Abstract
The feedback-inhibited form of Bacillus subtilis glutamine synthetase regulates the activity of the TnrA transcription factor through a protein-protein interaction that prevents TnrA from binding to DNA. Five mutants containing feedback-resistant glutamine synthetases (E65G, S66P, M68I, H195Y, and P318S) were isolated by screening for colonies capable of cross-feeding Gln− cells. In vitro enzymatic assays revealed that the mutant enzymes had increased resistance to inhibition by glutamine, AMP, and methionine sulfoximine. The mutant proteins had a variety of enzymatic alterations that included changes in the levels of enzymatic activity and in substrate Km values. Constitutive expression of TnrA- and GlnR-regulated genes was seen in all five mutants. In gel mobility shift assays, the E65G and S66P enzymes were unable to inhibit TnrA DNA binding, while the other three mutant proteins (M68I, H195Y, and P318S) showed partial inhibition of TnrA DNA binding. A homology model of B. subtilis glutamine synthetase revealed that the five mutated amino acid residues are located in the enzyme active site. These observations are consistent with the hypothesis that glutamine and AMP bind at the active site to bring about feedback inhibition of glutamine synthetase.
Glutamine is a key compound in nitrogen metabolism that serves as the nitrogen donor for the synthesis of 25% of the nitrogen-containing compounds in cells (33). Glutamine synthetase (GS) is a metalloenzyme that catalyzes the ATP-dependent synthesis of glutamine from glutamate and ammonium. Although Mg2+ or Mn2+ can be used to assay GS in vitro, the Mg2+-dependent reaction is the physiologically relevant enzymatic activity (34, 48).
To ensure that adequate supplies of glutamine are available for growth under all nutritional conditions, the expression and activity of GS are tightly regulated. GS levels are low when cells are growing with excess nitrogen but increase when nitrogen becomes limiting for growth. Nitrogen regulation in Bacillus subtilis is mediated by two closely related transcriptional factors, GlnR and TnrA (13). The genes encoding GlnR (glnR) and GS (glnA) are located together within the dicistronic glnRA operon (40). GlnR represses expression of glnRA, tnrA, and the genes encoding the urea-degradative enzymes (4, 36, 45, 53). TnrA controls the expression of multiple genes that include the catabolic enzymes and transport systems for nitrogen-containing compounds, which are utilized during nitrogen-limited growth (2, 12, 13, 17, 46, 52). GlnR and TnrA bind to sites with similar consensus sequences but are active under different growth conditions (21, 29, 49, 53). GlnR represses gene expression during growth with excess nitrogen, while TnrA represses and activates gene expression when nitrogen is limiting. Genetic analysis indicates that GS is required for the regulation of the activities of both TnrA and GlnR (36, 46, 48). Although the precise role of GS in controlling GlnR activity has not yet been determined, the feedback-inhibited form of GS forms a protein-protein complex with TnrA and prevents TnrA from binding to DNA (51).
The enzymatic activity of microbial GS is controlled posttranslationally by a variety of mechanisms. In Escherichia coli, GS activity is regulated by adenylylation, the covalent attachment of AMP to a specific tyrosine residue (34). The unmodified form of E. coli GS requires Mg2+ for optimal enzymatic activity and is relatively insensitive to feedback inhibition, while the adenylylated enzyme requires Mn2+ for activity and is subject to feedback inhibition by AMP and eight other end products of glutamine metabolism (34). Glutamine is not a feedback inhibitor of either form of the E. coli enzyme (43). Thus, under normal physiological conditions, where Mg2+ levels are relatively high and only trace amounts of Mn2+ are present, the activity of the E. coli GS is controlled by adenylylation (34). Although the tyrosine residue that is the site of adenylylation in E. coli GS is conserved in B. subtilis GS (40), posttranslational modification of B. subtilis GS has not been observed (16). Instead, the physiologically relevant Mg2+-dependent enzymatic activity of B. subtilis GS is subject to feedback inhibition by glutamine and AMP (9). Interestingly, although glutamine acts as a feedback inhibitor for only the B. subtilis GS enzyme, the amino acid sequences of the E. coli and B. subtilis GS proteins are highly similar (43% identical and 65% similar residues) (40, 47). The amino acid and structural differences that allow glutamine to bind with sufficient affinity to regulate the activity of the B. subtilis GS enzyme, but not the activity of the E. coli GS enzyme, are not understood.
The model in which the feedback-inhibited form of the B. subtilis GS regulates TnrA activity in vivo is supported by the properties of a mutant encoding the feedback-resistant S186F GS (48). Cells synthesizing the S186F enzyme express both TnrA- and GlnR-regulated genes constitutively. Although the feedback-resistant property of S186F GS results from reduced affinity for glutamine and AMP, no differences between the kinetic properties of the S186F and wild-type GS enzymes could be detected. The Ser-186 residue is located on a β-strand that lines the active site. Since the side chain of Ser-186 is oriented so that it points away from the active site, the phenylalanine substitution at this position indirectly confers resistance to glutamine and AMP feedback inhibition.
To identify additional residues in B. subtilis GS required for feedback inhibition, mutants encoding feedback-resistant GS enzymes were isolated using a novel plate screen. Interestingly, the five new feedback-resistant GS mutants isolated in this study were also found to be defective in regulating the activities of GlnR and TnrA.
MATERIALS AND METHODS
Bacterial strains, cell growth, and media.
Table 1 lists the B. subtilis strains used in this study. Derivatives of the feedback-resistant mutant strains containing lacZ fusions were constructed as previously described (44). The methods used for bacterial cultivation in the minimal medium of Neidhardt et al. (30) have been described previously (1). BSS minimal medium agar plates were prepared as previously described (5). Glucose was added to all media at a final concentration of 0.5%. All nitrogen sources were added to a final concentration of 0.2%. Casamino Acids, an acid hydrolysate of casein, was obtained from Difco Laboratories. 5-Bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) was added to agar plates to give a final concentration of 40 μg/ml.
TABLE 1.
Strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| 168 | trpC2 | This laboratory |
| SF300 | amtB::Tn917-lacZ trpC2 | 44 |
| SF300G7 | amtB::Tn917-lacZ ΔglnA207::spc trpC2 | SF300 × pGLN207 DNA |
| SF416G | amyE::[(amtB-lacZ)416 neo)] ΔglnA14::spc trpC2 | 14 |
| SF17GT | [amyE::(glnRA-lacZ)17 neo)] ΔglnA14::spc tnrA62::Tn917 trpC2 | 14 |
B. subtilis genotype symbols are those of Biaudet et al. (3). The amtB-glnK operon was formerly called nrgAB.
Plasmid construction.
Plasmid pGLN207, which is a clone of the glnRA region in which the entire glnA coding sequence has been replaced with a spectinomycin resistance cassette, was constructed in three steps. A 382-bp DNA fragment containing sequences located immediately downstream of the glnA gene was prepared by PCR amplification with primers GSDEL3 (5′-GTC ACT GCA GGT AAT AAA TTA TGG AGC GGC) and GSDEL4 (5′-ACT GAA GCT TTA AGA GTC ATC TAT AAC CGC) and inserted into pJDC9 (6) as a PstI-HindIII fragment to construct pGLN205. A 405-bp DNA fragment containing sequences located immediately upstream of the glnA gene was prepared by PCR amplification with primers GSDEL1 (5′-AGT CTC TAG ATA GTT ATC AGC AAG ACA AAT TCG) and GSDEL2 (5′-GTC ACT GCA GAA AAG GTA AAA ATG TAT ATA GC) and inserted into pGLN205 as an XbaI-PstI fragment to give pGLN206. Plasmid pGLN207 was constructed by inserting a spectinomycin resistance gene cassette (20) into the PstI site of pGLN206. Plasmid pGLN209 contains a neomycin resistance gene cassette (20) cloned into the BamHI site of pSF42 (15).
Mutant isolation.
pGLN209 plasmid DNA, which contains the entire glnA gene, was mutagenized by propagation in XL1-Red, an E. coli mutator strain (Stratagene). Mutagenized pGLN209 DNA was used to transform strain SF300G7 (ΔglnA207::spc amtB::Tn917-lacZ) cells to Gln+ on glucose minimal BSS plates containing X-Gal and an excess nitrogen source that lacked glutamine. A previously isolated mutant encoding a feedback-resistant GS (S186F) was shown to grow poorly with glutamate plus ammonium as the nitrogen source but had no defect when grown with Casamino Acids plus glutamate plus ammonium as the nitrogen source (48). Therefore, both glutamate plus ammonium and Casamino Acids plus glutamate plus ammonium were used as excess nitrogen sources during the mutant screen in order to avoid any growth-dependent bias. Gln+ transformants resulting from double-crossover events were identified by the absence of plasmid-encoded neomycin resistance.
Enzyme assays.
β-Galactosidase activity was assayed in crude extracts prepared from cells grown to mid-log growth phase (70 to 90 Klett units) as previously described (1). β-Galactosidase levels were corrected for the endogenous activity present in B. subtilis cells containing the promoterless lacZ gene from pSFL6 integrated at the amyE site.
The biosynthetic and reverse (transferase) enzymatic activities of glutamine synthetase were measured by the production of γ-glutamylhydroxamate as previously described (14). The kinetic constants for the Mg2+-dependent biosynthetic reaction were determined as previously described (48). The glutamine, AMP, and methionine sulfoximine concentrations necessary to reduce enzymatic activity by 50% (IC50) were determined with the Mg2+-dependent biosynthetic reaction, where the glutamate and ATP concentrations were 150 and 18 mM, respectively (48).
DNA and protein methods.
DNA sequencing of the feedback-resistant mutations and construction of mutant GS overexpression plasmids were performed as previously described (14). Purification of TnrA and GS were done by published procedures (50, 51). The concentrations of TnrA and GS were determined by measuring their absorbances at 280 nm. The molar absorption coefficients of the proteins were calculated from their amino acid sequences (31). Gel mobility shift experiments to examine the abilities of wild-type and mutant GS proteins to inhibit DNA binding by TnrA were performed as previously described (51).
Bioinformatics.
To identify amino acid residues uniquely conserved in GS proteins subject to feedback inhibition, a multiple-sequence alignment was used to compare 30 Bacilli GS proteins with 28 GS proteins from Proteobacteria. The rationale used for selection of the GS sequences in this alignment was as follows. Since feedback inhibition of B. subtilis GS is required for the regulation of GlnR and TnrA activity (48), a set of GS proteins that are presumably subject to feedback inhibition was obtained by selecting GS sequences from Bacilli species that encode orthologs of the B. subtilis GlnR or TnrA protein. The physiologically significant regulation of E. coli GS activity is mediated by adenylylation rather than by feedback inhibition (34). A set of GS proteins that presumably have the same pattern of regulation as the E. coli enzyme was obtained by selecting GS sequences from Proteobacteria species that encode orthologs of E. coli GS adenyltransferase, the enzyme that catalyzes the adenylylation of GS. None of the Proteobacteria species encode orthologs of GlnR or TnrA. None of the Bacilli species encode orthologs of the E. coli GS adenyltransferase. The sequence alignment was prepared with ClustalW (41) and manually edited with GeneDoc (http://www.psc.edu/biomed/genedoc/) to remove gaps within secondary-structural elements. The accession numbers for the protein sequences used in this alignment are listed in Table S1 in the supplemental material. The multiple-sequence alignment is presented in Fig. S1 in the supplemental material. Consensus sequence logos were prepared with WebLogo (7). A homology model of B. subtilis GS complexed with the reaction products glutamine, ADP, and phosphate was constructed using DeepView software and the SWISS-MODEL web server (37). The X-ray crystal structure of Salmonella enterica serovar Typhimurium GS complexed with ADP and the glutamine analogue phosphinothricin (18) was used as the template. The alignment of the B. subtilis and S. enterica serovar Typhimurium sequences used for modeling was guided by the multiple-sequence alignment described above and differs in the carboxyl-terminal region from the alignment used for a previous homology model (14). ADP and magnesium ions were placed into the active site of the homology model by structurally aligning Mycobacterium tuberculosis GS complexed with methionine sulfoximine phosphate and ADP (22) with the homology model. Similarly, glutamine was positioned in the active site by using the glutamate-bound structure of S. enterica serovar Typhimurium GS (23) as a template. Using the results of the structural model of S. enterica serovar Typhimurium GS complexed with the enzymatic reaction products (23) as a guide, a phosphate anion was manually docked into the active site and positioned so that it formed hydrogen bonds with the amide nitrogen of glutamine and the side chains of Arg-316 and Arg-335.
Protein structure accession number.
The coordinates for the homology model were deposited in the Protein Data Bank (accession number 2FWX).
RESULTS
Isolation and characterization of mutants that encode feedback-resistant GS.
A plate screen was developed that allowed direct visual identification of mutants with feedback-resistant glnA mutations. This screen was based on our previous observation that strain SF186, which synthesizes a feedback-resistant GS enzyme, excretes sufficient glutamine to cross-feed glnA null mutants (48). As a result, when the SF186 cells are plated on a lawn of Gln− cells in the absence of glutamine, the SF186 colonies become surrounded by a halo of Gln− cells. This cross-feeding phenotype was used to identify additional mutants that encode feedback-resistant GS enzymes.
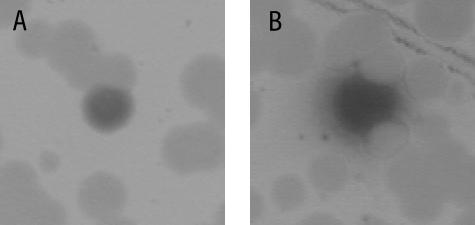
Mutagenized glnA DNA was used to transform strain SF300G7 (ΔglnA amtB-lacZ) to Gln+. Transcription from the TnrA-dependent amtB-lacZ fusion was visualized by including X-Gal, a chromogenic substrate for β-galactosidase, in the selection plates. Two types of blue colonies were found among the Gln+ transformants: colonies with sharp borders and colonies surrounded by a halo of blue Gln− cells (Fig. 1). Mutants that synthesize GS enzymes that are feedback sensitive but are unable to regulate the activity of TnrA give rise to blue Gln+ colonies with sharp borders (14). Blue Gln+ colonies with halos secrete glutamine due to the presence of a feedback-resistant GS and thus cross-feed the surrounding Gln− cells on the transformation plate. The Gln− cells in the halo are blue because TnrA-regulated genes are expressed constitutively in glnA null mutants (46). All of the colonies observed in the mutant screening that had halos were also colored blue. The nucleotide lesions in 15 independently isolated mutants with the cross-feeding phenotype were determined by PCR amplification and sequencing of the glnRA operon. In addition to the previously described glnA186 mutation (48), five new mutations in the glnA gene were identified (Table 2).
FIG. 1.
Colony morphology of Gln+ transformants. Gln+ transformants were selected on glucose X-Gal minimal medium containing Casamino Acids, glutamate, and ammonium as the nitrogen sources after transformation of strain SF300G7 with mutagenized glnA DNA. (A) A blue Gln+ colony with sharp borders. (B) A blue Gln+ colony with a halo of blue Gln− cells.
TABLE 2.
TnrA- and GlnR-dependent regulation in wild-type and glnA mutant strains
| Relevant genotype | Amino acid change | Codon change | TnrA-dependent regulation (β-galactosidase sp act [U/mg protein]) in amtB-lacZ fusion strain grown ona:
|
GlnR-dependent regulation (β-galactosidase sp act [U/mg protein]) in a glnRA-lacZ fusion strain grown ona:
|
||||
|---|---|---|---|---|---|---|---|---|
| Casamino Acids + glutamate | Glutamine | Glutamate | Casamino Acids + glutamate | Glutamine | Glutamate | |||
| Wild type | 6.2 | 0.05 | 111 | 21 | 0.3 | 42 | ||
| glnA(E65G) | Glu65 → Gly | GAG → GGG | 137 | NGb | 146 | 39 | NG | 54 |
| glnA(S66P) | Ser66 → Pro | TCA → CCA | 159 | NG | 125 | 42 | NG | 37 |
| glnA(M68I) | Met68 → Ile | ATG → ATA | 106 | 14 | 118 | NDc | 17 | 38 |
| glnA(H195Y) | His195 → Tyr | CAC → TAC | 80 | 7.2 | 105 | ND | 26 | 46 |
| glnA(P318S) | Pro318 → Ser | CCG → TCG | 94 | 12 | 90 | ND | 46 | 50 |
Cells were grown in glucose minimal medium containing the indicated nitrogen sources. Values are the average of two or more determinations and did not vary by more than 20%. Strains contained either the (amtB-lacZ)416 fusion or the (glnRA-lacZ)17 fusion. Strains with the glnRA-lacZ fusion also contained a tnrA null mutation.
NG, no growth.
ND, not determined.
When the growth properties of these five glnA mutants were examined on glucose minimal medium plates containing various nitrogen sources, the glnA(M68I) and glnA(H195Y) mutants were found to exhibit growth phenotypes that were identical to that of the wild-type strain. In contrast, the glnA(E65G) and glnA(S66P) mutants were unable to form colonies on plates containing ammonium as the sole nitrogen source but formed small colonies on plates with glutamate plus ammonium as the nitrogen source. Additionally, the glnA(P318S) mutant formed significantly smaller colonies than wild-type cells on plates containing ammonium as the sole nitrogen source, although the wild-type and glnA(P318S) strains formed similar-size colonies with glutamate plus ammonium as the nitrogen source. The growth defects with ammonium as the nitrogen source that were seen with the glnA(E65G), glnA(S66P), and glnA(P318S) mutants and the ability of glutamate supplementation to at least partially suppress this growth defect are most likely consequences of the enzymatically defective GS proteins that these mutants synthesize (see below). Strains with the glnA(E65G) and glnA(S66P) mutations were also unable to grow on media containing glutamine. This glutamine-sensitive growth phenotype was observed with the previously described glnA186 mutant (48).
TnrA- and GlnR-dependent regulation in the feedback-resistant mutants.
The effects of these glnA mutations on the expression of a TnrA-regulated gene were analyzed by measuring β-galactosidase production from an amtB-lacZ fusion in wild-type and mutant cells. β-Galactosidase levels were 2,000-fold higher in wild-type cells grown in minimal medium containing the limiting nitrogen source glutamate than in cells grown in minimal medium with the excess nitrogen source glutamine (Table 2). In wild-type cells, β-galactosidase levels were 18-fold higher in cultures grown with limiting nitrogen (glutamate) than in cultures grown with the excess nitrogen source Casamino Acids plus glutamate (Table 2).
Since the glnA(E65G) and glnA(S66P) mutants have a glutamine-sensitive growth phenotype and are unable to grow on minimal medium containing glutamine, Casamino Acids plus glutamate was used as the excess nitrogen source in these experiments. In contrast to the wild-type strain, similar levels of β-galactosidase were present in these two glnA mutants grown with limiting and excess nitrogen sources (Table 2). Since growth of the glnA(M68I), glnA(H195Y), and glnA(P318S) mutants is not inhibited by glutamine, transcription of the amtB promoter was examined in minimal medium containing either glutamine or Casamino Acids plus glutamate as the excess nitrogen source. When grown with glutamine as the excess nitrogen source, β-galactosidase levels were 140- to 280-fold higher in the glnA(M68I), glnA(H195Y), and glnA(P318S) cells than in wild-type cells (Table 2). In contrast amtB was expressed constitutively in glnA(M68I), glnA(H195Y), and glnA(P318S) cells grown with Casamino Acids plus glutamate as the nitrogen source (Table 2).
Since GS regulates the activity of GlnR (36), the effects of the feedback-resistant glnA mutations on the GlnR-dependent regulation of a glnRA-lacZ fusion were also examined. The expression of glnRA is repressed by both GlnR and TnrA (46). To eliminate the TnrA-dependent regulation of the glnRA promoter, the expression of this promoter was examined in a tnrA genetic background. In wild-type cells, β-galactosidase levels were 140-fold higher in nitrogen-limited (glutamate) cultures than in cells grown with the excess nitrogen source glutamine (Table 2). The GlnR-dependent repression of glnRA expression was relieved in the glnA(M68I), glnA(H195Y), and glnA(P318S) mutants, where the β-galactosidase levels in glutamine-grown cultures were 57- to 153-fold higher in the glnA mutants than in the wild-type strain (Table 2). When Casamino Acids plus glutamate was used as the excess nitrogen source for examining glnRA regulation in the glnA(E65G) and glnA(S66P) mutants, constitutive glnRA expression was observed (Table 2).
Enzymatic properties of the feedback-resistant enzymes.
To characterize the kinetic and feedback properties of the mutant enzymes, the proteins were overexpressed and purified to homogeneity. The results of enzymatic assays for the wild-type and mutant proteins are shown in Table 3. Because the Mg2+-dependent biosynthetic activity is the major route for glutamine synthesis in vivo (48), the kinetic constants for this enzymatic reaction were determined. The maximal velocities for the E65G and P318S enzymes were 6.5- and 3-fold lower, respectively, than for the wild-type enzyme (Table 3). Thus, the growth defects of these two glnA mutants on minimal medium containing ammonium as the sole nitrogen source most likely result from the synthesis of GS enzymes with low biosynthetic activities. The glutamate and ATP Km values for the S66P enzyme are five- and ninefold higher, respectively, than that of the wild-type enzyme, indicating that S66P GS does not bind these reaction substrates as tightly as the wild-type enzyme. These observations suggest that under in vivo conditions, where glutamate and ATP levels are 143 and 3.5 mM, respectively (16), lower levels of glutamine would be synthesized in the glnA(S66P) mutant than in wild-type cells. This impairment in glutamine synthesis most likely explains the inability of glnA(S66P) cells to grow with ammonium as the sole nitrogen source. Several of the kinetic parameters for the M68I and H195Y enzymes were slightly higher than the values for the wild-type enzyme (Table 3).
TABLE 3.
Kinetic parameters of wild-type and mutant glutamine synthetases
| Enzyme | Mg2+-dependent biosynthetic reactiona
|
Mn2+-dependent biosynthetic reaction (μmol/min/mg)a | Transferase reaction (μmol/min/mg)a | ||
|---|---|---|---|---|---|
| Km glutamate (mM) | Km ATP (mM) | Vmax (μmol/min/mg) | |||
| Wild type | 27 ± 2.2 | 2.4 ± 0.1 | 2.7 ± 0.2 | 2.4 ± 0.2 | 78 ± 8 |
| E65G | 7.3 ± 0.9 | 3.6 ± 0.4 | 0.35 ± 0.02 | 2.5 ± 0.7 | 73 ± 5 |
| S66P | 130 ± 11 | 23 ± 7 | 2.4 ± 0.2 | 11.0 ± 0.2 | 51 ± 4 |
| M68I | 56 ± 8 | 4.1 ± 0.5 | 2.4 ± 0.2 | 3.9 ± 0.4 | 57 ± 1 |
| H195Y | 30 ± 3 | 7.0 ± 0.6 | 4.7 ± 0.2 | 1.8 ± 0.1 | 43 ± 6 |
| P318S | 3.3 ± 0.5 | 0.81 ± 0.02 | 0.86 ± 0.03 | 0.29 ± 0.02 | 33 ± 3 |
Values are the average of at least two determinations ± the sample standard deviation.
Although the levels of the Mn2+-dependent biosynthetic specific activity varied significantly among the mutant GS proteins (Table 3), there was no correlation between this enzymatic activity and the growth phenotype or levels of transcriptional regulation in the mutants encoding these enzymes. Only a twofold variation in the levels of transferase (reverse) activity was observed among the mutant enzymes (Table 3).
Sensitivities of mutant enzymes to inhibitors.
The Mg2+-dependent biosynthetic reaction of B. subtilis GS is inhibited by glutamine and AMP (9). Structural and enzymatic data have shown that glutamine and AMP are competitive inhibitors that bind to the substrate sites for glutamate and ATP, respectively (23, 24, 48). While all of the mutant enzymes were highly resistant to inhibition by glutamine (Table 4), IC50s could be determined for only two of the proteins (M68I and H195Y) due to the limited solubility of glutamine. The previously reported S186F enzyme also has a glutamine IC50 that is greater than 140 mM (data not shown). Although all five mutant enzymes were more resistant to inhibition by AMP than the wild-type enzyme, there was a wide range in the AMP IC50s for the mutant proteins, and only the P318S enzyme was completely resistant to inhibition by AMP (Table 4).
TABLE 4.
Sensitivities of wild-type and mutant glutamine synthetases for inhibitors
| Enzyme | IC50 (mM) for Mg2+-dependent biosynthetic reactiona
|
||
|---|---|---|---|
| Glutamine | AMP | MetSox | |
| Wild type | 2.4 ± 0.1 | 0.5 ± 0.02 | 0.13 ± 0.01 |
| E65G | >140 | 12 ± 2 | 1.6 ± 0.2 |
| S66P | >140 | 2.0 ± 0.1 | 3.9 ± 0.3 |
| M68I | 139 ± 3 | 7.3 ± 0.6 | 0.38 ± 0.01 |
| H195Y | 115 ± 5 | 2.9 ± 0.1 | 0.24 ± 0.01 |
| P318S | >140 | >30 | 0.23 ± 0.01 |
Values are the average of at least two determinations ± the sample standard deviation.
The glutamate analogue l-methionine-s-sulfoximine (MetSox) inhibits GS enzymatic activity by a different mechanism than glutamine or AMP. MetSox is a substrate for GS and is phosphorylated in the presence of ATP (42). Phosphorylated MetSox is a transition state mimic that binds tightly to the enzyme, resulting in irreversible inhibition (42). The feedback-resistant GS proteins had MetSox IC50s that were 2- to 30-fold higher than those of the wild-type enzyme (Table 4). Curiously, the E65G and S66P mutant enzymes had the most significant increases in their MetSox IC50s, 12- and 30-fold, respectively, and were from the two mutant strains with the glutamine-sensitive growth phenotype (Table 2). This correspondence is most likely coincidental in that the previously characterized S186F mutant GS had only a 1.8-fold increase in resistance to MetSox but was also sensitive to glutamine (48).
Inhibition of TnrA in vitro DNA binding.
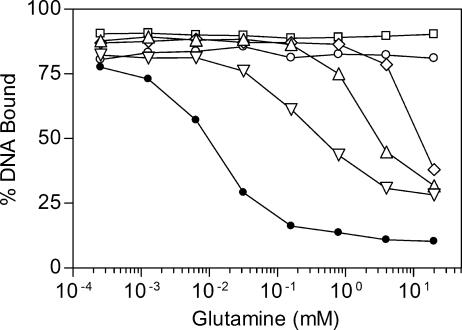
The ability of the feedback-resistant and wild-type enzymes to inhibit the binding of TnrA to the amtB promoter was examined using an in vitro DNA gel mobility shift assay. Glutamine is significantly more effective than AMP in promoting the ability of wild-type GS to impede TnrA DNA binding (48, 51). In addition, because all five mutants exhibited major defects in glutamine feedback inhibition but had variable alterations in their inhibition by AMP (Table 4), only glutamine was used as the feedback inhibitor in these assays.
In the first set of experiments, fixed amounts of GS and TnrA were incubated with various amounts of glutamine. Compared to the wild-type enzyme, all five feedback-resistant GS proteins were defective in the ability to suppress TnrA DNA binding in the presence of glutamine. No inhibition of TnrA DNA binding was observed with the E65G and S66P mutant enzymes (Fig. 2). In contrast, when the reaction mixture contained 20 mM glutamine, a concentration that matches the intracellular levels in wild-type B. subtilis cells grown with excess nitrogen (16), TnrA DNA binding was partially blocked by the M68I, H195Y, and P318S enzymes (Fig. 2). These in vitro results correlate with the observation that growth of the glnA(E65G) and glnA(S66P) mutants, but not of the glnA(M68I), glnA(H195Y), and glnA(P318S) mutants, is glutamine sensitive.
FIG. 2.
Effects of glutamine on the ability of wild-type and mutant GS proteins to inhibit TnrA DNA binding. Binding of TnrA to amtB promoter DNA was determined in a gel mobility shift assay. The abilities of wild-type (•), E65G (○), S66P (□), M68I (⋄), H195Y (▵), and P318S (▿) proteins to inhibit the DNA binding activity of TnrA were determined in the presence of various concentrations of glutamine. The TnrA dimer concentration was 30 nM, and the GS subunit concentration was 1 μM in all binding reactions. Each data point is the mean of at least two independent experiments and is reproducible to with ±10%.
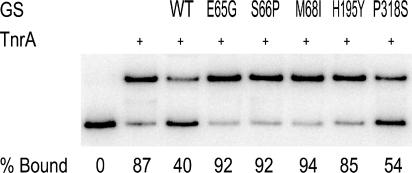
To determine whether the feedback-resistant enzymes had any inherent defect in the ability to interact with TnrA, high concentrations of mutant and wild-type proteins were incubated with TnrA in the absence of the feedback inhibitor glutamine. Under these reaction conditions, TnrA DNA binding is partially inhibited by wild-type GS (Fig. 3) (48, 51). Under the same experimental conditions, no inhibition of TnrA DNA binding was observed with the E65G, S66P, M68I, and H195Y enzymes. These results indicate that in the absence of glutamine, these four proteins have altered conformations that reduce their ability to interact with TnrA. In contrast, P318S GS was able to inhibit TnrA DNA binding to a level that was similar to that of the wild-type protein (Fig. 3).
FIG. 3.
Abilities of wild-type and mutant GS proteins to inhibit TnrA DNA binding in the absence of the feedback inhibitor glutamine. A gel mobility shift assay was used to examine the binding of TnrA to amtB promoter DNA. TnrA (30 nM dimer) was present in the binding mixtures shown in all but the far-left lane. The abilities of high concentrations (100 μM subunit) of wild-type (WT), E65G, S66P, M68I, H195Y, and P318S GS proteins to inhibit TnrA binding were determined in the absence of feedback inhibitors. The percentage of bound amtB DNA indicated below each lane is the mean of four independent measurements. The sample standard deviation was less than 15% for all values.
DISCUSSION
The observation that all the feedback-resistant glnA mutants isolated in this study have pleiotropic alterations in both TnrA- and GlnR-mediated gene regulation supports the model for the control of TnrA activity by feedback-inhibited GS and argues that the activity of GlnR is also controlled by feedback-inhibited GS. Based on their ability to grow in the presence of glutamine, the previously reported S186F mutant and the five feedback-resistant GS mutants described here fall into two groups. The E65G, S66P, and S186F mutants have a glutamine-sensitive growth phenotype, while the M68I, H195Y, and P318S mutants grew like wild-type cells in the presence of glutamine. This in vivo growth phenotype correlates well with the results of in vitro experiments showing that in the presence of glutamine, the M68I, H195Y, and P318S proteins partially inhibit TnrA binding, while TnrA binding is essentially unaffected by the presence of the E65G, S66P, and S186F proteins (Fig. 2) (48).
One explanation for the correlation between the glutamine growth phenotype and the in vitro levels of regulation of TnrA activity is that the E65G, S66P, and S186F GS proteins are more resistant to feedback inhibition by glutamine than the M68I, H195Y, and P318S proteins. The differences between these two groups of mutants do not appear to result from differences in their resistance to AMP inhibition, because there is no correlation between the in vivo phenotypes and the AMP IC50s. Although specific glutamine IC50s could not be determined for all of the mutant proteins due to limitations of the in vitro assay, the M68I and H195Y enzymes have IC50s that are lower than those of the E65G, S66P, S186F, and P318S enzymes. Interestingly, all the mutant proteins had glutamine IC50s that were significantly higher than 20 mM, the intracellular glutamine concentration in wild-type cells grown in the presence of excess nitrogen (16). Since all six feedback-resistant mutants, unlike the wild-type cells, can cross-feed Gln− cells, these mutants are likely to have significantly higher intracellular glutamine pools than wild-type cells. If the glutamine-sensitive phenotype results from the production of high intracellular levels of glutamine by mutant GS enzymes that are completely resistant to glutamine inhibition, then the E65G, S66P, and S186F enzymes would be more resistant to glutamine feedback inhibition than the M68I, H195Y, and P318S proteins.
The feedback-resistant glnA mutations could not only affect the level of feedback resistance, but also produce structural alterations that affect the TnrA binding site on GS. Indeed, compared to the wild-type enzyme, four of the feedback-resistant proteins had reduced affinity for TnrA in the absence of feedback inhibitors (Fig. 3) (48). Only the P318S and S186F proteins interacted with TnrA, as well as wild-type GS, under these conditions. However, since the abilities of the mutant proteins to bind TnrA in the absence of inhibitors do not correlate with the in vivo regulation of TnrA, this property cannot fully account for the difference in the level of TnrA regulation seen between these two groups of mutants.
A straightforward explanation for the feedback-resistant properties of the mutant enzymes is that they have lower affinities for the feedback inhibitors than the wild-type enzyme. Indeed the previously isolated S186F mutant was shown to be defective in the binding of both glutamine and AMP (48). However, the enzymatic properties of the S186F GS protein are similar to those of the wild-type GS, while all five feedback-resistant GSs reported here have a variety of enzymatic alterations that could contribute to their feedback-resistant phenotype. For example, since glutamine and AMP are competitive inhibitors that bind to the substrate sites for glutamate and ATP, respectively (23, 24, 48), mutant enzymes with higher substrate affinities than the wild-type enzyme would have increased resistance to the inhibitors as a result of competition with the substrates. Two of the mutant enzymes, E65G and P318S, have glutamate Km values that are lower than those of wild-type GS (Table 3). This observation implies that these two enzymes have a higher affinity for glutamate than the wild-type enzyme, and thus, their resistance to glutamine inhibition could result, at least in part, from the reduced ability of glutamine to bind to the active site in the presence of glutamate. Similarly, the high level of resistance to AMP inhibition by the P318S enzyme may be related to the fact that the P318S enzyme has an ATP Km value that is lower than that of wild-type GS (Tables 3 and 4).
In addition to having some substrate Km values that are lower than that of the wild-type enzyme, the E65G and P318S mutant enzymes have maximal velocity levels that are lower than that of wild-type GS (Table 3). The lower maximal velocity levels of the E65G and P318S mutant enzymes may directly result from their lower substrate Km values. According to the transition state model of catalysis, the level of enzymatic activity is inversely related to the difference in the energy level between the enzyme-substrate complex and the transition state complex (11). All other things being equal, an amino acid substitution that stabilized the enzyme-substrate complex would increase this energy difference and reduce the level of catalysis. Thus, the apparently higher affinities that these two mutant enzymes have for their substrates may stabilize the enzyme-substrate complex and contribute to the low levels of enzymatic activity observed with these mutant enzymes.
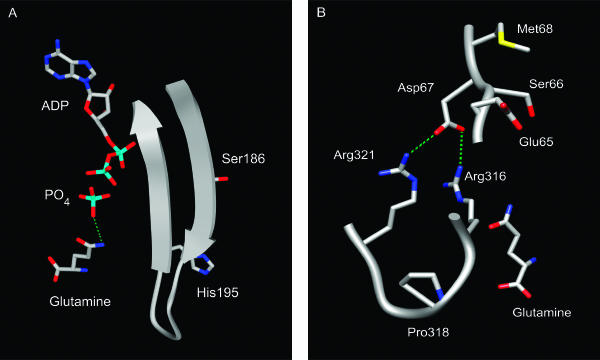
A structural approach was used to obtain a better understanding of how these glnA mutations might affect the feedback inhibition and activity of GS. Since the crystal structure of B. subtilis GS is not available, a homology model of this protein was prepared. The crystal structures of the GS proteins from S. enterica serovar Typhimurium and M. tuberculosis have been determined (10, 18, 19, 22, 23). These enzymes are dodecamers composed of two hexameric rings stacked on top of one another. The active sites are located at the subunit-subunit interfaces within the hexameric ring. Because B. subtilis GS is also a dodecamer and has significant sequence similarity to the proteins from S. enterica serovar Typhimurium and M. tuberculosis (8, 40), these proteins can be used as templates to produce an accurate model.
The structural model of B. subtilis GS revealed that all of the substituted residues in the feedback-resistant enzymes were located in the active site (Fig. 4). This observation is consistent with the hypothesis that glutamine and AMP act as feedback inhibitors by binding at the active site and not at an allosteric regulatory site (48). From structural studies of S. enterica serovar Typhimurium and M. tuberculosis GS (22, 23, 24), it is clear that none of the residues altered in the feedback-resistant B. subtilis enzymes directly contact the reaction substrates or enzymatic inhibitors. This finding indicates that these amino acid substitutions indirectly affect enzymatic activity and feedback inhibition. This result is not unprecedented. Studies of other enzymes have shown that amino acid substitutions that alter substrate specificity are often located in residues that do not make contact with the substrate (28). The mutant isolation procedure used to isolate the feedback-resistant enzymes reported here required that the mutant proteins both retain enough enzymatic activity to confer a Gln+ phenotype and simultaneously produce high levels of glutamine to cross-feed Gln− cells. As a result, glnA mutations that resulted in either defective substrate binding or catalysis would not have been isolated using this screen. Structural studies of Salmonella enterica serovar Typhimurium GS have revealed that the amino acid residues that interact with glutamine and AMP are also used to contact glutamate and ATP (23, 24). Since these same residues are conserved in B. subtilis GS, substitutions that altered residues directly involved in inhibitor binding would have simultaneously reduced substrate binding.
FIG. 4.
Locations of mutated residues in the homology model of GS. (A) The backbone of residues 183 to 199 is shown as a ribbon diagram. (B) The backbones for residues 65 to 68 and 316 to 321 are shown as smoothed tubes. Hydrogen bonds are shown as dashed lines. Color coding: carbon, gray; oxygen, red; nitrogen, blue; sulfur, yellow; phosphorus, cyan; hydrogen bonds, green. The diagrams were produced with UCSF Chimera (32).
The residues altered in the feedback-resistant enzymes are located in two regions of the active site. The His-195 and Ser-186 residues are located on adjacent β-strands that line the active site (Fig. 4A) (48). The side chains of these two residues are positioned so that they are directed away from the active-site chamber. This observation is consistent with the idea that these side chains do not directly contact the feedback inhibitors and suggests that the substitutions in these residues confer resistance to feedback inhibition by an indirect mechanism. Residues Glu-65, Ser-66, Met-68, and Pro-318 are located near a subunit-subunit interface where the side chain of Asp-67 from one subunit hydrogen bonds with the side chains of Arg-316 and Arg-321 from an adjacent subunit (Fig. 4B). The mutational substitutions in residues Glu-65, Ser-66, Met-68, and Pro-318 may alter this subunit-subunit interaction and thus change the conformation of the active site so that the enzymatic and feedback inhibition properties of the mutant enzymes are altered (26).
B. subtilis GS is functionally different from its E. coli homolog in that glutamine binds to the B. subtilis enzyme with sufficient affinity to inhibit catalytic activity. Therefore, the B. subtilis active site must contain unique amino acid residues that stabilize the binding of glutamine to the active site. Mutational and structural studies of a variety of different proteins have shown that evolutionarily conserved residues are important for binding specificity (25, 27, 39). To identify residues critical for glutamine feedback inhibition, a multiple-sequence alignment of GS proteins from Bacilli and Proteobacteria strains was constructed. This analysis assumes that the physiologically significant forms of the GS enzymes from these two groups of bacteria differ in that only the Bacilli enzymes are subject to feedback inhibition by glutamine (see Materials and Methods). The rationale for this approach is the fact that within any given protein family the amino acid residues critical for structure and function are highly conserved and that any subclass of a protein family with a different functional property or ligand-binding specificity will contain a unique set of conserved residues required for the distinguishing feature of that subclass (27, 38). Thus, the amino acid residues critical for glutamine feedback inhibition should be uniquely conserved within the Bacilli GS proteins.
The results of the genetic studies support this proposal. Three of the six residues altered in the feedback-resistant B. subtilis enzymes, His-195, Ser-186, and Pro-318, are completely conserved among the Bacilli GS proteins but have sequence divergences at the corresponding positions in the enzymes from Proteobacteria (Fig. 5). The fact that the other three residues, Glu-65, Ser-66, and Met-68, are highly conserved in both the Bacilli and Proteobacteria GS enzymes most likely reflects the fact that these amino acids are located in a protein region involved in subunit-subunit interactions. The observation that the Bacilli and Proteobacteria amino acid consensus sequences diverge at positions 69, 182, 198, 199, and 319 within the active site raises the possibility that these residues may also be critical for feedback inhibition by glutamine (Fig. 5). Further mutational analysis will be required to confirm this hypothesis.
FIG. 5.
Consensus sequence logos for regions of the GS active site. The residue numbers correspond to the B. subtilis enzyme. The heights of the letters indicate the information content at each position (35). Bac corresponds to the Bacilli sequences; Pro corresponds to the Proteobacteria sequences.
Supplementary Material
Acknowledgments
This research was supported by Public Health Service research grant GMA51127 from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Atkinson, M. R., L. V. Wray, Jr., and S. H. Fisher. 1990. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J. Bacteriol. 172:4758-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belitsky, B. R., L. V. Wray, Jr., S. H. Fisher, D. E. Bohannon, and A. L. Sonenshein. 2000. Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression. J. Bacteriol. 182:5939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biaudet, V., F. Samson, C. Anagnostopoulos, S. D. Erlich, and P. Bessières. 1996. Computerized map of Bacillus subtilis. Microbiology 142:2669-2729. [DOI] [PubMed] [Google Scholar]
- 4.Brown, S. W., and A. L. Sonenshein. 1996. Autogenous regulation of the Bacillus subtilis glnRA operon. J. Bacteriol. 178:2450-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chasin, L. A., and B. Magasanik. 1968. Induction and repression of the histidine-degrading enzymes of Bacillus subtilis. J. Biol. Chem. 243:5165-5178. [PubMed] [Google Scholar]
- 6.Chen, J. D., and D. A. Morrison. 1987. Cloning of Streptococcus pneumoniae DNA fragments in Escherichia coli requires vectors protected by strong transcriptional terminators. Gene 55:179-187. [DOI] [PubMed] [Google Scholar]
- 7.Crooks, G. E., G. Hon, J.-M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deul, T. F., A. Ginsburg, J. Yeh, E. Shelton, and E. R. Stadtman. 1970. Bacillus subtilis glutamine synthetase: purification and physical characterization. J. Biol. Chem. 245:5195-5205. [PubMed] [Google Scholar]
- 9.Deul, T. F., and S. Prusiner. 1974. Regulation of glutamine synthetase from Bacillus subtilis by divalent cations, feedback inhibitors, and l-glutamine. J. Biol. Chem. 249:257-264. [PubMed] [Google Scholar]
- 10.Eisenberg, D., H. S. Gill, G. M. U. Pfluegel, and S. H. Rotstein. 2000. Structure-function relationships of glutamine synthetases. Biochim. Biophys. Acta 1477:122-145. [DOI] [PubMed] [Google Scholar]
- 11.Fersht, A. 1999. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. W. H. Freeman and Company, New York, N.Y.
- 12.Ferson, A. E., L. V. Wray, Jr., and S. H. Fisher. 1996. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol. Microbiol. 22:693-701. [DOI] [PubMed] [Google Scholar]
- 13.Fisher, S. H. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la différence! Mol. Microbiol. 32:223-232. [DOI] [PubMed] [Google Scholar]
- 14.Fisher, S. H., J. L. Brandenburg, and L. V. Wray, Jr. 2002. Mutations in Bacillus subtilis glutamine synthetase that block its interaction with transcription factor TnrA. Mol. Microbiol. 45:627-635. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, S. H., M. S. Rosenkrantz, and A. L. Sonenshein. 1984. Glutamine synthetase gene of Bacillus subtilis. Gene 32:427-438. [DOI] [PubMed] [Google Scholar]
- 16.Fisher, S. H., and A. L. Sonenshein. 1984. Bacillus subtilis glutamine synthetase mutants pleiotropically altered in glucose catabolite repression. J. Bacteriol. 157:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher, S. H., and L. V. Wray, Jr. 2002. Bacillus subtilis 168 contains two differentially regulated genes encoding l-asparaginase. J. Bacteriol. 184:2148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill, H. S., and D. Eisenberg. 2001. The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition. Biochemistry 40:1903-1912. [DOI] [PubMed] [Google Scholar]
- 19.Gill, H. S., G. M. U. Pfleugl, and D. Eisenberg. 2002. Multicopy crystallographic refinement of a relaxed glutamine synthetase from Mycobacterium tuberculosis highlights flexible loops in the enzymatic mechanism and its regulation. Biochemistry 41:9863-9872. [DOI] [PubMed] [Google Scholar]
- 20.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. 167:335-336. [DOI] [PubMed]
- 21.Gutowski, J. C., and H. J. Schreier. 1992. Interaction of the Bacillus subtilis glnRA repressor with operator and promoter regions in vivo. J. Bacteriol. 174:671-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krajewski, W. W., T. A. Jones, and S. L. Mowbray. 2005. Structure of Mycobacterium tuberculosis glutamine synthetase in complex with a transition-state mimic provides functional insights. Proc. Natl. Acad. Sci. USA 102:10499-10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liaw, S.-H., and D. Eisenberg. 1994. Structural model for the reaction mechanism of glutamine synthetase, based on five crystal structures of enzyme-substrate complexes. Biochemistry 33:675-681. [DOI] [PubMed] [Google Scholar]
- 24.Liaw, S.-H., G. Jun, and D. Eisenberg. 1994. Interactions of nucleotides with fully unadenylylated glutamine synthetase from Salmonella typhimurium. Biochemistry 33:11184-11188. [DOI] [PubMed] [Google Scholar]
- 25.Lockless, S. W., and R. Ranganathan. 1999. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286:295-299. [DOI] [PubMed] [Google Scholar]
- 26.MacArthur, M. W., and J. M. Thornton. 1991. Influence of proline residues on protein conformation. J. Mol. Biol. 218:397-412. [DOI] [PubMed] [Google Scholar]
- 27.Madabushi, S., A. K. Gross, A. Philippi, E. C. Meng, T. G. Wensel, and O. Lichtarge. 2004. Evolutionary trace of G protein-coupled receptors reveals clusters of residues that determine global and class-specific functions. J. Biol. Chem. 279:8126-8132. [DOI] [PubMed] [Google Scholar]
- 28.Morely, K. L., and R. J. Kazlauskas. 2005. Improving enzyme properties: when are closer mutations better? Trends Biotechnol. 23:231-237. [DOI] [PubMed] [Google Scholar]
- 29.Nakano, M. M., F. Yang, P. Hardin, and P. Zuber. 1995. Nitrogen regulation of nasA and nasB operon, which encode genes required for nitrate assimilation in Bacillus subtilis. J. Bacteriol. 177:573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 33.Reitzer, L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57:155-176. [DOI] [PubMed] [Google Scholar]
- 34.Rhee, S. G., B. Chock, and E. R. Stadtman. 1989. Regulation of Escherichia coli glutamine synthetase. Adv. Enzymol. Relat. Areas Mol. Biol. 62:37-92. [DOI] [PubMed] [Google Scholar]
- 35.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schreier, H. J., S. W. Brown, K. D. Hirschi, J. F. Nomellini, and A. L. Sonenshein. 1989. Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J. Mol. Biol. 210:51-63. [DOI] [PubMed] [Google Scholar]
- 37.Schwede, T., J. Koop, N. Guex, and M. Peitsch. 2003. SWISS_MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shackelford, G. S., C. A. Regni, and L. J. Beamer. 2004. Evolutionary trace analysis of the α-d-phosphohexomutase superfamily. Protein Sci. 13:2130-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sowa, M. E., W. He, K. C. Slep, M. A. Kercher, O. Lichtarge, and T. G. Wensel. 2001. Prediction and confirmation of a site critical for effector regulation of RGS domain activity. Nat. Struct. Biol. 8:234-237. [DOI] [PubMed] [Google Scholar]
- 40.Strauch, M. A., A. I. Aronson, S. W. Brown, H. J. Schreier, and A. L. Sonenshein. 1988. Sequence of the Bacillus subtilis glutamine synthetase gene region. Gene 71:257-265. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weisbrod, R. E., and A. Meister. 1973. Studies on glutamine synthetase from Escherichia coli. Formation of pyrrolidone carboxylate and inhibition by methionine sulfoximine. J. Biol. Chem. 248:3997-4002. [PubMed] [Google Scholar]
- 43.Woolfolk, C. A., and E. R. Stadtman. 1967. Regulation of glutamine synthetase III. Cumulative feedback inhibition of glutamine synthetase from Escherichia coli. Arch. Biochem. Biophys. 118:736-755. [DOI] [PubMed] [Google Scholar]
- 44.Wray, L. V., Jr., M. R. Atkinson, and S. H. Fisher. 1994. The nitrogen-regulated Bacillus subtilis nrgAB operon encodes a membrane protein and a protein highly similar to the Escherichia coli glnB-encoded PII protein. J. Bacteriol. 176:108-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors, including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wray, L. V., Jr., A. E. Ferson, K. Rohrer, and S. H. Fisher. 1996. TnrA, a transcriptional factor required for global nitrogen regulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93:8841-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wray, L. V., Jr., and S. H. Fisher. 1988. Cloning and nucleotide sequence of the Streptomyces coelicolor gene encoding glutamine synthetase. Gene 71:247-256. [DOI] [PubMed] [Google Scholar]
- 48.Wray, L. V., Jr., and S. H. Fisher. 2005. A feedback-resistant mutant of Bacillus subtilis glutamine synthetase with pleiotropic defects in nitrogen-regulated gene expression. J. Biol. Chem. 280:33298-33304. [DOI] [PubMed] [Google Scholar]
- 49.Wray, L. V., Jr., J. M. Zalieckas, A. E. Ferson, and S. H. Fisher. 1998. Mutational analysis of the TnrA-binding sites in the Bacillus subtilis nrgAB and gabP promoter regions. J. Bacteriol. 180:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wray, L. V., Jr., J. M. Zalieckas, and S. H. Fisher. 2000. Purification and in vitro activities of the Bacillus subtilis TnrA transcription factor. J. Mol. Biol. 300:29-40. [DOI] [PubMed] [Google Scholar]
- 51.Wray, L. V., Jr., J. M. Zalieckas, and S. H. Fisher. 2001. Bacillus subtilis glutamine synthetase controls gene expression through a protein-protein interaction with transcription factor TnrA. Cell 107:427-435. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida, K., H. Yamaguchi, M. Kinehara, Y. H. Ohki, Y. Nakaura, and Y. Fujita. 2003. Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box. Mol. Microbiol. 49:157-165. [DOI] [PubMed] [Google Scholar]
- 53.Zalieckas, J. M., L. V. Wray, Jr., and S. H. Fisher. 2006. Cross-regulation of the Bacillus subtilis glnRA and tnrA genes provides evidence for DNA binding site discrimination by GlnR and TnrA. J. Bacteriol. 188:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.