EBV latency and replication may be the result of the collaborative actions of antiapoptotic signaling by LMP1 and the negative regulation of proapoptotic gene expression by BATF. Certainly the observations that BATF is induced following NF-κB activation (41) and that AP-1 activity is associated with apoptosis in some cell types (reviewed in reference 64) are consistent with this role. While validation of the model will rely on future studies aimed at characterizing the composition of BATF protein complexes and identifying the genetic targets of these complexes in B cells, we are encouraged by the potential of cellular proteins like BATF to control the in vivo progression of EBV-associated human disease.
FIG. 9.
Proposed function of BATF as a positive modulator of EBV latency and as a deterrent to lytic-cycle entry. The EBNA2 protein is expressed immediately following infection and utilizes its transactivator function to trigger cell proliferation as well as the direct induction of BATF, which functions in the context of a BATF: Jun heterodimer to restrict cell growth, inhibit AP-1 target gene expression, and, together with LMP1, block apoptosis. Exposure to an appropriate lytic-cycle inducer (e.g., TPA) enhances intracellular AP-1 activity through new protein synthesis (Fos) and the posttranslational modification of resident AP-1 family members (Jun). The resultant sustained increase in AP-1 activity is sufficient to overcome the BATF inhibition of multiple AP-1 target genes (including BZLF1) and promote production of the immediate-early transactivators required for lytic-gene expression.
Acknowledgments
We thank Al Zullo for technical assistance with the Notch experiments and Gerhard Laux for his generous exchange of unpublished information.
L.M.J. was supported by a predoctoral fellowship from the American Heart Association. C.D.D. and S.E.H. are predoctoral trainees of PHS T32 CA09634 and GM08298, respectively. This work was supported by National Institutes of Health grants AI01537 (J.M.M.), CA64610 (J.M.M.), and CA78264 (E.J.T.).
REFERENCES
- 1.Abbot, S. D., M. Rowe, K. Cadwallader, A. Ricksten, J. Gordon, F. Wang, L. Rymo, and A. B. Rickinson. 1990. Epstein-Barr virus nuclear antigen 2 induces expression of the virus-encoded latent membrane protein. J. Virol. 64:2126-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 3.Baichwal, V. R., and B. Sugden. 1989. Transformation of Balb/c 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2:461-467. [PubMed] [Google Scholar]
- 4.Ben-Bassat, H., N. Goldblum, S. Mitrani, T. Goldblum, J. M. Yoffey, M. M. Cohen, Z. Bentwich, B. Ramot, E. Klein, and G. Klein. 1977. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt-like” malignant lymphoma (line D.G.-75). Int. J. Cancer 19:27-33. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, J., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 86:9558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, J., and E. Kieff. 1991. An Epstein-Barr virus nuclear protein 2 domain essential for transformation is a direct transcriptional activator. J. Virol. 65:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, J., F. Wang, and E. Kieff. 1991. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J. Virol. 65:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordier, M., A. Calender, M. Billaud, U. Zimber, G. Rousselet, O. Pavlish, J. Banchereau, T. Tursz, G. W. Bornkamm, and G. M. Lenoir. 1990. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J. Virol. 64:1002-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorsey, M. J., H.-J. Tae, K. G. Sollenberger, N. T. Mascarenhas, L. M. Johansen, and E. J. Taparowsky. 1995. B-ATF: a novel human bZIP protein that associates with members of the AP-1 transcription factor complex. Oncogene 11:2255-2265. [PubMed] [Google Scholar]
- 10.Echlin, D., H.-J. Tae, N. Mitin, and E. J. Taparowsky. 2000. B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene 19:1752-1763. [DOI] [PubMed] [Google Scholar]
- 11.Erickson, K. D., and J. M. Martin. 1997. Early detection of the lytic LMP-1 protein in EBV-infected B-cells suggests its presence in the virion. Virology 234:1-13. [DOI] [PubMed] [Google Scholar]
- 12.Fahraeus, R., A. Jansson, A. Ricksten, A. Sjoblom, and L. Rymo. 1990. Epstein-Barr virus-encoded nuclear antigen 2 activates the latent membrane protein promoter by modulating the activity of a negative regulatory element. Proc. Natl. Acad. Sci. USA 87:7390-7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrell, P. J., D. T. Rowe, C. M. Rooney, and T. Kouzarides. 1989. Epstein-Barr virus BZLF1 transctivator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 8:127-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flemington, E., and S. H. Speck. 1990. Identification of phorbol ester response elements in the promoter of the Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuentes-Panana, E. M., R. Peng, G. Brewer, J. Tan, and P. D. Ling. 2000. Regulation of the Epstein-Barr C promoter by AUF1 and the cyclic AMP protein kinase A signaling pathway. J. Virol. 74:8166-8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordadze, A. V., R. Peng, J. Tan, G. Liu, R. Sutton, B. Kempkes, G. W. Bornkamn, and P. D. Ling. 2001. Notch1IC partially replaces EBNA2 function in B cells immortalized by Epstein-Barr virus. J. Virol. 75:5899-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grossman, S., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear protein 2 transactivator is directed to response elements by the Jk recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammerschmidt, W., and B. Sugden. 1989. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340:393-397. [DOI] [PubMed] [Google Scholar]
- 19.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 71:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasegawa, H., Y. Utsunomiya, K. Kishimoto, Y. Tange, M. Yasukawa, and S. Fujita. 1996. SFA-2, a novel bZIP transcription factor induced by human T-cell leukemia virus type 1, is highly expressed in mature lymphocytes. Biochem. Biophys. Res. Commun. 222:164-170. [DOI] [PubMed] [Google Scholar]
- 21.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA-2 transactivation by recombination signal-binding protein Jκ. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 22.Höfelmayr, H., L. J. Strobl, G. Marschall, G. W. Bornkamn, and U. Zimber-Strobl. 2001. Activated Notch1 can transiently substitute for EBNA2 in the maintenance of proliferation of LMP1-expressing immortalized B cells. J. Virol. 75:2033-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Höfelmayr, H., L. J. Strobl, C. Stein, G. Laux, G. Marschall, G. W. Bornkamm, and U. Zimber-Strobl. 1999. Activated mouse Notch1 transactivates Epstein-Barr virus nuclear antigen 2-regulated viral promoters. J. Virol. 73:2770-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holley-Guthrie, E. A., E. B. Quinlivan, E.-C. Mar, and S. Kenney. 1990. The Epstein-Barr virus (EBV) BMRF 1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J. Virol. 64:3753-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh, J. J.-D., and S. D. Hayward. 1995. Masking of the CBF1/RBP-Jκ transcriptional repression domain by Epstein-Barr virus EBNA2. Science 268:560-563. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh, J. J.-D., T. Henkel, P. Salmon, E. Robey, M. G. Peterson, and S. D. Hayward. 1996. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16:952-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh, J. J.-D., D. E. Nofziger, G. Weinmaster, and S. D. Hayward. 1997. Epstein-Barr virus immortalization: Notch2 interacts with CBF1 and blocks differentiation. J. Virol. 71:1938-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh, J. J., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeffries, S., and A. J. Capobianco. 2000. Neoplastic transformation by Notch requires nuclear localization. Mol. Cell. Biol. 20:3928-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jochner, N., D. Eick, U. Zimber-Strobl, M. Pawlita, G. W. Bornkamm, and B. Kempkes. 1996. Epstein-Barr virus nuclear antigen 2 is a transcriptional suppressor of the immunoglobulin μ gene: implications for the expression of the translocated c-myc gene in Burkitt's lymphoma cells. EMBO J. 15:375-382. [PMC free article] [PubMed] [Google Scholar]
- 31.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and E. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaiser, C., G. Laux, D. Eick, N. Jochner, G. W. Bornkamm, and B. Kempkes. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73:4481-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaiser, C., O. vonStein, G. Laux, and M. Hoffmann. 1999. Functional genomics in cancer research: identification of target genes of the Epstein-Barr virus nuclear antigen 2 by subtractive cDNA cloning and high-throughput differential screening using high-density agarose gels. Electrophoresis 20:261-268. [DOI] [PubMed] [Google Scholar]
- 34.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 1109-1162. In Fundamental virology, B. N. Fields, D. M. Knipe, and P. M. Howley (ed.). Lippincott-Raven, Philadelphia, Pa.
- 36.Klein, G., T. Lindahl, M. Jondal, W. Leibold, J. Menezes, K. Nilsson, and C. Sundstrom. 1974. Continuous lymphoid cell lines with characteristics of B cells (bone-marrow-derived), lacking the Epstein-Barr virus genome and derived from three human lymphomas. Proc. Natl. Acad. Sci. USA 71:3283-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knecht, H., C. Berger, A. S. Al-Homsi, C. McQuain, and P. Brousset. 1997. Epstein-Barr virus oncogenesis. CRC Crit. Rev. Oncol.-Hematol. 26:117-135. [DOI] [PubMed] [Google Scholar]
- 38.Knutson, J. C. 1990. The level of c-fgr RNA is increased by EBNA2, an Epstein-Barr virus gene required for B-cell immortalization. J. Virol. 64:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lassar, A. B., R. L. Davis, W. E. Wright, T. Kadesch, C. Murre, A. Voronova, D. Baltimore, and H. Weintraub. 1991. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66:305-315. [DOI] [PubMed] [Google Scholar]
- 40.Laux, G., B. Adam, L. J. Strobl, and F. Moreau-Gachelin. 1994. The Spi1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-Jκ interact with an Epstein-Barr nuclear antigen 2 responsive cis-element. EMBO J. 13:5624-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, J., G. W. Peet, D. Balzarano, X. Li, P. Massa, R. W. Barton, and K. Marcu. 2001. Novel NEMO/IκB kinase and NF-κB target genes at the pre-B to immature B cell transition. J. Biol. Chem. 276:18579-18590. [DOI] [PubMed] [Google Scholar]
- 42.Ling, P. D., D. R. Rawlins, and S. D. Hayward. 1993. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. USA 90:9237-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling, P. D., J. J.-D. Hsieh, I. K. Ruf, D. R. Rawlins, and S. D. Hayward. 1994. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J. Virol. 68:5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, P., S. Liu, and S. H. Speck. 1998. Identification of a negative cis-element within the ZII domain of the Epstein-Barr virus lytic switch BZLF1 gene promoter. J. Virol. 72:8230-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, F. M., and S. E. Lux. 1996. Constitutively active human Notch1 binds to the transcription factor CBF 1 and stimulates transcription through a promoter containing a CBF 1-responsive element. Proc. Natl. Acad. Sci. USA 93:5663-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacCallum, P., L. Karimi, and L. J. Nicholson. 1999. Definition of the transcription factors which bind the differentiation responsive element of the Epstein-Barr virus BZLF1 Z promoter in human epithelial cells. J. Gen. Virol. 80:1501-1512. [DOI] [PubMed] [Google Scholar]
- 47.Martin, J., and B. Sugden. 1991. Transformation by the oncogenic latent membrane protein correlates with its rapid turnover, membrane localization, and cytoskeletal association. J. Virol. 65:3246-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer, N. P., L. M. Johansen, H.-J. Tae, P. P. Budde, K. L. Williams, and E. J. Taparowsky. 1998. Genomic organization of human B-ATF, a target for regulation by EBV and HTLV-1. Mamm. Genome 9:849-852. [DOI] [PubMed] [Google Scholar]
- 49.Nitsche, F., A. Bell, and A. Rickinson. 1997. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: role for the W1W2 repeat domain. J. Virol. 71:6619-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nye, J. S., R. Kopan, and R. Axel. 1994. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development 120:2421-2430. [DOI] [PubMed] [Google Scholar]
- 51.Osborne, B., and L. Miele. 1999. Notch and the immune system. Immunity 11:653-663. [DOI] [PubMed] [Google Scholar]
- 52.Peng, R., J. Tan, and P. D. Ling. 2000. Conserved regions in the Epstein-Barr virus leader protein define distinct domains required for nuclear localization and transcriptional cooperation with EBNA2. J. Virol. 74:9953-9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peters, M. A., K. G. Sollenberger, T.-L. Kao, and E. J. Taparowsky. 1997. A minimal regulatory region maintains constitutive expression of the max gene. Mol. Cell. Biol. 17:1037-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price, T., J. Altken, and E. R. Simpson. 1992. Relative expression of aromatase cytochrome p450 in human fetal tissues as determined by competitive polymerase chain amplification. J. Clin. Endocrinol. Metab. 74:879-883. [DOI] [PubMed] [Google Scholar]
- 55.Quinlivan, E. B., E. A. Holley-Guthrie, M. Norris, D. Gutsch, S. L. Bachenheimer, and S. C. Kenney. 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 21:1999-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rabson, M., L. Heston, and G. Miller. 1983. Identification of a rare Epstein-Barr virus variant that enhances early antigen expression in Raji cells. Proc. Natl. Acad. Sci. USA 80:2762-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramocki, M. B., S. E. Johnson, M. A. White, C. L. Ashendel, S. F. Konieczny, and E. J. Taparowsky. 1997. Signaling through MAP kinase and Rac/Rho does not duplicate the effects of activated Ras on skeletal myogenesis. Mol. Cell. Biol. 17:3547-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reedman, B. M., and G. Klein. 1973. Cellular localization of an Epstein-Barr virus (EBV)-associated complement-fixing antigen in producer and non-producer lymphoblastoid cell lines. Int. J. Cancer 11:499-520. [DOI] [PubMed] [Google Scholar]
- 59.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p.2397-2446. In Fields virology, B. N. Fields, D. M. Knipe, and P. M. Howley (ed.). Lippincott-Raven, Philadelphia, Pa.
- 60.Ruf, I. K., and D. R. Rawlins. 1995. Identification and characterization of ZIIBC, a complex formed by cellular factors and the ZII site of the Epstein-Barr virus BZLF1 promoter. J. Virol. 69:7648-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakai, T., Y. Taniguchi, K. Tamura, S. Minoguchi, T. Fukuhara, L. J. Strobl, U. Zimber-Strobl, G. W. Bornkamm, and T. Honjo. 1998. Functional replacement of the intracellular region of the Notch1 receptor by Epstein-Barr virus nuclear antigen 2. J. Virol. 72:6034-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sample, J., L. Young, B. Martin, T. Chatman, E. Kieff, A. Rickinson, and E. Kieff. 1990. Epstein-Barr viruses types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 64:4084-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Severson, C. D., D. L. Burg, D. E. Lafrenz, and T. L. Feldbush. 1987. An alternative method of panning for rat B lymphocytes. Immunol. Lett. 15:291-295. [DOI] [PubMed] [Google Scholar]
- 64.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell. Biol. 4:131-136. [DOI] [PubMed] [Google Scholar]
- 65.Shimizu, Y., D. E. Geraghty, B. H. Koller, H. T. Orr, and R. DeMars. 1988. Transfer and expression of three cloned human non-HLA-A,B,C class I major histocompatibility complex genes in mutant lymphoblastoid cells. Proc. Natl. Acad. Sci. USA 85:227-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinclair, A. J., and P. J. Farrell. 1992. Epstein-Barr virus transcription factors. Cell Growth Diff. 3:557-563. [PubMed] [Google Scholar]
- 67.Sinclair, A. J., I. Palmero, G. Peters, and P. J. Farrell. 1994. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 13:3321-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Speck, S. H., T. Chatila, and E. Flemington. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399-405. [DOI] [PubMed] [Google Scholar]
- 69.Spender, L. C., G. H. Cornish, B. Rowland, B. Kempkes, and P. J. Farell. 2001. Direct and indirect regulation of cytokine and cell cycle proteins by EBNA-2 during Epstein-Barr virus infection. J. Virol. 75:3537-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strobl, L. J., H. Höfelmayr, C. Stein, G. Marschall, M. Brielmeier, G. Laux, G. W. Bornkamm, and U. Zimber-Strobl. 1997. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-Jκ. Immunobiology 198:299-306. [DOI] [PubMed] [Google Scholar]
- 71.Strobl, L. J., H. Höfelmayr, G. Marschall, M. Brielmeier, G. W. Bornkamm, and U. Zimber-Strobl. 2000. Activated Notch modulates gene expression in B cells similarly to Epstein-Barr viral nuclear antigen 2. J. Virol. 74:1727-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamura, K., Y. Taniguchi, S. Minoguchi, T. Sakai, T. Tun, T. Furukawa, and T. Honjo. 1995. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-Jκ/Su(H). Curr. Biol. 5:1416-1423. [DOI] [PubMed] [Google Scholar]
- 73.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waltzer, L., C. Logeat, C. Brou, A. Israel, A. Sergeant, and E. Manet. 1994. The human J kappa recombination signal sequence binding protein (RBP-J kappa) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 13:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]
- 76.Wang, F., S. Tsang, M. Kurilla, J. Cohne, and E. Kieff. 1990. Epstein-Barr virus nuclear antigen 2 transactivates the latent membrane protein (LMP1). J. Virol. 64:3407-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang, Y.-C. J., J.-M. Huang, and E. A. Montalvo. 1997. Characterization of proteins binding to the ZII element in the Epstein-Barr virus BZLF1 promoter: transactivation by ATF1. Virology 227:323-330. [DOI] [PubMed] [Google Scholar]
- 78.Weinmaster, G., V. J. Roberts, and G. Lemke. 1992. Notch2: a second mammalian Notch gene. Development 116:931-941. [DOI] [PubMed] [Google Scholar]
- 79.Williams, K. L., I. Nanda, G. E. Lyons, C. T. Kuo, M. Schmid, J. M. Leiden, M. H. Kaplan, and E. J. Taparowsky. 2001. Characterization of murine BATF: a negative regulator of activator protein-1 activity in the thymus. Eur. J. Immunol. 31:1620-1627. [DOI] [PubMed] [Google Scholar]
- 80.Yalamanchili, R., X. Tong, S. Grossman, E. Johannsen, G. Mosialos, and E. Kieff. 1994. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology 204:634-641. [DOI] [PubMed] [Google Scholar]
- 81.Zhang, Q., D. Gutsch, and S. Kenney. 1994. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol. Cell. Biol. 14:1929-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou, S., M. Fujimuro, J. J. Hseih, L. Chen, and S. D. Hayward. 2000. A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J. Virol. 74:1939-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zimber-Strobl, U., L. J. Strobl, C. Meitinger, R. Hinrichs, T. Sakai, T. Furukawa, T. Honjo, and G. W. Bornkamm. 1994. Eppstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 13:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]