Abstract
The ability to induce broadly neutralizing antibodies should be a key component of any forthcoming vaccine against human immunodeficiency virus type 1. One potential vaccine candidate, monomeric gp120, has generally failed to elicit such antibodies. We postulated that gp120 might be a better immunogen if it could be engineered to preferentially bind known broadly neutralizing antibodies. In a first study, we found that four alanine substitutions on the perimeter of the so-called Phe-43 cavity of gp120 could reduce binding of weakly neutralizing CD4-binding site antibodies (R. Pantophlet, E. O. Saphire, P. Poignard, P. W. H. I. Parren, I. A. Wilson, and D. R. Burton, J. Virol. 77:642-658, 2003), while slightly enhancing binding of the potent, broadly neutralizing antibody b12. In the present study, we sought to reduce or abolish the binding of a wider range of nonneutralizing antibodies, by incorporating extra N-glycosylation motifs at select positions into the hypervariable loops and the gp120 core. A hyperglycosylated mutant containing seven extra glycosylation sequons (consensus sequences) and the four alanine substitutions described above did not bind an extensive panel of nonneutralizing and weakly neutralizing antibodies, including a polyclonal immunoglobulin preparation (HIVIG) of low neutralizing potency. Binding of b12, at lowered affinity, and of four antibodies to the C1 and C5 regions was maintained. Removal of N- and C-terminal residues in the C1 and C5 regions, respectively, reduced or abolished binding of the four antibodies, but this also adversely affected b12 binding. The hyperglycosylated mutant and its analogues described here are novel antigens that may provide a new approach to eliciting antibodies with b12-like neutralizing properties.
Global efforts to obtain an effective vaccine against human immunodeficiency virus type 1 (HIV-1) have thus far failed. The induction of antibodies with broad antiviral activity, considered a highly beneficial feature of a future vaccine (16, 17, 53, 68, 92, 115, 117), has proven particularly problematic. The use of soluble monomeric gp120, the major component of the viral envelope spike, has yielded antibodies that bind solely to monomeric gp120 or only to a narrow range of HIV-1 isolates (6, 23, 49). The crystal structures of the gp120 core in complex with CD4 and an antibody Fab fragment (47, 48, 115) have shed light on why it may be difficult to elicit antibodies that are capable of recognizing gp120 as presented on the virion surface. Conserved sequences, such as are found in the CD4-binding domain, lie recessed within the core and are partially occluded by (hyper)variable loops, which then reduces antibody recognition (48, 115, 117). Furthermore, although other conserved regions, such as the interface between gp120 and the transmembrane unit glycoprotein gp41 (48), may be readily exposed on monomeric gp120, these epitopes are most likely occluded on the envelope spike (115, 117).
Because of the disappointing results with monomeric gp120, new approaches are being explored for eliciting broadly neutralizing antibodies. Two main approaches are currently being investigated by using HIV envelope glycoproteins. One strategy focuses on the preservation or reconstruction of the trimeric envelope spike. For example, virions have been chemically inactivated by modification of the zinc finger domains of the nucleocapsid region while maintaining the native envelope structure (2, 89). In another approach, soluble gp140 oligomers containing the ectodomain of gp41 covalently linked to gp120 have been generated by fusing GCN4 trimerization domains or T4 bacteriophage fibritin trimeric motifs to the C terminus of soluble, uncleaved gp140 glycoproteins (118-120). In other studies, cysteine residues have been incorporated into gp120 and gp41 (8, 9, 90) to prevent dissociation of the two subunits through the formation of an intersubunit disulfide bridge upon expression of cleaved gp140. More recently, proteoliposomes have been generated containing native, trimeric uncleaved gp160ΔCT (with the cytoplasmic tail deleted) glycoproteins (39). All of these approaches appear promising. However, such attempts to mimic native HIV envelope trimers have the limitation that key cross-neutralizing epitopes may be of relatively low immunogenicity on the trimer (115, 117).
A second strategy for obtaining broadly neutralizing antibodies with recombinant envelope glycoproteins focuses on the use of monomeric, but slightly modified, gp160 or gp140 glycoproteins. For example, various envelope glycoproteins have been generated in which the V2 loop has been deleted, with the aim of increasing the exposure of neutralizing epitopes (102). In other studies, partially deglycosylated recombinant gp160 (10) or recombinant viruses expressing gp120 glycosylation mutants have been generated (82). Unfortunately, all of these approaches have thus far failed to provide immunogens that elicit the desired level of neutralizing antibodies (20, 82), most likely because the elicited antibodies are unable to recognize their cognate epitopes on wild-type virus particles.
Logic suggests that neutralizing antibodies should target conserved regions on the HIV-1 envelope because such antibodies are most likely to be cross-reactive and useful in protection against HIV. The CD4-binding site (CD4bs) on gp120 of HIV-1 is a particularly attractive target for vaccine design since (i) it displays a high degree of conservation (48) and (ii) it is accessible to neutralizing monoclonal antibodies (MAbs) on the surface of primary HIV-1 isolates prior to CD4 binding (86). One antibody in particular is useful as a model for the design of a vaccine capable of inducing potently neutralizing antibodies targeted to the CD4bs. This antibody, b12, was isolated from a combinatorial phage display library developed from bone marrow donated by an individual who had been HIV positive for more than 6 years but who had not yet developed clinical symptoms (18). MAb b12 effectively neutralizes a broad range of primary isolates from various HIV-1 clades, as well as T-cell-line-adapted viruses (19, 28, 45). It has been inferred that this capability stems from b12 being able to bind functional oligomeric gp120 with comparable affinity to monomeric gp120 (76, 91, 96); nonneutralizing MAbs bind with substantially lower affinity, or not at all, to functional oligomeric gp120, presumably due to epitope occlusion or steric hindrance by vicinal gp120 protomers on the viral surface (79, 115, 117).
Monomeric gp120 thus contains all of the antigenic determinants to elicit a broadly neutralizing antibody, such as b12. However, the use of monomeric gp120 as such is jeopardized by the exposure of nonneutralizing epitopes that are normally occluded on oligomeric gp120 or that reside in the variable regions, in particular the V2 and V3 loops (77, 79, 117). These antigenic determinants are immunologically dominant over more conserved neutralizing epitopes (55, 70). Furthermore, they may induce antibodies that interact with gp120 in a manner that is not permitted on native envelope spikes. Therefore, we postulated that if monomeric gp120 can be engineered to restrict antibody binding to broadly neutralizing antibodies, such as b12, it might represent a more interesting immunogen. As a first step, we identified, by alanine scanning mutagenesis, residues on gp120 that are important for binding of b12, CD4, and the nonneutralizing CD4bs MAbs b3 and b6 (75). Although these three antibodies and CD4 bind monomeric gp120 with a similar footprint, we identified four residues on the perimeter of the Phe-43 cavity (48) that, when replaced with alanine, abolish binding of the two nonneutralizing antibodies and CD4 (75). The binding of another nonneutralizing MAb, F105, was also abolished, whereas that of two further nonneutralizing CD4bs MAbs, F91 and 15e, was reduced. In contrast, the binding of MAb b12 relative to wild-type monomeric gp120 was slightly enhanced (75).
Thus, these modified gp120 proteins already show promise as prospective immunogens. However, considering that the variable loops on monomeric gp120 may serve as immune decoys (29, 55, 70, 97), we surmised that it might be necessary to further alter gp120 so as to focus the antibody response on the CD4bs. The concept of refocusing B-cell responses has been postulated by Delves et al. (26). They suggested that epitope-specific molecules could be elicited by selectively mutating “undesired” epitopes, while preserving the overall fold of the protein and, hence, the desired B-cell (or T-cell) epitope(s). In a recent report (21), the β-chain of human chorionic gonadotrophin was used as a model system to show that this strategy is indeed feasible. That study showed that a single amino acid substitution, which does not disrupt the overall conformation of the protein, is sufficient to shift the immune response away from an unwanted epitope and toward a weakly immunogenic determinant (21).
Rather than inserting single mutations, we chose to adopt a previously described approach (33), namely, the masking of undesired epitopes through the incorporation of extra N-linked glycans. We show here that this strategy indeed blocks the binding of nonneutralizing and weakly neutralizing antibodies, whereas binding of the broadly neutralizing antibody b12 is retained. These reengineered gp120 molecules are potential immunogens as part of an HIV-1 vaccine and may serve as a basis for alternative approaches to the development of candidate vaccines against other viral pathogens.
MATERIALS AND METHODS
Plasmids and mutagenesis.
The generation of plasmid pCMV-Tag4A-tpaJR-FLgp120wt has been described recently (75). This plasmid, which is derived from plasmid pCMV-Tag4A (Stratagene), contains a tissue plasminogen activator leader sequence immediately upstream of the env gene to ensure secretion of gp120 envelope glycoprotein into the culture supernatant. The env gene, here of JR-FL, was obtained by PCR amplification by using as a template a plasmid (pSyngp140JR-FL) encoding the codon-optimized gp140 gene (1, 40) of this HIV isolate. Site-directed mutagenesis to substitute wild-type residues for alanine and to incorporate N-glycosylation sequence motifs was performed by using Quikchange (Stratagene).
The generation of gp120 containing deletions of residues in the N and C termini (52 and 19 residues, respectively) was performed by PCR. The primers flCORE-5 (5′-GGAGGTCAACAGCACCGCGCGCGAGGTGGTGCTGGAGAATGTGAC-3′), which contains a BssHII restriction site, and flCORE-3 (5′-GGAGGTCAACAGCACCCTCGAGTTAATTAATTAACTCAATCTTCACCACCTTGTA-3′), which contains an XhoI site, were used. Restriction sites are underlined. Plasmids encoding wild-type or mutant gp120 were used as templates. The PCR products were cloned into pCMV-Tag4A-tpa by using restriction enzymes BssHII and XhoI according to standard protocols. DNA sequencing prior to use verified that the plasmids and mutations generated in the present study were correct.
Antibodies.
A total of 22 MAbs and one polyclonal immunoglobulin preparation were used in the present study. MAbs b3, b6, b12, X5, and loop 2 were from our laboratory (4, 5, 18, 19, 30, 86), whereas the remaining antibodies were obtained from their respective sources or from the National Institutes of Health AIDS Research and Reference Reagent Program (NIH ARRRP). Primary citations for these MAbs are as follows: C11, 212A, 19b, 17b, 48d, F91, 15e, and A32 (J. Robinson [42, 60-62, 65, 66, 87, 88, 98, 105, 106, 116]); 2G12 (H. Katinger [14, 108]); G3-4 and G3-136 (M. Fung [32, 41, 104]); 447-52D (S. Zolla-Pazner [15, 36, 37]); F105 (NIH ARRRP [80, 81, 107]); M90 (F. di Marzo Veronese [27]); 133/290 and 133/237 (M. Niedrig [56, 66, 71]); and 522-149 (G. Robey [64]). The epitopes recognized by the antibodies are summarized in the HIV sequence database at http://resdb.lanl.gov/ABDB/antibody_id.htm, as well as in the cited literature. Human immunoglobulin G (IgG) purified from pooled plasma obtained from healthy asymptomatic seropositive individuals (HIVIG) was obtained from the NIH ARRRP. This manufactured product (NABI, Boca Raton, Fla.) is a 50-mg/ml solution that contains 98% monomeric IgG (24). An affinity-purified polyclonal antibody preparation against the C5 region of gp120, APTKAKRRVVQREKR sequence, was purchased from Cliniqa (Fallbrook, Calif.).
Transient expression of recombinant HIV glycoproteins.
Monomeric wild-type gp120 and mutant glycoproteins were obtained by transiently transfecting 293T cells, as reported earlier (75). Culture supernatants containing recombinant glycoproteins were stored at −20°C until needed.
MAb biotinylation.
MAb 2G12 was conjugated to biotin by using EZ-Link biotinylation reagents (Pierce) according to the manufacturer's instructions. Biotin-labeled MAb was tested in an enzyme-linked immunosorbent assay (ELISA) to assure that the conjugation did not adversely affect binding to gp120.
ELISAs.
ELISAs were performed essentially as described previously (75). Glycoproteins were captured onto ELISA plate wells by using the anti-C5 polyclonal antibody preparation unless otherwise indicated. Antibodies to CD4-induced epitopes were tested in the absence of soluble CD4. MAb 2G12, or in some cases HIVIG, was used to ensure that similar amounts of envelope proteins were captured in each experiment. In general, plates were developed with p-nitrophenyl phosphate (Sigma), and the absorbance was measured at 405 nm. When peroxidase-conjugated secondary antibody (Pierce) was used, plates were developed with 3,3′,5,5′-tetramethyl benzidine-hydrogen peroxide substrate (TMB; Pierce), and the absorbance was measured at 450 nm. In this case, the color reaction was stopped with sulfuric acid (2 M) prior to measurement. For detection of biotinylated 2G12, peroxidase-conjugated streptavidin (Jackson Immunoresearch) was used in combination with the TMB system. ELISAs were performed in duplicate and repeated on at least two different occasions.
SDS-PAGE and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot assays were performed essentially as described elsewhere (74, 111). For electrophoresis, supernatants from 293T cells transiently expressing wild-type or mutant glycoproteins (typically one six-well plate for each glycoprotein) were pooled and concentrated ∼10 times by using YM-50 concentrators (Amicon). The supernatants were diluted 1:2 with concentrated sample buffer (125 mM Tris-HCl, 4% SDS, 20% glycerol, 0.02% bromphenol blue; pH 6.8) and vortexed briefly. Of this mixture, 10 μl was loaded onto a 5% separating gel. After electrophoresis, the proteins were transferred to a polyvinylidene difluoride membrane by tank blotting (Bio-Rad). Blots were incubated overnight with primary antibody (1 or 5 μg/ml, as indicated) and then with alkaline phosphatase-conjugated secondary antibody (Pierce) and were developed by use of BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium as substrates (Sigma).
RESULTS
Effect of multiple alanine substitutions on the perimeter of the Phe-43 cavity of gp120 on MAb binding.
In a recent study (75), Pantophlet et al. reported on the effects of multiple alanine substitutions on the perimeter of the Phe-43 cavity (positions 473 to 476) of gp120JR-FL on the binding of a panel of seven CD4bs antibodies. One mutant, in which all four residues were replaced with alanine, proved to be of particular interest. Binding of MAbs b3, b6, CD4-IgG2, and F105 to this mutant, termed GDMR, was essentially abolished, whereas the apparent binding affinity of MAbs F91 and 15e was reduced relative to the wild type. In contrast, the binding affinity of MAb b12 was unaffected or slightly enhanced.
In the present study, a larger panel of antibodies was tested with this mutant. A total of 14 anti-gp120 antibodies were selected, including the seven antibodies described above. One of the antibodies, MAb A32, binds to a discontinuous epitope involving the C1-C4 domains on gp120 and competes with CD4bs antibodies for binding (13, 64, 116). The affinity of this antibody is increased in the presence of soluble CD4, thus qualifying it nominally as a “CD4-induced” antibody. However, MAb A32 does not compete with other CD4-induced MAbs, e.g., 17b and 48d, for binding to gp120 (116). The other antibodies bind to continuous and discontinuous epitopes in the C1 and C5 domains of gp120 (MAb C11), the V2 loop (MAbs G3-4 and G3-136), and the V3 loop (MAbs loop 2, 19b, and 447-52D). To determine apparent binding constants, cell culture supernatants containing recombinant glycoproteins were captured onto ELISA plate wells with a polyclonal antibody preparation to the C5 region of gp120. The specificity of this polyclonal antibody is similar to that of MAb D7324 and does not compete for binding with any of the antibodies tested here (64). Captured glycoproteins were then probed with various concentrations of antibody to generate binding curves. Apparent affinities were determined from the antibody concentration at half-maximal binding and related to that determined for wild-type gp120.
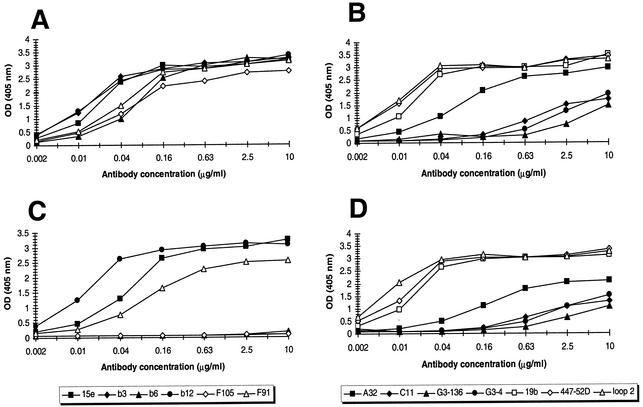
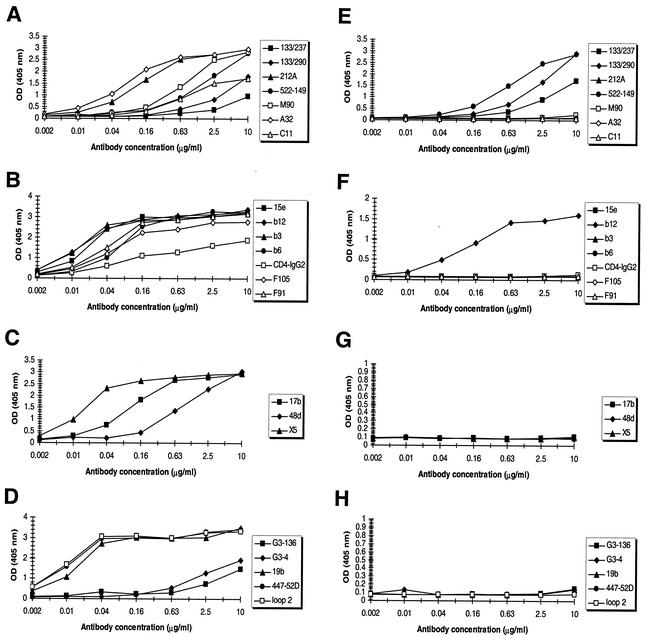
Most antibodies bound with high affinity to wild-type gp120JR-FL (Fig. 1A and B). However, MAbs G3-4 and G3-136, which recognize similar conformational epitopes in the V2 loop, and MAb C11 bound with only moderate affinity to monomeric gp120JR-FL. The moderate binding affinity of MAbs G3-4 and G3-136 to gp120JR-FL, in contrast to binding to gp120IIIB, is most probably due to sequence differences between the two gp120s. The sequence STSIRGKVKEYAFFYKLDI, which is believed to be involved in antibody binding to gp120IIIB (78, 121), is replaced by the sequence TTSIRDEVQKEYALFYKLDV in gp120JR-FL (residues in JR-FL that differ from those in IIIB are underlined). The indicated residues most likely cause slight changes in the epitope recognized by these two antibodies, thus diminishing antibody binding. The same may hold true for MAb C11, although it must be noted that the epitope of MAb C11 has not been clearly defined.
FIG. 1.
Binding of CD4bs MAbs (A and C) and non-CD4bs MAbs (B and D) to gp120JR-FL and mutant GDMR in ELISA. (A and B) Antibody binding to wild-type gp120. (C and D) Antibody binding to gp120 of mutant GDMR. Supernatants containing monomeric gp120 were captured onto ELISA plate wells and probed with various concentrations of MAb, starting at 10 μg/ml. Bound antibody was detected with alkaline phosphatase-conjugated secondary antibody. Absorbance was measured at 405 nm.
As expected, MAbs b3, b6, and F105 did not bind to gp120 of mutant GDMR, whereas the binding of MAbs F91 and 15e was reduced relative to that of the wild type (55 and 45%, respectively; Fig. 1). The binding of MAb A32 was slightly reduced (70% relative to wild-type gp120 [Fig. 1D]). In contrast, binding of the other antibodies, including b12, was largely unaffected by the alanine substitutions (Fig. 1C and D). Based on these results, it was apparent that, although introduction of the four alanine mutations is sufficient to abolish or reduce binding to gp120 of nonneutralizing or weakly neutralizing MAbs against epitopes close to or overlapping the CD4bs, further mutations would be required to eliminate the reactivity of undesired antibodies against other gp120 epitopes.
Effect of the introduction of an N-linked glycan in the V3 loop on reactivity with V3 loop antibodies.
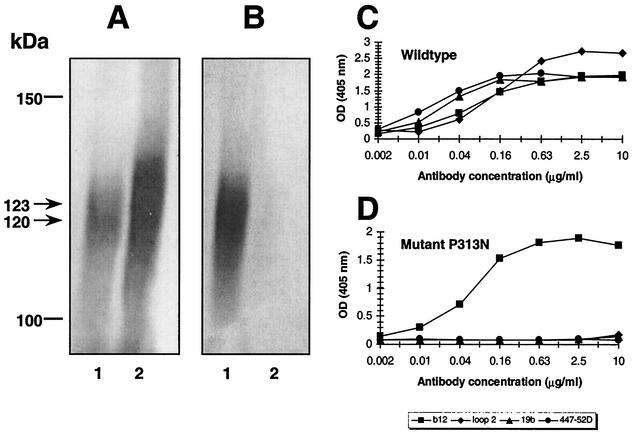
First, we sought to determine whether introduction of an N-glycan in the V3 loop could inhibit reactivity by V3 loop MAbs. For this purpose, a mutant, termed P313N, was generated. In this mutant, Arg and Thr substitute for Pro and Arg at positions 313 and 315, respectively, in the “tip” of the V3 loop (-Gly-Pro-Gly-Arg-Ala-Phe- segment). This N-glycosylation consensus sequence (sequon; NXT) thus permits addition of a potential N-linked glycan at position 313. SDS-PAGE and Western blotting were used to investigate the presence of the extra glycan (Fig. 2A); a slight increase in average molecular weight was observed for the mutant glycoprotein in comparison to wild-type gp120, suggesting the presence of an additional glycan in the mutant.
FIG. 2.
Binding of V3 loop MAbs to wild-type gp120JR-FL and glycoprotein of mutant P313N in Western blot and in ELISA. (A) Wild-type gp120 (lane 1) and mutant glycoprotein (lane 2) were reacted with MAb b12 (1 μg/ml). (B) Glycoproteins were reacted with MAbs 19b, 447-52D, and loop 2 (pooled mixture; 1 μg of each per ml). Molecular mass indicators (bars) and the average molecular masses of wild-type and mutant glycoproteins (arrows) are shown on the left in kilodaltons. (C and D) MAb binding to wild-type gp120 (C) and mutant P313N (D) was determined by ELISA.
The effect of the mutation on antibody reactivity was determined for three V3 loop MAbs, namely, loop 2, 19b, and 447-52D. These three antibodies pooled together were unable to react with glycoprotein of mutant P313N in Western blot (Fig. 2B). ELISA, with individual MAbs, confirmed these results (Fig. 2C and D). In contrast, the apparent affinity of MAb b12, relative to wild type, was unaffected by the introduced mutation. We do note that the glycoprotein of mutant P313N was stained much more strongly by b12 in comparison to wild-type gp120 (Fig. 2A). However, this may be due to a difference in the amount of glycoprotein present in the respective preparations, which were not standardized prior to SDS-PAGE. To determine whether the inability of the three V3 loop antibodies to bind mutant P313N is due solely to incorporation of an N-linked glycan, we replaced the Asn at position 313 with Gln, leaving the Arg→Thr substitution at position 315 unchanged. The three antibodies were also unable to react with this mutant (data not shown). This finding suggests that, even though the V3 loop MAbs may be blocked from binding to their respective epitopes by the introduced glycan, reactivity is probably also abolished due to substitution of the proline residue alone or in combination with the arginine residue. The mutations most likely disrupt the β-turn at the apex of the V3 loop comprising the -Gly-Pro-Gly-Arg- segment (35, 103).
Effect of the introduction of N-linked glycans in gp120 on reactivity with anti-gp120 MAbs.
Next, we sought to block other epitopes that might be recognized by nonneutralizing antibodies by using the same glycosylation strategy. First, we introduced N-glycosylation sequons individually at various positions in gp120, primarily on the nonneutralizing face (115) and in the V1 and V2 loops, to determine the effect on b12 binding. The NXT glycosylation sequon (where “X” is any amino acid except proline) was chosen because the asparagine in this motif is twofold more likely to be glycosylated than the asparagine in the NXS motif (34, 44, 57, 99). In some cases two substitutions, at the N and T positions, were required to introduce the glycosylation motif, whereas in other instances only one of the two positions needed to be substituted by the appropriate residue. The mutations incorporated were Q114N/L116T and Q246N (masking of the nonneutralizing face and the putative gp41-gp120 interface), S143T and E150N/G152T (masking of the V1 loop), K171N/Y173T (masking of the V2 loop), S365T (masking of potential epitopes on the outer perimeter of the CD4bs), and R440N/Q442T (masking of potential epitopes close to the base of the V3 loop, adjacent to the silent face). The S143T and S365T mutations were introduced to enhance the likelihood of glycan attachment at residues N141 and N363, respectively. Binding of MAb b12 was not significantly affected by mutations Q114N/L116T and S143T, whereas binding was somewhat decreased by the E150N/G152T mutation (Fig. 3). Binding was more strongly affected by the mutation in the V2 loop (K171N/Y173T) and by the Q246N mutation. With the two remaining mutants, S365T and R440N, no reactivity with b12 was observed. Notably, expression of the mutant with the R440N/Q442T substitution was very poor (not shown), suggesting that introduction of a glycan at position 440 may interfere with the proper folding or intracellular processing of gp120.
FIG. 3.
Binding of MAb b12 to gp120 with added N-glycosylation sequons. Wild-type gp120 and mutant glycoproteins (labeled I to VII) were captured onto ELISA wells and probed with MAb b12 (10 ng/ml). HIVIG (1 μg/ml) was used to ensure that equivalent amounts of protein were captured. Bound antibody was detected with peroxidase-conjugated secondary antibody, and the absorbance was measured at 450 nm.
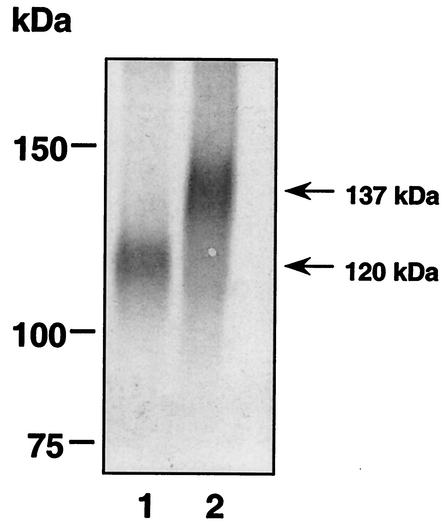
Glycosylation motifs that did not affect b12 binding, or affected it only slightly, i.e., mutations Q114N/L116T, S143T, and E150N/G152T, were then combined. The Pro→Asn and Arg→Thr substitutions at positions 313 and 315 (mutant P313N), respectively, and the alanine substitutions at positions 473 to 476 (mutant GDMR) were also incorporated to generate a hyperglycosylated mutant (Fig. 4). We decided to also insert the K171N/Y173T mutation, despite its negative effect on b12 binding, since this was the only mutation in the V2 loop that could potentially hinder the binding of V2 loop antibodies. Extra N-glycosylation sites were also introduced at positions 92 and 423, without prior testing with b12. Residue H92 lies on the nonneutralizing face of gp120, and residue I423 lies in the coreceptor-binding site. Western blots were performed to confirm that the glycoprotein of this mutant, termed mCHO*-GDMR, was indeed hyperglycosylated. The molecular mass of the modified glycoprotein was observed to be ∼137 kDa (Fig. 5), suggesting that most, if not all, extra potential glycosylation sites are likely to be occupied in this mutant. The largest oligosaccharide that can be incorporated at each site is a fully sialylated, fucosylated, and galactosylated tetraantennary complex glycan (∼3.5 kDa), which would increase the average molecular mass of wild-type gp120 by ∼25 kDa. However, this complex glycan type is observed in fewer than 1% of the total glycans on gp120JR-FL (94). Rather, recombinant gp120JR-FL expressed in mammalian cells appears to contain predominantly galactosylated, fucosylated biantennary complex glycans (sialylated and nonsialylated) and high-oligomannose glycans (Man-8) (94). Such glycans would add an average of ∼2 kDa per glycosylation site, thus increasing the molecular mass of wild-type gp120 to ∼134 kDa, which approximates the average mass determined from the SDS gel for the mutant mCHO*-GDMR. However, we cannot exclude the possibility that some sites are not glycosylated; mass spectrometric analyses of the mutagenized protein (122) would be required to address this issue in more detail.
FIG. 4.
Location of introduced N-glycan attachment sites and alanine substitutions, mapped onto the gp120 core. N-linked oligosaccharides (yellow) were modeled onto the core structure (48) according to the most likely glycoforms, as inferred from a previous study (122). The putative locations of the V1 and V2 loops are also indicated in each panel. The 20-Å bar represents the average width of a typical antibody-combining site. (A) Depiction of the gp120 core is shown from the perspective of CD4. The attachment sites of the extra glycans are labeled and colored dark blue; dark blue spheres indicate those in the V1 and V2 loops. The alanine substitutions at positions 473 to 476 (GDMR) are labeled and colored red. The perspectives shown in panels B and C are also indicated by arrows. (B) View of the gp120 outer domain (48, 116). The spatial location of the V3 and V4 loops, which are proposed to extend from the protein surface, are also indicated. (C) View of the proposed gp41-gp120 interface (48, 116). Figures were made with RasMol (93) and modified by using Adobe Photoshop.
FIG. 5.
Determination of the molecular mass of mutant mCHO*-GDMR by Western blot. Wild-type gp120JR-FL and mutant glycoprotein were separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and probed with a polyclonal antibody prearation generated against the C5 region of gp120 (5 μg/ml). Lane 1, wild type; lane 2, mutant mCHO*-GDMR. Molecular mass indicators are shown on the left, and the average molecular mass of each glycoprotein is denoted on the right in kilodaltons.
To investigate the antigenic properties of this glycoprotein mutant, binding affinities were determined for b12 and for a large panel of nonneutralizing anti-gp120 MAbs. Most MAbs bound wild-type gp120JR-FL with high affinity (Fig. 6A to D). Saturation levels were not obtained with certain MAbs against the C1 domain (133/237, 133/290, and 522-149) and, as already observed, by MAbs C11, G3-4, and G3-136. MAbs 133/237 and 133/290 bind better to denatured gp120 than to native glycoprotein (63) and here display moderate affinity for gp120 in ELISA. Whether this is also the case for MAb 522-149 is not known. It is also possible the moderate binding affinity stems from sequence differences between the homologous antigen and gp120JR-FL. Antibodies 17b and 48d also bound with lower affinity to wild-type gp120 due to the absence of sCD4 in these experiments. When tested with mutant mCHO*-GDMR, binding of virtually all MAbs was abolished, whereas binding of MAb b12 was reduced in comparison to that of wild-type gp120. Unfortunately, but not unexpectedly, the binding of only two of the five C1 antibodies was inhibited. In the present study, no glycan attachment motifs were introduced in the C1 region because glycosylation of the N terminus is reported to be rare (34). Surprisingly, binding of the three other C1 antibodies to mutant mCHO*-GDMR was nearly twofold higher than binding to wild-type gp120 (Fig. 6E). Removal of N-terminal residues in the C1 region reduced or abolished the ability of these antibodies to bind gp120, and deletion of residues in the C terminus of the C5 region reduced the binding of the anti-C5 polyclonal antibody preparation used to capture gp120 (Fig. 7). Surprisingly, b12 binding was also severely diminished (Fig. 7), indicating that the combined removal of these N- and C-terminal residues dramatically influences the conformation of the b12 epitope in mutant mCHO*-GDMR.
FIG. 6.
Binding of anti-gp120 MAbs to wild-type (A to D, left) and mutant mCHO*-GDMR (E to H, right) glycoproteins. (A and E) Binding of MAbs to the C1, C5, C1-C4, and C1-C5 domains; (B and F) binding MAbs to the CD4bs; (C and G) binding of MAbs to the coreceptor-binding site; (D and H) binding of MAbs to the V2 and V3 loops. Captured glycoproteins were probed with MAb concentrations indicated on the x axis. The absorbance was measured at 405 nm.
FIG. 7.
Glycoproteins of mutants mCHO*-GDMR and mCHO*-GDMR ΔN/C (N and C termini deleted), captured onto ELISA plate wells with indicated antibodies (10 μg/ml) and detected with biotinylated MAb 2G12 (1μg/ml). Gray bars represent mutant mCHO*-GDMR; dark bars represent mutant mCHO*-GDMR ΔN/C. The absorbance was measured at 450 nm.
Epitope masking by introduction of N-glycans abolishes reactivity with a polyclonal antibody preparation.
To test the degree to which our approach was able to mask nonneutralizing epitopes on gp120, we also determined the effects of the introduced glycans on binding of a preparation of polyclonal antibodies. This preparation, HIVIG, which is derived from pooled plasma of asymptomatic individuals that was selected based on high antibody titers to the HIV structural protein p24, neutralizes primary HIV-1 isolates poorly (24). When tested with mutant GDMR, a twofold reduction in relative binding affinity was observed, in comparison to wild-type gp120JR-FL (Fig. 8). However, binding was much more reduced to the mutant with an extra glycosylation site in the V3 loop, thus suggesting the presence of a large percentage of V3 loop antibodies in this preparation. No binding of HIVIG to mutant mCHO*-GDMR was observed, supporting the notion that we may have indeed succeeded in masking a number of epitopes on gp120 that have the potential to induce nonneutralizing antibodies.
FIG. 8.
Reactivity of HIVIG (open symbols) and MAb b12 (solid symbols) in ELISA with wild-type gp120 and mutant glycoproteins. wt, wild-type gp120; GDMR, mutant GDMR in which residues at positions 473 to 476 in gp120 have been replaced with alanine; GDMR-P313N, similar to mutant GDMR with the addition of an N-glycosylation site in the V3 loop; mCHO*-GDMR, similar to mutant GDMR-P313N, with the addition of six glycosylation motifs (two in the V1 loop, one in the V2 loop, and three on the gp120 core).
Influence of the N-glycosylation motif in the V2 loop on b12 binding.
We suspected that the lowered affinity of MAb b12 for mutant mCHO*-GDMR may be caused by the K171N/Y173T mutation in the V2 loop. To test this postulate, the mutation was reverted to wild-type sequence. Surprisingly, b12 binding affinity for this mutant, termed K171Nx, was similar to that for mutant mCHO*-GDMR (data not shown). Therefore, the lower affinity of b12 for mutant mCHO*-GDMR in comparison to wild-type glycoprotein most probably stems from the introduction of a particular combination of glycans in mutant mCHO*-GDMR rather than from steric hindrance by a specific glycan. Interestingly, the V2 loop antibody G3-4 did not react with mutant K171Nx (data not shown), indicating that the extra V2 loop-glycan may not be required for masking antigenic determinants in this loop. Rather, the result with MAb G3-4 suggests that neighboring glycans that were inserted additionally may be sufficient to block the binding of V2 loop antibodies.
Influence of alanine substitution at positions 473 to 476 in mutant mCHO*-GDMR on binding of MAbs to epitopes overlapping the CD4bs.
Although we had been successful in masking the epitopes of various nonneutralizing and weakly neutralizing antibodies, it was unclear whether blockage of CD4bs antibodies and MAb A32 could be accomplished by the glycans alone, without the need for the introduced alanine substitutions at positions 473 to 476. Therefore, another variant of mutant mCHO*-GDMR was generated. In this variant, termed mCHO*, the alanine substitutions at positions 473 to 476 were reverted to wild type. As shown in Fig. 9, the binding of MAbs 15e, F91, F105, and A32 was significantly reduced or completely abolished, indicating that the added glycans are indeed sufficient to block binding of many nonneutralizing or weakly neutralizing CD4bs antibodies. However, the weakly neutralizing antibodies b3 and b6 were still able to bind mutant glycoprotein, albeit with lower affinity than that of MAb b12. Thus, the extra glycans appear not to have completely blocked access to the CD4bs but, rather, may have confined the space available to antibodies for interaction with this site.
FIG. 9.
Binding of CD4bs MAbs and MAb A32 to glycoprotein of mutant mCHO*. This mutant contains all of the glycosylation sequons that are also present in mutant mCHO*-GDMR but lacks the alanine substitutions on the edge of the Phe-43 cavity.
DISCUSSION
A major concern for HIV-1 vaccine design is that at present no immunogen or combination of immunogens is capable of eliciting the levels of broadly neutralizing antibodies that are likely needed to contribute to significant protection against infection. Nevertheless, the anti-gp120 and anti-gp41 MAbs that have been isolated from natural infection and from immunization studies serve as valuable tools to screen prospective antigenic formulations for their suitability as candidate immunogens (17, 117); antigens that preferentially bind neutralizing antibodies, but not nonneutralizing ones, would naturally be most desirable. In a recent report, we observed that binding of the broadly and potently neutralizing human antibody b12 was unaffected or slightly enhanced by the introduction of four alanine mutations on the perimeter of the Phe-43 cavity on gp120 (75). In contrast, binding of CD4 and five weakly neutralizing CD4bs antibodies was abolished or reduced. The aim of the current study was to further evaluate and improve the antigenic properties of this gp120 mutant, termed GDMR.
As a first step, mutant GDMR was tested in ELISA with a selection of MAbs against various linear and discontinuous epitopes on gp120. Only nonneutralizing or weakly neutralizing CD4bs MAbs were significantly affected by the alanine substitutions (Fig. 1). Whether this correlates with how these antibodies interact with gp120 and their lack of neutralizing potency is uncertain. However, considering that antibodies often form salt bridges or hydrogen bonds with available polar groups on the antigen, most probably because this compensates for the loss in entropy upon antigen interaction (7, 11, 12, 100, 109, 110), it is noteworthy that MAbs b3, b6, and F105 are particularly sensitive to replacement of residues at positions 474 (Asp) and 476 (Arg) with alanine (75). The results obtained here with mutant GDMR suggest that these aspartate and arginine residues may be ideal contact residues for many nonneutralizing antibodies. Although it is speculative at this point, these residues may be more accessible on monomeric gp120 than on the trimeric envelope spikes of primary HIV isolates, thus explaining why these antibodies are unable to neutralize virions potently.
The observation that non-CD4bs antibodies were not inhibited from binding to mutant GDMR prompted us to pursue further means of blocking epitopes recognized by nonneutralizing antibodies. The concept of diverting B-cell immune responses away from undesired epitopes has been discussed previously (26). A recent study (21) showed that, by introducing a single amino acid mutation in the β-chain of human chorionic gonadotrophin, cross-reactive antibodies to luteinizing hormone, which are normally elicited upon immunization with wild-type human chorionic gonadotrophin β-chain, could be eliminated. The immune response was thus refocused to epitopes that are normally only weakly immunogenic.
In the case of HIV, such a strategy is likely to be tedious and time-consuming, particularly considering the vast genetic diversity that is manifested among HIV isolates. Therefore, we chose to insert N-glycosylation sequons into the gp120 sequence with the aim of selectively incorporating additional N-glycans onto the glycoprotein to mask undesired epitopes. Masking epitopes in the (hyper)variable loops of HIV was of particular importance, since antibodies to such sites are often induced upon immunization with gp120 or during natural infection (22, 29, 38, 52, 54, 55, 59, 93, 97, 101, 112). During infection, these antibodies are detected relatively early, suggesting the presence of immunodominant epitopes within the loops (29, 55, 70, 97). The masking of epitopes by glycans is in itself not novel but, in fact, a strategy employed by HIV to avoid facile recognition by the host immune system (3, 25, 73, 82, 83, 95, 115). For example, a large portion of gp120 is covered by N-glycans, thus rendering those regions of the antigen immunologically “silent” (115). Furthermore, changes in the number and placement of N-linked glycans in gp120 are known to modulate the exposure of antigenic determinants. For example, elimination of an N-glycan in the V3 loop by site-directed mutagenesis increases viral sensitivity to neutralizing antibodies (3). Also, when virus lacking an N-linked glycan in the V3 loop is grown in the presence of a V3 loop antibody it rapidly reverts to a variant in which a glycan is reincorporated at that position (95). However, in terms of vaccine design, the approach of epitope masking by the incorporation of additional N-linked glycans is not widespread. In fact, this particular approach has been applied in only one previous study (33). In that report, incorporation of an N-glycosylation site in the V3 loop resulted in a shift in the immune response toward epitopes in the V1 loop (33).
Here, the approach was expanded. N-linked glycans were introduced based primarily, but not solely, on the following criteria: (i) the side chain of the residue to be mutated should be solvent exposed; (ii) the introduced glycan should block a region on gp120 that might elicit nonneutralizing antibodies; (iii) the site selected for introduction of the N-glycan should contain residues that might tolerate mutation, yet still retain a similar conformation; and (iv) the potential glycosylation site should not be hindered by neighboring sequence elements. For incorporation of the N-glycans, an NXT glycosylation sequence was chosen over an NXS motif because studies have shown that the asparagine in the former sequon is more likely to be glycosylated (34, 44, 57, 99). Furthermore, in an NXT glycosylation motif, more residues are tolerated at position X in terms of glycosylation efficiency than in an NXS sequence (44).
First, we established whether introduction of an N-glycan in the V3 loop could abolish binding of V3 loop antibodies. Considering that a glycan at the apex of V3 might mask a substantial portion of the loop on both sides, a glycosylation site was incorporated at position 313 (Pro). Indeed, none of the three V3 loop MAbs tested were able to bind to this mutant. However, antibody binding to a second mutant, in which the Asn in the glycosylation sequon was replaced by Gln, was also abrogated. These results suggest that incorporation of the extra glycosylation sequon may serve two functions. First, incorporation of the glycosylation motif allows potential antibody epitopes to be masked due to the presence of the glycan. Whether the glycan indeed masks the entire loop is uncertain, since there are currently no antibodies available that are reactive with residues downstream or upstream from the introduced glycosylation site in the V3 loop of JR-FL. The second effect of replacing the proline and arginine residues is that this most likely eliminates or modifies the β-turn and β-type hairpin, which are characteristic of the apex of the V3 loop and confer immunodominance to the loop (35, 84). Lowering this predominant characteristic of the V3 loop may further increase the potential to obtain antibodies against epitopes on gp120 that are normally only weakly immunogenic.
Next, the glycosylation strategy was extended to other epitopes that might elicit nonneutralizing antibodies. Seven sites were chosen for the incorporation of N-glycosylation motifs (Fig. 3). The mutations were generated in individual mutants first and tested with b12 to ensure that none severely compromised gp120 folding as detected by antibody binding. Of the seven mutations, three were eliminated from subsequent studies because they indeed abolished b12 binding. The added glycosylation site in the V2 loop also reduced b12 binding but was still selected for further studies since no other glycosylation motifs had been incorporated in the V2 loop for epitope masking. This mutation and the others that did not significantly affect b12 binding were then combined together with the glycosylation sequon in the V3 loop and the alanine substitutions in the Phe-43 cavity to generate a hyperglycosylated mutant glycoprotein. Two additional glycosylation motifs were introduced at positions 92 and 423 to further mask the nonneutralizing face on gp120 and the coreceptor-binding site, respectively. This hyperglycosylated gp120 mutant, termed mCHO*-GDMR, blocked the binding of virtually all nonneutralizing and weakly neutralizing antibodies used in the present study, including a polyclonal antibody preparation (HIVIG) of low neutralizing potency. Importantly, b12 binding was maintained, albeit at lowered affinity. However, this reduced binding was not caused by the N-glycan that was incorporated in the V2 loop, since reversion to the wild-type sequence did not alter b12 affinity. Binding to mutant mCHO*-GDMR was also still observed with three MAbs against the C1 region and a polyclonal antibody against the C5 region. Although the binding of these antibodies could be reduced or abolished by removal of residues in C1 and C5, respectively, b12 binding was also severely reduced (Fig. 7). Thus, in the mutant mCHO*-GDMR, it appears that b12 binding cannot be maintained at the expense of the remaining C1- and C5-reactive antibodies by truncating the N and C termini because of the apparent negative effect on the conformation of the b12 epitope.
Thus, we have generated a series of mutant gp120s that diminish or abolish the binding of nonneutralizing or weakly neutralizing antibodies to various degrees, but retain, to some extent, b12 binding. Whether the reduced b12 affinity for mutant mCHO*-GDMR translates into difficulties in using this mutant to elicit b12-like antibodies is unpredictable. However, one of the remarkable features of the humoral immune response is its plasticity (50, 51, 58, 69, 113). This flexibility, which is represented by the variable regions of immunoglobulins, in particular within the paratope, may enable antibodies with disparate sequences to recognize gp120 equivalent to b12 and neutralize viral isolates with comparable efficacy.
The primary goal of the present study has been to generate gp120 molecules that have the potential to induce broadly neutralizing antibodies, particularly those targeted to the CD4bs. It could be argued that, although the CD4bs shows a high degree of amino acid conservation, the coreceptor binding site is even more conserved (48, 85) and, therefore, is an equally important target. The reason for not directing our efforts to this site relates to recent observations which indicate that Fab and single-chain antibody fragments of CD4-induced antibodies, such as 17b and X5, are more effective in neutralizing primary isolates than the corresponding full-length antibody (A. F. Labrijn, P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. M. Binley, V. Vivona, C. Grundner, C.-C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton, unpublished observations), presumably due to spatial constraints between the target membrane and the CD4-bound virus. The coreceptor-binding site may thus be a target that is more suitable for the design of small molecules to inhibit viral entry rather than a viable epitope to elicit neutralizing antibodies.
Although the approach taken is this study is novel, one caveat may be that, because of the extra glycans, gp120 has assumed a different conformation from that presumably present on the viral surface. Considering that b12 binding to mutant mCHO*-GDMR is maintained, it is unlikely that the extra oligosaccharide moieties significantly alter the overall gp120 core structure, including the CD4bs. Since the majority of broadly neutralizing antibodies appear to bind equally well to monomeric and oligomeric gp120 (31, 76, 91), antibodies that recognize the CD4bs on gp120 in a b12-like manner should also be reactive with oligomeric gp120. In fact, with the introduction of the extra N-glycans, we may have also succeeded in creating spatial constraints around the CD4 binding domain that antibodies are likely to encounter with native, virion-associated oligomeric gp120.
Besides epitope masking, the extra glycans may also serve an additional function. Recent studies suggest that monomeric gp120 contains unusually high intrinsic entropy, which might undermine efforts to obtain broadly neutralizing antibodies (46, 67). Considering that asparagine-linked glycosylation is known to influence protein structure and folding (43, 44, 72, 114) and that the addition of carbohydrate moieties may minimize the conformational flexibility of proteins (72, 114), it is possible that the extra glycans incorporated into the gp120 mutants described here reduce the overall flexibility of monomeric gp120. If so, this could further promote the induction of antibodies with neutralizing properties superior to those obtained with unmodified gp120.
In conclusion, we have presented here a panel of novel HIV glycoprotein mutants that, based on their antigenic properties, are promising immunogens. The antigenic characteristics of the hyperglycosylated mutant mCHO*-GDMR in particular are unparalleled. It will now be interesting to determine whether these modified glycoproteins are capable of inducing broadly cross-reactive neutralizing antibodies, the results of which may provide useful information for the further development of an effective HIV vaccine.
Acknowledgments
We are grateful to J. Robinson (Tulane University) for MAbs C11, 212A, 17b, 48d, 19b, F91, 15e, and A32; H. Katinger (Institute of Applied Microbiology, University of Agriculture, Vienna, Austria) for MAb 2G12; M. Fung (Tanox Biosystems, Inc.) for MAbs G3-4 and G3-136; and S. Zolla-Pazner (New York University School of Medicine) for MAb 447-52D. We also thank E. O. Saphire for help selecting potential sites on the gp120 core for the introduction of N-glycosylation sequons and acknowledge the contribution of plasmid pSyngp140JR-FL to the NIH ARRRP by E.-C. Park and B. Seed (Department of Molecular Biology, Massachusetts General Hospital). The technical assistance of R. Kelleher for the production of antibody Fab fragments and the fruitful discussions with M. Zwick and J. Binley are also acknowledged.
The present study was supported by NIH grants GM46192 and AI33292 to I. A. Wilson and D. R. Burton, respectively, and by the International AIDS Vaccine Initiative through the Neutralizing Antibody Consortium.
REFERENCES
- 1.Andre, S., B. Seed, J. Eberle, W. Schraut, A. Bultmann, and J. Haas. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, L. O., J. W. Bess, Jr., E. N. Chertova, J. L. Rossio, M. T. Esser, R. E. Benveniste, L. E. Henderson, and J. D. Lifson. 1998. Chemical inactivation of retroviral infectivity by targeting nucleocapsid protein zinc fingers: a candidate SIV vaccine. AIDS Res. Hum. Retrovir. 14(Suppl. 3):S311-S319. [PubMed] [Google Scholar]
- 3.Back, N. K. T., L. Smit, J.-J. de Jong, W. Keulen, M. Schutten, J. Goudsmit, and M. Tersmette. 1994. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199:431-438. [DOI] [PubMed] [Google Scholar]
- 4.Barbas, C. F., III, E. Bjorling, F. Chiodi, N. Dunlop, D. Cababa, T. M. Jones, S. L. Zebedee, M. A. Persson, P. L. Nara, E. Norrby, and D. R. Burton. 1992. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. USA 89:9339-9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbas, C. F., III, T. A. Collet, W. Amberg, P. Roben, J. M. Binley, D. Hoekstra, D. Cababa, T. M. Jones, R. A. Williamson, G. R. Pilkington, N. L. Haigwood, E. Cabezas, A. C. Satterthwait, I. Sanz, and D. R. Burton. 1993. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J. Mol. Biol. 230:812-823. [DOI] [PubMed] [Google Scholar]
- 6.Belshe, R. B., G. J. Gorse, M. J. Mulligan, T. G. Evans, M. C. Keefer, J. L. Excler, A. M. Duliege, J. Tartaglia, W. I. Cox, J. McNamara, K. L. Hwang, A. Bradney, D. Montefiori, and K. J. Weinhold. 1998. Induction of immune responses to HIV-1 by canarypox virus (ALVAC) HIV-1 and gp120 SF-2 recombinant vaccines in uninfected volunteers. AIDS 12:2407-2415. [DOI] [PubMed] [Google Scholar]
- 7.Bhat, T. N., G. A. Bentley, G. Boulot, M. I. Greene, D. Tello, W. Dall'Acqua, H. Souchon, F. P. Schwarz, R. A. Mariuzza, and R. J. Poljak. 1994. Bound water molecules and conformational stabilization help mediate an antigen-antibody association. Proc. Natl. Acad. Sci. USA 91:1089-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binley, J. M., R. W. Sanders, A. Master, C. S. Cayanan, C. L. Wiley, L. Schiffner, B. Travis, S. Kuhmann, D. R. Burton, S.-L. Hu, W. Olson, and J. P. Moore. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 76:2606-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolmstedt, A., S. Sjolander, J.-E. S. Hansen, L. Akerblom, A. Hemming, S.-L. Hu, B. Morein, and S. Olofsson. 1996. Influence of N-linked glycans in V4-5 region of human immunodeficiency virus type 1 glycoprotein gp160 on induction of a virus-neutralizing humoral response. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12:213-220. [DOI] [PubMed] [Google Scholar]
- 11.Braden, B. C., B. A. Fields, and R. J. Poljak. 1995. Conservation of water molecules in an antibody-antigen interaction. J. Mol. Recognit. 8:317-325. [DOI] [PubMed] [Google Scholar]
- 12.Braden, B. C., and R. J. Poljak. 1995. Structural features of the reactions between antibodies and protein antigens. FASEB J. 9:9-16. [DOI] [PubMed] [Google Scholar]
- 13.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauder, and H. Katinger. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359-369. [DOI] [PubMed] [Google Scholar]
- 14.Buchacher, A., R. Predl, C. Tauer, M. Purtscher, G. Gruber, R. Heider, F. Steindl, A. Trkola, A. Jungbauer, and H. Katinger. 1992. Human monoclonal antibodies against gp41 and gp120 as potential agents for passive immunization, p. 191-194. In F. Brown, R. Chanock, H. S. Ginsberg, and R. Lerner (ed.), Vaccines '92: modern approaches to new vaccines including prevention of AIDS. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 15.Buchbinder, A., S. Karwowska, M. K. Gorny, S. T. Burda, and S. Zolla-Pazner. 1992. Synergy between human monoclonal antibodies to HIV extends their effective biological activity against homologous and divergent strains. AIDS Res. Hum. Retrovir. 8:425-427. [DOI] [PubMed] [Google Scholar]
- 16.Burton, D. R. 1997. A vaccine for HIV type 1: the antibody perspective. Proc. Natl. Acad. Sci. USA 94:10018-10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burton, D. R. 2002. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2:706-713. [DOI] [PubMed] [Google Scholar]
- 18.Burton, D. R., C. F. Barbas III, M. A. A. Persson, S. Koenig, R. M. Chanock, and R. A. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 88:10134-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. H. I. Parren, L. S. W. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garratty, E. R. Stiehm, Y. J. Bryson, Y. Cao, J. P. Moore, D. D. Ho, and C. F. Barbas III. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 20.Cherpelis, S., I. Shrivastava, A. Gettie, X. Jin, D. D. Ho, S. W. Barnett, and L. Stamatatos. 2001. DNA vaccination with the human immunodeficiency virus type 1 SF162ΔV2 envelope elicits immune responses that offer partial protection from simian/human immunodeficiency virus infection to CD8+-T-cell-depleted rhesus macaques. J. Virol. 75:1547-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiesa, M. D., P. M. Martensen, C. Simmons, N. Porakishvili, J. Justesen, G. Dougan, I. M. Roitt, P. J. Delves, and T. Lund. 2001. Refocusing of B-cell responses following a single amino acid substitution in an antigen. Immunology 103:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clements-Mann, M. L., K. Weinhod, T. J. Matthews, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, R. H. Hsieh, J. Mestechy, S. Zolla-Pazner, R. Belshe, R. Dolin, S. Jackson, S. Xu, P. Fast, M. C. Walker, D. Stablein, J. L. Excler, and J. Tartaglia. 1998. Immune responses to human immunodeficiency virus (HIV) type 1 induced by canarypox expressing HIV-1MN gp120, HIV-1SF2 recombinant gp120, or both vaccines in seronegative adults. J. Infect. Dis. 177:1230-1246. [DOI] [PubMed] [Google Scholar]
- 23.Connor, R. I., B. T. M. Korber, B. S. Graham, B. H. Hahn, D. D. Ho, B. D. Walker, A. U. Neumann, S. H. Vermund, J. Mestecky, S. Jackson, E. Fenamore, Y. Cao, F. Gao, S. Kalams, K. J. Kunstman, D. McDonald, N. McWilliams, A. Trkola, J. P. Moore, and S. M. Wolinsky. 1998. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J. Virol. 72:1552-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cummins, L. M., K. J. Weinhold, T. J. Matthews, A. J. Langlois, C. F. Perno, R. M. Condie, and J. P. Allain. 1991. Preparation and characterization of an intravenous solution of IgG from human immunodeficiency virus-seropositive donors. Blood 77:1111-1117. [PubMed] [Google Scholar]
- 25.Davis, D., D. M. Stephens, C. Willers, and P. J. Lachmann. 1990. Glycosylation governs the binding of antipeptide antibodies to regions of hypervariable amino acid sequence within recombinant gp120 of human immunodeficiency virus type 1. J. Gen. Virol. 71:2889-2898. [DOI] [PubMed] [Google Scholar]
- 26.Delves, P. J., T. Lund, and I. M. Roitt. 1997. Can epitope-focused vaccines select advantageous immune responses? Mol. Med. Today 3:55-60. [DOI] [PubMed] [Google Scholar]
- 27.di Marzo Veronese, F. R., R. Rahman, R. Pal, C. Boyer, J. Romano, V. S. Kalyanaraman, B. C. Nair, R. C. Gallo, and M. G. Sarngadharan. 1992. Delineation of immunoreactive, conserved regions in the external envelope glycoprotein of the human immunodeficiency virus type I. AIDS Res. Hum. Retrovir. 8:1125-1132. [DOI] [PubMed] [Google Scholar]
- 28.D'Souza, M. P., D. Livnat, J. A. Bradac, S. Bridges, et al. 1997. Evaluation of monoclonal antibodies to HIV-1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 29.Ebenbichler, C., T. McNearney, H. Stoiber, J. Most, R. Zangerle, W. Vogetseder, J. R. Patsch, L. Ratner, and M. P. Dierich. 1995. Sera from HIV-1 infected individuals in all stages of disease preferentially recognize the V3 loop of the prototypic macrophage-tropic glycoprotein gp120 ADA. Mol. Immunol. 32:1039-1045. [DOI] [PubMed] [Google Scholar]
- 30.Finnegan, C. M., W. Berg, G. K. Lewis, and A. L. DeVico. 2001. Antigenic properties of the human immunodeficiency virus envelope during cell-cell fusion. J. Virol. 75:11096-11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fung, M. S. C., C. R. Y. Sun, W. L. Gordon, R.-S. Liou, T. W. Chang, W. N. C. Sun, E. S. Daar, and D. D. Ho. 1992. Identification and characterization of a neutralization site within the second variable region of human immunodeficiency virus type 1 gp120. J. Virol. 66:848-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garrity, R. R., G. Rimmelzwaan, A. Minassian, W.-P. Tsai, G. Lin, J.-J. de Jong, J. Goudsmit, and P. L. Nara. 1997. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J. Immunol. 159:279-289. [PubMed] [Google Scholar]
- 34.Gavel, Y., and G. von Heijne. 1990. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 3:433-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghiara, J. B., E. A. Stura, R. L. Stanfield, A. T. Profy, and I. A. Wilson. 1994. Crystal structure of the principal neutralization site of HIV-1. Science 264:82-85. [DOI] [PubMed] [Google Scholar]
- 36.Gorny, M. K., A. J. Conley, S. Karwowska, A. Buchbinder, J.-Y. Xu, E. A. Emini, S. Koenig, and S. Zolla-Pazner. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorny, M. K., J. Y. Xu, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1993. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J. Immunol. 150:635-643. [PubMed] [Google Scholar]
- 38.Graham, B. S., M. C. Keefer, M. J. McElrath, G. J. Gorse, D. H. Schwartz, K. Weinhold, T. J. Matthews, J. R. Esterlitz, F. Sinangil, P. E. Fast, et al. 1996. Safety and immunogenicity of a candidate HIV-1 vaccine in healthy adults: recombinant glycoprotein (rgp) 120: a randomized, double-bind trial. Ann. Intern. Med. 125:270-279. [DOI] [PubMed] [Google Scholar]
- 39.Grundner, C., T. Mirzabekov, J. Sodroski, and R. Wyatt. 2002. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J. Virol. 76:3511-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 41.Ho, D. D., M. S. Fung, Y. Z. Cao, X. L. Li, C. Sun, T. W. Chang, and N. C. Sun. 1991. Another discontinuous epitope on glycoprotein gp120 that is important in human immunodeficiency virus type 1 neutralization is identified by a monoclonal antibody. Proc. Natl. Acad. Sci. USA 88:8949-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho, D. D., J. A. McKeating, X. L. Li, T. Moudgil, E. S. Daar, N. C. Sun, and J. E. Robinson. 1991. Conformational epitope on gp120 important in CD4 binding and human immunodeficiency virus type 1 neutralization identified by a human monoclonal antibody. J. Virol. 65:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imperiali, B., and K. W. Rickert. 1995. Conformational implications of asparagine-linked glycosylation. Proc. Natl. Acad. Sci. USA 92:97-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasturi, L., H. Chen, and S. H. Shakin-Eshleman. 1997. Regulation of N-linked core glycosylation: use of a site-directed mutagenesis approach to identify Asn-Xaa-Ser/Thr sequons that are poor oligosaccharide acceptors. Biochem. J. 323:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kessler, J. A., P. M. McKenna, E. A. Emini, C. P. Chan, M. D. Patel, S. K. Gupta, G. E. Mark, C. F. Barbas III, D. R. Burton, and A. J. Conley. 1997. Recombinant human monoclonal antibody IgG1 b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retrovir. 13:575-581. [DOI] [PubMed] [Google Scholar]
- 46.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. M. Chaiken, M. Fung, H. Katinger, P. W. H. I. Parren, J. Robinson, D. van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 47.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure Fold Des. 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 48.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Letvin, N. L., D. H. Barouch, and D. C. Montefiori. 2002. Prospects for vaccine protection against HIV-1 infection and AIDS. Annu. Rev. Immunol. 20:73-99. [DOI] [PubMed] [Google Scholar]
- 50.Manivel, V., F. Bayiroglu, Z. Siddiqui, D. M. Salunke, and K. V. Rao. 2002. The primary antibody repertoire represents a linked network of degenerate antigen specificities. J. Immunol. 169:888-897. [DOI] [PubMed] [Google Scholar]
- 51.Manivel, V., N. C. Sahoo, D. M. Salunke, and K. V. Rao. 2000. Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immunity 13:611-620. [DOI] [PubMed] [Google Scholar]
- 52.Markham, R. B., J. Coberly, A. J. Ruff, D. Hoover, J. Gomez, E. Holt, J. Desormeaux, R. Boulos, T. C. Quinn, and N. A. Halsey. 1994. Maternal IgG1 and IgA antibody to V3 loop consensus sequence and maternal-infant HIV-1 tranmission. Lancet 343:390-391. [DOI] [PubMed] [Google Scholar]
- 53.Mascola, J. R., S. Schlesinger Frankel, and K. Broliden. 2000. HIV-1 entry at the mucosal surface: role of antibodies in protection. AIDS 14(Suppl. 3):S167-S174. [PubMed] [Google Scholar]
- 54.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, and D. S. Burke. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 55.McKeating, J. A., C. Shotton, S. Jeffs, C. Palmer, A. Hammond, J. Lewis, K. Oliver, J. May, and P. Balfe. 1996. Immunogenicity of full length and truncated forms of the human immunodeficiency virus type I envelope glycoprotein. Immunol. Lett. 51:101-105. [DOI] [PubMed] [Google Scholar]
- 56.McKeating, J. A., M. Thali, C. Furman, S. Karwowska, M. K. Gorny, J. Cordell, S. Zolla-Pazner, J. Sodroski, and R. A. Weiss. 1992. Amino acid residues of the human immunodeficiency virus type 1 gp120 critical for the binding of rat and human neutralizing antibodies that block the gp120-sCD4 interaction. Virology 190:134-142. [DOI] [PubMed] [Google Scholar]
- 57.Mellquist, J. L., L. Kasturi, S. L. Spitalnik, and S. H. Shakin-Eshleman. 1998. The amino acid following an Asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 37:6833-6837. [DOI] [PubMed] [Google Scholar]
- 58.Mian, I. S., A. R. Bradwell, and A. J. Olson. 1991. Structure, function, and properties of antibody binding sites. J. Mol. Biol. 217:133-151. [DOI] [PubMed] [Google Scholar]
- 59.Montefiori, D. C., B. S. Graham, J. Zhou, D. H. Schwartz, L. A. Cavacini, and M. R. Posner. 1993. V3-specific neutralizing antibodies in sera from HIV-1 gp160-immunized volunteers block virus fusion and act synergistically with human monoclonal antibody to the conformation-dependent CD4 binding site of gp120. J. Clin. Investig. 92:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moore, J. P., and D. D. Ho. 1993. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J. Virol. 67:863-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117-S136. [PubMed] [Google Scholar]
- 62.Moore, J. P., F. E. McCutchan, S. W. Poon, J. Mascola, J. Liu, Y. Cao, and D. D. Ho. 1994. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J. Virol. 68:8350-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moore, J. P., Q. J. Sattentau, R. Wyatt, and J. Sodroski. 1994. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J. Virol. 68:469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moore, J. P., A. Trkola, B. Korber, L. J. Boots, J. A. I. Kessler, F. E. McCutchan, J. Mascola, D. D. Ho, J. Robinson, and A. J. Conley. 1995. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J. Virol. 69:122-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moore, J. P., R. L. Willey, G. K. Lewis, J. Robinson, and J. Sodroski. 1994. Immunological evidence for interactions between the first, second, and fifth conserved domains of the gp120 surface glycoprotein of human immunodeficiency virus type 1. J. Virol. 68:6836-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nabel, G. J., and N. J. Sullivan. 2000. Antibodies and resistance to natural HIV infection. N. Engl. J. Med. 343:1263-1265. [DOI] [PubMed] [Google Scholar]
- 69.Nair, D. T., K. Singh, Z. Siddiqui, B. P. Nayak, K. V. Rao, and D. M. Salunke. 2002. Epitope recognition by diverse antibodies suggests conformational convergence in an antibody response. J. Immunol. 168:2371-2382. [DOI] [PubMed] [Google Scholar]
- 70.Nara, P. L., R. R. Garrity, and J. Goudsmit. 1991. Neutralization of HIV-1: a paradox of humoral proportions. FASEB J. 5:2437-2455. [DOI] [PubMed] [Google Scholar]
- 71.Niedrig, M., H. P. Harthus, J. Hinkula, M. Broker, H. Bickhard, G. Pauli, H. R. Gelderblom, and B. Wahren. 1992. Inhibition of viral replication by monoclonal antibodies directed against human immunodeficiency virus gp120. J. Gen. Virol. 73:2451-2455. [DOI] [PubMed] [Google Scholar]
- 72.O'Connor, S. E., and B. Imperiali. 1996. Modulation of protein structure and function by asparagine-linked glycosylation. Chem. Biol. 3:803-812. [DOI] [PubMed] [Google Scholar]
- 73.Overbaugh, J., and L. M. Rudensey. 1992. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J. Virol. 66:5937-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pantophlet, R., L. Brade, L. Dijkshoorn, and H. Brade. 1998. Specificity of rabbit antisera against lipopolysaccharide of Acinetobacter. J. Clin. Microbiol. 36:1245-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pantophlet, R., E. O. Saphire, P. Poignard, P. W. H. I. Parren, I. A. Wilson, and D. R. Burton. 2003. Fine mapping of the interaction of neutralizing and non-neutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parren, P. W. H. I., I. Mondor, D. Naniche, H. J. Ditzel, P. J. Klasse, D. R. Burton, and Q. J. Sattentau. 1998. Neutralization of HIV-1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parren, P. W. H. I., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13(Suppl. A):S137-S162. [PubMed] [Google Scholar]
- 78.Poignard, P., T. Fouts, D. Naniche, J. P. Moore, and Q. J. Sattentau. 1996. Neutralizing antibodies to human immunodeficiency virus type-1 gp120 induce envelope glycoprotein subunit dissociation. J. Exp. Med. 183:473-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poignard, P., E. O. Saphire, P. W. H. I. Parren, and D. R. Burton. 2001. GP120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 80.Posner, M. R., L. A. Cavacini, C. L. Emes, J. Power, and R. Byrn. 1993. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 6:7-14. [PubMed] [Google Scholar]
- 81.Posner, M. R., H. S. Elboim, T. Cannon, L. Cavacini, and T. Hideshima. 1992. Functional activity of an HIV-1 neutralizing IgG human monoclonal antibody: ADCC and complement-mediated lysis. AIDS Res. Hum. Retrovir. 8:553-558. [DOI] [PubMed] [Google Scholar]
- 82.Quiñones-Kochs, M. I., L. Buonocore, and J. K. Rose. 2002. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J. Virol. 76:4199-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 84.Rini, J. M., R. L. Stanfield, E. A. Stura, P. A. Salinas, A. T. Profy, and I. A. Wilson. 1993. Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen. Proc. Natl. Acad. Sci. USA 90:6325-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 86.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robinson, J. E., D. Holton, S. Pacheco-Morell, J. Liu, and H. McMurdo. 1990. Identification of conserved and variant epitopes of human immunodeficiency virus type 1 (HIV-1) gp120 by human monoclonal antibodies produced by EBV-transformed cell lines. AIDS Res. Hum. Retrovir. 6:567-579. [DOI] [PubMed] [Google Scholar]
- 88.Robinson, J. E., H. Yoshiyama, D. Holton, S. Elliott, and D. D. Ho. 1992. Distinct antigenic sites on HIV gp120 identified by a panel of human monoclonal antibodies. J. Cell. Biochem. 50(Suppl. 16E):71. [Google Scholar]
- 89.Rossio, J. L., M. T. Esser, K. Suryanarayana, D. K. Schneider, J. W. Bess, Jr., G. M. Vasquez, T. A. Wiltrout, E. Chertova, M. K. Grimes, Q. Sattentau, L. O. Arthur, L. E. Henderson, and J. D. Lifson. 1998. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72:7992-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sanders, R. W., L. Schiffner, A. Master, F. Kajumo, Y. Guo, T. Dragic, J. P. Moore, and J. M. Binley. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J. Virol. 74:5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sattentau, Q. J., M. Moulard, B. Brivet, F. Botto, J. C. Cuillemot, I. Mondor, P. Poignard, and S. Ugolini. 1999. Antibody neutralization of HIV-1 and the potential for vaccine design. Immunol. Lett. 66:143-149. [DOI] [PubMed] [Google Scholar]
- 93.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374-376. [DOI] [PubMed] [Google Scholar]
- 94.Scanlan, C. N., R. Pantophlet, M. R. Wormwald, E. O. Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schønning, K., B. Jansson, S. Olofsson, and J. E. S. Hansen. 1996. Rapid selection for an N-linked oligosaccharide by monoclonal antibodies directed against the V3 loop of human immunodeficiency virus type 1. J. Gen. Virol. 77:753-758. [DOI] [PubMed] [Google Scholar]
- 96.Schønning, K., O. Lund, O. S. Lund, and J. E. S. Hanssen. 1999. Stochiometry of monoclonal antibody neutralization of T-cell-line-adapted human immunodeficiency virus type 1. J. Virol. 73:8364-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schreiber, M., H. Petersen, C. Wachsmuth, H. Müller, F. T. Hufert, and H. Schmitz. 1994. Antibodies of symptomatic human immunodeficiency virus type 1-infected individuals are directed to the V3 domain of noninfectious and not of infectious virions present in autologous serum. J. Virol. 68:3908-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scott, C. F., Jr., S. Silver, A. T. Profy, S. D. Putney, A. Langlois, K. Weinhold, and J. E. Robinson. 1990. Human monoclonal antibody that recognizes the V3 region of human immunodeficiency virus gp120 and neutralizes the human T-lymphotropic virus type IIIMN strain. Proc. Natl. Acad. Sci. USA 87:8597-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shakin-Eshleman, S. H., S. L. Spitalnik, and L. Kasturi. 1996. The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. J. Biol. Chem. 271:6363-6366. [DOI] [PubMed] [Google Scholar]
- 100.Slagle, S. P., R. E. Kozack, and S. Subramaniam. 1994. Role of electrostatics in antibody-antigen association: anti-hen egg lysozyme/lysozyme complex (HyHEL-5/HEL). J. Biomol. Struct. Dyn. 12:439-456. [DOI] [PubMed] [Google Scholar]
- 101.Spear, G. T., D. M. Takefman, S. Sharpe, M. Ghassemi, and S. Zolla-Pazner. 1994. Antibodies to the HIV-1 V3 loop in serum from infected persons contribute a major proportion of immune effector functions including complement activation, antibody binding and neutralization. Virology 204:609-615. [DOI] [PubMed] [Google Scholar]
- 102.Stamatatos, L., and C. Cheng-Mayer. 1998. An envelope modification that renders a primary neutralization resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stanfield, R. L., T. M. Fiester, R. A. Lerner, and I. A. Wilson. 1990. Crystal structures of an antibody to a peptide and its complex with peptide antigen at 2.8 Å. Science 248:712-719. [DOI] [PubMed] [Google Scholar]
- 104.Sullivan, N., M. Thali, C. Furman, D. D. Ho, and J. Sodroski. 1993. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J. Virol. 67:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thali, M., C. Furman, D. D. Ho, J. Robinson, S. Tilley, A. Pinter, and J. Sodroski. 1992. Discontinuous, conserved neutralization epitopes overlapping the CD4 binding region of the HIV-1 gp120 envelope glycoprotein. J. Virol. 66:5635-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thali, M., U. Olshevshy, C. Furman, D. Gabuzda, M. Posner, and J. Sodroski. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120-epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 65:6188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsumoto, K., K. Ogasahara, Y. Ueda, K. Watanabe, K. Yutani, and I. Kumagai. 1996. Role of salt bridge formation in antigen-antibody interaction. Entropic contribution to the complex between hen egg white lysozyme and its monoclonal antibody HyHEL10. J. Biol. Chem. 271:32612-32616. [DOI] [PubMed] [Google Scholar]
- 110.Tsumoto, K., Y. Ueda, K. Maenaka, K. Watanabe, K. Ogasahara, K. Yutani, and I. Kumagai. 1994. Contribution to antibody-antigen interaction of structurally perturbed antigenic residues upon antibody binding. J. Biol. Chem. 269:28777-28782. [PubMed] [Google Scholar]
- 111.Vinogradov, E. V., R. Pantophlet, L. Dijkshoorn, L. Brade, O. Holst, and H. Brade. 1996. Structural and serological characterisation of two O-specific polysaccharides of Acinetobacter. Eur. J. Biochem. 239:602-610. [DOI] [PubMed] [Google Scholar]
- 112.Vogel, T., R. Kurth, and S. Norley. 1994. The majority of neutralizing abs in HIV-1-infected patients recognize linear V3 loop sequences: studies using HIV-1MN multiple antigenic peptides. J. Immunol. 153:1895-1904. [PubMed] [Google Scholar]
- 113.Wilson, I. A., and L. K. Jolliffe. 1999. The structure, organization, activation, and plasticity of the erythropoietin receptor. Curr. Opin. Struct. Biol. 9:696-704. [DOI] [PubMed] [Google Scholar]
- 114.Wormald, M. R., and R. A. Dwek. 1999. Glycoproteins: glycan presentation and protein-fold stability. Struct. Fold Des. 7:R155-R160. [DOI] [PubMed] [Google Scholar]
- 115.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 116.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 118.Yang, X., M. Farzan, R. Wyatt, and J. Sodroski. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 74:5716-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang, X., L. Florin, M. Farzan, P. Kolchinsky, P. D. Kwong, J. Sodroski, and R. Wyatt. 2000. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J. Virol. 74:4746-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang, X., J. Lee, E. M. Mahony, P. D. Kwong, R. Wyatt, and J. Sodroski. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76:4634-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yoshiyama, H., H. Mo, J. P. Moore, and D. D. Ho. 1994. Characterization of mutants of human immunodeficiency virus type 1 that have escaped neutralization by a monoclonal antibody to the gp120 V2 loop. J. Virol. 68:974-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhu, X., C. Borchers, R. J. Bienstock, and K. B. Tomer. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 39:11194-11204. [DOI] [PubMed] [Google Scholar]