Abstract
Herpesvirus saimiri (HVS), a T-lymphotropic tumor virus of neotropical primates, and the Kaposi's sarcoma-associated human herpesvirus 8 (KSHV) belong to the gamma-2-herpesvirus (Rhadinovirus) subfamily and share numerous features of genome structure and organization. The KSHV latency-associated nuclear antigen (LANA) protein appears to be relevant for viral persistence, latency, and transformation. It binds to DNA, colocalizes with viral episomal DNA, and presumably mediates efficient persistence of viral genomes. LANA further represses the transcriptional and proapoptotic activities of the p53 tumor suppressor protein. Here we report on the ORF73 gene of HVS strain C488, which is the positional and structural homolog of KSHV LANA. The ORF73 gene in OMK cells can encode a 62-kDa protein that localizes to the nucleus in a pattern similar to that of LANA. We show that the ORF73 gene product can regulate viral gene expression by acting as a transcriptional modulator of latent and lytic viral promoters. To define the HVS ORF73 function in the background of a replication-competent virus, we constructed a viral mutant that expresses ORF73 under the transcriptional control of a mifepristone (RU-486)-inducible promoter. The HVS ORF73 gene product efficiently suppresses lytic viral replication in permissive cells, indicating that it defines a critical control point between viral persistence and lytic replication.
Herpesvirus saimiri (HVS) is the prototype of the herpesvirus subfamily Rhadinoviridae (gamma-2-herpesviruses). Thus, it is closely related to Kaposi's sarcoma-associated human herpesvirus 8 (KSHV), Rhesus rhadinovirus, and Murine gammaherpesvirus type 68 (MHV-68), but it also has clear homologs to most genes of the Lymphocryptovirus Epstein-Barr virus (EBV). HVS asymptomatically persists in its natural host, the squirrel monkey (Saimiri sciureus), without overt disease, while infection of other New World primates such as Saguinus spp. and Callithrix spp. usually results in fulminant T-cell lymphomas. Based on sequence variations at the left end of the nonrepetitive L-DNA (5, 38, 39), HVS strains can be classified into three subgroups, A to C. Subgroup C strains are capable of transforming human lymphocytes to continuous growth in vitro (4, 15). The two oncoproteins StpC and Tip of strain C488 were shown previously to be essential for transformation of human and simian T lymphocytes and pathogenicity in common marmosets (11). The HVS genome persists as a stable nonintegrated circular episome in transformed human and simian T cells. A cluster of genes, ORF71, ORF72, and ORF73, is localized at the right end of the L-DNA; they are expressed as a polycistronic mRNA (14). Previous analysis has shown that both KSHV and HVS ORF71 encode the antiapoptotic FLICE inhibitory protein (vFLIP) (55), but the HVS ORF71 is not essential for viral replication, transformation, and pathogenicity (19). ORF72 encodes a v-Cyclin D homolog which is also dispensable for transformation of human T lymphocytes (12). The function of HVS ORF73 has remained unknown up to now.
The ORF73 of KSHV encodes the latency-associated nuclear antigen (LANA). LANA is a large (222- to 234-kDa) nuclear protein with three distinct domains: a proline-rich N-terminal domain, a long glutamic-acid-rich internal repeat domain, and a carboxy-terminal domain that includes a putative nuclear localization signal (NLS) (30, 46). It has recently been shown that the KSHV ORF73 gene product interacts with cellular p53 (17), pRb (18), a homolog of Drosophila melanogaster female sterile homeotic (fsh) gene Ring3 (37, 44), the mSin3 corepressor complex (32), ATF4/CREB2 (34), and CBP (33). LANA is able to modulate the transcriptional activity of the human immunodeficiency virus type 1 long terminal repeat (28, 47). The effects on latent promoters of EBV are more controversial: in BJAB and 293 cells transactivation of LMP-1 and Cp (22) was observed, while another group described downmodulation of EBNA-1 Cp and Qp promoters in cells derived from primary effusion lymphoma (PEL) (32). The transcriptional activity of the LANA on its own promoter is also controversial. It was reported elsewhere that LANA binds a 20-bp palindrome within the terminal repeats of the KSHV genome and, when this sequence is cloned upstream of a Gal-4 recognition motif, the transcriptional activity is repressed by binding of LANA (18). While no binding of LANA on its promoter was seen (18), there is evidence that LANA is able to modulate its own promoter (29, 47). A major function of LANA has been suggested to be maintaining episomal persistence of the KSHV genome in latently infected cells (2, 3, 43). Hereby it tethers the viral episome to cellular mitotic chromosomes via an interaction with histone H1 (8). The ORF73 proteins of KSHV and HVS share an amino acid homology of 33% and have similar structural features including (i) an internal glutamic-acid-rich repetitive region, (ii) a DNA-binding domain (DBD) in the C terminus with structural homology to EBV EBNA-1, and (iii) NLSs. While the NLS of LANA is localized within the DBD at the C terminus (43), the two NLSs of HVS ORF73 are localized at the N terminus (24), and ORF73 of HVS, like that of MHV-68, lacks the proline-rich region at the amino terminus.
Herpesviruses are characterized by an initial lytic replication, usually followed by establishment of a lifelong persistent infection in which the viral genome is maintained episomally in specific tissue compartments and occasionally reactivates from a dormant state to lytic replication. The lytic cycle of EBV is controlled by transactivating proteins encoded by the ORFs BZLF-1 (Zta), BRLF-1 (Rta), and BMLF-1 (M) that trigger the switch between latency and lytic replication (16). The transactivator Rta seems to be a downstream target of Zta transactivation (52). However, Rta overexpression alone can reactivate EBV in the epithelial cell model of latency (63). The KSHV homologs to the lytic EBV transactivators are K8 (Zta), ORF50 (Rta), and ORF57 (M) (48). In KSHV infection, the Rta homolog (ORF50) can induce markers of lytic replication in B-cell models for KSHV latency (36, 53), and it was shown previously that transient expression of Rta is able to disrupt the latent state and induce the lytic replication cycle in B-cell lines latently infected with KSHV (21, 36, 53) and MHV-68 (62), respectively. It was demonstrated previously that the ORF50 gene product is a nuclear protein that can directly transactivate KSHV delayed-early, but not late, promoters and that the ORF50 carboxy terminus contains the major transactivating domain (36). Although multiple interactions of the K8 protein have been described previously (27, 42, 45), no clear-cut function could be assigned, and yet the transcription of both K8/Zta and ORF57/M seems to be controlled by the ORF50 gene product (35), suggesting that the ORF50 gene product may be a key switch protein in KSHV lytic reactivation.
In HVS strain A11, gene expression during the lytic replication cycle is controlled by the products of the two major transcriptional regulatory genes encoded by ORF50/Rta and ORF57/M; no homolog of the Z protein has been identified in the HVS genome (1). It has been shown previously that the ORF50 immediate-early gene product (41) can directly activate delayed-early gene transcription (40). The ORF50 locus encodes an ORF50A protein, derived from a longer spliced mRNA, and ORF50B, derived from a shorter, unspliced coterminal transcript (59). Both proteins are able to transactivate the promoters of viral delayed-early genes like the major single-stranded DNA (ssDNA)-binding protein (ORF6); this promoter contains the specific ORF50-responsive element CCN9GG (60). Expression of ORF50A causes strong transactivation of the ORF6 promoter, while the shorter ORF50B is less proficient in transactivating promoters of other HVS genes. A carboxy-terminal activation domain has been identified in ORF50 transactivation which is required for the interaction with a basal cellular transcription factor, TATA-binding protein (25). However, considerable divergence has been observed between the ORF50 genes of HVS strains A11 and C488, which may lead to altered transactivation properties (56). While it seems that ORF50A is the relevant transactivator for HVS strain A11, a dominant regulatory function is provided by ORF50B in strain C488 (56). In the human lung carcinoma cell line A549, which is semipermissive for HVS lytic replication, transfection of the HVS ORF50/Rta initiates a gene cascade that switches on expression of early viral replication genes and increases viral lytic replication (20). There is no recognizable homolog to the EBV Zta or KSHV K8 in HVS, and it is unknown which viral or cellular gene product regulates the transcription of ORF50.
In this study we performed a functional analysis of the ORF73 gene of HVS strain C488. Our data indicate that ORF73 downregulates the viral ORF50A and ORF50B promoters in permissive OMK cells and that it can inhibit the ORF50-mediated expression of viral early replication genes. Studies with a recombinant HVS C488 virus that contains a mifepristone-inducible ORF73 showed that lytic gene expression and viral lytic replication of HVS could be completely blocked by overexpression of ORF73 protein in OMK cells. Taken together, these data suggest that ORF73 can block the transition from the latent to the lytic phase of the viral life cycle; by inhibiting the expression of the ORF50 R-transactivator homolog, ORF73 appears to prevent the activation of lytic genes by the herpesvirus regulatory gene expression cascade.
MATERIALS AND METHODS
Cell culture and virus propagation.
OMK cells (ATCC CRL1556) were used for transfection assays and for propagation of HVS (9). The cells were cultivated in Dulbecco's modified Eagle's medium supplemented with glutamine (350 μg/ml), gentamicin (100 μg/ml), and 10% heat-inactivated fetal calf serum (FCS; Invitrogen/LifeTechnologies, Groningen, The Netherlands). Virus stocks were generated by infection of confluent OMK cells in 175-cm2 tissue culture flasks at a low multiplicity (∼0.1) of infection. When lysis was complete, supernatant was cleared from cell debris by centrifugation at 2,000 × g for 10 min, and cell-free supernatants were stored at −80°C.
Plasmids.
The effector plasmid pcDNA-HA73MH, which was used for expression studies, contains viral sequences from nucleotide (nt) 105809 to nt 107176. It contains the HVS ORF73 that was cloned by simultaneous insertion of two fragments into NotI-EcoRI-digested pcDNA3.1(−)MycHisB (Invitrogen). A 1.5-kb HindIII-AspHI fragment including ORF73 was subcloned from right-terminal cosmid Dc5 into the plasmid pJ3H (50), adding the 10-amino-acid hemagglutinin (HA) epitope tag (YPYDVPDYAS) to the amino terminus of ORF73. A HindIII-SmaI fragment from pJ3H-73 was then subcloned into pGeneV5-HisA (Invitrogen) to generate the mifepristone-inducible HA-tagged ORF73 expression construct pGene-HA73. The HA-tagged ORF73 5′ fragment was excised as an EagI-AgeI fragment from plasmid pGene-HA73. The 3′ fragment of ORF73 was PCR amplified from cosmid Dc5 with primers ORF73noStop (5′-TCCGGAATTCTATGGGCAAGCTTTTGC-3′), which contained an EcoRI site, and ark61 (5′-CTAAAAATGCAGCATCGTCACC-3′), which lies directly adjacent to the AgeI site within ORF73. PCR was done in a 50-μl reaction mixture that contained 10 ng of template cosmid Dc5, 0.2 μM (each) deoxynucleoside triphosphates, 10 μM (each) primer, and 2.5 U of AmpliTaq polymerase (Perkin-Elmer) in 1× PCR buffer. After an initial 1-min denaturation step at 96°C, 25 cycles of 15 s at 96°C, 20 s at 55°C, and 40 s at 70°C were performed in an MJ Research PTC-200 thermal cycler (Biozym, Oldendorf, Germany), followed by a 1-min final extension step at 70°C. The resulting ORF73-3′ was digested with AgeI and EcoRI to generate compatible ends. The complete insert of the plasmid pcDNA-HA73MH was sequenced. The control plasmid pcDNA-HA-MH was generated by deleting the ORF73 coding sequence from pcDNA-HA73MH by digestion with BamHI and religation. The expression plasmids for ORF73 deletion mutants that lack amino acids 4 to 56 (pcDNA-HA73MHd3), amino acids 53 to 334 (pcDNA-HA73MHd8), and amino acids 342 to 501 (pcDNA-HA73MHd15) were generated from the original plasmid pcDNA-HA73MH by PCR-directed mutagenesis with the Expand High Fidelity PCR kit (Roche Molecular Biochemicals, Mannheim, Germany). The following oligonucleotides were designed to delete the respective coding sequences: for pcDNA-HA73MHd3, 73dHA (5′-TCCTCTAGAAGCGTAATCTGGAAC-3′) and 73d3 (5′-TCCCCAACAGAATACGAACAACGTG-3′); for pcDNA-HA73MHd8, ark164 (5′-GAGCGTTGCGTCAATGTCATC-3′) and 73d4 (5′-TCCGGACCAAGTGCCCAACGTTTACC-3′); and for pcDNA-HA73MHd15, 73d5 (5′-ACGTTGGGCACTTGGTCCTGC-3′) and 73d8-MH (5′-AGCTTTCTAGAACAAAAACTC-3′). The PCR fragments spanning the complete plasmids were blunted with T4 DNA polymerase, residual template DNA was removed by DpnI digestion, and the PCR products were phosphorylated with T4 polynucleotide kinase and religated with T4 DNA ligase. All enzymes were purchased from New England Biolabs (Frankfurt-am-Main, Germany), except T4 DNA ligase (Takara Biowhittaker, Apen, Germany). The correct in-frame religation of the constructs and absence of PCR-derived errors were verified by sequencing of the complete ORF73 inserts and most of the adjacent cytomegalovirus immediate-early (CMVIE) promoter and the bovine growth hormone-derived poly(A) regions of the vector backbone.
Reporter gene assays.
The firefly luciferase expression plasmids containing the different promoter sequences of the HVS genome were constructed by PCR amplification of the respective viral regions and insertion of the PCR fragments into the pGL3-Basic luciferase reporter plasmid (Invitrogen). pGL3-StpC corresponds to nt 2073 to nt 3000, pGL3-ORF6 corresponds to nt 11260 to nt 12285, pGL3-ORF15 corresponds to nt 28623 to nt 29068, pGL3-ORF44 corresponds to nt 62053 to nt 62703, pGL3-ORF50A corresponds to nt 68230 to nt 69348, pGL3-ORF50B corresponds to nt 69917 to nt 70820, pGL3-ORF70 corresponds to nt 102240 to nt 103182, and pGL3-ORF73 and pGL3-ORF74 correspond to nt 107119 to nt 107675 of HVS C488. The following oligonucleotides were designed to amplify the respective 5′ upstream regions of the HVS genes: for pGL3-StpC, 304582 (5′-GGTTCGGTTAGCTTGCCAATTTTTTC-3′) and 280600 (5′-TACGTAGTAAACACGCAAATGCACAAG-3′); for pGL3-ORF15, 14XhoI (5′-AGCTCGAGTTGCAAATGAAATGAGAATCTGG-3′) and 15XhoI (5′-AGCTCGAGCAATGAGACAAGAATCAAG-3′); for pGL3-ORF44, HF263 (5′-AAGCTTTCTACTGCTATATGCTTG-3′) and HF264 (5′-TCTAGATAGTTCTGCCATTATTCG-3′); for pGL3-ORF50A, AR48r (5′-GACACGTTCAAAACTGGTTGG-3′) and 50Aluc3 (5′-ACAAGCTTGTGTGTCATTGTTG-3′); for pGL3-ORF50B, 50Bluc5 (5′-TTTTAGCACAATAAGCTTAGG-3′) and 50Bluc3 (5′-TTAAGCTTCTAGTCCCATTAACACGC-3′); for pGL3-ORF70, 204408 (5′-CGTGCCGTTCTTCAGTGTG-3′) and 204406 (5′-CAGCTAAAAATTACTTGCTTG-3′); and for pGL3-ORF73 and pGL3-ORF74, ark181 (5′-TGCACTCAGAACGCAGAGTGTGCTTCCTTC-3′) and ark221 (5′-CTACTGAAGTCCAGCTTGACCTCC-3′). For construction of pGL3-ORF6, an SstI fragment of construct Orf6prok1ta containing the ORF6 promoter sequence was cloned into the SstI-linearized pGL3-Basic luciferase reporter plasmid (56). The correct insertion and sequence of the reporter gene constructs were verified by DNA sequencing of the complete PCR-derived fragments in the reporter constructs. The promoterless pGL3-Basic vector and plasmid pGL3-PGK containing the cellular phosphoglycerate kinase promoter were used as negative and positive transfection controls, respectively. The plasmids containing the genomic fragments of ORF50A (pCR2.1-50Agen) and ORF50B (pCR2.1-50Bgen) and the cDNA expression construct of CMVIE promoter-driven spliced ORF50A (pcDNA-50) were provided by Mathias Thurau and Helmut Fickenscher (20, 56). For luciferase reporter assays 2.5 × 105 OMK cells were plated onto 24-well dishes the day before transfection. Liposome-mediated transfection was performed by combining 1 μl of Lipofectamine 2000 (Invitrogen) per 1 μg of plasmid DNA in Optimem-I (Invitrogen). Unless indicated otherwise, 0.5 μg of each luciferase reporter plasmid, 1 μg of the effector plasmid pcDNA-HA73MH or the control plasmid pcDNA-HA-MH, and 1 μg of the indicated ORF50 constructs were transfected. The total amount of transfected DNA was kept constant by adding appropriate amounts of the control plasmid pcDNA-HA-MH in order to replace the missing effector plasmid. At 24 h after transfection, cells were harvested in lysis buffer (50 mM Tris-H3PO4 [pH 7.8], 50 mM trans-1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 2% Triton X-100, 4 mM dithiothreitol, 20% glycerol), and standard luciferase assays were performed. Luciferase activity in the supernatant was determined with a single-tube luminometer (Bertholt, Freiburg, Germany). Each transfection was performed in triplicate and was repeated at least three times.
RNA isolation and cDNA synthesis.
OMK cells were cultured and infected as described above. At day 7 postinfection (p.i.), when the first cytopathic effects (CPEs) were detectable in the control cultures (infected with C488_switch virus) and in the ORF73 uninduced culture, cells were harvested and used for mRNA isolation. RNA was isolated (Nucleospin RNA II kit; Macherey-Nagel, Düren, Germany), and 1.5 μg of RNA was digested in a volume of 12 μl with 10 U of RNase-free DNase I (Roche Diagnostics) in the presence of 1 U of RNaseOUT RNase inhibitor (Invitrogen) and 1 mM dithiothreitol at 37°C for 30 min, followed by a heat inactivation step of 10 min at 70°C. The samples were then divided into two parallel reaction mixtures and processed with the ThermoScript reverse transcription PCR (RT-PCR) system (Invitrogen) in 20-μl reaction mixtures with or without reverse transcriptase according to the supplier's protocol. The reaction mixtures were stored at −20°C. A total of 2 μl of the reaction mixtures was used for RT-PCR analysis. PCR conditions were as follows: a 5-min initial denaturation step at 94°C; 30 cycles of 15 s at 94°C, 30 s at 64°C, and 30 s at 72°C; a final extension of 5 min at 72°C; and a 12°C hold. Primers were specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (GAPDH1c, 5′-GCAGGGGGGAGCCAAAAGGG-3′, and GAPDH2n, 5′-GCCTCAAGATCATCAGCAATGCC-3′), for HVS C488 ORF50A (50aSGc, 5′-GCCTCAAGATCATCAGCAATGCC-3′, and 50aSGn, 5′-CAATGACACACAAGCCTGTTAAGGAG-3′), and for HVS C488 ORF50A and ORF50B (50bSGc, 5′-TTGAGAGGAAATCCTCCAATTCGTG-3′, and 50bSGn, 5′-CATGGACTGTTTGGTGACGTGTTTC-3′). Reaction products (5 μl) were resolved on 1.5% agarose gels in 1× Tris-borate-EDTA buffer.
Real-time quantitative PCR.
The PCR mixtures (50 μl) contained 0.3 μM ROX (6-carboxy-X-rhodamine), 0.4× PCR buffer, 3.5 mM MgCl2, 0.5 mM deoxynucleoside triphosphates, 0.3× SYBR Green I (from 10,000× stock solution; Molecular Probes, Eugene, Oreg.), 200 μM (each) primer (see above), 2.5 U of Taq polymerase (Perkin-Elmer), and 10 μl of 1:10-diluted cDNA sample as template. Quantitative real-time PCR was performed on an ABI Prism 7700 sequence detection system (Applied Biosystems) with identical cycle conditions (5 min at 94°C and 50 cycles of 15 s at 94°C, 30 s at 64°C, and 30 s at 72°C) for all fragments. Amplification threshold cycle values were analyzed with the sequence detection system software (version 1.6.3), and copy numbers were calculated by comparison to 10-fold dilution series (10−9 to 10−2 copies) of standards processed in parallel. Each sample was analyzed in parallel, and the PCR was repeated once with identical results.
Western blot analysis and immunofluorescence.
OMK cells were infected and transfected as described above. For Western blot analysis, OMK cells were lysed in 1× RIPA buffer (10 mM Tris-Cl [pH 8], 150 mM NaCl, 1% NP-40, sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], and 1% aprotinin-leupeptin) 24 h after transfection. The total protein concentration was determined by the bicinchoninic acid assay (Pierce, Bonn, Germany). Cell lysates (20 μg per lane) were separated by electrophoresis on SDS-10% polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Immobilon; Millipore, Schwalbach, Germany). The immobilized proteins were then probed with the following primary antibodies: a monoclonal anti-HA antibody (HA-11; Babco, Mississauga, Ontario, Canada) at a dilution of 1:1,000, a monoclonal anti-β-actin antibody (clone AC-15; Abcam, Cambridge, United Kingdom) at a dilution of 1:5,000, or specific rabbit polyclonal antiserum directed against ORF73 (1:500) or against the viral protease-minor capsid scaffold protein ORF17 (1:500). Primary antibodies were detected with murine or rabbit immunoglobulin-specific, horseradish peroxidase-coupled secondary antibodies (Dako, Hamburg, Germany) diluted 1:5,000 and an enhanced chemoluminescence substrate (ECL; Amersham, Freiburg, Germany) by a Fuji LAS-1000 chemoluminescence detection system (Raytest, Straubenhart, Germany).
For immunofluorescence analysis transfected or infected OMK cells were fixed in 4% paraformaldehyde. After two washing steps in phosphate-buffered saline (PBS), cells were permeabilized for 5 min with 0.1% Triton X-100 in PBS. Unspecific binding of cells was blocked with 1% FCS for 1 h at room temperature. The HA epitope tag was visualized with the monoclonal HA antibody (HA-11; Babco) at a dilution of 1:1,000 in blocking diluent (1% FCS in PBS) and incubated on cells for 1 h at room temperature. Three 5-min washes with PBS were performed before anti-mouse Cy3-conjugated antibody (Sigma, Taufkirchen, Germany) was used at a dilution of 1:100 in the same buffer. Cells were incubated with secondary antibody for 1 h at room temperature. After three to five washes in PBS at room temperature, cells were mounted in Moviol mounting medium containing 1 μg of the intercalating nuclear dye Hoechst 33358 (Sigma)/ml and visualized by fluorescence microscopy on a Zeiss Axiophot-2 microscope (Göttingen, Germany) equipped with a SPOT cooled charge-coupled device camera (INTAS, Göttingen, Germany).
Recombinant viruses and infection studies.
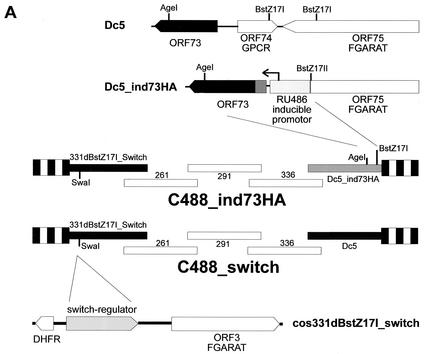
The recombinant viruses were constructed by cotransfection of overlapping cosmids covering the complete HVS C488 genome into permissive OMK cells (13). All cloning procedures were performed according to standard protocols. The SspI-AgeI fragment from pGene-HA73 containing the mifepristone-inducible promoter and the HA-tagged ORF73 gene was reinserted into AgeI-BstZ17I-digested cosmid Dc5, resulting in cosmid Dc5_indHA73. An MbiI fragment of pSwitch (Invitrogen) containing the expression cassette for the mifepristone-dependent transcriptional activator protein (GeneSwitch) was cloned into the SwaI site of cosmid 331dBstZ17I in a noncoding region between the viral ORF02 (encoding dihydrofolate reductase) and ORF03 (viral homolog of FGARAT, EC 6.3.5.3).
The correct insertions into the cosmids were verified by restriction enzyme mapping and sequencing. Recombinant viruses C488_ind73HA and C488_switch were generated by liposome-mediated cotransfection of a set of five overlapping cosmids, including cosmids 331dBstZ17I_switch and Dc5_indHA73, into permissive OMK cells. The cosmids were linearized by restriction with NotI before transfection, which also removed the pWE15 cloning vector, which contains two NotI sites flanking the BamHI site that was used to clone the viral DNA. For 80 to 90% confluent OMK cells in 25-cm2 flasks, 0.6 μg of each cosmid (total, 3 μg) was combined with 15 μl of Lipofectamine (Invitrogen). The correct genotype of the recombinant viruses was verified by PCR analysis; 1 ml of virus-containing supernatant from completely lysed cultures was harvested by centrifugation (90 min at 35,000 × g at 4°C). The virions in the pellet were lysed in 100 μl of PCR buffer containing 100 μg of proteinase K (Roche Diagnostics)/ml and 0.5% Tween 20 for 1 h at 56°C. The proteinase K was then heat inactivated for 15 min at 95°C. An aliquot of 2 μl was used for PCR analysis.
OMK cells were seeded in six-well plates 24 h before infection, which was performed in parallel with a C488 wild-type control, C488_switch control virus containing only the regulator protein, and C488_ind73HA. Infected cultures were induced with 500 nM RU-486 (mifepristone [Sigma], 1 mM stock solution in ethanol) to induce expression of HA-tagged ORF73; infected control cultures were kept in parallel without induction. The cultures were reinduced every 5 days by addition of RU-486 to a final concentration of 500 nM. Induction was either kept on constantly or switched during the experiment by withdrawal-new addition of RU-486. To withdraw RU-486, cells were washed twice with medium without supplements and fed fresh medium without RU-486. During infection, the infected cell cultures were observed for development of CPE and documented with an Olympus IMT-2 inverted microscope (Hamburg, Germany) equipped with an OM-2 camera body. For immunoblot analysis, infected OMK cultures that were kept under identical conditions in parallel were harvested after 14 days. Cell lysates were prepared in RIPA buffer, and 20 μg of cellular protein per lane was separated on SDS-10% polyacrylamide gels and detected as described above. For immunofluorescence detection of induced HA-ORF73, cells were infected and induced for 48 h with 5 nM RU-486, fixed with 4% phosphonoformic acid, and visualized with the anti-HA antibody and anti-murine Cy3 as described above.
RESULTS
The LANA homolog HVS ORF73 encodes a nuclear protein that can modulate the transcriptional activity of viral promoters.
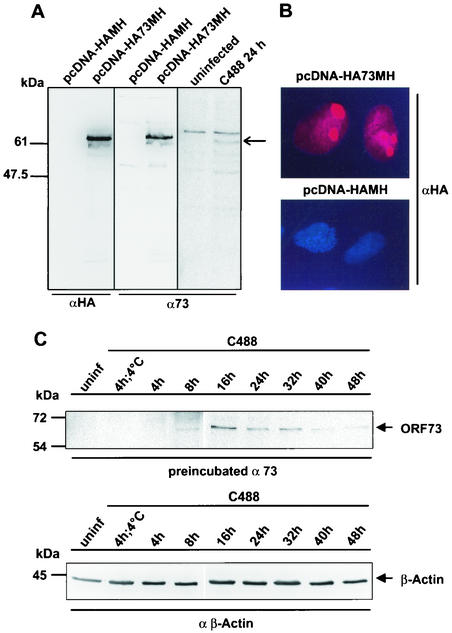
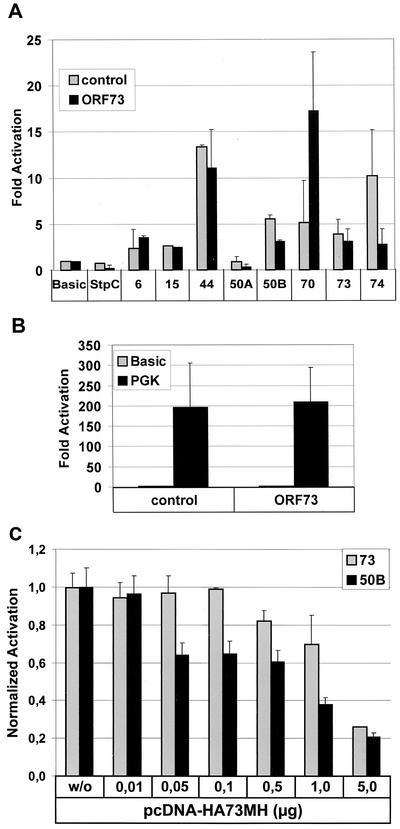
The HVS ORF73 positional and structural homolog KSHV LANA has been associated with multiple functions, including modulation of cellular or viral transcription. To test the hypothesis that the ORF73-encoded LANA homolog of HVS can modulate viral promoter activity, we constructed an epitope-tagged ORF73 expression vector where the coding sequence of the ORF73 was linked at the amino terminus to an epitope derived from the influenza virus HA (HA tag), while the carboxy-terminal end was bearing an epitope derived from the cellular myc oncogene linked to six consecutive histidine residues (MycHis tag). The construct HA73MH was expressed under the transcriptional control of the commonly used human CMVIE (6). This expression construct was verified in transiently transfected OMK cells, where a functional fusion protein of 64 kDa was detectable by a monoclonal anti-HA antibody as well as by a rabbit polyclonal ORF73-specific antiserum that was derived by immunization with the C-terminal domain of the protein expressed in Escherichia coli (Fig. 1A). The subcellular localization of ORF73 was determined by immunofluorescence studies performed on OMK cells that were seeded in four-chamber slides and transfected with pcDNA-HA73MH or empty control vector pcDNA-HA-MH. In the transfected cells, ORF73 was localized in the nucleus with a predominantly nucleolar accumulation (Fig. 1B) or a more speckled pattern at lower expression levels, while no staining was observed in the control plasmid-transfected culture. Detection of ORF73 with a specific rabbit polyclonal antiserum was compromised due to nonspecific reactivity with cellular antigens that could not be removed by absorption (data not shown). In addition, the localization of the HVS C488 ORF73 is very similar to that of the related ORF73 protein of the HVS strain A11, which has an amino acid sequence that is 75% identical to the C488 protein (24). In HVS C488-infected OMK cells, the polyclonal antiserum detected a slightly faster migrating protein of approximately 62 kDa. This size difference is attributable to the missing epitope tags in the viral protein. After infection of OMK cells, the ORF73 protein was first detectable at 8 h p.i., reached the highest level at 16 h and declined thereafter. At 40 to 48 h, when lytic CPE was apparent in the culture, only weak ORF73 expression was seen (Fig. 1C, upper panel), while the cellular β-actin expression was largely unchanged (Fig. 1C, bottom panel). The ORF73 expression construct was then transfected into the permissive OMK cell line, which is the cell line commonly used to propagate the virus and study lytic replication, together with a series of firefly luciferase reporter constructs. These reporter constructs contained the promoter regions of viral genes representing the regulatory-immediate-early EBV R-transactivator homolog ORF50A and ORF50B, delayed-early-DNA replication-associated genes (ORF6 and ORF44), the latently transcribed StpC oncoprotein, and other proteins (ORF15/vCD59, ORF70/vTS, ORF73, and ORF74/vGPCR). The cellular phosphoglycerate kinase promoter was included as a control. To test if ORF73 operates as a transcriptional modulator of viral promoters, we constructed several luciferase reporter vectors. For the latent phase, reporter vectors were cloned to contain the promoter sequences of the oncogene StpC, ORF73 (LANA homolog), and ORF74 (GPCR), respectively. The promoters of the two major HVS regulatory proteins ORF50A and ORF50B were used as immediate-early reporter plasmids. The promoter sequences of ORF6 (major ssDNA-binding protein), ORF15 (CD59 homolog), ORF44 (helicase), and ORF70 (thymidylate synthase) served as reporter constructs of delayed-early and late genes. For luciferase assays, OMK cells were transiently transfected by lipofection with 0.5 μg of reporter plasmid and 1 μg of either pcDNA-HA73MH or pcDNA-HA-MH. After incubation for 24 h, cell extracts were prepared and analyzed for amount of luciferase activity as previously described. Figure 2A shows normalized luciferase activities of the reporter constructs in OMK cells cotransfected with pcDNA-HA73MH compared to pcDNA-HA-MH control plasmid. There was virtually no influence of ORF73 on the promoter of latently expressed oncogene StpC or on the promoters of the delayed-early genes ORF6 and ORF44 or ORF15. There is a considerably increased transcriptional activity of the ORF70 promoter in the presence of ORF73. In contrast, the activity of ORF73 is slightly downregulated by its own gene product (by 25%), possibly hinting at a negative feedback regulation; the activity of the same promoter element in the reverse orientation, which drives transcription of ORF74 (vGPCR), is decreased more significantly to 25% of basal activity. Both promoter sequences of the immediate-early genes ORF50A and ORF50B were repressed at least twofold in the presence of ORF73. The phosphoglycerate kinase control construct is not significantly influenced by ORF73 (Fig. 2B). Taken together, these results suggest that the ORF73 gene product has distinct modulating effects on the transcription of several HVS promoters. ORF50 has been shown elsewhere to induce lytic virus replication in HVS A11-infected A549 cells (20), most likely by transactivation of viral promoters (56, 60).
FIG. 1.
HVS ORF73 expression in infected and transfected OMK cells. (A) OMK cells transfected with vector alone or with pcDNA-HA73MH or infected with HVS C488 were electrophoresed on an SDS-10% polyacrylamide gel and analyzed by Western blotting. An ORF73-specific signal is detected with the anti-HA-tag monoclonal antibody and the polyclonal antiserum; the double-epitope-tagged protein migrates at approximately 64 kDa, while the virus-encoded protein migrates at approximately 62 kDa (arrow). (B) OMK cells were transfected in chamber slides with pcDNA-HA73MH or with vector alone and analyzed by HA-tag-specific immunofluorescence microscopy 24 h later. (C) Time course of ORF73 expression in permissive OMK cells. Cells were infected with HVS C488 at 4°C for 4 h and at 37°C for 4, 8, 16, 24, 32, 40, and 48 h. ORF73 was detected with a specific polyclonal antiserum that was preabsorbed with uninfected OMK cell lysate (upper panel). As a control, cellular β-actin was detected with a specific monoclonal antibody (clone AC-15) (bottom panel). Uninfected OMK cells are shown as controls.
FIG. 2.
Transcriptional activity of HVS ORF73 on HVS gene promoters in OMK cells. (A) Luciferase reporter constructs (0.5 μg) were cotransfected with pcDNA-HA73MH (black columns) or the control vector pcDNA-HA-MH (gray columns, 1 μg each) into OMK cells in 24-well plates. Basic, pGL3-Basic; StpC, 6, 15, 44, 50A, 50B, 70, 73, 74, pGL reporter constructs containing the respective HVS gene promoters. (B) Cotransfections of the pGL3-PGK (phosphoglycerate kinase promoter) together with pcDNA-HA-MH or pcDNA-HA73MH were done as a positive control, respectively. After 24 h equal amounts of lysates were assayed for luciferase activity. Luciferase activity is expressed as fold activation relative to that obtained with pGL3-Basic alone. The summary of three independent experiments, each with cultures done in triplicate, is shown. (C) Differential susceptibilities of ORF50B and ORF73 promoters to downregulation by ORF73/LANA. Luciferase reporter constructs of the ORF50B (black columns) promoter and ORF73 promoter (gray columns) (each 0.5 μg) were cotransfected in OMK cells either with control vector (pcDNA-HA-MH;1 μg; “w/o”) or with increasing concentrations of pcDNA-HA73MH (from 0.01 to 5 μg of DNA). After 24 h equal amounts of cell lysates were assayed for luciferase activity. Luciferase activity is here expressed as fold activation relative to that obtained with ORF50B and ORF73 promoter alone, respectively. The summary of three independent experiments, each with cultures done in triplicate, is shown.
Under the hypothesis that ORF73 may regulate viral latency by preventing the expression of the transactivator protein ORF50B, the promoter repression of ORF50 and ORF73 was studied in further detail. Increasing concentrations of the ORF73 expression construct were cotransfected with constant amounts of either ORF50B or ORF73 promoter constructs in OMK cells, and lysates were measured for luciferase reporter activity. The results were normalized to the transcriptional activity of either the ORF50B or ORF73 promoter alone (relative activity of promoters alone = 1). As shown in Fig. 2C, the ORF50B promoter activity is more sensitive to repression by expression of ORF73 than is that of the ORF73 promoter. While the ORF50B promoter responds at 0.05 μg, at least 10-fold-more ORF73 expression plasmid is necessary to observe a similar effect on the ORF73 promoter. This supports the hypothesis that low expression levels of ORF73 suppress expression of the major HVS strain C488 transcriptional activator ORF50B (56), while higher expression levels of ORF73 may shut down the ORF73 expression by autoregulation and allow lytic replication to proceed.
Two ORF73 domains are necessary for the repression of the ORF50B promoter.
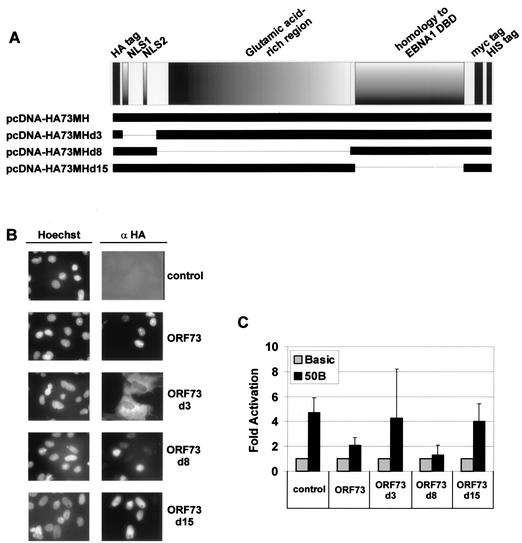
ORF73 deletion mutants were constructed to determine which domains of ORF73 protein are responsible for inhibition of ORF50B promoter. Deletion mutant ORF73d3 lacks the N terminus with the two putative NLSs, deletion mutant ORF73d8 lacks the whole glutamine-rich repeat region, and ORF73d15 lacks the C terminus containing a putative DBD with homology to the EBV EBNA-1 DBD (Fig. 3A). The expression of mutant proteins of the expected size was confirmed by Western blot analysis of transfected cells (data not shown). Immunofluorescence studies were then performed to examine the subcellular localization of the respective deletion mutants. While expressed at approximately equal levels, ORF73d3 was found in the cytoplasm, whereas ORF73d8 and ORF73d15 were localized in the nucleus, as was the full-length protein (Fig. 3B). In order to determine the transcriptional influence of these ORF73 deletions on the ORF50B promoter, OMK cells were cotransfected with the ORF50B luciferase reporter construct and the ORF73 deletion mutant, full-length, or empty control plasmids (Fig. 3C). ORF73d3, which lacks the N terminus and is not nucleus localized, shows no influence on the ORF50B promoter in comparison to wild-type ORF73. While both the full-length and the glutamine-rich-repeat-deletion mutant ORF73d8 were still able to repress the ORF50B promoter, neither the amino-terminal ORF73d3 nor the carboxy-terminal ORF73d15 deletion construct was able to suppress the approximately fourfold stimulation of transcription by the ORF50B promoter, similar to the empty control vector. This suggested that the transcriptional activity of ORF50B promoter is modulated by the ORF73 protein. Since the cytoplasmic localization of ORF73d3 may prevent it from repressing transcription in the nucleus, this could indicate that the responsible repressor domain is localized in the C terminus of ORF73.
FIG. 3.
ORF73 domains necessary for the transactivation of the ORF50B promoter. (A) Graphical representation of the ORF73 deletion mutants used in this experiment. (B) The subcellular localization of each ORF73 deletion mutant was determined in OMK cells that were grown and transfected in chamber slides with the respective expression constructs according to panel A (1 μg): pcDNA-HA-MH (negative control), pcDNA-HA73MH (ORF73), pcDNA-HA73MHd3 (ORF73d3), pcDNA-HA73MHd8 (ORF73d8), and pcDNA-HA73MHd15 (ORF73d15). Cells were fixed 24 h later and stained for expression of the HA-tagged proteins as described in the text. (C) Transcriptional activity of the ORF73 deletion mutants of the ORF50B promoter. OMK cells were cotransfected with 0.5 μg of luciferase reporter plasmid pGL3-ORF50B and with 1 μg of expression constructs for either pcDNA-HA-MH (negative control), pcDNA-HA73MH (ORF73), pcDNA-HA73MHd3 (ORF73d3), pcDNA-HA73MHd8 (ORF73d8), or pcDNA-HA73MHd15 (ORF73d15). Equal amounts of cell lysates were assayed for luciferase activity after 24 h, and values were normalized to that obtained with pGL3-Basic alone. The black and gray bars indicate the relative activities of the ORF50B and pGL3-Basic promoter in the presence of the respective pcDNA-HA73MH constructs and pcDNA-HA-MH control. The mean values obtained from three independent experiments, again performed in triplicate, are shown.
ORF73 downregulates ORF6 gene expression by modulating ORF50 promoters.
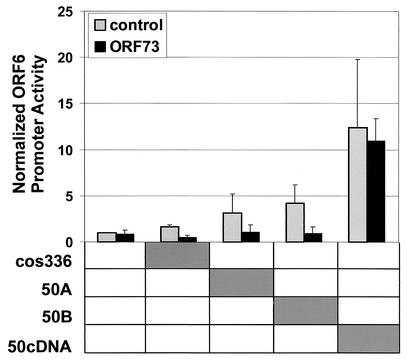
It was previously shown that ORF50A in HVS strain A11 and ORF50B in HVS strain C488 are important transactivators of the ORF6 gene promoter (56, 60). The reporter construct pGL3-ORF6 was used to determine the ability of ORF73 to influence ORF50 gene expression and to modulate the transactivating function of ORF50 in vitro. Constructs containing the ORF50 gene in the context of its own transcriptional regulatory elements (cosmid 336, which encompasses the HVS genome from ORF46 to ORF65.5, and plasmids pCR2.1-50A, containing the genomic ORF50A and ORF50B genes including the respective promoters, and plasmid pCR2.1-50B, containing only ORF50B with its own promoter) and, as a positive control, the CMVIE promoter-driven pcDNA-50cDNA construct were cotransfected with the ORF6 reporter construct and either pcDNA-HA73MH or empty vector pcDNA-HA-MH (Fig. 4). The responsiveness of the ORF6 promoter regions to the ORF50 expression constructs and the expression of ORF50 protein from these expression constructs were shown previously (56). Luciferase activity was normalized to the ORF6 promoter activity in the absence of ORF50. There was a clear increase in the ORF6 promoter activity detectable when the reporter construct was cotransfected with cos336 and ORF50A; the strongest increase was observed with ORF50B, which enhanced the activity of the ORF6 promoter by fourfold. In each case, the activation was completely and significantly repressed in the presence of ORF73. There was no influence of ORF73 expression when the ORF6 promoter was transactivated by the CMVIE promoter-driven ORF50 cDNA. These data support the hypothesis that ORF73 represses ORF6 promoter activity via modulation of ORF50A and ORF50B.
FIG. 4.
Transcriptional activity of ORF6 promoter in the presence of ORF50 and ORF73. OMK cells (2.5 × 105) were cotransfected with 0.5 μg of ORF6 reporter construct, with 1 μg of each genomic ORF50 construct containing the respective viral promoter (50A or 50B), and with 1 μg of pcDNA-HA73MH and pcDNA-HA-MH alone, respectively. A pcDNA-ORF50 construct (50cDNA) under the control of a constitutive CMV promoter serves as a positive control. In addition, cosmid 336, which includes the genomic region from ORF46 to ORF67.5 of HVS C488, including both loci, ORF50A and ORF50B, was transfected. After 24 h equal amounts of cell lysates were assayed for luciferase activity. Luciferase activity is expressed as fold activation over that obtained with pGL3-ORF50B or pGL3-ORF73 promoter alone. The mean values of triplicates of three independent experiments are shown.
Construction of mifepristone-inducible recombinant viruses.
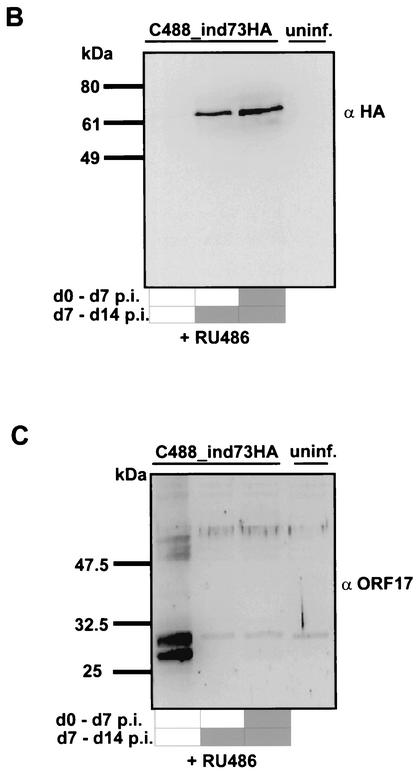
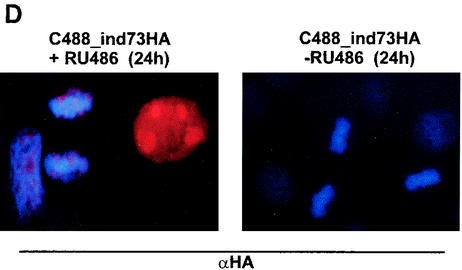
The regulatory function of the ORF73 protein was verified in the context of the viral genome by constructing a recombinant HVS where the ORF73 expression was tightly controlled by a conditional RU-486-inducible promoter. Recombinant HVS expressing the regulatory GeneSwitch protein either alone or together with ORF73 driven by the conditional RU-486-inducible promoter was created by a cosmid-based approach (Fig. 5A) (13). The right-terminal cosmid Dc5 was modified by insertion of RU-486-inducible ORF73. By this insertion, the natural promoter of ORF73, the nonessential ORF74, and the carboxy-terminal residues of ORF75 are deleted. The correct insertion was verified by restriction mapping and sequencing (data not shown). The switch regulator protein was cloned into the SwaI site of cos331dBstZ17I, a noncoding region that is dispensable for viral replication and transformation (13). Recombinant viruses were then constructed by cotransfection of linearized cosmids into OMK cells. The recombinant C488_ind73HA was generated from cosmids 331dBstZ17I_switch, 261, 291, 336, and Dc5_indHA73. The control virus C488_NFκB-switch was generated from cosmids 331dBstZ17I_switch, 261, 291, 336, and Dc5 (Fig. 5A). The induction of ORF73 expression was studied by Western blot analysis from infected and RU-486-induced OMK cell extracts, as well as with control cell lysates from uninduced and uninfected cells (Fig. 5B). Cell extracts were separated by SDS-10% polyacrylamide gel electrophoresis and detected with anti-HA-tag antibody. OMK cells infected with C488_ind73HA that were RU-486 induced showed high expression of ORF73 (lane 3). ORF73 expression is also detectable in cells which were induced between day 7 and day 14 p.i. (lane 2). ORF73 was not visible in uninduced cultures (lane 1) and in uninfected control cultures (lane 4). HVS ORF17 was used as a marker of lytic virus replication. ORF17 encodes the minor scaffold protein and the viral protease, which digests itself by autoproteolytic cleavage and is involved in capsid protein maturation; it is expressed during lytic herpesvirus replication and is considered a typical herpesvirus late gene. ORF17 was detected by a polyclonal anti-ORF17 rabbit antiserum in the same OMK cell extracts from the conditional ORF73 expression experiment. ORF17 was detectable only in the infected and uninduced cell extract (Fig. 5C, lane 1). Upper bands at 52 and 50 kDa correspond to immature, not fully cleaved, precursors of the protein, while the two major lower bands (25 and 23 kDa) represent the mature ORF17 protein after autocleavage at two sites. In the infected and RU-486-induced cell lysates, no protease proteins were identifiable (lanes 2 and 3). These data indicate that overexpression of ORF73 during virus infection inhibits the transit into late gene expression and the lytic life cycle of HVS. Immunofluorescence studies were done to study subcellular localization of induced HA-ORF73. OMK cells were infected with C488_ind73HA and cultured with and without RU-486 for 24 h, respectively. Induced ORF73 was seen in the nucleoli with a similar pattern as that observed in transient-transfection assays (Fig. 1B). There was no ORF73 detectable in infected but uninduced control cultures (Fig. 5D), confirming the Western blot analysis (Fig. 5B).
FIG. 5.
Induction of ORF73 suppresses late gene expression in permissive OMK cells. (A) Construction of the recombinant HVS C488_NFκB-switch and C488_ind73HA by cotransfection of linearized overlapping cosmids into permissive OMK cells. The regulatory GeneSwitch protein was inserted into the cosmid 331dBstZ17I representing the left end of the genome, and the HA-tagged ORF73 under the transcriptional control of the RU-486-inducible promoter was inserted into BstZ17I-AgeI-digested cosmid Dc5. DHFR, dihydrofolate reductase. (B) RU-486-inducible expression of ORF73. Shown are the results of immunoblot analysis of OMK cells that were infected with C488_ind73HA and were cultured for 2 weeks. Twenty micrograms of each cell lysate was electrophoresed on a 10% polyacrylamide-SDS gel and analyzed by Western blotting. One infected culture was uninduced the whole time (lane 1). A second culture was RU-486 induced (500 nM) only for the second week of infection (lane 2), and a third culture was RU-486 induced from the beginning (lane 3). Uninfected OMK cells are shown as negative controls (lane 4). (C) The same extracts were analyzed for expression of late antigens, encoded by ORF17, the viral protease and minor scaffold protein. Several bands around 30 kDa are visible due to autoproteolytic processing of the 53-kDa precursor protein. (D) HA-tagged ORF73 is RU-486 inducible and localized in the nuclei of C488_ind73HA-infected OMK cells. There is a noticeable colocalization of the HA-ORF73 to chromosomal host cell DNA in mitosis. OMK cells were infected with C488_ind73HA, and ORF73 expression was induced by 2 μM RU-486 for 24 h. An infected but uninduced culture is shown as a control. Cells were fixed, and the HA-ORF73 protein was visualized by immunofluorescence.
Replication of recombinant virus C488_ind73HA in OMK cells.
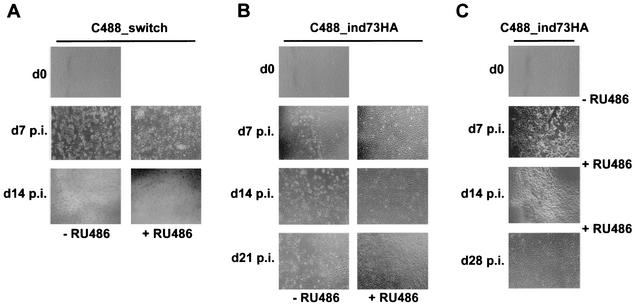
For replication studies, OMK cells were cultivated in six-well plates for 24 h and then infected with C488_ind73HA or the control virus C488_switch. The amount of inoculum virus was identical in the respective parallel cultures. The C488_switch virus grew to similar titers (106 to 107) as the wild-type virus; due to a slower progress of CPE in the C488_ind73HA virus, titration of this virus was not possible by conventional limiting dilution or plaque assays. In this case, the amount of C488_ind73HA virus was chosen so that >90% of cells showed LANA-specific signal by immunofluorescence microscopy. To investigate the effect of induced expression of ORF73 during virus replication, cells were observed in parallel cultures induced with RU-486 or not induced. At time points day 0, day 7 p.i., day 14 p.i., day 21 p.i., and day 28 p.i., infected cell cultures were documented by photography (Fig. 6). In the control virus C488_switch-infected cells, CPE progressed independently of the addition of RU-486 to the culture to complete lysis within 14 days (Fig. 6A). At time point day 7 p.i. initial CPEs were detectable in the uninduced C488_ind73HA-infected culture, while there were no CPEs visible in the RU-486-induced culture. While the CPE progressed to complete lysis of OMK cells in the uninduced cultures, there was still no CPE apparent at time points day 14 p.i. and day 21 p.i. in ORF73-expressing OMK cells. The uninduced culture infected with C488_ind73HA had fully lysed at day 28 p.i., in contrast to the RU-486-induced cell cultures, which showed no CPE (Fig. 6B). In a further experiment, RU-486 was added to OMK cells that were infected with C488_ind73HA and cultured for 7 days in the absence of RU-486 until the first CPEs began to develop. The CPE in these cultures began to decrease at time point day 14 and had completely disappeared at day 28 p.i. (Fig. 6C). Cultures which were RU-486 induced from day 0 to day 14 p.i., and then cultured without RU-486 began to show initial CPEs at day 21 p.i. (data not shown). Again, C488_switch-infected OMK cultures served as controls and were treated under the same conditions. The first indications of CPE were detectable at time point day 7 p.i. in RU-486-induced and uninduced cultures; at day 14 p.i., CPE had progressed to complete lysis.
FIG. 6.
Induction of ORF73 suppresses lytic replication in permissive OMK cells. OMK cells were grown on six-well plates and infected at comparable multiplicities of infection with C488_switch control (A) or C488_ind73HA (B and C) viruses. (A) The addition of 500 nM RU-486 to infected cells could not prevent lytic replication of the control virus; CPE was apparent at 1 week p.i., and lysis was complete at 2 weeks p.i. (B) C488_ind73HA-infected cultures were RU-486 induced and observed in parallel to uninduced but otherwise identically treated cultures for the indicated period. The induction of ORF73 expression by 500 nM RU-486 completely prevented the formation of CPE in C488_ind73HA-infected cells, whereas CPE developed in the uninduced infected cells. (C) Lytic replication can be switched by RU-486-induced ORF73 expression. A C488_ind73HA-infected culture was RU-486 induced for the first 7 days of infection and then switched to medium without RU-486. Another infected culture was uninduced during the first week of infection, and when development of initial CPEs became apparent, RU-486 was added to the medium to induce ORF73 expression from the second week of infection onward.
Regulated expression of ORF73 in infected OMK cells inhibits lytic virus replication by direct downregulation of HVS C488 lytic transactivators ORF50A and ORF50B.
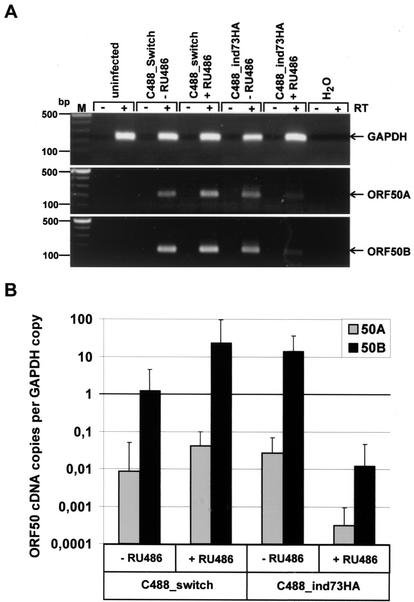
Parallel cultures of OMK cells were infected with C488_switch and C488_ind73HA viruses, respectively; one culture was treated with mifepristone, while the other was not induced. The amount of inoculum virus was identical in the respective parallel cultures and in the case of C488_ind73HA was chosen so that >90% of cells showed LANA-specific signal by immunofluorescence microscopy. After 1 week, when CPE became apparent in uninduced and control cultures, total RNA was isolated. RT-PCR for ORF50A, ORF50B, and GAPDH was performed, and reaction products were analyzed by agarose gel electrophoresis (Fig. 7A). RT-PCR for GAPDH (228-bp fragment) shows that similar amounts of cellular RNA were used for cDNA synthesis (Fig. 7A, top panel). A 195-bp fragment specific for expression of the spliced ORF50A (Fig. 7A, middle panel) and a 170-bp fragment derived from both unspliced ORF50B and spliced ORF50A RNA (Fig. 7A, bottom panel) were detectable from all infected cultures; since this transcript was found to be far more abundant than the ORF50A transcript (see below), it is referred to below as ORF50B specific. The signal of both ORF50A and ORF50B in ORF73-induced infected cells is clearly decreased compared to that in the infected control cultures and the C488_ind73HA-infected cells cultured without RU-486.
FIG. 7.
ORF50A and ORF50B transcription is regulated by ORF73 in infected OMK cells. (A) OMK cells were infected with either C488_switch or C488_ind73HA in parallel cultures and maintained in the presence or absence of RU-486. At day 7 p.i., when CPE became apparent in uninduced or control virus-infected cultures, cells were harvested for RNA isolation. Total RNA of each culture was used for ORF50A- and ORF50B-specific RT-PCR. The top panel shows corresponding RNAs as detected by RT-PCR for GAPDH (resulting fragment of 228 bp). The middle panel shows amplified ORF50A transcripts (spliced fragment of 195 bp), and the bottom panel shows ORF50B transcripts (fragment of 170 bp) detected in the infected OMK cells. +RT, RT-PCR from first-strand cDNA; −RT, PCR from parallel control reaction mixture, where the reverse transcriptase was omitted. (B) For quantitative analysis, the same samples were diluted 1:10 and analyzed by SYBR Green real-time PCR. The diagram shows the relative number of ORF50A (gray columns) and ORF50B (black columns) cDNA copies that were normalized to the number of GAPDH cDNA copies in each sample.
In order to obtain a quantitative estimate of the cDNA copies of ORF50A and ORF50B in the infected cultures, the same cDNAs were analyzed by quantitative real-time PCR along with corresponding standards (Fig. 7B). The ABI Prism sequence detection system software (version 1.6.3) was used to calculate cDNA copy numbers from threshold amplification values of each sample. The established values were normalized to cDNA copies of GAPDH for every analyzed sample. As shown in Fig. 7B, ORF50A is expressed in C488_switch control cultures and in uninduced C488_ind73HA cultures at similar levels (gray columns). However, the ORF50A cDNA copy number in ORF73-induced cultures is about 100-fold lower than in the C488_ind73HA-infected OMK cells that were cultured without RU-486. The copy number of the amplified fragment derived from both the spliced ORF50A and the unspliced ORF50B (black columns) was about 100- to 1,000-fold higher than the ORF50A copy number alone; thus, more than 99% of this detected transcript was derived from ORF50B. This further supports the data from Thurau et al. that ORF50B is the major transactivator of HVS strain C488 (56). The expression of ORF50B in ORF73-induced cultures was significantly decreased by 1,000-fold compared to that in uninduced control cultures. These data indicate that expression of ORF73 represses both ORF50A and ORF50B expression in the viral context similarly to that in the reporter gene assays and that this effect is most likely responsible for the inhibition of viral lytic replication in infected OMK cells.
DISCUSSION
In latent infection with KSHV, a cluster of genes is expressed that includes vFLIP (ORF71), the v-Cyclin D homolog (ORF72), and LANA (ORF73) (10, 31, 49, 54). KSHV LANA is a 222- to 234-kDa protein that is expressed during the latent phase of infection and that is localized in the nucleus (30, 31). It has been shown elsewhere that, by tethering the viral episome to host cell chromosomes, LANA may play a role in plasmid maintenance and segregation during latent infection (2, 3, 8). There are several aspects of this function of LANA that remind us of the EBNA-1 protein of the Lymphocryptovirus EBV, and EBNA-1 is also involved in transcriptional regulation via the family of repeat sites (61). Within the Rhadinovirus subfamily, all viruses encode an ORF73 protein with homology to KSHV LANA, but their function has not been defined yet. Here we report on a functional aspect of ORF73 in downregulating viral lytic replication.
The subcellular distribution of HVS strain A11 ORF73 has been studied previously with OMK cells, revealing a nuclear localization pattern similar but not identical to that of KSHV LANA (24). In the human lung cancer-derived epithelial cell line A549, the strain A11 ORF73 promoter is able to drive strong expression of the reporter gene construct, and ORF73 expression constructs are able to transactivate the homologous ORF73 promoter (23). However, the transcribed mRNAs in A549 cells differ from the pattern that has been found in strain C488-transformed human and simian lymphocytes and from OMK cells (14). In this paper, we addressed the putative function of HVS strain C488 ORF73 as a transcriptional modulator in the permissive OMK cell line, the standard culture for propagation and production of infectious HVS (9). HVS strain C488 encodes an ORF73 that is considerably larger than the A11 ORF73 (501 versus 407 amino acids); the amino acid sequence is conserved by 70% over the full ORF and by 75% in the nonrepetitive regions flanking the glutamic-acid-rich central repeat, including the NLS and DBD. In transient-transfection assays, and in C488-infected OMK cells, a protein of approximately 62 kDa was detectable by using epitope-tagged proteins or specific antiserum, respectively (Fig. 1A and C), and the nuclear distribution was similar to that for the A11 protein (Fig. 1B) (24). By performing transient-transcription assays, it could be shown that ORF73 expression can modulate the transcriptional activity of various HVS gene promoter-driven reporter constructs in permissive OMK cells (Fig. 2). Specifically, we found that expression of ORF73 downregulates the promoter activity of the immediate-early transactivators ORF50A and ORF50B. Although it cannot be excluded that other mechanisms might also contribute, the difference in ORF50B and ORF73 promoter susceptibilities to ORF73 suppression (Fig. 2C) and the reduction in ORF73 protein levels as lytic replication proceeds (Fig. 1C) further support the hypothesis that ORF73 can regulate its own gene expression by an autoregulatory loop. Thus, different expression levels of ORF73 may determine the choice between latency or lytic growth in the viral life cycle.
The downregulation of the ORF50A and ORF50B promoters was particularly relevant in the context that these are the main transactivators for the induction of HVS lytic replication. Transcription of the HVS strain C488 ORF50 gene locus has been extensively mapped by rapid amplification of cDNA ends, Northern blotting, and RNase protection experiments (56). In the same paper, expression of ORF50 protein from the expression constructs was shown by in vitro translation, and the responsiveness of the ORF6 promoter regions to the ORF50 expression constructs that were also used in this study was shown. ORF50B is believed to be the main effector for induction of the lytic replication cycle (56). When the ORF73 domains that are essential for ORF50B promoter repression were mapped by luciferase reporter assays with ORF73 deletion constructs (Fig. 3A), the N- and C-terminal domains were necessary for transcriptional repression (Fig. 3C). Since the cytoplasmic localization of ORF73d3 may prevent it from activating transcription in the nucleus, it could be argued that the responsible repressor domain is localized in the C terminus of ORF73. An attempt to delineate the ORF73-responsive elements by studying nested deletion constructs of the ORF50B promoter did not reveal specific elements (data not shown); this may be due to complex interactions along larger parts of the promoter. Taken together, these findings indicate that ORF73 is a transcriptional regulator, although the precise mechanism of how it represses the expression of the Rta homologs ORF50A and ORF50B remains to be clarified. This led us to the hypothesis that ORF73 may prevent the expression of early viral replication genes such as the major ssDNA-binding protein ORF6. The ORF6 promoter is known to include an ORF50-responsive element and is transactivated by both ORF50A and ORF50B (25, 40, 56, 60). When ORF73 was cotransfected to constructs where ORF50A and ORF50B were under the control of their own promoters, the normalized ORF6 promoter activity was reduced to its baseline activity, but not when ORF50 expression was driven by the heterologous CMVIE promoter (Fig. 4). These data suggest that ORF73 can inhibit the expression of delayed-early viral replication genes via transcriptional repression of the Rta homologs ORF50A and ORF50B.
We then extended this by constructing a recombinant HVS C488 virus with an inducible ORF73 by using the RU-486-inducible GeneSwitch system (7, 58). The insertion of the fragment containing the RU-486-inducible promoter in front of an HA-tagged ORF73 was possible only by simultaneous deletion of the open reading frame of ORF74 (GPCR) and the 3′ end of ORF75 (FGARAT) (Fig. 5A). However, these elements are dispensable for lytic viral replication (unpublished data), and since the sequence for ORF73 and ORF74 promoters is bidirectional (Fig. 2), it would have been impossible to generate ORF73 promoter mutants that would not affect ORF74 expression. This system allowed us to study the effects of controlled ORF73 expression in the viral context, and it could be shown that lytic viral replication in OMK cells could be controlled by regulated ORF73 expression (Fig. 5 and 6). Western blot analysis showed that ORF73 is expressed only in RU-486-induced C488_ind73HA-infected cultures, not in the uninduced cultures (Fig. 5B), while expression of the lytic viral protease gene (ORF17) was detectable only in the infected and uninduced culture (Fig. 5C). The expression of the ORF50 protein under the same conditions could not be studied directly due to a lack of specific reagents (five polyclonal sera were tested; data not shown). Therefore, we studied the downregulation of the ORF50A and ORF50B transcripts by RT-PCR (Fig. 7A) and SYBR Green real-time quantitative RT-PCR (Fig. 7B). Both methods clearly revealed that the ORF50 transcription is downregulated in the infected cells by induction of ORF73 expression.
There is evidence that overexpression of ORF73 in the infected cultures downregulates the transcriptional activity of the Rta promoter ORF50; thereby the ORF50-driven expression of early viral replication genes like ORF6 and the M homolog ORF57 (60) is inhibited, and thus, the initiation of the lytic replication cycle is prevented by ORF73. In KSHV, three differentially spliced mRNAs encoding the vFLIP homolog (ORF71), the v-Cyclin D homolog (ORF72), and LANA (ORF73) are latently expressed in PEL cells from a common ORF73 promoter (10). Immunoblotting for ORF72 was performed to test if the v-Cyclin is also overexpressed by the inducible promoter, from cells that were infected with the ORF73-inducible virus and C488 control virus, but no significant differences in the expression levels of v-Cyclin in the infected cell cultures could be detected, excluding the possibility that the observed effects are due to overexpression of the HVS v-Cyclin (data not shown). Furthermore, the assays were performed in nondividing, resting OMK cells, while the main contribution of the HVS v-Cyclin gene may be in reactivation of viral infection from latently infected lymphocytes (12), similar to results from the MHV-68 system (26, 57).
The data presented in this paper would be consistent with the following model. The Rta homologs ORF50A and ORF50B initiate lytic viral replication by transactivation of early viral replication genes (Fig. 8A). Our results expand this model by showing that the ORF50 promoters are repressed by ORF73, that the ORF50-mediated transactivation of delayed-early genes can be prevented by ORF73, and that lytic replication of HVS can be blocked by overexpression of ORF73 in a recombinant viral system (Fig. 8B). This shows how rhadinovirus replication could be regulated upstream of the lytic R transactivators. Although the recombinant viruses allow us to compare the effects of a regulated gene (in this case, ORF73) with a very low background expression to those of effects of expression in the induced state (Fig. 5B), the mifepristone-inducible gene expression system employed in this study does not allow us to titrate the amount of ORF73 expression by varying the concentration of the inducing drug. The levels of RU-486-induced ORF73 protein that achieve suppression of lytic replication are higher than in naturally infected cells, but this is the likely reason why lytic replication is efficiently suppressed in this experimental system. It is also difficult to estimate what effects local accumulation and subcellular distribution of a protein may have in an infected cell. Furthermore, the ORF50B promoter was sensitive to low amounts of ORF73, while higher quantities of ORF73 expression plasmid were required to shut down the ORF73 gene promoter (Fig. 2C). Thus, high levels of ORF73 can shut down the ORF73 promoter, which may be consistent with the hypothesis that ORF73 protein levels may then become low enough to allow expression of the ORF50-encoded lytic transactivators; ORF50A and ORF50B then initiate the cascade of gene expression, and the lytic phase of replication starts. The reduction in ORF73 levels as lytic replication proceeds (Fig. 1C) is also compatible with this model. Although it was not the focus of this study, our model could help to explain why HVS lytic replication is suppressed in the lung carcinoma cell line A549 (23, 24) and other human cell lines (51). In A549 cells, a high level of activity of the ORF73 promoter was detected, and ORF73 can activate this promoter (24), presumably resulting in relatively high amounts of ORF73 protein that could block the ORF50 promoters and prevent efficient lytic replication in these cells. Consequently, expression of ORF50 from heterologous promoters has been able to partially overcome the block of lytic replication in A549 cells (23).
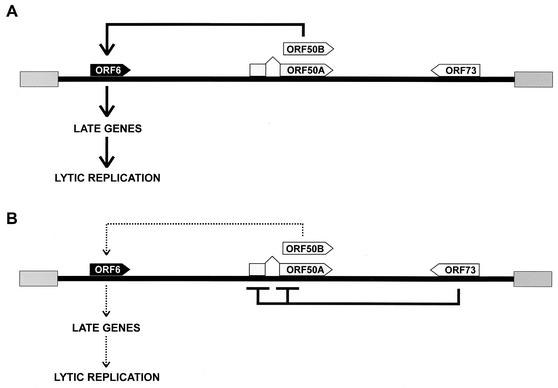
FIG. 8.
A model for ORF73-mediated regulation of HVS lytic replication. (A) In the absence of ORF73, both forms of ORF50 protein are expressed and activate expression of delayed-early genes involved in DNA replication, leading to expression of late viral genes. (B) ORF73 expression downregulates the transcriptional activity of ORF50 promoters and prevents ORF50 expression and the initiation of the lytic viral replication cycle via delayed-early gene expression.
Our studies revealed an important function of ORF73 in the control of lytic replication of HVS in the standard permissive cell culture system. Further studies will be necessary to elucidate the putative role of ORF73 in episomal maintenance and segregation of viral genomes and to identify viral or cellular molecules that interact with ORF73. It is notable that the punctate ORF73 staining associated with mitotic chromosomes (Fig. 5D) is highly reminiscent of data published previously for KSHV LANA (2). Moreover, such studies may provide insight into a possible function of HVS ORF73 in latent infection of transformed simian and human T lymphocytes, also with respect to transformation. Although it cannot be excluded that repression of cellular promoters or viral promoters other than ORF50 may contribute to the block of lytic replication by ORF73, we believe that HVS ORF73 controls the lytic replication cascade upstream of the immediate-early transactivators and that it regulates viral latency via a feedback mechanism.
Acknowledgments
We thank Mathias Thurau and Helmut Fickenscher for the generous gift of plasmids and cell lines, Karin Metzner for help and advice with real-time PCR, and Ingrid Müller-Fleckenstein and Monika Schmidt for technical assistance.
This project was supported by the Deutsche Forschungsgemeinschaft (SFB 466, Lymphoproliferation und virale Immundefizienz), IZKF (Entzündungsprozesse: Genese, Diagnostik und Therapie, 01 KS 9601/1), and Akademie der Wissenschaften und der Literatur, Mainz, Germany.
Footnotes
This article is dedicated to Harald zur Hausen on the occasion of his retirement as chairman of the executive board of the German Cancer Research Center (Deutsches Krebsforschungzentrum), Heidelberg, Germany.
REFERENCES
- 1.Albrecht, J. C., J. Nicholas, D. Biller, K. R. Cameron, B. Biesinger, C. Newman, S. Wittmann, M. A. Craxton, H. Coleman, B. Fleckenstein, and R. W. Honess. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 66:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biesinger, B., I. Müller-Fleckenstein, B. Simmer, G. Lang, S. Wittmann, E. Platzer, R. C. Desrosiers, and B. Fleckenstein. 1992. Stable growth transformation of human T lymphocytes by herpesvirus saimiri. Proc. Natl. Acad. Sci. USA 89:3116-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biesinger, B., J. J. Trimble, R. C. Desrosiers, and B. Fleckenstein. 1990. The divergence between two oncogenic herpesvirus saimiri strains in a genomic region related to the transforming phenotype. Virology 176:505-514. [DOI] [PubMed] [Google Scholar]
- 6.Boshart, M., F. Weber, G. Jahn, K. Dorsch-Hasler, B. Fleckenstein, and W. Schaffner. 1985. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 41:521-530. [DOI] [PubMed] [Google Scholar]
- 7.Burcin, M. M., G. Schiedner, S. Kochanek, S. Y. Tsai, and B. W. O'Malley. 1999. Adenovirus-mediated regulable target gene expression in vivo. Proc. Natl. Acad. Sci. USA 96:355-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotter, M. A., and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 9.Daniel, M. D., D. Silva, and N. Ma. 1976. Establishment of owl monkey kidney 210 cell line for virological studies. In Vitro 12:290. [Google Scholar]
- 10.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duboise, S. M., J. Guo, S. Czajak, R. C. Desrosiers, and J. U. Jung. 1998. STP and Tip are essential for herpesvirus saimiri oncogenicity. J. Virol. 72:1308-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensser, A., D. Glykofrydes, H. Niphuis, E. M. Kuhn, B. Rosenwirth, J. L. Heeney, G. Niedobitek, I. Müller-Fleckenstein, and B. Fleckenstein. 2001. Independence of herpesvirus-induced T cell lymphoma from viral cyclin D homologue. J. Exp. Med. 193:637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ensser, A., A. Pfinder, I. Müller-Fleckenstein, and B. Fleckenstein. 1999. The URNA genes of herpesvirus saimiri (strain C488) are dispensable for transformation of human T cells in vitro. J. Virol. 73:10551-10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fickenscher, H., B. Biesinger, A. Knappe, S. Wittmann, and B. Fleckenstein. 1996. Regulation of the herpesvirus saimiri oncogene stpC, similar to that of T-cell activation genes, in growth-transformed human T lymphocytes. J. Virol. 70:6012-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fickenscher, H., C. Bökel, A. Knappe, B. Biesinger, E. Meinl, B. Fleischer, B. Fleckenstein, and B. M. Bröker. 1997. Functional phenotype of transformed human αβ and γδ T cells determined by different subgroup C strains of herpesvirus saimiri. J. Virol. 71:2252-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 18.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glykofrydes, D., H. Niphuis, E. M. Kuhn, B. Rosenwirth, J. L. Heeney, J. Bruder, G. Niedobitek, I. Müller-Fleckenstein, B. Fleckenstein, and A. Ensser. 2000. Herpesvirus saimiri vFLIP provides an antiapoptotic function but is not essential for viral replication, transformation, or pathogenicity. J. Virol. 74:11919-11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodwin, D. J., M. S. Walters, P. G. Smith, M. Thurau, H. Fickenscher, and A. Whitehouse. 2001. Herpesvirus saimiri open reading frame 50 (Rta) protein reactivates the lytic replication cycle in a persistently infected A549 cell line. J. Virol. 75:4008-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groves, A. K., M. A. Cotter, C. Subramanian, and E. S. Robertson. 2001. The latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus activates two major essential Epstein-Barr virus latent promoters. J. Virol. 75:9446-9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall, K. T., M. S. Giles, D. J. Goodwin, M. A. Calderwood, I. M. Carr, A. J. Stevenson, A. F. Markham, and A. Whitehouse. 2000. Analysis of gene expression in a human cell line stably transduced with herpesvirus saimiri. J. Virol. 74:7331-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall, K. T., M. S. Giles, D. J. Goodwin, M. A. Calderwood, A. F. Markham, and A. Whitehouse. 2000. Characterization of the herpesvirus saimiri ORF73 gene product. J. Gen. Virol. 81:2653-2658. [DOI] [PubMed] [Google Scholar]
- 25.Hall, K. T., A. J. Stevenson, D. J. Goodwin, P. C. Gibson, A. F. Markham, and A. Whitehouse. 1999. The activation domain of herpesvirus saimiri R protein interacts with the TATA-binding protein. J. Virol. 73:9756-9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoge, A. T., S. B. Hendrickson, and W. H. Burns. 2000. Murine gammaherpesvirus 68 cyclin D homologue is required for efficient reactivation from latency. J. Virol. 74:7016-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang, S., Y. Gwack, H. Byun, C. Lim, and J. Choe. 2001. The Kaposi's sarcoma-associated herpesvirus K8 protein interacts with CREB-binding protein (CBP) and represses CBP-mediated transcription. J. Virol. 75:9509-9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyun, T. S., C. Subramanian, M. A. Cotter, R. A. Thomas, and E. S. Robertson. 2001. Latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus interacts with Tat and activates the long terminal repeat of human immunodeficiency virus type 1 in human cells. J. Virol. 75:8761-8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 1:19-29. [PubMed] [Google Scholar]
- 32.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim, C., Y. Gwack, S. Hwang, S. Kim, and J. Choe. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 276:31016-31022. [DOI] [PubMed] [Google Scholar]
- 34.Lim, C., H. Sohn, Y. Gwack, and J. Choe. 2000. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J. Gen. Virol. 81:2645-2652. [DOI] [PubMed] [Google Scholar]
- 35.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattsson, K., C. Kiss, G. M. Platt, G. R. Simpson, E. Kashuba, G. Klein, T. F. Schulz, and L. Szekely. 2002. Latent nuclear antigen of Kaposi's sarcoma herpesvirus/human herpesvirus-8 induces and relocates RING3 to nuclear heterochromatin regions. J. Gen. Virol. 83:179-188. [DOI] [PubMed] [Google Scholar]
- 38.Medveczky, M. M., E. Szomolanyi, R. Hesselton, D. DeGrand, P. Geck, and P. G. Medveczky. 1989. Herpesvirus saimiri strains from three DNA subgroups have different oncogenic potentials in New Zealand White rabbits. J. Virol. 63:3601-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medveczky, P., E. Szomolanyi, R. C. Desrosiers, and C. Mulder. 1984. Classification of herpesvirus saimiri into three groups based on extreme variation in a DNA region required for oncogenicity. J. Virol. 52:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicholas, J., L. S. Coles, C. Newman, and R. W. Honess. 1991. Regulation of the herpesvirus saimiri (HVS) delayed-early 110-kilodalton promoter by HVS immediate-early gene products and a homolog of the Epstein-Barr virus R trans activator. J. Virol. 65:2457-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholas, J., E. P. Smith, L. Coles, and R. Honess. 1990. Gene expression in cells infected with gammaherpesvirus saimiri: properties of transcripts from two immediate-early genes. Virology 179:189-200. [DOI] [PubMed] [Google Scholar]
- 42.Park, J., T. Seo, S. Hwang, D. Lee, Y. Gwack, and J. Choe. 2000. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J. Virol. 74:11977-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polson, A. G., L. Huang, D. M. Lukac, J. D. Blethrow, D. O. Morgan, A. L. Burlingame, and D. Ganem. 2001. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 75:3175-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russo, J. J., R. A. Bohenzky, M.-C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi's sarcoma associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sells, M. A., and J. Chernoff. 1995. Epitope-tag vectors for eukaryotic protein production. Gene 152:187-189. [DOI] [PubMed] [Google Scholar]
- 51.Simmer, B., M. Alt, I. Buckreus, S. Berthold, B. Fleckenstein, E. Platzer, and R. Grassmann. 1991. Persistence of selectable herpesvirus saimiri in various human haematopoietic and epithelial cell lines. J. Gen. Virol. 72:1953-1958. [DOI] [PubMed] [Google Scholar]
- 52.Sinclair, A. J., M. Brimmell, F. Shanahan, and P. J. Farrell. 1991. Pathways of activation of the Epstein-Barr virus productive cycle. J. Virol. 65:2237-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 55.Thome, M., P. Schneider, K. Hofmann, H. Fickenscher, E. Meinl, F. Neipel, C. Mattmann, K. Burns, J. L. Bodmer, M. Schroter, C. Scaffidi, P. H. Krammer, M. E. Peter, and J. Tschopp. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386:517-521. [DOI] [PubMed] [Google Scholar]
- 56.Thurau, M., A. Whitehouse, S. Wittmann, D. Meredith, and H. Fickenscher. 2000. Distinct transcriptional and functional properties of the R transactivator gene orf50 of the transforming herpesvirus saimiri strain C488. Virology 268:167-177. [DOI] [PubMed] [Google Scholar]
- 57.van Dyk, L. F., H. W. Virgin IV, and S. H. Speck. 2000. The murine gammaherpesvirus 68 v-cyclin is a critical regulator of reactivation from latency. J. Virol. 74:7451-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, Y., B. W. O'Malley, Jr., S. Y. Tsai, and B. W. O'Malley. 1994. A regulatory system for use in gene transfer. Proc. Natl. Acad. Sci. USA 91:8180-8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitehouse, A., I. M. Carr, J. C. Griffiths, and D. M. Meredith. 1997. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J. Virol. 71:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitehouse, A., A. J. Stevenson, M. Cooper, and D. M. Meredith. 1997. Identification of a cis-acting element within the herpesvirus saimiri ORF 6 promoter that is responsive to the HVS.R transactivator. J. Gen. Virol. 78:1411-1415. [DOI] [PubMed] [Google Scholar]
- 61.Wu, H., P. Kapoor, and L. Frappier. 2002. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J. Virol. 76:2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, T. T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zalani, S., G. E. Holley, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]