Abstract
Enterobacterial common antigen (ECA) is a polysaccharide found on the outer membrane of virtually all gram-negative enteric bacteria and consists of three sugars, N-acetyl-d-glucosamine, N-acetyl-d-mannosaminuronic acid, and 4-acetamido-4,6-dideoxy-d-galactose, organized into trisaccharide repeating units having the sequence →3)-α-d-Fuc4NAc-(1→4)-β-d-ManNAcA-(1→4)-α-d-GlcNAc-(1→. While the precise function of ECA is unknown, it has been linked to the resistance of Shiga-toxin-producing Escherichia coli (STEC) O157:H7 to organic acids and the resistance of Salmonella enterica to bile salts. The final step in the synthesis of 4-acetamido-4,6-dideoxy-d-galactose, the acetyl-coenzyme A (CoA)-dependent acetylation of the 4-amino group, is carried out by TDP-fucosamine acetyltransferase (WecD). We have determined the crystal structure of WecD in apo form at a 1.95-Å resolution and bound to acetyl-CoA at a 1.66-Å resolution. WecD is a dimeric enzyme, with each monomer adopting the GNAT N-acetyltransferase fold, common to a number of enzymes involved in acetylation of histones, aminoglycoside antibiotics, serotonin, and sugars. The crystal structure of WecD, however, represents the first structure of a GNAT family member that acts on nucleotide sugars. Based on this cocrystal structure, we have used flexible docking to generate a WecD-bound model of the acetyl-CoA-TDP-fucosamine tetrahedral intermediate, representing the structure during acetyl transfer. Our structural data show that WecD does not possess a residue that directly functions as a catalytic base, although Tyr208 is well positioned to function as a general acid by protonating the thiolate anion of coenzyme A.
Enterobacterial common antigen (ECA) was first identified as a common hapten associated with the gram-negative family of Enterobacteriaceae (77). The name enterobacterial common antigen was coined by Mäkel ä and Mayer in their review paper (36). It was later shown that ECA is a glycolipid (30). The biological role of ECA is not yet well understood, although its presence in all members of the Enterobacteriaceae suggests a role unique to these organisms (53). It has been shown that ECA contributes, at least partially, to virulence (47, 68) and to the resistance of enteric bacteria to bile salts in the digestive tract of their host (15, 51). The polysaccharide portion of ECA is heterogeneous in length and is formed by a trisaccharide repeat unit composed of amino sugars linked as follows: [4-acetamido-4,6-dideoxy-d-galactose (α-d-Fuc4NAc)]-(1→4)-[N-acetyl-d-mannosaminuronic acid (β-d-ManNAcA)]-(1→4)-[N-acetyl-d-glucosamine (α-d-GlcNAc)] (35). In most Enterobacteriaceae, the ECA polysaccharide chain is covalently bound to a phospholipid molecule in the outer membrane. The reducing end of the terminal GlcNAc residue is linked via a phosphodiester linkage to a phosphoglyceride aglycone. However, a few members of this family instead have ECA bound to the lipopolysaccharide core of the outer membrane (29). ECA is also found in a lipid-free cyclic form of several repeating units (19, 30, 73), and the crystal structure of ECACYC composed of four repeating units has been recently determined (21).
ECA synthesis occurs in three steps, at the cytoplasmic side of the inner membrane, through successive addition of nucleotide diphosphate-activated sugars onto the lipid carrier undecaprenol phosphate (50). The synthesis of the repeat is initiated by the transfer of GlcNAc from UDP-GlcNAc to undecaprenol phosphate, yielding GlcNAc-P-P-undecaprenol (lipid I), the first lipid intermediate. Second, ManNAcA is transferred from UDP-ManNAcA onto lipid I, resulting in ManNAcA-lipid I (lipid II). Next, Fuc4NAc is transferred from TDP-Fuc4NAc to form Fuc4NAc-lipid II (lipid III). Lipid III molecules are then translocated across the membrane to the periplasmic side via a Wzx “flippase” (55), are polymerized into longer chains by a Wzy-dependent block polymerization mechanism (53), bind to phospholipids, and finally localize at the outer leaflet of the outer membrane (3, 52, 54, 56).
The genes responsible for ECA biosynthesis are mostly located in the wec gene cluster, formerly known as the rfe-rff gene cluster (39). The genes wecA (rfe), wecB (rffE), wecC (rffD), and wecG (rffM) are required for the synthesis of lipid I and lipid II. In lipid III synthesis, the wecE (rffA) gene encodes a transaminase that catalyzes the transfer of an amino group from glutamate to TDP-fucose, yielding TDP-Fuc4N, which then receives an acetyl group from acetyl-coenzyme A (AcCoA), a reaction catalyzed by TDP-fucosamine acetyltransferase encoded by wecD (rffC), to yield TDP-Fuc4NAc. Finally, a wecF (rffT)-encoded fucosyltransferase transfers Fuc4NAc to lipid II. Several genes, including wecA, rmlA2 (TDP-glucose pyrophosphorylase 2), and rffG (rmlB, TDP-glucose dehydratase), essential for ECA synthesis, are also required for O-antigen synthesis in the Enterobacteriaceae (38, 39).
The synthesis of TDP-Fuc4NAc in the presence of TDP-Fuc4NH and AcCoA was first analyzed by Matsuhashi and Strominger (37) in Escherichia coli cell extracts. The gene encoding the protein, now known as wecD, was cloned and its nucleotide sequence determined together with those of other wec genes from E. coli (16). The amino acid sequences of WecD are well conserved across enterobacteria, with sequence identities of 40% or more. WecD orthologs are also found in other gammaproteobacteria, such as Actinobacillus pleuropneumoniae, Haemophilus ducreyi, Vibrio vulnificus, and Vibrio cholerae, and show 25 to 35% sequence identity to the E. coli enzyme. The acetyltransferase activity of WecD is essential for the formation of lipid III, and it was shown that a mutation in the wecD gene in Salmonella enterica causes sensitivity of this bacterium to deoxycholate (51).
Sequence analysis of WecD from E. coli identifies the C-terminal segment (residues 98 to 171; PFAM entry PF00583; http://www.sanger.ac.uk/Software/Pfam/) as belonging to the GCN5-related N-acetyltransferase (GNAT) superfamily. This superfamily was first noted by Shaw et al. (60), who identified four conserved amino acid motifs present in these proteins. The GNAT superfamily of enzymes catalyzes transfer of an acetyl group from acetyl coenzyme A to a primary amine. Structures of more than 20 enzymes containing this domain are known (SCOP database; http://scop.berkeley.edu/ [44]) and include, among others, histone N-acetyltransferase, aminoglycoside N-acetyltransferase, serotonin N-acetyltransferase, protein N-myristoyltransferase, mycothiol synthase, glucosamine-6-phosphate N-acetyltransferase, and the Fem family of amino acyl transferases. Together, these structures clearly show that the primary function of the GNAT domain is to bind AcCoA and that somewhat different mechanisms have evolved to catalyze this reaction (18, 72). The compounds accepting the acetyl group from AcCoA vary substantially between these enzymes, with the specificity of each enzyme determined by the presence of additional secondary structural elements or separate domains. Most if not all of these enzymes form dimers.
We describe here the structure of the WecD acetyltransferase from uropathogenic E. coli CFT073, as well as its complex with AcCoA, and compare these structures with those of other members of the GNAT superfamily. We have used computational flexible docking to obtain further insights into substrate binding. The experimental and modeled structures allow us to propose WecD residues that may be involved in TDP-Fuc4NH binding and acetyl transfer. The structural information presented here serves to demonstrate further the diversity of proteins that belong to the GNAT superfamily.
MATERIALS AND METHODS
Gene cloning, protein expression, and purification.
The wecD gene was amplified by PCR from Escherichia coli CFT073 genomic DNA (75) and was cloned into a modified pET15b vector (Amersham Biosciences) to obtain an in-frame N-terminal fusion protein with a noncleavable His6 tag. E. coli BL21(DE3) cells were transformed with the vector DNA, the cells were grown in 1 liter of Circle Grow medium (QBiogene, Irvine, CA), and the culture was induced by adding 0.1 mM isopropyl-1-thio-β-d-galactopyranoside (Sigma Chemical Co.) followed by culturing at 22°C for 5 h. Cells were harvested by centrifugation (4,000 × g, 4°C, 20 min), resuspended in 30 ml of lysis buffer (50 mM Tris-Cl, pH 7.5, 0.4 M NaCl, 5% [wt/vol] glycerol, 20 mM imidazole, 10 mM β-mercaptoethanol, one tablet of Complete protease inhibitors [Roche Diagnostics, Laval, Canada]), and lysed by sonication on ice for a total of five 30-s cycles with 45 s between cycles. The lysate was clarified by centrifugation (100,000 × g, 4°C, 30 min). The clarified cell lysate was passed through an equilibrated, 3-ml DEAE-Sepharose column (Amersham Biosciences), and the eluted protein loaded on a 2-ml nickel-nitrilotriacetic acid column (QIAGEN). Bound protein was eluted in buffer containing 200 mM imidazole. Fractions containing purified protein were pooled and further purified on a Superdex 200 gel filtration column (Amersham Biosciences), using a buffer consisting of 20 mM Tris-Cl, pH 8.0, 450 mM NaCl, 5 mM dithiothreitol (DTT). Protein purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and pure fractions were pooled and dialyzed overnight in a final buffer consisting of 20 mM Tris-Cl, pH 8.0, 50 mM NaCl, and 5 mM DTT. The homogeneity of the protein was further analyzed by native polyacrylamide gel electrophoresis and dynamic light scattering using a DynaPro MASPRII molecular sizing instrument (Proterion Corp., Piscataway, N.J.) running Dynamics V6 software.
Crystallization.
WecD was concentrated to 7.1 mg/ml in a buffer containing 20 mM Tris-Cl, pH 8.0, 450 mM NaCl, 5 mM DTT. The best native crystals were obtained in one day at 20°C from a hanging drop containing 1 μl of protein in buffer mixed with 1 μl of reservoir solution (100 mM Na-acetate, pH 4.5, 10 mM ZnSO4). The crystals belong to the space group P43212 with unit cell dimensions a = b = 84.8, c = 155.6 Å, and diffract to a 1.95-Å resolution. There are two molecules in the asymmetric unit (Z = 16). Crystals of the AcCoA complex of WecD were obtained under similar conditions, but with the protein buffer containing 20 mM Tris, pH 8.0, 200 mM NaCl, 5 mM DTT, and the addition of 2 mM AcCoA and 5 mM TDP (Fluka Chemical Co.) to the reservoir solution. The crystals belong to space group P43212, have unit cell dimensions a = b = 84.9, c = 154.1 Å, and diffract to 1.65-Å resolution. For cryoprotection, crystals were transferred to a reservoir solution with the addition of 25% (vol/vol) glycerol, picked up with a nylon loop, and flash cooled in the nitrogen cold stream at 100 K. Diffraction data were collected at beamlines X8C (for the WecD crystal), and X29 (for the AcCoA complex). Data were recorded using a Quantum-4 or Quantum-315 charge-coupled-device detector (Area Detector Systems Corp., Poway, CA) at beamlines X8C or X29, respectively, at the National Synchrotron Light Source, Brookhaven National Laboratory. Integration and scaling of diffraction data were performed using HKL2000 (48).
Structure determination and refinement.
The three-dimensional structure of apo-WecD was solved using a two-wavelength MAD experiment about the Zn K edge (E = 9.6586 keV). One Zn site per asymmetric unit was found, and phases were calculated using the program ShelxD/E (59), having a figure of merit of 0.47. These phases were improved by density modification, giving a FOM of 0.77, and a partial model was built using RESOLVE (65). The structure was completed through further rounds of manual model fitting using the program O (28), alternating with refinement of the model with REFMAC (43) of the CCP4 suite (78). Final refinement statistics are summarized in Table 1. The apo-WecD model contains residues Val3 to Arg224 in each subunit and one Zn atom, coordinated to the side chains of Asp136, Glu157, and three water molecules. The electron density is overall well defined, except for the solvent-exposed loops between β5 and β6 and between α5 and β7, where the side chains of residues Asn78, Asn79, and Val80 in one subunit and Ser81 and Phe135 in another subunit are not well ordered. The model of the WecD-AcCoA complex contains two AcCoA molecules per dimer. No electron density corresponding to TDP was visible in the maps for the AcCoA complex, although TDP was present in the crystallization.
TABLE 1.
X-ray crystallographic data
| Parametera | Value for model
|
|||
|---|---|---|---|---|
| WecD-Zn
|
WecD-Zn | WecD-AcCoA | ||
| Peak | Inflection | |||
| Data collection (unit cell) | ||||
| a (Å) | 84.8 | 84.8 | 84.8 | 84.9 |
| b (Å) | 84.8 | 84.8 | 84.8 | 84.9 |
| c (Å) | 155.5 | 155.5 | 155.6 | 154.1 |
| Z | 16 | 16 | 16 | 16 |
| Resolution (Å) | 50-2 | 50-2 | 50-1.95 | 50-1.66 |
| Wavelength | 1.282 | 1.283 | 1.1 | 1.1 |
| Observed hkl | 198,351 | 197,147 | 318,786 | 407,076 |
| Unique hkl | 37,288 | 37,234 | 40,825 | 65,489 |
| Redundancy | 5.3 | 5.3 | 7.8 | 6.2 |
| Completeness (%) | 95.2 | 95.1 | 96.6 | 97.3 |
| Rsym | 0.043 | 0.045 | 0.042 | 0.053 |
| I/σ(I) | 21.6 | 20.4 | 24.1 | 18.0 |
| Refinement | ||||
| Resolution (Å) | 50-1.95 | 50-1.66 | ||
| Rwork (no. of hkl) | 0.181 (38,592) | 0.20 (62,158) | ||
| Rfree (no. of hkl) | 0.223 (2,012) | 0.22 (3,259) | ||
| B-factor (Å)2/(no. of atoms) | ||||
| Protein | 22.0 (3,390) | 23.6 (3,390) | ||
| Solvent | 36.9 (627) | 36.1 (463) | ||
| Ligands | 18.3 (1) | 42.9 (102) | ||
| Ramachandran | ||||
| Allowed (%) | 99.7 | 99.5 | ||
| Generous (%) | 0.3 | 0.3 | ||
| Disallowed (%) | 0.0 | 0.3 | ||
| rms deviation | ||||
| Bond length (Å) | 0.012 | 0.009 | ||
| Angle (°) | 1.31 | 1.28 | ||
Rsym = (∑|Iobs − Iavg|)/ΣIavg; Rwork = (∑|Fobs − Fcalc|)/∑Fobs; Rfree = Rwork for a subset of 5% of reflections not used in refinement.
Modeling of TDP-d-Fuc4N:acetyl-CoA transition state bound to WecD.
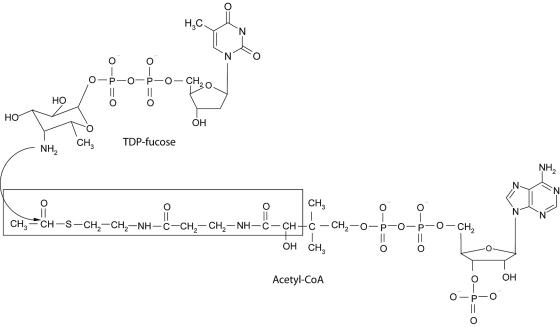
The putative transition state (tetrahedral intermediate) formed between AcCoA and TDP-β-d-fucosamine (TDP-Fuc4N) was modeled into the active site of chain A of the WecD dimer. All crystallographic water molecules were removed, and hydrogen atoms were added explicitly. The protonation state at physiological pH was adopted, except for the Glu70 side chain, which was treated in neutral form due to its close proximity to the Glu68 carboxylate group. The polar hydrogen atoms of Ser, Thr, and Tyr side chains were manually oriented to maximize hydrogen-bonding interactions. The positions of all hydrogen atoms were then refined by energy minimization using the AMBER all-atom molecular mechanics force field (13) in the absence of the AcCoA ligands and with the nonhydrogen atoms fixed at their crystallographic positions. The structure of the transition state ligand modeled at the WecD active site, shown schematically in Fig. 1, consists of the tetrahedral intermediate formed by the attack of the amino group of TDP-Fuc4N to the acetyl carbonyl group of AcCoA, followed by zwitterion neutralization. Geometric constraints lead to the (R) absolute configuration at the chiral center formed following the attack. Only the acetyl-proximal half of the AcCoA molecule was retained for transition state modeling (see Fig. 1), in its crystallographically determined geometry bound to monomer A of the WecD homodimer. The TDP-Fuc4N part was built starting from the crystallographic structure of dTDP-β-d-glucose (1), replacing β-d-glucose by β-d-fucosamine. The latter was built in the chair conformation bearing both the C-4 amino and the C-1 phosphate substituents in axial orientations, in agreement with the nuclear magnetic resonance spectra of TDP-Fuc4N produced by WecE (27), the aminotransferase immediately upstream of WecD in the ECA biosynthetic pathway. This ring conformation is also consistent with the crystallographic structure of β-d-fucose (66). All structural manipulations and visualizations were done with SYBYL 6.9 (Tripos, Inc., St. Louis, MO). Partial charges of the transition state ligand (bearing a net charge of −2e) were calculated by a two-stage restrained fitting procedure (5, 14) to the single-point HF 6-31G* electrostatic potential calculated with GAMESS (58).
FIG. 1.
Enzymatic reaction catalyzed by TDP-fucosamine acetyltransferase (WecD). The portion of the CoA cofactor utilized in computational molecular modeling is boxed.
The Monte Carlo minimization (MCM) approach (32), adapted to protein-ligand flexible docking (9, 33, 61), was used to generate and rank feasible conformations of the TDP-Fuc4N portion of the constructed transition state bound to WecD. In each MCM cycle, the conformation of the TDP-Fuc4N fragment was first perturbed by introducing random changes in one or more of its dihedral angles and then was conjugate gradient energy minimized using the AMBER force field (13, 74), a distance-dependent dielectric (4rij), and an 8-Å nonbonded cutoff, up to a root mean square (rms) gradient of 0.05 kcal/(mol · Å). A predefined set of protein residues in the region expected to be explored by the conformationally sampled ligand was allowed to move during energy minimization, while the rest of the WecD homodimer, as well as the more distal portion of the retained AcCoA fragment (see Fig. 1), were fixed at their crystallographic positions. The set of mobile protein residues included Leu10 to Glu13, Ile23 to Arg25, Gln45 to Ile48, Glu68 to Leu73, Phe104, Ser107 to Phe109, Phe122, Tyr123, Trp126, Asn129, His137, Ser154 to Leu158, Arg164 to Leu168, Arg194 to Gln198, Tyr208, and Ser217 to Trp221 from monomer A in the WecD homodimer. In a first docking run, 5,000 MCM cycles were carried out to sample all 10 acyclic rotatable bonds of the TDP-Fuc4N moiety. In order to increase the sampling efficiency in the solvent-exposed region, the lowest-energy conformation obtained after the first docking run was submitted to a second docking run consisting of 2,000 MCM cycles, in which only the four terminal rotatable bonds of the TDP moiety were included in conformational sampling. The lowest-energy conformation obtained after the second docking run was selected as the final model of the WecD-transition state complex.
Protein structure accession numbers.
Protein Data Bank codes for WecD-Apo and WecD-AcCoA are 2FS5 and 2FT0, respectively.
RESULTS AND DISCUSSION
Overall structure of WecD.
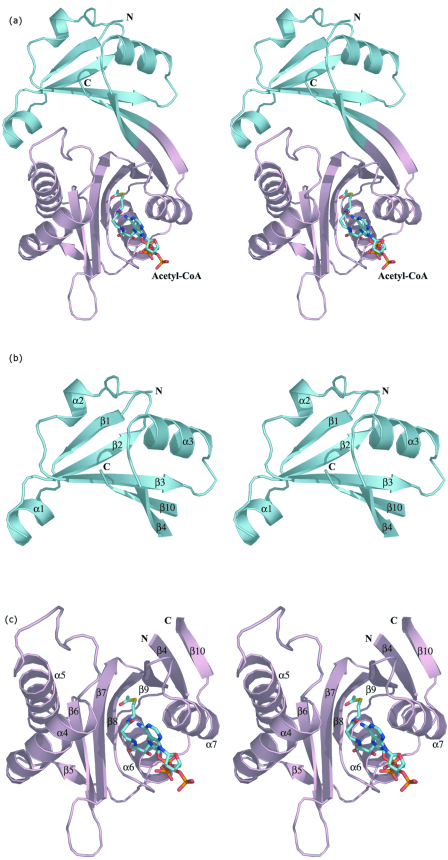
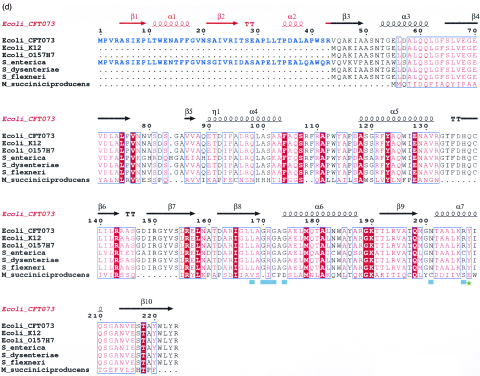
The three-dimensional structure of the WecD monomer is shown in Fig. 2a. The protein belongs to the α/β class and is composed of two domains. The N-terminal domain consists of residues Val3 to Gly69 and Ala219 to Arg224 from the C terminus. It has a central, five-stranded mixed β-sheet with two α-helices, α2 and α3, on one side of the sheet and helix α1 following after the first strand β1 (Fig. 2b). The strands are arranged in the order β↓β2↑β3↑β10↓β4↑. The C-terminal domain is comprised of residues Glu70 to Thr218 and is formed from a seven-stranded mixed β-sheet with four α-helices, two of them (α6 and α7) on one side of the sheet and the other two (α4 and α5) within a long loop on the side of the domain. The strands β4 and β10 are long and extend across both domains (Fig. 2). The order of strands in the C-terminal domain is β5↑β6↓β7↑β8↓β9↓β4↑β10↓. The sequence predicted to belong to the GNAT family is localized within this domain. In many other WecD-like sequences, the N-terminal 43 residues forming β1, α1, β2, and α2 are absent, including those from E. coli K12 and O157:H7 (Fig. 2d).
FIG.2.
Three-dimensional structure of E. coli WecD. (a) Stereo view of the WecD monomer. The domains are differently colored; N-terminal domain is shown in cyan and the C-terminal GNAT domain in light pink. (b) The N-terminal domain with secondary structure elements labeled. (c) The C-terminal domain with secondary structure elements labeled. These and subsequent figures were prepared using PyMol (www.pymol.org). (d) Sequence alignment of selected WecD sequences, highlighting the N-terminal extended sequence present in WecD from E. coli CFT073 and Salmonella enterica (blue). The potential general acid, Tyr 208, is highlighted as a green star, and other AcCoA-binding residues are highlighted by cyan squares. Sequence alignment was carried out using ClustalW (7) and formatted using ESPript (23).
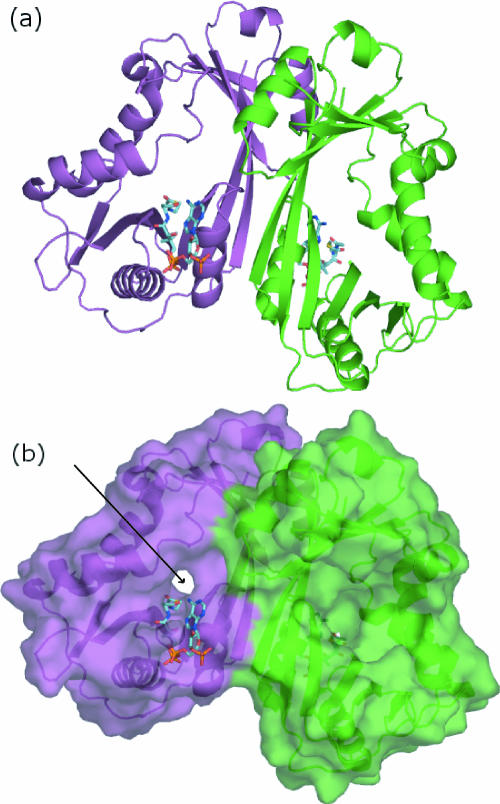
Although the two domains are distinct in the WecD structure, their β-strands extend towards each other to form a continuous, highly concave 10-stranded β-sheet with all α-helices, except α4 and α5, lining the outside of this sheet. Two polypeptide segments, Leu10 to Asn20 from the N-terminal domain, which includes α1, and Gln89 to Thr134 from the C-terminal domain, which includes α4 and α5, embrace each other on one side of the β-sheet, creating a tunnel through the molecule with a wide funnel-like entrance on one side and a narrow exit on the opposite side (Fig. 3).
FIG. 3.
Structure of the WecD dimer. (a) Ribbon representation of the WecD dimer with chain A colored green and chain B colored magenta. (b) Surface representation of the WecD dimer, with acetyl-CoA shown in stick representation. The arrow defines the location of the narrow side of the tunnel in monomer B, while the wide funnel-like side of the tunnel is towards the viewer in monomer A.
Unexpectedly, the three-dimensional structure of the WecD N-terminal domain shows remarkable similarity to a part of the C-terminal domain. Of 68 Cα atoms of the N-terminal domain, 42 superimpose with Cα atoms of the C-terminal domain with an rms deviation of 1.2 Å. In addition, two segments that are distant in sequence from the superposing regions, namely residues 219 to 224, which are part of the N-terminal domain, and residues 70 to 75 of the C-terminal domain, superimpose almost exactly. Structure-based sequence alignment of the N and C domains shows that 10 out of 75 residues (13%) are identical, suggestive of an early gene duplication event in the evolution of this protein.
Dimer formation.
Gel filtration chromatography and dynamic light scattering indicate that WecD forms dimers in solution. In the crystal structure, the two WecD monomers within the asymmetric unit associate into a tight dimer (Fig. 3a). The two monomers of the dimer are virtually identical, giving an rms deviation of 0.45 Å for all main-chain atoms upon superposition. The dimer is created through interactions on the convex side of the β-sheet along strands β4 and β10 from each monomer that face each other. Helices α3 and α7 are also part of the dimer interface. In addition to hydrophobic contacts, there are several hydrogen bonds, predominantly between the side chains of one monomer (Asp57, Asn214, Thr218, and Tyr220) and the backbone of the other. There are no water molecules in the dimer interface and no cavities, indicating very good shape complementarity of the two interacting chains. The surface area buried by dimer formation calculated using a 1.4-Å probe is 1,153 Å2 per monomer, corresponding to 10.5% of the total surface area of the monomer.
Comparison with other structures.
Sequence comparison methods employed by various public databases (InterPro [http://www.ebi.ac.uk/interpro/] [41] and PFAM [http://www.sanger.ac.uk/Software/Pfam/] [4]) identified the presence of an acetyltransferase (GNAT) superfamily domain (PFAM PF00583) (46, 72). This is one of the largest sequence superfamilies, with more than 10,000 proteins from available sequenced genomes associated with this fold. The three-dimensional structure of WecD confirms the presence of this domain and localizes it to residues 70 to 219. A search for structurally similar proteins performed using DALI (26) identified a number of proteins containing a GNAT domain (72) that superimposed very well on the corresponding C-terminal domain of WecD. The closest structural relatives identified by DALI with a Z-score of >9 include Mycobacterium tuberculosis mycothiol synthase MshD (Rv0819; PDB code 1OZP [70]), an aminoglycoside acetyltransferase [AAC(6′)-ly] from Enterococcus faecium (PDB code 1S3Z [71]), an aminoglycoside 6′-N-acetyltransferase from Salmonella enterica (PDB code 1B87 [80]), a histone acetyltransferase (Hpa2) from Saccharomyces cerevisiae (PDB code 1QSM [2]), an acetyltransferase from Bacillus subtilis (PDB code 1TIQ; unpublished), the aminoglycoside 2′-N-acetyltransferase [AAC(2′)-Ic] from Mycobacterium tuberculosis (PDB code 1M44 [(69]), N-myristoyl transferase from Candida albicans (PDB code 1NMT [76]), and the FemX transferase from Weissella viridescens (PDB code 1NE9 [6]). As described above, the N-terminal domain shares clear structural similarity to a part of the C-terminal domain corresponding to a partial GNAT fold, missing the first two α-helices and two β-strands. It is not surprising, then, that the N-terminal domain of WecD can be aligned with parts of the GNAT domains of other proteins, albeit with rather low Z-values of 4 and less.
Four sequence motifs, designated C, D, A, and B (45, 46), are shared by all members of the GNAT superfamily. The typical N-acetyltransferase protein fold resembles β1-α1-α2-β2-β3-β4-α3-β5-α4-β6 (72) with variation mostly associated with the lengths of the loops linking β1 and α1, α1, and α2, the position of α2, and the length of β6. Motifs A and B are the most conserved segments and are critical to cofactor binding (12, 34, 64).
As is the case with most proteins from the GNAT superfamily, WecD associates into dimers which form the biologically functional unit. Mycothiol synthase MshD from Mycobacterium tuberculosis (70) and N-myristoyl transferase from S. cerevisiae (20) and C. albicans (76) are exceptions to this observation, as both proteins are active as monomers. However, these two proteins contain two repeats of the full-length GNAT domain, and while they do not form dimers, their GNAT domains associate in a manner resembling dimeric forms of other GNAT proteins.
Acetyl-CoA binding site.
In crystals of WecD cocrystallized with AcCoA, we observed electron density that corresponded well to this ligand. The electron density is well defined for the β-mercaptoethylamine and pantothenate portions of CoA, as well as the central pyrophosphate, but the adenosine and the 3′-phosphate have weaker and less well defined density, indicating substantial mobility at this end of the CoA cofactor (Fig. 4a). Both the substrate and cofactor binding sites are located within the tunnel that traverses the entire width of each WecD molecule. The AcCoA cofactor binds at the narrow side of this tunnel, with the acetyl group reaching the midpoint of the tunnel and the nucleotide portion extending outside the tunnel entrance and largely exposed to bulk solvent (Fig. 3). The AcCoA binding site is formed by residues from strands β8 and β9, the loop linking α4 and α5 as well as contributions from the loop β8-α6 and from helix α7. The loop between α4 and α5 is longer in WecD than in other GNAT proteins and affects the size and shape of the substrate-binding site. In most GNAT members, the active site resembles a canyon with AcCoA bound at the bottom, whereas the longer loop linking α4 and α5 in WecD closes up the canyon, converting it into a channel. Examination of the electrostatic potential near the AcCoA binding site reveals a particularly basic region near the pyrophosphate and adenosine 3′-phosphate groups.
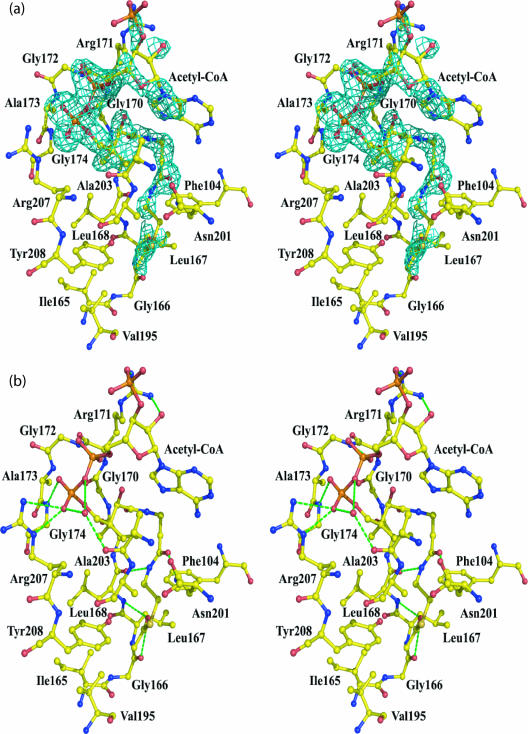
FIG. 4.
WecD acetyl-CoA binding site. (a) Stereo view of the Fo-Fc (omit) electron density around AcCoA from subunit A contoured at the 2.5 σ level. Residues interacting with AcCoA are indicated. (b) Interactions between AcCoA and WecD. Residues within 4 Å of CoA are shown. Hydrogen bonds are marked by dashed lines.
The variation in the quality of the electron density for CoA is reflected by a greater number of van der Waals and hydrogen-bonding interactions between the protein and the mercaptoethylamine and pantothenate moieties and by the pyrophosphate group, with very few contacts made by the adenosine (Fig. 4a). Each molecule of AcCoA adopts a bent conformation, with the bend occurring at the P2A atom of the pyrophosphate group. The linear part of CoA binds along strand β8, following the β-bulge produced by Gly166 and Leu167, and wedges between this strand and strand β9. It makes three hydrogen bonds to the main-chain atoms of Leu168 and Gly170 and to the side chain of Asn201 (Fig. 4b). The side chain of Tyr208 is 3.0 Å away from the sulfur atom of AcCoA and may also provide a hydrogen bond interaction. In addition to hydrogen-bonding interactions, Phe104, Leu168, Ala169, and Ala204 contribute to formation of a hydrophobic pocket suitable for binding the pantetheine moiety. The acetyl methyl group is oriented toward a hydrophobic pocket formed by residues Ile165 to Leu168 and Val195, and the carbonyl oxygen of the acetyl group is hydrogen bonded to the main chain amide of Leu168.
The pyrophosphate makes hydrogen bonds to the backbone NH groups of Gly172 and Gly174 and to the side chain of Arg207. These two glycine residues, located at the loop region linking strand β8 and helix α6, are part of the Arg/Gln-X-X-Gly-X-Gly/Ala segment of the GNAT motif A (46). In WecD, the first position of this motif is atypically an Ala instead of the usual Arg or Gln. The adenosine moiety at the end of the CoA molecule is less well defined in electron density, consistent with its few interactions with the protein. The adenosine folds back on the pantothenate portion of CoA with the formation of an intramolecular hydrogen bond between the pantothenate hydroxyl and the oxygen of the 5′-phosphate (Fig. 4b). Arg171 interacts with the adenosine segment, but these interactions differ in the two molecules. In subunit A, Arg171 forms a hydrogen bond to the terminal phosphate, while in subunit B, this hydrogen bond is not formed, and instead this side chain is positioned closer to the adenosine moiety. Several water molecules participate in bridging interactions between the CoA and protein.
Comparison with other GNAT family proteins shows that the mode of binding of the mercaptoethylamine and pantothenate segments of AcCoA found in WecD is similar to that observed for other GNAT proteins. However, binding of the ADP segment differs from that observed for other proteins. Indeed, the position of this segment of the CoA varies between different proteins and originates from differences in the binding mode of the diphosphate. In WecD, the loop that binds the diphosphate is five to six residues shorter than in other proteins. This shorter loop causes a different conformation of the CoA diphosphate and induces a sharp bend in the CoA structure, resulting in exposure of the adenine ring and the 3′-phosphate to the solvent and therefore greater mobility than in other GNAT proteins, where stronger interactions with the protein restrict their mobility.
Superposing the two subunits of AcCoA-bound WecD showed that the two monomers are very similar, except for a loop (residues Gly133 to His137) from subunit B which is flipped, such that the Cα of Phe135 moves ∼11 Å. Since TDP was included in the crystallization but was not clearly visible in the map, this reorientation of the loop could be the result of weak TDP binding. A second data set collected from a crystal in which TDP was omitted showed an identical conformation for the two loops, the same as chain A from WecD cocrystallized with TDP (not shown). The differences in structure between apo and AcCoA bound forms of WecD are minor. Two notable differences are observed in the orientation of side chains that are involved in binding AcCoA. The side chain of Arg171 bends toward and stabilizes the 3′-phosphate, while the side chain of Arg207 turns away from the binding site in order to avoid steric clashes with the cofactor.
Predicted TDP-Fuc4N binding site.
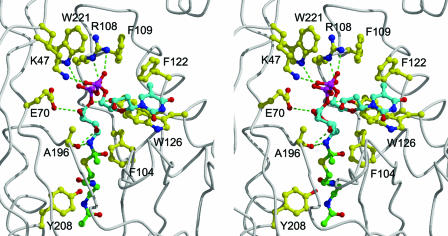
The three-dimensional structure reported here reveals that the acetyl group of AcCoA is located approximately halfway in a tunnel that traverses the WecD molecule. This tunnel leads to the AcCoA binding site on one face of the protein. On the opposite face of the WecD molecule, the tunnel opens into a relatively well-shaped pocket that can be regarded as the most probable binding site for the TDP-β-d-fucosamine substrate. The positions of the AcCoA sulfur atom and the tunnel surrounding the acetyl group help define the possible binding location of the substrate. In order to better define putative enzyme-substrate interactions in this region, we carried out flexible docking of TDP-Fuc4N, which was covalently linked as a tetrahedral intermediate transition state adduct to the acetyl group of WecD-bound AcCoA. A chemically meaningful anchor was thus introduced at the 4-nitrogen atom of the fucosamine moiety, which in turn allowed docking of the TDP-Fuc4N substrate by conformational sampling of its rotatable bonds using the Monte Carlo minimization approach (see Materials and Methods). The lowest-energy binding mode of TDP-Fuc4N for the WecD protein is shown in Fig. 5. All residues predicted to interact with TDP-Fuc4N are absolutely conserved in the sequences of the 14 known homologs of WecD.
FIG. 5.
Structural model of the WecD tetrahedral intermediate resulting from nucleophilic attack of the 4-amino group of TDP-fucosamine at the acetyl-CoA thioester carbon atom (stereo view). The backbone of the WecD protein is rendered as a tube. Select WecD residues lining the putative binding site for TDP-Fuc4N, together with the putative general acid Tyr208, are displayed. Carbon atoms are colored in cyan for the TDP-Fuc4N part, green for the AcCoA part, and yellow for the displayed WecD residues. The other atom types are colored as follows: N, blue; O, red; S, yellow; P, magenta. Intermolecular hydrogen bonds are indicated by green dashed lines. Hydrogen atoms are omitted for clarity.
In this model, the fucosamine moiety binds towards the end of the tunnel traversing the WecD molecule, where it establishes hydrogen bond and nonpolar contacts with residues Ala196, Glu70, Tyr123, and Phe104. The fucosamine pyranose ring adopts a chair conformation with the C-1 and C-4 substituents in the axial orientation and C-2, C-3, and C-5 substituents in the equatorial orientation, according to experimental structural data (27, 66).
The diphosphate group of the modeled TDP-Fuc4N binds at the opening of the tunnel, where it establishes salt bridge and hydrogen bond interactions with three WecD residues located on one side of the binding pocket. Therefore, the “in-register” disposition of the Arg108, Trp221, and Lys47 side chains, with the indole ring sandwiched between the two positively charged moieties, creates a hydrogen-bonding capability poised to accept and neutralize the negatively charged diphosphate group of the substrate. In the present crystal structure, a number of relatively well-structured water molecules were identified interacting with this hydrogen-bonding donor area. We also note the conformational restriction on the Arg108 side chain, which is sandwiched between the aromatic rings of Trp221 and Phe109 and also anchored by hydrogen bonds to the Gln45 and Tyr223 side chains.
The β-d-2-deoxyribose fragment is located completely outside the tunnel and is largely exposed to the solvent. In this lowest-energy model of the complex, the thymidine fragment adopts a conformation that is typically found in nucleotides, i.e., with the deoxyribose 5′ oxygen atom oriented on top of the deoxyribose ring and facing the thymine C-6—hydrogen atom (1, 8, 10, 22, 42).
The modeled thymine moiety interacts with the Phe122 side chain but mostly contacts a flat region of the WecD surface through parallel stacking with the indole ring of Trp126. Structural studies have repeatedly revealed parallel stacking interactions between the thymine base of nucleotides and aromatic rings of proteins (1, 8, 10, 22, 42). In our model, the other face of the thymine ring is completely exposed to solvent. We note, however, that when TDP was added to the crystallization mixture, we observed an alternate conformation for the loop Gly133 to His137 in one monomer in the dimer. Not only is the alternate conformation of this loop compatible with our model of the bound substrate, but it would potentially provide additional stabilization of the complex. Specifically, the alternate geometry packs the Phe135 side chain directly against the available face of the thymine ring (not shown), which would effectively sandwich the thymine ring between the Trp126 and Phe135 aromatic rings.
Implications for catalysis.
Analysis of the WecD-AcCoA complex, as well as the model of the tetrahedral intermediate with TDP-Fuc4N, offers insight into the mode of catalysis by this enzyme. Acetyl transfer likely occurs by direct nucleophilic attack of the TDP-fucose 4-amino group at the AcCoA thioester carbon, involving a ternary enzyme-substrate complex, as previously shown for various GNAT family members (11, 17, 31, 62, 63). While the kinetic mechanism of WecD has not yet been investigated, formation of an enzyme-substrate ternary complex is entirely reasonable, given the clear accessibility of the active sites, suggesting that both substrates can bind simultaneously. A ping-pong kinetic mechanism involving a covalent enzyme intermediate for WecD is chemically unlikely, as there are no residue side chains proximal to the acetyl moiety of AcCoA that could act as an acetyl acceptor.
Given that the pKa's for alkyl-amines are in the basic range (>9.5), the 4-amino group of the substrate likely requires deprotonation in order to act as an effective nucleophile. Various strategies have been put forward for how the substrate amino group is deprotonated in members of the GNAT family, including direct proton abstraction by an Asp or Glu residue (67), via an activated water molecule (24), or through a series of hydrogen-bonded water molecules that together form a “proton wire” (25, 69). In some instances, as with serotonin acetyltransferase (57, 79), clear evidence for a catalytic base has not yet been obtained. Inspection of the model of the WecD-bound adduct does not indicate any nearby residue that could directly function as a general base. The closest acidic residue, Glu68, is 7.5 Å from the 4-amino group in the tetrahedral intermediate model. Several water molecules are present in this region that could potentially serve as a base if deprotonated. The region surrounding the 4-amino group is predominantly nonpolar, which should favor deprotonation by lowering its pKa. That is, extensive burial of a protonated amino group in the middle of the tunnel traversing the WecD molecule would likely be accompanied by a significant desolvation cost, and no salt bridge interaction can be established within the tunnel to offset this. A single hydrogen bond is observed in the modeled tetrahedral intermediate, between the carbonyl O of Ala196 and the 4-amino group of the sugar. Unlike the case of a salt bridge interaction, this hydrogen bond interaction is not expected to favor the positively charged form of the bound amine.
A structurally conserved active site tyrosine in many GNAT enzymes, Tyr208 in WecD, is positioned 3.0 Å from the sulfur atom of CoA. This residue has been suggested to stabilize and protonate the thiolate anion of the leaving CoA and to assist in correctly orienting the acetyl group for transfer (24, 25, 49, 57). The GNA1 mutant enzyme Tyr143Ala has severely diminished activity, consistent with an important role for this residue in activity (40). Similarly, mutation of Tyr168 to Phe in serotonin N-acetyltransferase significantly reduced Vmax (25, 57). Studies of the pH-versus-rate profile for the latter enzyme established that Tyr168 is responsible for the basic limb of the activity profile, with an apparent pKa of 8.5 (57). The relatively nonpolar environment of Tyr208 in WecD may contribute to lowering its pKa such that it can function to protonate the thiolate anion leaving group.
Acknowledgments
We thank Martin McMillan and Leon Flaks for assistance in data collection. Data for this study were measured at beamlines X8C and X29 of the National Synchrotron Light Source. We also thank P. Iannuzzi for assistance in cloning, Stephane Raymond for maintenance of the computing environment, and Mirek Cygler for support and encouragement.
This work was supported in part by a CIHR grant 200103GSP-90094-GMX-CFAA-19924. Financial support of PXRR beamlines comes principally from the Office of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy and the National Center for Research Resources of the National Institutes of Health.
REFERENCES
- 1.Allard, S. T., W. W. Cleland, and H. M. Holden. 2004. High resolution X-ray structure of dTDP-glucose-4,6-dehydratase from Streptomyces venezuelae. J. Biol. Chem. 279:2211-2220. [DOI] [PubMed] [Google Scholar]
- 2.Angus-Hill, M. L., R. N. Dutnall, S. T. Tafrov, R. Sternglanz, and V. Ramakrishnan. 1999. Crystal structure of the histone acetyltransferase Hpa2: a tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J. Mol. Biol. 294:1311-1325. [DOI] [PubMed] [Google Scholar]
- 3.Barr, K., and P. D. Rick. 1987. Biosynthesis of enterobacterial common antigen in Escherichia coli. In vitro synthesis of lipid-linked intermediates. J. Biol. Chem. 262:7142-7150. [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayly, C. I., P. Cieplak, W. D. Cornell, and P. A. Kollman. 1993. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 97:10269-10280. [Google Scholar]
- 6.Biarrotte-Sorin, S., A. P. Maillard, J. Delettré, W. Sougakoff, M. Arthur, and C. Mayer. 2004. Crystal structures of Weissella viridescens FemX and its complex with UDP-MurNac-pentapeptide: insights into FemABX family substrates recognition. Structure 12:257-267. [DOI] [PubMed] [Google Scholar]
- 7.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherfils, J., S. Morera, I. Lascu, M. Vernon, and J. Janin. 1994. X-ray structure of nucleoside diphosphate kinase complexed with thymidine diphosphate and Mg2+ at 2-Å resolution. Biochemistry 33:9062-9069. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhury, S. F., J. Sivaraman, J. Wang, G. Devanathan, P. Lachance, H. Qi, R. Menard, J. Lefebvre, Y. Konishi, M. Cygler, T. Sulea, and E. O. Purisima. 2002. Design of noncovalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides. J. Med. Chem. 45:5321-5329. [DOI] [PubMed] [Google Scholar]
- 10.Christendat, D., V. Saridakis, A. Dharamsi, A. Bochkarev, E. F. Pai, C. H. Arrowsmith, and A. M. Edwards. 2000. Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP. J. Biol. Chem. 275:24608-24612. [DOI] [PubMed] [Google Scholar]
- 11.Clements, A., J. R. Rojas, R. C. Trievel, L. Wang, S. L. Berger, and R. Marmorstein. 1999. Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO J. 18:3521-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman, C. S., H. Huang, and A. E. Pegg. 1996. Structure and critical residues at the active site of spermidine/spermine-N1-acetyltransferase. Biochem. J. 316:697-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornell, W. D., P. Cieplak, C. L. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117:5179-5197. [Google Scholar]
- 14.Cornell, W. D., P. Cieplak, C. I. Bayly, and P. A. Kollman. 1993. Application of RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J. Am. Chem. Soc. 115:9620-9631. [Google Scholar]
- 15.Danese, P. N., G. R. Oliver, K. Barr, G. D. Bowman, P. D. Rick, and T. J. Silhavy. 1998. Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli. J. Bacteriol. 180:5875-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniels, D. L., G. Plunkett III, V. Burland, and F. R. Blattner. 1992. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science 257:771-778. [DOI] [PubMed] [Google Scholar]
- 17.De Angelis, J., J. Gastel, D. C. Klein, and P. A. Cole. 1998. Kinetic analysis of the catalytic mechanism of serotonin N-acetyltransferase (E.C. 2.3.1.87). J. Biol. Chem. 273:3045-3050. [DOI] [PubMed] [Google Scholar]
- 18.Dyda, F., D. C. Klein, and A. B. Hickman. 2000. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 29:81-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erbel, P. J., K. Barr, N. Gao, G. J. Gerwig, P. D. Rick, and K. H. Gardner. 2003. Identification and biosynthesis of cyclic enterobacterial common antigen in Escherichia coli. J. Bacteriol. 185:1995-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farazi, T. A., G. Waksman, and J. I. Gordon. 2001. Structures of Saccharomyces cerevisiae N-myristoyltransferase with bound myristoylCoA and peptide provide insights about substrate recognition and catalysis. Biochemistry 40:6335-6343. [DOI] [PubMed] [Google Scholar]
- 21.Farnback, M., L. Eriksson, S. Senchenkova, K. Zych, Y. A. Knirel, Z. Sidorczyk, and G. Widmalm. 2003. Crystal structure of a cyclic enterobacterial common antigen. Angew. Chem. 42:2543-2546. [DOI] [PubMed] [Google Scholar]
- 22.Fioravanti, E., A. Haouz, T. Ursby, H. Munier-Lehmann, M. Delarue, and D. Bourgeois. 2003. Mycobacterium tuberculosis thymidylate kinase: structural studies of intermediates along the reaction pathway. J. Mol. Biol. 327:1077-1092. [DOI] [PubMed] [Google Scholar]
- 23.Gouet, P., X. Robert, and E. Courcelle. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He, H., Y. Ding, M. Bartlam, F. Sun, Y. Le, X. Qin, H. Tang, R. Zhang, A. Joachimiak, J. Liu, N. Zhao, and Z. Rao. 2003. Crystal structure of tabtoxin resistance protein complexed with acetyl coenzyme A reveals the mechanism for β-lactam aceylation. J. Mol. Biol. 325:1019-1030. [DOI] [PubMed] [Google Scholar]
- 25.Hickman, A. B., M. A. A. Namboodiri, D. C. Klein, and F. Dyda. 1999. The structural basis of ordered substrate binding by serotonin N-acetyltransferase: enzyme complex at 1.8 Å resolution with a bisubstrate analog. Cell 97:361-389. [DOI] [PubMed] [Google Scholar]
- 26.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 27.Hwang, B. Y., H. J. Lee, Y. H. Yang, H. S. Joo, and B. G. Kim. 2004. Characterization and investigation of substrate specificity of the sugar aminotransferase WecE from E. coli K12. Chem. Biol. 11:915-925. [DOI] [PubMed] [Google Scholar]
- 28.Jones, T. A., J.-Y. Zou, S. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:100-119. [DOI] [PubMed] [Google Scholar]
- 29.Kiss, P., J. Rinno, G. Schmidt, and H. Mayer. 1978. Structural studies on the immunogenic form of the enterobacterial common antigen. Eur. J. Biochem. 88:211-218. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn, H. M., U. Meier-Dieter, and H. Mayer. 1988. ECA, the enterobacterial common antigen. FEMS Microbiol. Rev. 3:195-222. [DOI] [PubMed] [Google Scholar]
- 31.Lau, O. D., A. D. Courtney, A. Vassilev, L. A. Marzilli, R. J. Cotter, Y. Nakatani, and P. A. Cole. 2000. p300/CBP-associated factor histone acetyltransferase processing of a peptide substrate. J. Biol. Chem. 275:21953-21959. [DOI] [PubMed] [Google Scholar]
- 32.Li, Z., and H. A. Scheraga. 1987. Monte Carlo-minimization approach to the multiple-minima problem in protein folding. Proc. Natl. Acad. Sci. USA 84:6611-6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, L. Y., T. Sulea, R. Szittner, V. Vassilyev, E. O. Purisima, and E. A. Meighen. 2001. Modeling of the bacterial luciferase-flavin mononucleotide complex combining flexible docking with structure-activity data. Prot. Sci. 10:1563-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, L., K. A. Berkey, and R. A. Casero, Jr. 1996. RGFGIGS is an amino acid sequence required for acetyl coenzyme A binding and activity of human spermidine/spermine N1acetyltransferase. J. Biol. Chem. 271:18920-18924. [DOI] [PubMed] [Google Scholar]
- 35.Lugowski, C., E. Romanowska, L. Kenne, and B. Lindberg. 1983. Identification of a trisaccharide repeating unit in the enterobacterial common antigen. Carbohydr. Res. 118:173-181. [DOI] [PubMed] [Google Scholar]
- 36.Makela, P. H., and H. Mayer. 1976. Enterobacterial common antigen. Bacteriol. Rev. 40:591-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuhashi, M., and J. L. Strominger. 1964. Thymidine diphosphate 4-acetamido-4,6-dideoxyhexoses. J. Biol. Chem. 239:2454-2463. [PubMed] [Google Scholar]
- 38.Meier-Dieter, U., R. Starman, K. Barr, H. Mayer, and P. D. Rick. 1990. Biosynthesis of enterobacterial common antigen in Escherichia coli. Biochemical characterization of Tn10 insertion mutants defective in enterobacterial common antigen synthesis. J. Biol. Chem. 265:13490-13497. [PubMed] [Google Scholar]
- 39.Meier-Dieter, U., K. Barr, R. Starman, L. Hatch, and P. D. Rick. 1992. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. Molecular cloning of the rfe-rff gene cluster. J. Biol. Chem. 267:746-753. [PubMed] [Google Scholar]
- 40.Mio, T., T. Yamada-Okabe, M. Arisawa, and H. Yamada-Okabe. 1999. Saccharomyces cerevisiae GNA1, an essential gene encoding a novel acetyltransferase involved in UDP-N-acetylglucosamine synthesis. J. Biol. Chem. 274:424-429. [DOI] [PubMed] [Google Scholar]
- 41.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, A. Bateman, D. Binns, P. Bradley, P. Bork, P. Bucher, L. Cerutti, R. Copley, E. Courcelle, U. Das, R. Durbin, W. Fleischmann, J. Gough, D. Haft, N. Harte, N. Hulo, D. Kahn, A. Kanapin, M. Krestyaninova, D. Lonsdale, R. Lopez, I. Letunic, M. Madera, J. Maslen, J. McDowall, A. Mitchell, A. N. Nikolskaya, S. Orchard, M. Pagni, C. P. Ponting, E. Quevillon, J. Selengut, C. J. A. Sigrist, V. Silventoinen, D. J. Studholme, R. Vaughan, and C. H. Wu. 2005. InterPro, progress and status in 2005. Nucleic Acids Res. 33:D201-D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulichak, A. M., W. Lu, H. C. Losey, C. T. Walsh, and R. M. Garavito. 2004. Crystal structure of vancosaminyltransferase GtfD from the vancomycin biosynthetic pathway: interactions with acceptor and nucleotide ligands. Biochemistry 43:5170-5180. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov, G. N., A. A. Vagin, A. Lebedev, K. S. Wilson, and E. J. Dodson. 1999. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. Sect. D Biol Crystallogr. 55:247-255. [DOI] [PubMed] [Google Scholar]
- 44.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536-540. [DOI] [PubMed] [Google Scholar]
- 45.Neuwald, A. F., J. S. Liu, D. J. Lipman, and C. E. Lawrence. 1997. Extracting protein alignment models from the sequence database. Nucleic Acids Res. 25:1665-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neuwald, A. F., and D. Landsman. 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22:154-155. [DOI] [PubMed] [Google Scholar]
- 47.Nnalue, N. A., and B. A. Stocker. 1987. The effects of O-antigen character and enterobacterial common antigen content on the in vivo persistence of aromatic-dependent Salmonella sp. live-vaccine strains. Microb. Pathog. 3:31-44. [DOI] [PubMed] [Google Scholar]
- 48.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 49.Peneff, C., D. Mengin-Lecreulx, and Y. Bourne. 2001. The crystal structures of apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase. J. Biol. Chem. 276:16328-16334. [DOI] [PubMed] [Google Scholar]
- 50.Rahman, A., K. Barr, and P. D. Rick. 2001. Identification of the structural gene for the TDP-Fuc4NAc:lipid II Fuc4NAc transferase involved in synthesis of enterobacterial common antigen in Escherichia coli K-12. J. Bacteriol. 183:6509-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramos-Morales, F., A. I. Prieto, C. R. Beuzon, D. W. Holden, and J. Casadesus. 2003. Role for Salmonella enterica enterobacterial common antigen in bile resistance and virulence. J. Bacteriol. 185:5328-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rick, P. D., H. Mayer, B. A. Neumeyer, S. Wolski, and D. Bitter-Suermann. 1985. Biosynthesis of enterobacterial common antigen. J. Bacteriol. 162:494-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rick, P. D., and R. P. Silver. 1996. Enterobacterial common antigen and capsular polysaccharides, p. 104-122. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 54.Rick, P. D., G. L. Hubbard, M. Kitaoka, H. Nagaki, T. Kinoshita, S. Dowd, V. Simplaceanu, and C. Ho. 1998. Characterization of the lipid-carrier involved in the synthesis of enterobacterial common antigen (ECA) and identification of a novel phosphoglyceride in a mutant of Salmonella typhimurium defective in ECA synthesis. Glycobiology 8:557-567. [DOI] [PubMed] [Google Scholar]
- 55.Rick, P. D., K. Barr, K. Sankaran, J. Kajimura, J. S. Rush, and C. J. Waechter. 2003. Evidence that the wzxE gene of Escherichia coli K-12 encodes a protein involved in the transbilayer movement of a trisaccharide-lipid repeat intermediate in the assembly of enterobacterial common antigen. J. Biol. Chem. 278:16534-16542. [DOI] [PubMed] [Google Scholar]
- 56.Rinno, J., J. Gmeiner, J. R. Golecki, and H. Mayer. 1980. Localization of enterobacterial common antigen: Proteus mirabilis and its various L-forms. J. Bacteriol. 141:822-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheibner, K. A., J. De Angelis, S. K. Burley, and P. A. Cole. 2002. Investigation of the roles of catalytic residues in serotonin N-acetyltransferase. J. Biol. Chem. 277:18118-18126. [DOI] [PubMed] [Google Scholar]
- 58.Schmidt, M. W., K. K. Baldridge, J. A. Boatz, S. T. Elbert, M. S. Gordon, J. H. Jensen, S. Koseki, N. Matsunaga, K. A. Nguyen, S. Su., T. L. Windus, M. Dupuis, and J. A. Montgomery. 1993. General atomic and molecular electronic structure system. J. Comput. Chem. 14:1347-1363. [Google Scholar]
- 59.Schneider, T. R., and G. M. Sheldrick. 2002. Substructure solution with ShelxD. Acta Crystallogr. Sect. D Biol. Crystallogr. 58:1772-1779. [DOI] [PubMed] [Google Scholar]
- 60.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sivaraman, J., R. S. Meyers, L. Boju, T. Sulea, M. Cygler, J. Davisson, and J. D. Schrag. 2005. Crystal structure of Methanobacterium thermoautotrophicum phosphoribosyl-AMP cyclohydrolase HisI. Biochemistry 44:10071-10080. [DOI] [PubMed] [Google Scholar]
- 62.Tanner, K. G., M. R. Langer, Y. Kim, and J. M. Denu. 2000. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J. Biol. Chem. 275:22048-22055. [DOI] [PubMed] [Google Scholar]
- 63.Tanner, K. G., M. R. Langer, and J. M. Denu. 2000. Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry 39:11961-11969. [DOI] [PubMed] [Google Scholar]
- 64.Tercero, J. C., L. E. Riles, and R. B. Wickner. 1992. Localized mutagenesis and evidence for post-transcriptional regulation of MAK3. A putative N-acetyltransferase required for double-stranded RNA virus propagation in Saccharomyces cerevisiae. J. Biol. Chem. 267:20270-20276. [PubMed] [Google Scholar]
- 65.Terwilliger, T. C. 2000. Maximum-likelihood density modification. Acta Crystallogr. Sect. D Biol. Crystallogr. 56:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thoden, J. B., J. Kim, F. M. Raushel, and H. M. Holden. 2002. Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis. J. Biol. Chem. 277:45458-45465. [DOI] [PubMed] [Google Scholar]
- 67.Trievel, R. C., J. R. Rojas, D. E. Sterner, R. V. Venkataramani, L. Wang, J. Zhou, C. D. Allis, S. L. Berger, and R. Marmorstein. 1999. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl. Acad. Sci. USA 96:8931-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Valtonen, M. V., U. M. Larinkari, M. Plosila, V. V. Valtonen, and P. H. Makela. 1976. Effect of enterobacterial common antigen on mouse virulence of. Salmonella typhimurium. Infect. Immun. 13:1601-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vetting, M. W., S. S. Hegde, F. Javid-Majd, J. S. Blanchard, and S. L. Roderick. 2002. Aminoglycoside 2′-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates. Nat. Struct. Biol. 9:653-658. [DOI] [PubMed] [Google Scholar]
- 70.Vetting, M. W., S. L. Roderick, M. Yu, and J. S. Blanchard. 2003. Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases. Protein Sci. 12:1954-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vetting, M. W., S. Magnet, E. Nieves, S. L. Roderick, and J. S. Blanchard. 2004. A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones. Chem. Biol. 11:565-573. [DOI] [PubMed] [Google Scholar]
- 72.Vetting, M. W., L. P. S. de Carvalho, M. Yu, S. S. Hegde, S. Magnet, S. L. Roderick, and J. S. Blanchard. 2005. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433:212-226. [DOI] [PubMed] [Google Scholar]
- 73.Vinogradov, E. V., Y. A. Knirel, J. E. Thomas-Oates, A. S. Shashkov, and V. L. L'vov. 1994. The structure of the cyclic enterobacterial common antigen (ECA) from Yersinia pestis. Carbohydr. Res. 258:223-232. [DOI] [PubMed] [Google Scholar]
- 74.Wang, J., R. M. Wolf, J. W. Caldwell, P. A. Kollman, and D. A. Case. 2004. Development and testing of a general amber force field. J. Comput. Chem. 25:1157-1174. [DOI] [PubMed] [Google Scholar]
- 75.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic. Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weston, S. A., R. Camble, J. Colls, G. Rosenbrock, I. Taylor, M. Egerton, A. D. Tucker, A. Tunnicliffe, A. Mistry, F. Mancia, E. de la Fortelle, J. Irwin, G. Bricogne, and R. A. Pauptit. 1998. Crystal structure of the anti-fungal target N-myristoyl transferase. Nat. Struct. Biol. 5:213-221. [DOI] [PubMed] [Google Scholar]
- 77.Whang, H. Y., and E. Neter. 1962. Immunological studies of a heterogenetic enterobacterial antigen (Kunin). J. Bacteriol. 84:1245-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Winn, M. D., A. W. Ashton, P. J. Briggs, C. C. Ballard, and P. Patel. 2002. Ongoing developments in CCP4 for high-throughput structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1929-1936. [DOI] [PubMed] [Google Scholar]
- 79.Wolf, E., J. De Angelis, E. M. Khalil, P. A. Cole, and S. K. Burley. 2002. X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition. J. Mol. Biol. 317:215-224. [DOI] [PubMed] [Google Scholar]
- 80.Wybenga-Groot, L. E., K. Draker, G. D. Wright, and A. M. Berghuis. 1999. Crystal structure of an aminoglycoside 6′-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Structure 7:497-507. [DOI] [PubMed] [Google Scholar]