Abstract
Structural maintenance of chromosomes proteins (SMCs) bind to DNA and function to ensure proper chromosome organization in both eukaryotes and bacteria. Caulobacter crescentus possesses a single SMC homolog that plays a role in organizing and segregating daughter chromosomes. Approximately 1,500 to 2,000 SMC molecules are present per cell during active growth, corresponding to one SMC complex per 6,000 to 8,000 bp of chromosomal DNA. Although transcription from the smc promoter is induced during early S phase, a cell cycle transcription pattern previously observed with multiple DNA replication and repair genes, the SMC protein is present throughout the entire cell cycle. Examination of the intracellular location of SMC showed that in swarmer cells, which do not replicate DNA, the protein forms two or three foci. Stalked cells, which are actively engaged in DNA replication, have three or four SMC foci per cell. The SMC foci appear randomly distributed in the cell. Many predivisional cells have bright polar SMC foci, which are lost upon cell division. Thus, chromosome compaction likely involves dynamic aggregates of SMC bound to DNA. The aggregation pattern changes as a function of the cell cycle both during and upon completion of chromosome replication.
All organisms face the challenge of chromosome compaction because the contour length of DNA is vastly longer than the space assigned to it. Generally, bacteria have to compact their chromosome 1,000-fold for it to fit into the cell (20). Yet the compacted chromosomes must remain accessible for transcription, methylation, and DNA repair reactions, and the condensation must be reversible to allow DNA replication and proper separation of replicated chromosomes. Cytological studies of living and fixed bacterial cells have revealed that regions of the chromosomes have specific intracellular locations, suggesting that bacterial chromosomes are highly organized (reviewed in references 13 and 24).
Significant condensation of bacterial chromosomes is achieved by the supercoiling of the DNA. Supercoiling draws DNA in on itself, forming interwound superhelixes and superhelical branches, thereby decreasing the volume it occupies (20, 56). Negative supercoiling of DNA, which is observed in most bacteria, also aids cellular processes that require separation of the DNA strands, e.g., transcription and replication. Additionally, most bacteria possess a homolog of the structural maintenance of chromosomes protein (SMC), which appears to have a role in chromosome condensation and organization (15). SMCs are very large, containing 800 to 1,500 amino acids, and consist of five domains: globular N- and C-terminal domains connected by two long coiled-coil regions, which are separated by a flexible hinge region. The SMC protomers fold up into rod-shaped molecules, where the N- and C-terminal domains associate, forming an ATPase domain with structural similarity to ATP-binding cassette transport proteins (17, 19, 21, 31, 55). SMC forms dimers through interactions in the hinge region, forming symmetric molecules where both ends contain an ATPase and DNA binding domain. Electron microscopy shows that the hinge region is flexible, resulting in V-shaped molecules that when fully stretched are 100 to 150 nm long or headphone-like structures where the two globular domains come together (34). ATP binding and hydrolysis may control association of the SMC head domains from opposite ends of the molecule, perhaps condensing regions of DNA that are as much as 150 nm apart in a scissoring fashion.
SMCs are ubiquitous in eukaryotes and present in most eubacteria and archaea. Eukaryotes possess multiple SMCs that function in chromosome condensation, sister chromatid cohesion, DNA repair, and dosage condensation (reviewed in references 5 and 51). In eukaryotes, multiple SMC molecules and accessory proteins assemble into complexes, such as the condensin complex, which condenses the chromosomes during mitosis, and the cohesin complex, which mediates sister chromatid cohesion. In contrast, most bacteria possess only a single SMC. The gamma proteobacteria (such as Escherichia coli) possess the MukB protein instead of a typical bacterial SMC. Despite the lack of obvious sequence homology, MukB is distantly related to SMC, and the proteins are structurally and functionally similar (31, 34). MukB and many bacterial SMCs interact with accessory proteins, which are required for their activities (30, 33, 48).
SMC from Bacillus subtilis or Caulobacter crescentus and MukB from E. coli appear to have similar functions. The proteins are not essential, but disruption of smc or mukB results in temperature-sensitive growth (4, 23, 36, 38). The DNA is decondensed in the smc and mukB deletion strains, the origin-proximal region of the chromosome is mislocalized in a subpopulation of the cells, and there is a DNA segregation defect (4, 14, 23, 36, 38, 58). Thus, bacterial SMCs appear to function in condensing and organizing the chromosome. The defects of the E. coli mukB and B. subtilis smc mutants can be suppressed by mutations in topoisomerase I, resulting in DNA with more negative supercoiling (20, 29, 46). Since increased negative supercoiling makes the DNA more compact (20, 56), this suppression supports a function for SMC and MukB in condensing the chromosome. Analysis of the intracellular localization of SMC in B. subtilis and MukB in E. coli showed that the proteins bind to the chromosomal DNA and form specific foci (4, 7, 16, 30, 33, 39). In the absence of SMC, the origins in B. subtilis move away from each other normally after initiation of DNA replication but the termini are unable to separate. These observations suggest that SMC is not a part of the apparatus responsible for movement of the newly replicated chromosomal origins but rather functions later in the DNA compaction and segregation processes (14).
We showed previously that Caulobacter SMC is required for proper chromosome organization and the completion of chromosome segregation (23). In Caulobacter, many genes involved in cell-cycle-regulated events are differentially transcribed during the part of the cell cycle in which they function and many exhibit dynamic intracellular localization patterns (28, 47). We show here that transcription of smc is increased at the beginning of the S phase, coincident with the transcription of many genes involved in DNA replication and repair, although the SMC abundance does not change significantly during the cell cycle. There are between 1,500 and 2,000 SMC molecules per cell, sufficient for it to have a global role in organizing the chromosome. Instead of a stoichiometric distribution along the DNA, SMC forms a small number of foci, and the number increases at the G1-to-S transition. The predivisional cell accumulates bright SMC foci at the poles of the cell. Thus, either SMC complexes bind only to a few sites in the chromosome or aggregation of SMC molecules brings different regions of the chromosome into close proximity.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
C. crescentus strain CB15N (also named NA1000) (10) and derivatives were grown at 30°C in M2-glucose minimal medium or PYE medium (9). Synchronization was performed as described previously (10). The smc knockout strain CB15NΔsmc was described previously (23).
Low-copy-number plasmid pRBJ570 containing the entire smc region was constructed. The 3′ end of smc was amplified by PCR using the primers SMC23 (5′-CCAATGTCGACCGCTACTG-3′) and SMC24 (5′-ACAGGATCCCTGCTGGTGGTGATCTCGT-3′). An EcoRI-NcoI fragment from pGZ7 (61) containing the majority of the smc gene and the upstream region and the PCR fragment digested with NcoI and BamHI were ligated into pMR20 (44) digested with BamHI and EcoRI, resulting in pRBJ570. The plasmid contains the entire smc gene and ∼3.2 kb of DNA upstream of smc and 0.3 kb downstream.
The orfA deletion strain CB15NΔorfA was constructed by a two-step knockout technique. A DNA fragment containing an in-frame deletion of the majority of the orfA open reading frame was constructed by double PCR. For the first PCR round, the primers OrfA1 (5′-AATTGAATTCTGCTGAAGGACCTGGAAGAC-3′) and OrfA2 (5′-CGCCAGGGCGAAGATCTTGCTGGCCGCGAGGGCG-3′) or OrfA3 (5′-CGCCCTCGCGGCCAGCAAGATCTTCGCCCTGGCG-3′) and SMC27 (5′-AATTGGATCCTCTTCCAGACGCGAGAGATT-3′) were used. For the second round of PCR, we used these two PCR products as the template and the primers OrfA1 and SMC27. The BamHI- and EcoRI-digested PCR product was cloned into the sacB-containing integration plasmid pNPTS138 (M. R. K. Alley), resulting in plasmid pRBJ515. The plasmid was integrated by a single crossover event into the chromosome of CB15N or CB15N containing plasmid pRBJ570, expressing the OrfA gene product in trans. Next, cells were grown without selection to allow a second recombination event, removing the integrated plasmid and either the full-length orfA gene or the version of orfA with the in-frame deletion. Sucrose-resistant and kanamycin-sensitive clones were isolated to select for ΔorfA cells. PCR was used to identify clones with an in-frame deletion in the orfA gene. Clones with an orfA deletion were recovered only when the complementing plasmid pRBJ570 was present in the cells.
Plasmids containing transcriptional fusions between the smc promoter and a promoterless lacZ gene were constructed as follows. The NruI, ScaI-NruI, and NruI-ScaI fragments containing the region upstream of smc were inserted into the transcriptional fusion vector pRKLac290 (12), resulting in pRBJ591, pRBJ592, and pRBJ593, respectively. To construct transcriptional fusions containing shorter DNA fragments, regions upstream of smc were amplified by PCR using the following primer sets and cloned into pRKLac290: for pRBJ594, primers SMC30 (5′-ATATGAATTCAAGCTTGCCCCATGCTAGTTTCCTC-3′) and SMC31 (5′-ATATGGATCCGCGACGATGGTGACTCG-3′); for pRBJ595, SMC32 (5′-ATATGAATTCAAGCTTATCATCAAGGGCCGCAAC-3′) and SMC31; for pRBJ596, SMC33 (5′-ATATGAATTCAAGCTTCGCTGGAAAAGCAACTGG-3′) and SMC31; for pRBJ597, SMC34 (5′-ATATGAATTCAAGCTTCTTTTTCGCCGCCTAGAG-3′) and SMC31; for pRBJ598, SMC30 and SMC35 (5′-ATATGGATCCCTTGTCAATGGTCCGATCC-3′).
The pRBJ580 plasmid, used for overexpressing His6-SMC, was constructed by ligating the MluI-BamHI fragment from pRBJ570 and an NdeI- and MluI-digested PCR product made with primers SMC25 (5′-GGAATTCCATATGGTGCAGTTCCAGCGCCTC-3′) and SMC26 (5′-ACGACAGGCGCTTGTACTTC-3′) into NdeI- and BamHI-digested pET28a (Novagen). The plasmid was transformed into the E. coli strain BL21(DE3)/pLysS (Novagen).
The SMC-yellow fluorescent protein (YFP) fusion strain was constructed as follows. The 3′ end of smc was amplified by PCR using the primers SMC5 (5′-AAAACTGCAGATGGAGCCTGAGGAGCTG-3′) and SMC6 (5′-TTGGGATCCTCAGCCGCCACCAGCTTCTC-3′). The BamHI- and PstI-digested PCR product and the BamHI-EcoRI yfp fragment from pEYFP (Clontech) were cloned into the PstI- and EcoRI-digested integration vector pNPT228 (M. R. K. Alley), resulting in pRBJ600. The plasmid was integrated into the chromosome of CB15N by a single crossover event. We used PCR to confirm that the integration resulted in an SMC-YFP-expressing strain. Western blotting using anti-SMC or anti-green fluorescent protein (GFP) antibodies showed that a fusion protein of the expected size was expressed.
Transcription analysis.
The transcriptional start site of the smc promoter was mapped by primer extension analysis as described previously (43). The primer SMC29 (5′-GCGACGTAGGTGACTCG-3′) with sequence complementary to the 5′ end of the orfA gene upstream of smc was used. To examine the cell cycle transcription pattern of the orfA and smc promoter, cells containing a Psmc-lacZ transcriptional fusion (pRBJ593) were synchronized. At various times during the cell cycle, aliquots of cells were pulse-labeled for 5 min with [35S]methionine, the cells were lysed, and β-galactosidase and flagellin proteins were immunoprecipitated as described previously (43).
Antibody production and Western blotting.
His6-SMC was overproduced in E. coli from the plasmid pRBJ580 and purified with Talon resin (Clontech) as described by the manufacturer. Antibodies were produced in rabbits with the His6-SMC protein as the antigen. The antibodies were affinity purified as described previously (45) and used at a 1:2,000 dilution for Western blotting (8). For determining the relative abundance of SMC during the cell cycle, samples were normalized so equal amounts of total protein were loaded in all lanes. For quantitative Western analysis, cell lysates from a known number of cells were prepared as follows. Caulobacter cells were grown in M2-glucose medium at 30°C. Dilutions of the culture were plated to determine the number of cells per milliliter. Concentrated sodium dodecyl sulfate (SDS) loading buffer was added directly to an aliquot of the culture, and the mixture was immediately heated to 95°C for 5 min. The cell lysates and known quantities of purified His6-SMC protein were serially diluted in SDS loading buffer containing 50 μg of bovine serum albumin/ml and applied to an SDS-8% polyacrylamide gel. The SMC protein was detected by Western blotting. The films were scanned, and the relative signals from the bands were quantified with ImageQuant (Molecular Dynamics). The signals from the protein standard were used to make a standard curve, and the amounts of SMC in the cell lysates were determined. The signals were in the linear range, and the correlation coefficients for the standard curves were at least 0.98 for all determinations. The number of SMC molecules per cell reported is an average of four determinations.
Microscopy.
For immunofluorescence microscopy, synchronized CB15N cells were fixed with 3% formaldehyde or methanol at different stages of the cell cycle. The fixation solution was removed by filtration. Immunofluorescence microscopy was performed as described previously (8). Affinity-purified anti-SMC antibodies were used at a 1:100 dilution, and Alexa-488-labeled secondary antibodies (Molecular Probes) were used at a 1:200 dilution. For live-cell microscopy, SMC-YFP-expressing cells were immobilized by using a thin layer of agarose as described previously (22). Nomarski differential interference contrast (DIC) and fluorescence images were acquired with a Nikon E800 microscope with a 100× DIC objective and a 5-Mhz Micromax 5600 cooled charge-coupled device camera controlled through Metamorph (Universal Imaging Corp.). Images were processed with Metamorph and Photoshop (Adobe).
RESULTS
Mapping the smc promoter.
Analysis of the chromosomal region containing the smc gene showed that smc is directly downstream of a hypothetical open reading frame (named orfA) with homology to putative open reading frames of unknown function in other bacteria (24) (Fig. 1A). To determine if orfA is an essential gene, we attempted to construct an in-frame deletion where the majority of the putative open reading frame was removed. We were unable to obtain a ΔorfA strain unless the strain simultaneously contained a plasmid (pRBJ570) expressing the orfA gene product in trans. Additionally, the orfA-containing plasmid could not be cured from the chromosomal orfA deletion strain, whereas it was readily cured from the wild-type strain. Thus, orfA is an essential gene.
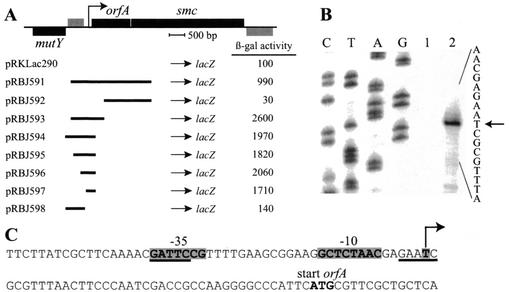
FIG. 1.
Mapping of the promoter transcribing the smc gene. (A) Schematic showing the organization of the chromosomal region containing the smc gene. Bars above the line, genes (black) and an open reading frame (grey) transcribed from left to right; bars below the line, gene and open reading frame transcribed in the opposite direction. Arrow, orfA and smc promoter mapped in this study. Below is shown the extent of the regions upstream of smc that were cloned in front of a promoterless lacZ gene, resulting in different transcriptional fusions. The β-galactosidase activities (in Miller units) of strains containing these plasmids are shown. All activities reported are averages of at least four independent measurements. (B) Mapping of the transcriptional start site of the orfA and smc promoter by primer extension analysis. A DNA sequencing ladder was generated with the oligonucleotide used for the primer extension analysis. Lane 1, yeast tRNA; lane 2, total RNA isolated from Caulobacter cells. Arrow, only major band within the DNA region present in the pRBJ597 plasmid. (C) Sequence of the orfA and smc promoter region. Arrow, mapped start site; grey shading, promoter −35 and −10 elements. GAnTC methylation sites are underlined.
Since there are only 23 bp between the stop site of orfA and the start site of smc, the promoter transcribing smc is probably located upstream of orfA. To map the orfA and smc promoter, we constructed a series of transcriptional fusions to lacZ containing different regions upstream of smc (Fig. 1A). Full promoter activity was observed with pRBJ597 containing a 309-bp fragment located immediately upstream of the orfA start site. A fragment further upstream (pRBJ598) exhibited no promoter activity. Furthermore, no promoter activity was observed with an internal orfA fragment (pRBJ592). Thus, orfA and smc appear to be in an operon. The lower β-galactosidase activity that was observed when the fusion point was within the smc gene (pRBJ591) was probably caused by the presence of a weak transcriptional terminator between orfA and smc.
The start site of the orfA and smc promoter was mapped by primer extension analysis. There was only one prominent band within the DNA region that gave full promoter activity with transcriptional fusions (pRBJ597), showing that the orfA and smc genes are transcribed from a single promoter, located immediately upstream of the start site of orfA (Fig. 1B and C). The −35 and −10 regions of the smc promoter show a reasonably good match to the published Caulobacter consensus σ70 promoter sequence (32).
Cell cycle regulation of smc transcription.
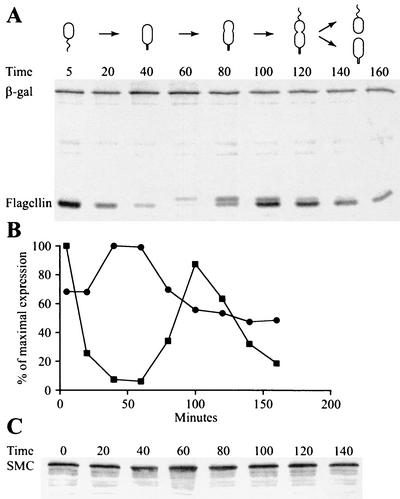
To determine if the transcription of the orfA-smc operon is under cell cycle control, swarmer cells of the strain CB15N/pRBJ593, containing a Psmc-lacZ transcriptional fusion, were isolated and allowed to progress synchronously through the cell cycle. The levels of transcription from the orfA and smc promoter at different time points during the cell cycle were measured by pulse-labeling newly synthesized proteins with [35S]methionine and immunoprecipitating the β-galactosidase protein (Fig. 2A). The radioactivity in the β-galactosidase band represents the activity of the smc promoter during the 5-min pulse-labeling. Synthesis of the 25-kDa flagellin protein was used as an internal control, since this protein is expressed specifically in swarmer and predivisional cells (11). In these experiments, the orfA and smc promoter was induced two- to threefold in stalked cells, during early S phase. The cell cycle timing of orfA and smc transcription and the level of induction are similar to the transcription pattern previously observed with several genes involved in DNA replication and repair (26, 28, 42, 43, 59).
FIG. 2.
Analysis of the smc transcription pattern and SMC level during the cell cycle. (A) Swarmer cells from strain CB15N/pRBJ593, containing a Psmc-lacZ transcriptional fusion, were isolated and allowed to progress synchronously through the cell cycle. At the indicated times (minutes), an aliquot of the cells was pulse-labeled for 5 min with [35S]methionine. The β-galactosidase and the 25-kDa flagellin proteins were immunoprecipitated from cell lysates, followed by SDS-polyacrylamide electrophoresis. Schematics show cell cycle progression of the strain. (B) The amounts of radioactivity in the different bands were quantified with a phosphorimager. Circles, radioactivity in the β-galactosidase band; squares, radioactivity in the flagellin bands. (C) The relative amount of the SMC during the cell cycle was analyzed by Western blotting using anti-SMC antibodies. Samples were withdrawn at the indicated time points (minutes), and equal amounts of total protein were loaded in all lanes.
The smc promoter contains two GAnTC sites overlapping essential promoter elements at the −35 region and at the transcription start site (Fig. 1C). These sites are known to be potential methylation sites for the essential CcrM methyltransferase (61). In Caulobacter, DNA methylation takes place only late in S phase, so GAnTC sites remain hemimethylated from the time the replication fork passes that region of the chromosome until the end of the cell cycle, when CcrM is expressed (49). For at least two Caulobacter genes, the methylation status of GAnTC sites in the promoter regions has roles in regulating gene expression (41, 50). To examine if the methylation state of the GAnTC sites in the orfA and smc promoter influenced promoter activity, we examined the transcript levels when the abundance of the CcrM methyltransferase was varied. No significant change in transcript levels was observed in ccrM mutants that either overexpress or deplete CcrM (data not shown), indicating that the methylation state of the GAnTC sites in the smc promoter does not play an important role in regulating the orfA and smc promoter activity.
Abundance of the SMC during the cell cycle.
To determine the abundance of the SMC at specific stages of the cell cycle, we produced antibodies to SMC. Western blots showed a band of the expected size (∼125 kDa) in extracts of the wild-type strain, a larger protein (∼150 kDa) in a smc-yfp fusion strain, and no signal in extracts of the smc deletion strain (data not shown). Thus, the antibodies are specific to SMC. Western blots of cell samples taken at different time points from a culture progressing synchronously through the cell cycle showed that SMC is present in approximately equal amounts at all times (Fig. 2C). Thus, SMC is probably a stable protein, so SMC molecules synthesized earlier remain in the cell, and the modest two- to threefold induction of orfA and smc transcription in early S phase does not result in a significant increase in the abundance of the SMC. Similar cell cycle regulation of transcription, but only very little variation in protein abundance, have previously been observed with other Caulobacter proteins (22). It is possible that the temporal regulation of transcription of the first gene in the operon, orfA, is important in modulating the concentration of its protein product at specific times in the cell cycle.
The number of SMC molecules present per cell has implications for the function of SMC. Therefore, we measured the number of SMC molecules per Caulobacter cell by quantitative Western blotting. Different dilutions of extracts from a known number of cells and known quantities of purified His6-SMC were fractionated using SDS-polyacrylamide gel electrophoresis. Western blots with anti-SMC antibodies were used to detect the protein (data not shown). The amount of SMC in the cell lysates was determined from a standard curve. We calculated that 1,690 ± 270 SMC molecules per cell are present in actively growing Caulobacter cells. Caulobacter SMC most likely forms a dimer, as is observed with B. subtilis SMC (34). Therefore, if the SMC complexes are evenly distributed throughout the chromosome, there would be approximately one SMC complex per 6,000 to 8,000 bp of chromosomal DNA in actively growing Caulobacter cells.
Intracellular localization of SMC during the cell cycle.
To carry out functions of DNA compaction and segregation, SMC is likely to interact with the DNA in an ordered pattern. If SMC binds to sites throughout the chromosome, SMC may be distributed throughout the cell. However, if SMC binds to a few sites in the chromosome or SMC molecules aggregate at specific sites in the cell, SMC foci should be observed. Thus, knowing the intracellular location of SMC may give information about the functions of SMC. Initially, we determined the intracellular localization of SMC by immunofluorescence microscopy with affinity-purified anti-SMC antibodies. Immunofluorescence microscopy using these antibodies gave a strong signal in more than 98% of the wild-type cells and no significant signal when Δsmc cells were used (data not shown), showing that the signal is specific for SMC. At different time points during the cell cycle, cells were fixed and the intracellular location of SMC was determined by immunofluorescence microscopy (Fig. 3). In all cell types SMC exhibited a punctate pattern, forming a limited number of small foci located throughout the cells. Weak background fluorescence in the cells, which could originate from noncomplexed SMC molecules, was observed.
FIG. 3.
Intracellular localization of the SMC in cells at different stages of the Caulobacter cell cycle as determined by indirect immunofluorescence microscopy. Swarmer cells were isolated and allowed to progress synchronously through the cell cycle. When the cells reached the indicated stages of the cell cycle (0, 60, 90, or 120 min, respectively), they were fixed and the intracellular location of SMC was visualized by indirect immunofluorescence microscopy using affinity-purified anti-SMC antibodies. Only a low level of background signal was observed with a Δsmc strain, showing that the signal is specific for SMC. Shown are Nomarski DIC microscopy images of the cells, immunofluorescence microscopy (IFM) images of the cells showing the intracellular localizations of SMC, and images of 4′,6′-diamidino-2-phenylindole (DAPI)-stained chromosomal DNA in the cells. Arrows, typical predivisional cells with bright SMC staining near the poles of the cells. Bar, 2 μm.
Approximately 30 to 40% of the predivisional cells showed large and bright polar SMC foci (Fig. 3), suggesting that SMCs aggregate near the poles at some point during the predivisional cell stage of the cell cycle. Since these polar foci are largely missing from swarmer and stalked cells, the polar SMC aggregates must dissociate upon cell division. The same localization pattern was observed when the cells were grown in a minimal medium (generation time, 150 min) and a rich growth medium (generation time, 70 min) (data not shown).
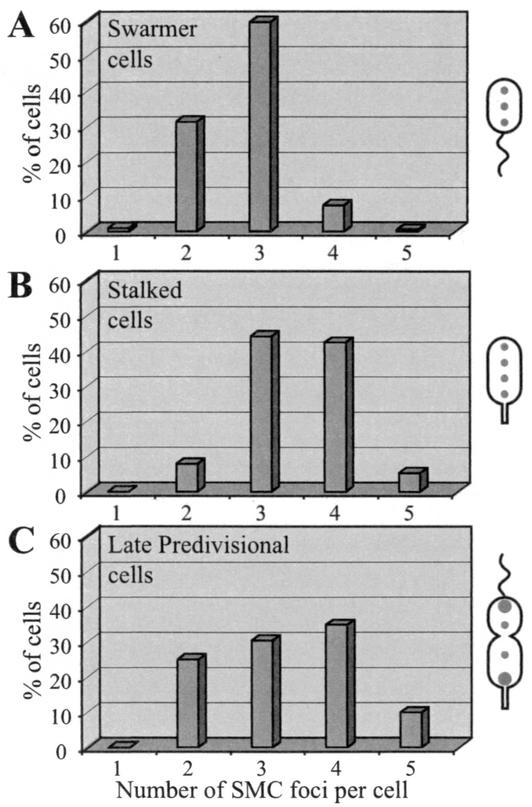
We quantified the number of SMC foci per cell at different stages of the cell cycle (Fig. 4). The number of SMC foci per cell varied, even among cells in the same stage of the cell cycle, with swarmer cells exhibiting predominantly two or three foci per cell, with a transition to three or four foci per cell coincident with the initiation of DNA replication in the stalked cell. The late predivisional cell, containing two fully replicated chromosomes, exhibited two to five SMC foci per cell.
FIG. 4.
Distribution of the numbers of SMC foci per cell in swarmer cells (A), stalked cells (B), and late predivisional cells (C). The numbers of SMC foci, visualized by immunofluorescence microscopy, per cell at each stage of the cell cycle were determined. The schematics to the right show cells at the respective stage of the cell cycle and typical SMC localization patterns. At least 200 cells were counted at each stage.
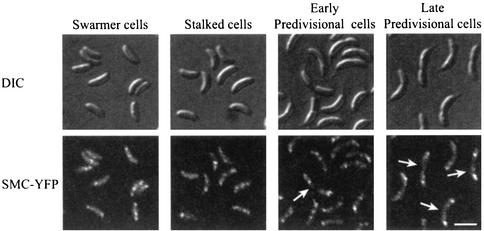
To confirm the intracellular localization pattern of SMC observed by immunofluorescence microscopy, we examined the subcellular localization of SMC in live cells by using fusions to GFP or YFP. A fusion of YFP to the C terminus of SMC was constructed, and the smc-yfp gene fusion replaced the wild type smc allele in single copy in the Caulobacter chromosome under the control of the endogenous smc promoter. The strain expressing SMC-YFP as its only source of SMC protein showed no growth or cell cycle defects, indicating that the fusion protein is fully functional. To examine the intracellular location of SMC during the Caulobacter cell cycle, we synchronized the smc-yfp fusion strain. At different times during the cell cycle, the intracellular location of SMC-YFP was examined by fluorescence microscopy (Fig. 5). There were several SMC-YFP foci per cell, as was observed by immunofluorescence microscopy of fixed cells. Many predivisional cells had large and bright polar SMC-YFP foci and faint foci located at other positions within the cells. Identical subcellular localization was obtained with a fusion of GFP to the N terminus of SMC (data not shown). Thus, the localization pattern observed in live cells with smc-yfp or gfp-smc fusions is identical to the subcellular localization of native SMC, determined by immunofluorescence microscopy.
FIG. 5.
Intracellular localization of SMC in live cells at different stages of the cell cycle. SMC-YFP-expressing cells were synchronized, and samples were taken for microscopy when the cells reached the indicated stages (0, 60, 90, or 120 min into the cell cycle, respectively). DIC microscopy images of the cells (top) and YFP fluorescence (bottom) are shown. Arrows, predivisional cells with bright polar SMC-YFP foci. Scale bar, 2 μm.
DISCUSSION
SMCs are ubiquitously present in eukaryotes, eubacteria, and archaea. Where examined, the SMCs function in organizing chromosomal DNA (5, 15, 51). Caulobacter possesses a single SMC homolog, and deletion of SMC results in irregular nucleoids. Visualization of the intracellular locations of the origin- and terminus-proximal regions of the chromosome shows that they are mislocalized in a subpopulation of the Δsmc cells. Additionally, these regions are less condensed in the Δsmc strain than in the wild-type strain. Thus, deletion of smc results in chromosome organization defects (23). At elevated temperatures, the Δsmc cells, like strains with defects in chromosome replication or segregation, arrest at the predivisional cell stage (6, 40, 57, 60). Thus, Caulobacter SMC plays a role in maintaining proper organization of the chromosomal DNA, and lack of SMC-mediated chromosome organization during rapid growth results in a chromosome segregation defect.
In exponentially growing Caulobacter cells, approximately 1,500 to 2,000 SMC molecules are present per cell, corresponding to approximately one SMC complex per 6,000 to 8,000 bp of chromosomal DNA. This density of SMC complexes is similar to the densities of condensin complexes in eukaryotes and SMC in B. subtilis (30, 52) but is much lower than the density of histone-like proteins in E. coli, which are present at one complex per 200 to 600 bp in exponentially growing cells (2). Thus, there are sufficient SMC molecules in Caulobacter to affect the global structure of the nucleoid but too few to compact the chromosome directly by wrapping DNA around them. Condensation of the DNA by a scissoring action that brings the two SMC head domains from opposite ends of the protein into contact is unlikely to occur in a stoichiometric manner because too few SMC molecules are present but may operate at selected regions of the chromosome. Relevant to these mechanisms is the question of how SMC is dynamically deployed within the cell.
Examination of the intracellular localization of SMC during the Caulobacter cell cycle showed a punctate pattern, with SMC forming two to five small foci distributed throughout the cell (Fig. 3 to 5). We cannot rule out the possibility that, in addition to the SMC molecules that form foci, nonaggregated SMC molecules may be distributed throughout the cell. Many predivisional cells had bright polar SMC foci, indicating that some SMCs aggregate near the poles during a period in the predivisional cell stage of the cell cycle. Since only a subpopulation of the predivisional cells (30 to 40%) had bright polar foci, SMCs may aggregate near the poles only during a brief period of the cell cycle. A similar pattern of SMC localization was observed in B. subtilis, with SMC localized in a punctate pattern as well as forming specific polar foci (16, 33). Similarly, MukB in E. coli forms multiple foci within the nucleoid (7). However, other studies report that SMC and MukB localize as discrete foci in positions similar to that of the replisome (30, 39). The differences in localization may reflect the use of different methods for examining the localization of the proteins or different growth conditions. The Caulobacter SMC localization pattern reported here was observed by immunofluorescence microscopy of fixed cells and smc-yfp or gfp-smc fusions in live cells and was observed in minimal as well as rich growth media. Since immunofluorescence microscopy and live-cell microscopy are very different methods, the consistent SMC localization pattern is most likely an accurate reflection of the localization of SMC within the Caulobacter cell. SMC localization clearly differs from replisome localization in Caulobacter (25).
SMCs possess a DNA binding motif, and DNA binding has been observed with both bacterial and eukaryotic SMC proteins (1, 3, 18, 19, 27, 37, 53). Therefore, the majority of the SMCs are expected to interact with the chromosome. The formation of discrete SMC foci at different positions in the cell most likely represents the association of multiple SMCs. If the SMCs simultaneously bind to multiple sites throughout the chromosome, different chromosomal regions could be brought into close proximity by SMC aggregation. Purified B. subtilis SMC can, in the presence of ATP, aggregate single-stranded DNA (18). It was suggested that aggregation of single-stranded DNA brings unpaired regions of the bacterial chromosome together, thereby compacting it. Alternatively, SMC could bind with high affinity to a small number of chromosomal regions, thereby concentrating SMC in regions of the cell where chromosomal domains with multiple SMC binding sites are located. This case would be similar to that of yeast cohesin, which binds to specific regions of the chromosome and forms multiple discrete foci in spread nuclei (3, 35, 53, 54).
Organization and condensation of the chromosomal DNA by SMC could take place when chromosome replication has been completed, or chromosomal domains could be formed and condensed during the process of DNA replication, shortly after a given region has been replicated. SMC appears to be deployed to more sites in stalked cells than in swarmer cells, since two or three SMC foci generally are observed in swarmer cells and three or four foci are present in stalked cells, even though a constant amount of SMC is present. Since the chromosome is being replicated in stalked cells, this increase in number of SMC foci may mean that SMC organizes and condenses the chromosome continuously during DNA replication, not only after completion of DNA replication. This is supported by the analysis of the smc cell cycle transcription pattern. The orfA and smc promoter is activated during early S phase, a cell cycle transcription pattern similar to that previously observed with several DNA replication and repair genes (26, 28, 42, 43, 59). It is clearly different from the cell cycle transcription pattern of known partitioning genes, which are expressed much later in the cell cycle (28).
In the predivisional cell stage of the cell cycle, many cells showed bright polar SMC foci and only faint foci in other parts of the cell. In those cells, the two new daughter chromosomes are separated from each other and the nucleoids are further condensed to form a DNA-free region at the place where septation takes place. If SMCs remain bound to sites throughout the chromosome while aggregating near the poles, they could have a role in the further condensation of the chromosomes that takes place at this time in the cell cycle. Thus, SMC may function both during DNA replication, by refolding and condensing newly replicated DNA, and after completion of DNA replication, by condensing the entire nucleoid.
Acknowledgments
We thank Ann Reisenauer for testing the role of methylation in regulating promoter activity.
R. B. Jensen was supported by postdoctoral fellowships from EMBO and the Carlsberg foundation. This work was supported by National Institutes of Health grants GM32506/5120MZ and GM51426 and Office of Naval Research grant N00014-96-1-0564.
REFERENCES
- 1.Akhmedov, A. T., C. Frei, M. Tsai-Pflugfelder, B. Kemper, S. M. Gasser, and R. Jessberger. 1998. Structural maintenance of chromosomes: protein C-terminal domains bind preferentially to DNA with secondary structure. J. Biol. Chem. 273:24088-24094. [DOI] [PubMed] [Google Scholar]
- 2.Azam, T. A., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blat, Y., and N. Kleckner. 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98:249-259. [DOI] [PubMed] [Google Scholar]
- 4.Britton, R. A., D. C. Lin, and A. D. Grossman. 1998. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 12:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cobbe, N., and M. M. Heck. 2000. Review: SMCs in the world of chromosome biology—from prokaryotes to higher eukaryotes. J. Struct. Biol. 129:123-143. [DOI] [PubMed] [Google Scholar]
- 6.Degnen, S. T., and A. Newton. 1972. Dependence of cell division on the completion of chromosome replication in Caulobacter. J. Bacteriol. 110:852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.den Blaauwen, T., A. Lindqvist, J. Lowe, and N. Nanninga. 2001. Distribution of the Escherichia coli structural maintenance of chromosomes (SMC)-like protein MukB in the cell. Mol. Microbiol. 42:1179-1188. [DOI] [PubMed] [Google Scholar]
- 8.Domian, I. J., K. C. Quon, and L. Shapiro. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415-424. [DOI] [PubMed] [Google Scholar]
- 9.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 10.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gober, J. W., and J. C. England. 2000. Regulation of flagellum biosynthesis and motility in Caulobacter, p. 319-339. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 12.Gober, J. W., and L. Shapiro. 1992. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol. Biol. Cell 3:913-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon, G. S., and A. Wright. 2000. DNA segregation in bacteria. Annu. Rev. Microbiol. 54:681-708. [DOI] [PubMed] [Google Scholar]
- 14.Graumann, P. L. 2000. Bacillus subtilis SMC is required for proper arrangement of the chromosome and for efficient segregation of replication termini but not for bipolar movement of newly duplicated origin regions. J. Bacteriol. 182:6463-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graumann, P. L. 2001. SMC proteins in bacteria: condensation motors for chromosome segregation? Biochimie 83:53-59. [DOI] [PubMed] [Google Scholar]
- 16.Graumann, P. L., R. Losick, and A. V. Strunnikov. 1998. Subcellular localization of Bacillus subtilis SMC, a protein involved in chromosome condensation and segregation. J. Bacteriol. 180:5749-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haering, C. H., J. Lowe, A. Hochwagen, and K. Nasmyth. 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9:773-788. [DOI] [PubMed] [Google Scholar]
- 18.Hirano, M., and T. Hirano. 1998. ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J. 17:7139-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirano, M., and T. Hirano. 2002. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 21:5733-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes, V. F., and N. R. Cozzarelli. 2000. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc. Natl. Acad. Sci. USA 97:1322-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopfner, K. P., A. Karcher, D. S. Shin, L. Craig, L. M. Arthur, J. P. Carney, and J. A. Tainer. 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101:789-800. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs, C., I. J. Domian, J. R. Maddock, and L. Shapiro. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97:111-120. [DOI] [PubMed] [Google Scholar]
- 23.Jensen, R. B., and L. Shapiro. 1999. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl. Acad. Sci. USA 96:10661-10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen, R. B., and L. Shapiro. 1999. Chromosome segregation during the prokaryotic cell division cycle. Curr. Opin. Cell Biol. 11:726-731. [DOI] [PubMed] [Google Scholar]
- 25.Jensen, R. B., S. C. Wang, and L. Shapiro. 2001. A moving DNA replication factory in Caulobacter crescentus. EMBO J. 20:4952-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keiler, K. C., and L. Shapiro. 2001. Conserved promoter motif is required for cell cycle timing of dnaX transcription in Caulobacter. J. Bacteriol. 183:4860-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura, K., and T. Hirano. 1997. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90:625-634. [DOI] [PubMed] [Google Scholar]
- 28.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 29.Lindow, J. C., R. A. Britton, and A. D. Grossman. 2002. Structural maintenance of chromosomes protein of Bacillus subtilis affects supercoiling in vivo. J. Bacteriol. 184:5317-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindow, J. C., M. Kuwano, S. Moriya, and A. D. Grossman. 2002. Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein. Mol. Microbiol. 46:997-1009. [DOI] [PubMed] [Google Scholar]
- 31.Lowe, J., S. C. Cordell, and F. van den Ent. 2001. Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled-coil inserted. J. Mol. Biol. 306:25-35. [DOI] [PubMed] [Google Scholar]
- 32.Malakooti, J., S. P. Wang, and B. Ely. 1995. A consensus promoter sequence for Caulobacter crescentus genes involved in biosynthetic and housekeeping functions. J. Bacteriol. 177:4372-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mascarenhas, J., J. Soppa, A. V. Strunnikov, and P. L. Graumann. 2002. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 21:3108-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melby, T. E., C. N. Ciampaglio, G. Briscoe, and H. P. Erickson. 1998. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 142:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 36.Moriya, S., E. Tsujikawa, A. K. Hassan, K. Asai, T. Kodama, and N. Ogasawara. 1998. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol. 29:179-187. [DOI] [PubMed] [Google Scholar]
- 37.Niki, H., R. Imamura, M. Kitaoka, K. Yamanaka, T. Ogura, and S. Hiraga. 1992. E. coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J. 11:5101-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niki, H., A. Jaffe, R. Imamura, T. Ogura, and S. Hiraga. 1991. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10:183-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohsumi, K., M. Yamazoe, and S. Hiraga. 2001. Different localization of SeqA-bound nascent DNA clusters and MukF-MukE-MukB complex in Escherichia coli cells. Mol. Microbiol. 40:835-845. [DOI] [PubMed] [Google Scholar]
- 40.Osley, M. A., and A. Newton. 1980. Temporal control of the cell cycle in Caulobacter crescentus: roles of DNA chain elongation and completion. J. Mol. Biol. 138:109-128. [DOI] [PubMed] [Google Scholar]
- 41.Reisenauer, A., and L. Shapiro. 2002. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 21:4969-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizzo, M. F., L. Shapiro, and J. Gober. 1993. Asymmetric expression of the gyrase B gene from the replication-competent chromosome in the Caulobacter crescentus predivisional cell. J. Bacteriol. 175:6970-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts, R. C., and L. Shapiro. 1997. Transcription of genes encoding DNA replication proteins is coincident with cell cycle control of DNA replication in Caulobacter crescentus. J. Bacteriol. 179:2319-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts, R. C., C. Toochinda, M. Avedissian, R. L. Baldini, S. L. Gomes, and L. Shapiro. 1996. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J. Bacteriol. 178:1829-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salamitou, S., M. Lemaire, T. Fujino, H. Ohayon, P. Gounon, P. Beguin, and J. P. Aubert. 1994. Subcellular localization of Clostridium thermocellum ORF3p, a protein carrying a receptor for the docking sequence borne by the catalytic components of the cellulosome. J. Bacteriol. 176:2828-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawitzke, J. A., and S. Austin. 2000. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc. Natl. Acad. Sci. USA 97:1671-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shapiro, L., and R. Losick. 2000. Dynamic spatial regulation in the bacterial cell. Cell 100:89-98. [DOI] [PubMed] [Google Scholar]
- 48.Soppa, J., K. Kobayashi, M. F. Noirot-Gros, D. Oesterhelt, S. D. Ehrlich, E. Dervyn, N. Ogasawara, and S. Moriya. 2002. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 45:59-71. [DOI] [PubMed] [Google Scholar]
- 49.Stephens, C., A. Reisenauer, R. Wright, and L. Shapiro. 1996. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc. Natl. Acad. Sci. USA 93:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephens, C. M., G. Zweiger, and L. Shapiro. 1995. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J. Bacteriol. 177:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strunnikov, A. V., and R. Jessberger. 1999. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur. J. Biochem. 263:6-13. [DOI] [PubMed] [Google Scholar]
- 52.Sutani, T., and M. Yanagida. 1997. DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature 388:798-801. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka, T., M. P. Cosma, K. Wirth, and K. Nasmyth. 1999. Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98:847-858. [DOI] [PubMed] [Google Scholar]
- 54.Toth, A., R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, and K. Nasmyth. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van den Ent, F., A. Lockhart, J. Kendrick-Jones, and J. Lowe. 1999. Crystal structure of the N-terminal domain of MukB: a protein involved in chromosome partitioning. Struct. Fold Design 7:1181-1187. [DOI] [PubMed] [Google Scholar]
- 56.Vologodskii, A. V., and N. R. Cozzarelli. 1994. Conformational and thermodynamic properties of supercoiled DNA. Annu. Rev. Biophys. Biomol. Struct. 23:609-643. [DOI] [PubMed] [Google Scholar]
- 57.Ward, D., and A. Newton. 1997. Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol. Microbiol. 26:897-910. [DOI] [PubMed] [Google Scholar]
- 58.Weitao, T., S. Dasgupta, and K. Nordstrom. 2000. Role of the mukB gene in chromosome and plasmid partition in Escherichia coli. Mol. Microbiol. 38:392-400. [DOI] [PubMed] [Google Scholar]
- 59.Winzeler, E., and L. Shapiro. 1996. A novel promoter motif for Caulobacter cell cycle-controlled DNA replication genes. J. Mol. Biol. 264:412-425. [DOI] [PubMed] [Google Scholar]
- 60.Wortinger, M., M. J. Sackett, and Y. V. Brun. 2000. CtrA mediates a DNA replication checkpoint that prevents cell division in Caulobacter crescentus. EMBO J. 19:4503-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zweiger, G., G. Marczynski, and L. Shapiro. 1994. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J. Mol. Biol. 235:472-485. [DOI] [PubMed] [Google Scholar]