Abstract
RNA helicase A (RHA) is a multifunctional protein involved in various nuclear processes such as transcription and RNA export. It is believed that the interacting factors play important roles in determining the functional specificity of RHA. Here we show that RHA directly interacts with double-stranded (ds) nucleic acids (NAs) and assembles complexes with topoisomerase IIα. First, electrophoresis mobility shift assays demonstrate that RHA interacts with dsDNAs of different lengths ranging from 15 to 104 bp. Secondly, the binding of RHA to closed circular dsDNA stimulates the relaxation reaction catalyzed by either calf thymus topoisomerase I or HeLa topoisomerase IIα. Thirdly, immunoprecipitation, coupled with western blot analysis using anti-RHA and anti-topoisomerase IIα antibodies, shows that RHA and topoisomerase IIα assemble a complex in the presence of as yet unknown RNA molecules and additional protein factors such as Ubc9. Our observation suggests physical and functional interaction between RHA and topoisomerase IIα, which, perhaps, play important roles in regulating chromatin structure. The putative role of RHA–topoisomerase IIα complex in RNA polymerase II-mediated transcription is discussed.
INTRODUCTION
RNA helicase A (RHA) belongs to the asp-glu-ala-his (DEAH) helicase family (1) and unwinds both double-stranded (ds) DNA and dsRNA (2–5). It has been shown previously that RHA plays a critical role in mammalian development (6) and that RHA mediates the interaction between CBP/p300 and RNA polymerase II (7). RHA interacts with various nuclear factors, including the tumor suppressor BRCA1 (8), HAP95, a newly identified protein with extensive homology to AKAP95, a member of the A-kinase anchoring protein family (9), and the survival motor neuron (SMN) complex (10). These findings suggest that, in addition to transcription, RHA participates in diverse nuclear processes and that the functional specificity is not inherent but a property endowed by its interacting factors. In particular, the latter possibility can be inferred from MLE (maleless), a Drosophila ortholog of RHA that is critical for dosage compensation (11–13). Although present in both female and male flies, MLE increases the transcription activity of X-linked genes only in male flies through the interaction with male-specific lethal (MSL) complexes and regulatory RNA molecules such as roX1 and roX2 (RNA on X 1 and 2) (14,15).
Recent studies, employing RHA and MLE carrying mutations in the ATP binding motif (GKT), have shown that ATPase activity is essential for both CREB-dependent pathway in mammals and dosage compensation in Drosophila (5,7). Loss of MLE ATPase activity decreases roX2 RNA levels both in the nucleoplasm and in MSL complexes (16), implying that ATPase activity is required either for the association of roX2 RNA with MSL complexes, for the stability of roX2 RNA, or both. Similar to the functional interaction of MLE with roX2 RNA, RHA is also regulated by RNA molecules such as constitutive transport element (CTE)-containing RNAs (17) or TAR encoded by type D retroviruses (18). Adenoviral VA RNAs suppress the helicase activity of RHA (19), but the functional significance of the interaction remains unknown.
Although mammalian homologs of Drosophila roX RNAs or virally encoded RNAs have yet to be found, the above observations show that RNA molecules can regulate the function of RHA. In addition to the functional interaction of RNA molecules with RHA and MLE, the importance of RNA molecules in the protein–protein interaction has been shown for topoisomerase IIα, which associates with as yet unidentified complexes in an RNase-sensitive but DNase I-resistant manner (20). This observation raises an intriguing possibility that, like MLE and MSL complexes, topoisomerase IIα forms multiprotein complexes containing RNA molecules. However, the identity of RNA molecules interacting with topoisomerase IIα and whether RNA binding activity is an intrinsic activity of topoisomerase IIα remain undetermined.
In this report, we show that RHA and topoisomerase IIα form complexes containing RNA molecules in the presence of Ubc9, a functional homolog of E2-type ubiquitin-conjugating enzyme (21,22). Like several non-coding (nc) RNA molecules with regulatory function, including XIST/Xist (23,24), SRA (25) and Air RNA (26), there can exist specific RNA molecules that play a role in the RHA–topoisomerase IIα interaction. Using purified RHA and topoisomerase IIα, we also show that the binding of RHA to dsDNA augments the relaxation activity of topoisomerase IIα, whereas topoisomerase IIα suppresses the helicase activity of RHA. Our observation suggests that RHA–topoisomerase IIα complexes might regulate both the superhelicity and helicity of DNA, potentially influencing the stability and transcription activity of chromatin.
MATERIALS AND METHODS
Proteins and plasmid DNAs
Recombinant RHA and MLE used in the present study were essentially the same as described previously (1,5). Calf thymus topoisomerase I was commercially obtained (Invitrogen). Topoisomerase IIα was purified as follows. In brief, nuclear extracts (2.3 g) prepared from HeLa cells (60 l), as described previously (2), were subjected to ammonium sulfate (AS) fractionation. AS 35–65% fraction was loaded onto DEAE Sepharose (Pharmacia), followed by heparin Sepharose (Pharmacia), P11 (Whatman), brown agarose (Sigma) and green agarose (Sigma). The final fraction (850 µg) was enriched in topoisomerase IIα, which constituted >90% of the protein detected on SDS–PAGE. The detailed purification procedure is available upon request. Supercoiled pBluescript IIKS– (pBS) was prepared using the Qiagen plasmid kit.
Electrophoresis mobility shift assay (EMSA)
The polylinker region (167 bp) of pBS was amplified by PCR using T7 and T3 primers. The PCR product was digested with either ClaI, EcoRI/NotI or EcoRI/XbaI to prepare a mixture of 32P-labeled DNA probes of (65 + 104 bp), (41 + 84 bp) or (34 + 57 bp), respectively. To prepare 10 and 15 bp DNA probes, partial dsDNAs were formed using two sets of synthetic primers, 5′-CGACGGTATC-3′/5′-GATACCG-3′ and 5′-CGACGGTATCGATAA-3′/5′-TTATCGATACG-3′. DNA fragments were end-labeled by filling-in reactions containing Klenow fragment and either [α-32P]dCTP or [α-32P]dATP, and subsequently purified using an 8% polyacrylamide (30:1) gel as described previously (2). However, 10 and 15 bp dsDNAs were purified by gel-filtration chromatography on G-50 column (5 ml) using a buffer containing 50 mM Tris–HCl, 0.1 mM ethylenediamine tetra-acetate (EDTA), 150 mM NaCl and 0.05% NP-40, followed by ethanol precipitation. A standard EMSA reaction (20 µl) contained 20 mM HEPES–NaOH, pH 7.4, 2 mM dithiothreitol (DTT), 3 mM MgCl2, 0.05% NP-40, 12 nM 32P-labeled DNA, and varying concentrations (10–40 nM) of RHA. After incubation for 30 min at 37°C, reaction mixtures were transferred on ice and mixed with 5 µl of gel-loading solution consisting of 0.1 M Tris–HCl, pH 7.4, 0.1% bromophenol blue, 0.1% xylene cyanol, 0.1% NP-40 and 50% glycerol. An aliquot (10 µl) was analyzed for the formation of RHA–DNA complexes on an 8% (30:1) polyacrylamide gel. Following autoradiography, RHA–DNA complexes were quantified using a Phosphor Imager (Molecular Dynamics).
Topoisomerase-mediated relaxation assays
Reaction mixtures (20 µl) contained 0.25–0.5 µg of pBS, 50 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 0.5 mM DTT, 0.5 mM ATP, 0.2 mg/ml bovine serum albumin (BSA) and indicated amounts of topoisomerase I or topoisomerase IIα. After incubation for 15 min at 37°C, 5 µl of termination buffer (0.1 M Tris–HCl, pH 7.4, 50 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol, 0.1% NP-40, 0.5% SDS, 1 mg/ml proteinase K) was added to each reaction. After incubation for an additional 15 min at 37°C, reaction mixtures were analyzed on a 1% agarose gel in 1× Tris–acetate/EDTA (TAE) buffer, pH 8.0. DNAs of different linking number were visualized by staining with ethidium bromide (0.5 µg/ml) for 20 min and quantified using a Gel Documentation System (Bio-Rad).
Helicase assay
Standard reaction was carried out for 30 min at 37°C in a mixture (20 µl) containing 20 mM HEPES–NaOH, pH 7.4, 2 mM DTT, 3 mM MgCl2, 1 mM ATP, 0.1 mg/ml BSA, 2 U RNasin, 50 fmol dsRNA or dsDNA substrate (2), and 10 ng RHA or MLE. Reactions were analyzed as described previously (2).
Construction of mammalian expression vector for the flag-tagged RHA
The N‐terminal 500 bp of RHA cDNA in pBSIISK– (1) was subcloned into pCRII (Invitrogen) subsequent to the PCR using two primers, 5′-CCCGGGTACCATGGGTGACGTT AAAAATTTTC-3′ and 5′-CTGATTCTAGAGTCGCTTGC ACTTC-3′. The EcoRI–ApaI fragment of RHA cDNA was recovered from pCRII vector and used to replace the corresponding region of a full-length RHA cDNA in pBSIISK– (1). A synthetic linker, containing coding sequences for both flag epitope and hexahistidine (underlined sequences), was made from two primers, 5′-CCGGGCTAGC ACCATGGATTACAAG(GAT)4AAG(CAC)6-3′ and 5′-CC G(GTG)6CTT(ATC)4CTTGTAATCCATGGTGCTAGC-3′, and inserted into the SmaI site upstream of ATG, the translation start codon of RHA gene. A full-length RHA cDNA, containing the synthetic linker at its 5′ end, was then subcloned into the EcoRI site of pCDNA3 (Invitrogen). The finally obtained expression vector was named pCDNA3-fhis-RHA.
A PCR-based site-directed mutagenesis was performed to introduce a substitution mutation in the ATP binding motif of RHA (GKT to GET) according to the procedure recommended by the manufacturer (Stratagene). In brief, using pCDNA3-fhis-RHA as template, a PCR was carried out for 12 cycles with a forward primer (nucleotide position 1318–1343 of RHA cDNA) (1), 5′-CTGGATGTGGGGAAACCACAC AGGTT-3′, and its reverse complementary primer. Positive clone carrying the GET mutation was confirmed by direct nucleotide sequencing in the Molecular Resources Core Facility at UMDNJ-New Jersey Medical School.
Preparation of cytoplasmic extract (CE) and nuclear extract (N450)
293T cells, grown in 90-mm culture plates at ∼20% confluence, were transfected with 4 µg of pCDNA3-fhis-RHA alone or in combination with 4 µg of CMV-Ubc9-V5 (provided by R. Hay) using the calcium phosphate precipitation method. After a 24 h incubation, cells were supplied with fresh DME media containing 10% fetal bovine serum and incubated for an additional 24 h. Cells, grown in five culture plates, were collected by centrifugation at 1000 g for 10 min and washed once with ice-cold 1× phosphate buffered saline (PBS). Cells were then resuspended in 1 ml of lysis buffer consisting of 10 mM potassium phosphate buffer, pH 7.4, 0.5% NP-40, 1% deoxycholate, 2 mM DTT and 5% glycerol. Cell slurries were cleared by centrifugation at 15 000 r.p.m. at 4°C, and the supernatants containing CE were directly stored at –70°C. To the remaining nuclear pellet, 0.45 ml of lysis buffer and 50 µl of 5 M NaCl were sequentially added. After a 30 min incubation on ice, the mixture was sonicated for 30 s and centrifuged at 20 000 r.p.m. for 30 min at 4°C. Cleared supernatant, N450, was stored at –70°C until use.
Immunoprecipitation (IP) and western blot analyses
One to 5 mg of HeLa nuclear extract (N450) was incubated with 20–50 µg of the indicated antibody for 2 h at 4°C. Subsequently, protein A–Sepharose (10–20 µl) was added to the above mixture and incubated for an additional 1 h. After centrifugation at 800 g for 2 min, the IP pellet was washed four times with 1× PBS containing 0.5% Tween-20 and 0.5% Triton X-100, and resuspended in 1× SDS sample buffer to a final volume of 60–100 µl. After boiling for 5 min, an aliquot (25%) was separated by 7.5% SDS–PAGE and proteins were visualized by Coomassie staining. Among the proteins stained, a 170 kDa protein was subjected to mass spectrometry analysis. Other aliquots (25%), resolved on a 7.5% SDS– polyacrylamide gel, were transferred to a nylon filter (Schleicher and Schuell) using the SemiDry system (Bio-Rad). Western blot analysis was performed using rabbit anti-RHA antibody, rabbit anti-Topo II antibody (Topogen) or mouse anti-V5 antibody (Invitrogen), as described previously (1).
RESULTS
RHA interacts with dsNAs
RHA is the only known cellular helicase that catalyzes the unwinding of dsRNA, dsDNA and RNA:DNA hybrid (2,3). Most helicases possess two functional domains, one for interaction with single-stranded nucleic acid (ssNA) and the other with double-stranded nucleic acid (dsNA) (27–30). Although RHA possesses two dsRNA binding motifs at the N-terminus (31), it has yet to be experimentally proven if RHA interacts directly with dsNA. Therefore, we determined whether RHA binds to dsNA of either linear or circular form. Consistent with its ability to unwind both dsRNA and dsDNA containing a ssNA region, RHA interacted stably with both dsRNA and dsDNA lacking a ssNA region, forming two discernible complexes (Fig. 1A). This result provides evidence that RHA directly interacts with dsNA.
Figure 1.
Binding of RHA to dsDNA and dsRNA. (A) Increasing concentrations of RHA (5, 10 and 20 nM) were incubated for 30 min at 37°C in reactions containing 36 nM of dsRNA (lanes 1–4) or dsDNA substrate of 34 bp (lanes 5–8) and subsequently analyzed by electrophoresis on an 8% polyacrylamide (30:1) gel. (B and C) Various dsDNAs (12.5 nM) of indicated length were analyzed for their association with varying concentrations (5, 10 and 20 nM) of RHA, as described in (A). dsNAs complexed with RHA were quantitated using a PhosphorImager.
We also explored whether the length of dsNA quantitatively influences the interaction with RHA. As shown in Figure 1B and C, RHA associated with dsNAs of different lengths (39–104 bp) with varying affinities. With dsDNA of 34 and 104 bp, Kds were estimated to be ∼5–18 nM. RHA formed a complex with a 15 bp dsDNA in a concentration-dependent manner but with almost 50-fold lower affinity. In contrast, a 10 bp dsDNA barely interacted with RHA. These results suggest that the longer the duplex region is, the more efficiently RHA interacts with it. Our observation also implies that the minimal length of dsDNA required for the RHA binding is ∼15 bp. Multiple complexes formed with 34 bp (Fig. 1A) or longer DNAs (Fig. 1B), therefore, might be due to the association of more than one RHA molecule.
RHA stimulates the relaxation of supercoiled DNAs catalyzed by topoisomerases
It has been shown previously that the SWI/SNF complex binds and imposes a structural constraint on supercoiled DNAs. In doing so, the SWI/SNF complex assists topoisomerase I in inducing a positive supercoiling in an ATP-independent fashion (32–34). We sought to address whether RHA similarly influences the topology of bound dsDNA and consequentially affects the reaction catalyzed by topoisomerases. We first examined whether superhelicity influences the binding of RHA to dsDNA. As shown in Figure 2A, varying amounts (0.25–1 µg) of RHA were incubated with 0.2 µg of either linear or supercoiled pBS, and the reaction mixtures analyzed on a native agarose gel. Both circular (lanes 3–5) and linear forms (lanes 6–8) of pBS exhibited retarded mobility with the increment of RHA. The smeared migration pattern of pBS complexed with RHA appears to be due to the association of varying numbers of RHA molecules (see below). Treatment of reaction mixtures with SDS and proteinase K abrogates the generation of RHA–pBS complexes (data not shown). These results suggest that superhelicity does not significantly influence the interaction of RHA with dsDNA and that RHA does not alter the topology of bound pBS.
Figure 2.
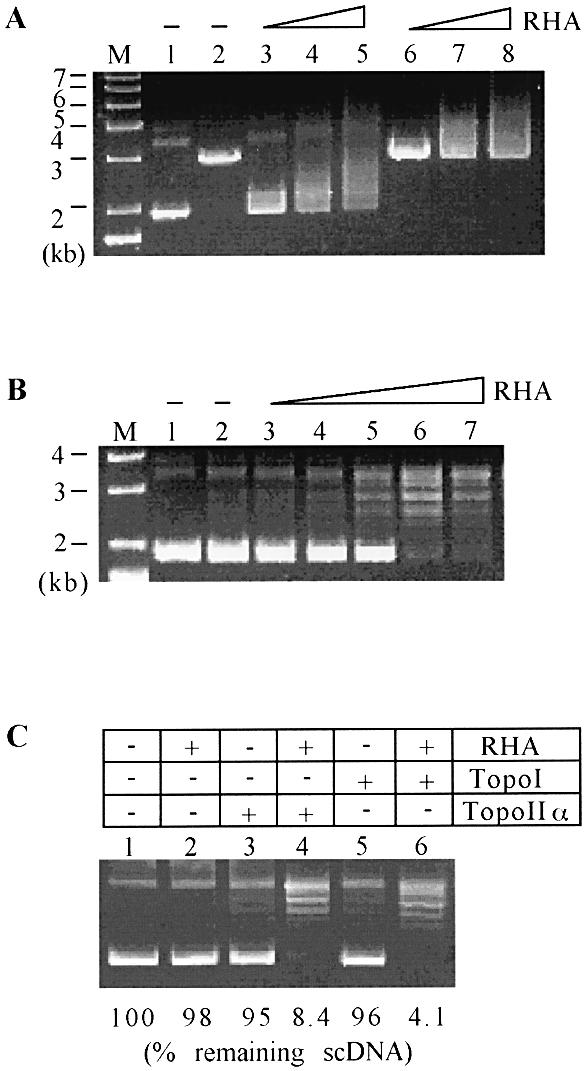
Binding of RHA to circular DNA. (A) Increasing amounts (0.25, 0.5 and 1 µg) of RHA were incubated for 30 min at 37°C in reactions (20 µl) containing 0.25 µg of either linear (lanes 2 and 6–8) or supercoiled circular pBS (lanes 1 and 3–5). Subsequently, reaction mixtures were resolved on a 0.7% agarose gel in 1× TAE, and DNA was visualized by ethidium bromide staining. (B) Increasing amounts (0.125, 0.25, 0.5, 1 and 2 µg) of RHA were added in reactions containing 0.25 µg of supercoiled pBS and 8 ng of topoisomerase IIα. After a 30 min incubation at 37°C, topoisomers were resolved on a 0.7% agarose gel. (C) RHA (1 µg) and topoisomerase IIα (8 ng) or calf thymus topoisomerase I (1 U) were added in the indicated reaction containing 0.25 µg of supercoiled pBS. After a 30 min incubation at 37°C, topoisomers were analyzed as described in (B) and quantified using the QuantityOne software. scDNA, supercoiled DNA.
Next, we addressed whether the binding of RHA to supercoiled DNA affects the relaxation reaction mediated by topoisomerases. Similar to SWI/SNF complex, RHA stimulated the relaxation reaction mediated by either calf thymus topoisomerase I (Promega) or HeLa topoisomerase IIα Increasing amounts (0.125–2.0 µg) of RHA progressively activated the relaxation of 3 kb plasmid DNA (pBS) catalyzed by topoisomerase IIα (Fig. 2B). It was estimated that in a reaction containing 1 µg of RHA and 0.25 µg of supercoiled pBS, RHA could be present at every 50–60 bp. Under this condition, the relaxation reaction by topoisomerase I or IIα was stimulated ∼10–20-fold, determined by the percent remaining supercoiled DNAs (Fig. 2C). Similar results were obtained with Escherichia coli topoisomerase I (data not shown). These results suggest that the relaxation reaction is stimulated not by the specific interaction between RHA and topoisomerases but rather by the presence of RHA itself on dsDNA. Similar to SWI/SNF, the stimulatory effect of RHA on the relaxation reaction might be due to its association with dsDNA. Unlike SWI/SNF complex, RHA did not lead to the formation of positively supercoiled DNA in the presence of either topoisomerase I or II (data not shown). Thus, there seems to be a mechanistic difference between RHA and SWI/SNF in activating topoisomerases, which might, in part, be attributed to the ability of RHA to form complexes with topoisomerase IIα (see below).
Topoisomerase IIα suppresses the helicase activity of RHA
The above results, together with the observation of RHA–topoisomerase IIα complexes (see below), prompted us to determine whether the catalytic function of RHA is influenced by topoisomerase IIα. For this purpose, unwinding reactions catalyzed by RHA were performed in the presence or absence of topoisomerase IIα. In contrast to the stimulatory effect of RHA on the relaxation reaction, topoisomerase IIα inhibited the unwinding of dsDNA catalyzed by either RHA or MLE (Fig. 3). Though less severe, topoisomerase IIα also interfered with the unwinding of dsRNA (Fig. 3B, closed symbols). The result that neither BSA nor HeLa SSB (single-stranded DNA binding protein) significantly influenced the unwinding of dsDNA or dsRNA by RHA (data not shown) suggests that the inhibitory effect of topoisomerase IIα on the helicase activity of RHA and MLE is not non-specific. In addition, it is important to note that the inhibitory effect of toposiomerase IIα on the helicase activity of RHA is functionally relevant to the activated relaxation of supercoiled DNAs, since both lead to the stabilization of duplex nature of NAs. The possible joint function of RHA and topoisomerase IIα during transcription is discussed below.
Figure 3.

Topoisomerase IIα suppresses the unwinding reaction mediated by RHA. (A) Indicated amounts of topoisomerase IIα were added in reactions containing 50 fmol of dsDNA and 10 ng of either MLE or RHA. After incubation for 30 min at 37°C, ssDNA was resolved on an 8% polyacrylamide (30:1) gel and visualized by autoradiography. (B) Unwinding reactions were carried with the indicated amount of HeLa topoisomerase IIα, dsRNA (closed symbols) or dsDNA (open symbols), and 10 ng of MLE (squares) or RHA (triangles). Reaction products, ssRNA or ssDNA were resolved on an 8% polyacrylamide (30:1) gel and quantified using a PhosphorImager.
RHA and topoisomerase IIα assemble a complex in vivo
In order to gain insights into the in vivo function of RHA, we identified RHA-interacting factors by performing a series of IP experiments using anti-RHA antibody and HeLa nuclear extract. The IP pellets were subjected to 7.5% SDS–PAGE. Coomassie staining revealed several proteins of 300, 170, 100 and 80 kDa (Fig. 4A, lane 2). Considering the stimulatory effect of RHA on the relaxation activity of topoisomerase IIα (Fig. 2), the presence of a 170 kDa protein among those co-immunoprecipitated with RHA was of great interest since the apparent molecular weight of topoisomerase IIα in human is 170 kDa. Subsequent mass spectrometry analysis identified that the 170 kDa protein is indeed topoisomerase IIα (data not shown).
Figure 4.

In vivo association of RHA with topoisomerase IIα. (A) Five milligrams of HeLa nuclear extract was incubated with 50 µg of preimmune (lane 1) or immune antibody specific for RHA (lane 2). Subsequently, proteins associated with antibodies were isolated using protein A–Sepharose and analyzed by 7.5% SDS–PAGE. Proteins were visualized by Coomassie staining, and the 170 kDa protein was subjected to the mass spectrometry analysis. ND, not determined. (B) IP was performed with 1 mg of HeLa nuclear extract and 20 µg of indicated antibody. The IP pellets obtained were incubated at room temperature for 30 min in the presence or absence of RNase A (0.1 mg/ml). Ten micrograms of nuclear extracts (lane 1) or 25% aliquots of the indicated IP pellet (lanes 2–6) were subjected to 7.5% SDS–PAGE, followed by western blot analysis using either anti-RHA or anti-topoisomerase IIα antibody. IB, immunoblotting.
To confirm the above result, IP was repeated using anti-RHA and anti-topoisomerase IIα antibodies, and the resulting pellets were analyzed by immunoblotting. As shown in Figure 4B, both topoisomerase IIα and RHA were present in the IP pellets obtained with anti-RHA (lane 3) and anti-topoisomerase IIα antibodies (lane 4), respectively. Densitometry analysis of western signals detected in the IP pellet and nuclear extract suggests that ∼15% of nuclear RHA and topoisomerase IIα is found in the IP pellets. However, it is presently unknown whether RHA and topoisomerase IIα are in direct physical contact. The identification of remaining proteins co-precipitated with RHA (NDs in Fig. 5A) might help address this issue.
Figure 5.
Influence of Ubc9 and ATPase activity on the interaction between RHA and topoisomerase IIα. (A) 293T cells were transfected with various vectors expressing the indicated proteins. Forty-eight hours later, cytoplasmic extracts (CE) and nuclear extract (N450) were prepared as described in Materials and Methods. Identity of ectopically expressed RHA or Ubc9 was confirmed by western blot analysis using aliquots of CE (20 µg) or N450 (10 µg) and anti-flag antibody (M2, Sigma) specific for flag-tagged RHA or anti-V5 antibody for V5-tagged Ubc9. The nylon filter used to detect flag-RHA, was treated with 0.2 M NaOH and reused for western blotting against anti-topoisomerase IIα. In addition to those obtained with anti-topoisomerase IIα antibody, incompletely removed anti-flag antibody also yielded a western signal for flag-tagged RHA. (B) Aliquots (0.5 mg) of the indicated N450 preparation were subjected to IP using 20 µg of anti-flag antibody, and the resulting IP pellet analyzed for the presence of topoisomerase IIα by western blotting using anti-toposomerase IIα antibody. (C) 293T cells were transfected with CMV-Ubc9-V5 in combination with a vector expressing wild-type RHA or a mutant RHA, carrying a substitution mutation (GKT to GET) in the ATP binding motif. As described in (B), nuclear extract (N450) was prepared and aliquot (0.5 mg) was subjected to IP. In (B) and (C), western blot analysis was performed with a 2 or 50% aliquot of the IP supernatant (SPNT) and the IP pellets (PPT), respectively. αFlag, anti-flag antibody; αTopoIIα, anti-topoisomerase IIα antibody; αV5, anti-V5 antibody; IB, immunoblotting.
Interestingly, treatment of the IP pellets with RNase A resulted in the dissociation of RHA or topoisomerase IIα from the IP pellet containing topoisomerase IIα or RHA complex, respectively (Fig. 4B, lanes 5 and 6). However, prolonged incubation of the IP pellets in the presence of ATP or DNase I did not disturb the interaction between RHA and topoisomerase IIα (data not shown). These results suggest that yet unknown but specific RNA molecules are involved in the assembly of the RHA–topoisomerase IIα complex.
Ubc9 induces the association of RHA with topoisomerase IIα
That only a fraction of nuclear RHA and topoisomerase IIα exists in a complex may indicate their instability. However, this is unlikely, since prolonged incubation (1 h at 37°C) of the IP pellet obtained with anti-RHA or anti-topoisomerase IIα antibody did not lead to the dissociation of topoisomerase IIα or RHA (see above). Alternatively, RHA–topoisomerase IIα complex may be formed in a cell cycle-dependent manner. It is also possible that additional factors involved in the formation of RHA–topoisomerase IIα complex may be present in limiting concentrations. We explored the above possibilities by determining whether ectopically expressed RHA (flag-tagged RHA) associates with endogenous topoisomerase IIα. If additional but limiting factors are required for the formation of RHA–topoisomerase IIα complex, it can be predicted that unless they are co-expressed, little or no complex will be formed between flag-tagged RHA and endogenous topoisomerase IIα. Otherwise, flag-tagged RHA is expected to associate with topoisomerase IIα to a certain degree. For this purpose, 293T cells were transfected with a vector expressing flag-tagged RHA and, after 48 h, CEs were prepared using a hypotonic buffer. The remaining nuclear pellet was extracted with 0.45 M NaCl, and the soluble fraction (N450) cleared by centrifugation, as described in Materials and Methods.
As shown in Figure 5A, both CE and N450 contained flag-RHA but endogenous topoisomerase IIα was present only in N450, as determined by western blotting using anti-flag (M2, Sigma) and anti-topoisomerase IIα antibodies (Topogen) (compare lanes 1 and 3). Thus, only the N450 fraction was subjected to IP using M2 antibody and the resulting pellet was examined for the presence of endogenous topoisomerase IIα. Unlike endogenous RHA, flag-RHA did not associate with endogenous topoisomerase IIα (Fig. 5B, lane 3). This result supports the possibility that additional factors are required for the formation of RHA–topoisomerase IIα complex in the nucleus, but their availability may be a limiting factor.
Recently, we have screened a HeLa cDNA library (Clontech) employing the yeast two-hybrid assay using full-length RHA as bait, and we have identified Ubc9, an E2-type enzyme specific for small ubiquitin-like molecule-1 (Sumo-1), as an RHA-interacting factor (K.Zhou, J.Argasinska, R.J.Donnelly, R.T.Hay and C.G.Lee, unpublished data). Our observation, together with the previous report describing a direct interaction between Ubc9 and topoisomerase IIα or IIβ (35), prompted us to explore whether Ubc9 is required for the association of ectopically expressed RHA with topoisomerase IIα. For this purpose, 293T cells were co-transfected with pCDNA3-fhis-RHA and CMV-Ubc9-V5. Western blot analysis using anti-V5 antibody indicated the distribution of V5-Ubc9 in both CE and N450 (Fig. 5A, lanes 2 and 4). When subjected to IP using N450 and M2 antibody, as described above, endogenous topoisomerase IIα was co-precipitated with flag-RHA (Fig. 5B, lane 4).
In addition to Ubc9, RHA interacts with CBP/p300 (7), BRCA1 (8) and RNA polymerase II (7,36). Thus, similar assays were performed with constructs expressing CBP or p300. In contrast to Ubc9, neither CBP nor p300 co-expression supported the interaction between flag-RHA and topoisomerase IIα (data not shown). These results indicate that Ubc9 is a limiting factor required for the interaction between RHA and topoisomerase IIα. As described above, the interaction between Ubc9 and topoisomerase IIα has been demonstrated by yeast two-hybrid and GST pull-down assays (35). However, we were unable to find any optimal condition for the detection of Ubc9 in the IP pellet obtained with anti-RHA or anti-topoisomerase IIα antibody (data not shown). Our result might suggest that the association of Ubc9 with RHA or topoisomerase IIα is too weak to be stably detected in vitro. Alternatively, Ubc9 might transiently interact with RHA and topoisomerase IIα to induce their interaction. The identification of additional factors required for the interaction between RHA and topoisomerase IIα will allow us to test the above possibilities.
The ATPase activity of RHA is dispensable for its interaction with topoisomerase IIα
RHA and MLE interact with topoisomerase IIα and MSL complexes, respectively, through the involvement of RNA molecules. A substitution mutation in the Walker motif A of MLE (GKT to GET) leads to its association not only with the male X chromosome but also with autosomes (31). In addition, such a mutation decreases cellular levels of roX2 RNA, eliminating both MLE–MSL and roX2 RNA–MSL complexes (16). These observations illustrate the importance of the ATPase activity of MLE in stabilizing roX2 RNA as well as in its activity to interact with MSL complexes. Therefore, we explored whether the ATPase activity of RHA is required for the interaction between RHA and topoisomerase IIα. For this purpose, 293T cells were transfected with a construct, expressing a mutant RHA (RHAGET), carrying GET in place of GKT in the Walker motif A, and the N450 extract was prepared and subjected to IP using anti-RHA antibody. As shown in Figure 5C, topoisomerase IIα was present in both RHA- and RHAGET-containing IP pellets (lanes 3 and 4). The association of RHAGET with topoisomerase IIα was also sensitive to RNase A (data not shown). These results indicate that the ATPase activity of RHA is not required for its association with topoisomerase IIα and with RNA molecules.
DISCUSSION
There are many parallels between RHA and MLE. Previously, we have shown that the biochemical properties of RHA and MLE are indistinguishable. The present study exhibits an additional similarity between RHA and MLE: the requirement of RNA molecules for their interaction with other proteins. The interaction between RHA and topoisomerase IIα resembles the assembly of MLE in MSL complexes containing roX1/2 RNAs (31). RNase A treatment dissociates MLE from the male X chromosome without affecting other MSL proteins or RNA polymerase II associated with the chromosome (31). In addition to MLE, at least two other MSL proteins, male-absent on the first (MOF) and MSL-3, are capable of interacting with roX2 RNA through the chromodomain (37). It is, therefore, possible that MLE interacts indirectly with MSL complexes on the X chromosome, bridged by roX2 RNA.
Based on a similar scenario, RHA and topoisomerase IIα may be brought together by RNA molecules. Obviously, the ability of topoisomerase IIα to interact with RNA molecules (20,38) would greatly facilitate this interaction. Alternatively, upon its binding, RNA molecules may induce a conformational change in RHA, which promotes RHA interaction with topoisomerase IIα or with other factors. To date, RNA-induced conformational changes have been shown for a few proteins such as the 40S ribosomal subunit interacting with hepatitis C virus IRES RNA (39) and PRK, a protein kinase activated by dsRNA (40).
The interaction between RHA and topoisomerase IIα requires protein factors in addition to RNA molecules. Our study shows that at least Ubc9 needs to be co-expressed to support the interaction between ectopically expressed flag-tagged RHA with topoisomerase IIα. A direct interaction has been demonstrated between Ubc9 and topoisomerase IIα or IIβ by yeast two-hybrid assay and GST pull-down assay (35). The N‐terminal 137 amino acids of RHA support the interaction of RHA with Ubc9 (K.Zhou, J.Argasinska, R.J.Donnelly, R.T.Hay and C.G.Lee, unpublished data). Thus, Ubc9 may bridge the interaction between RHA and topoisomerase IIα. The physical association with Ubc9 at the N‐terminal domain may activate RHA to recognize and bind both RNA molecules and topoisomerase IIα. Alternatively, the association with Ubc9 may enable topoisomerase IIα to form a complex with flag-tagged RHA. Identification of various factors associated with the RHA–topoisomerase IIα complex, including RNA molecules, will provide clues to the molecular nature of the RHA–topoisomerase IIα interaction.
The functional significance of RHA–topoisomerase IIα complex remains to be established. In Drosophila, topoisomerase II interacts with at least three factors such as supercoiling factor (SCF) (41), chromatin accessibility complex (CHRAC) (42) and histone deacetylases 1 and 2 (HDAC1/2) in nucleosome remodeling and deacetylase (NuRD) complex (43). SCF and topoisomerase II function jointly to introduce negative superhelical turns into DNA in vitro and co-localize to puffs on polytene chromosomes (2), implying a role for the SCF–topoisomerase II complex in gene expression. The role of topoisomerase II in transcription is also implied by the transcriptional repression of genes containing Polycomb group (PcG) target sequences in Drosophila. In general, the PcG proteins are required to preserve the transcriptionally silenced state of affected chromatin region and this process accompanies the interaction of the PcG protein Polyhomeotic with both topoisomerase II and Barren (44). The function of topoisomerase II as a co-repressor is suggested by its interaction with retinoblastoma protein (Rb) (45) and histone deacetylases HDAC1/2 in NuRD complex that mediates histone deacetylation and ATP-dependent nucleosome remodeling (46).
As seen with purified RHA and topoisomerase IIα, RHA in complex may stimulate the relaxation reaction either by facilitating the access of topoisomerase IIα to chromatin DNA or by directly binding to chromatin DNA. If complexed RHA and topoisomerase IIα function in a concerted manner, the affected chromatin region may undergo a significant topological alteration due to the reduction in superhelicity. Mechanistically a similar mode of action has been proposed to explain the stimulatory function of topoisomerase IIα in RNA polymerase II-mediated transcription on chromatin template (46). Interestingly, chromatin-dependent transcription is activated when topoisomerase IIα is provided during but not after chromatin assembly (46). This observation suggests that additional factors may be required for the interaction of topoisomerase IIα with nucleosomal DNA. If the access of topoisomerase IIα to chromatin is facilitated by complexed RHA, it can be predicted that, unlike topoisomerase IIα alone, the RHA–topoisomerase IIα complexes will activate RNA polymersae II-mediated transcription on pre-assembled chromatin template. Moreover, the DNA, once isolated post-transcriptionally, would be highly underwound if RHA–topoisomerase II complexes, operating in front of the transcription machinery, remove the positive supercoiling generated downstream of transcription site as a result of the movement of RNA polymerase II.
Several transcription factors such as ATF-2, CREB, c-Jun and p53 bind and stimulate the catalytic activity of topoisomerase IIα (47,48). In addition, there is increasing evidence indicating the conjoined function of topoisomerases and helicases. The RecQ family of asp-glu-x-his (DExH) DNA helicases act in concert with topoisomerase III (49). The interactions between two mammalian RecQ-like helicases, Bloom’s syndrome gene product (BLM) and Werner’s syndrome gene product (WRN), and topoisomerase III (50,51) suggest the conserved conjoined function of RecQ helicases and type I topoisomerases in eukaryotic species. Our observations show that RHA functionally and physically interacts with topoisomerase IIα, which suggests that a concerted function might be more general among topoisomerases and helicases. Quantitative isolation and thorough identification of RHA–topoisomerase IIα complex is underway in an effort to define their biochemical and biological functions in more detail.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Drs Mukund Modak and Andrew Parrott for their comments and critical reading of the manuscript. C.-G.L. was supported by a Foundation of UMDNJ research grant and UMD-NJMS Dean’s BRSG program.
REFERENCES
- 1.Lee C.G. and Hurwitz,J. (1993) Human RNA helicase A is homologous to the maleless protein of Drosophila. J. Biol. Chem., 268, 16822–16830. [PubMed]
- 2.Lee C.G. and Hurwitz,J. (1992) A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J. Biol. Chem., 267, 4398–4407. [PubMed]
- 3.Zhang S. and Grosse,F. (1994) Nuclear DNA helicase II unwinds both DNA and RNA. Biochemistry, 33, 3906–3912. [DOI] [PubMed]
- 4.Zhang S., Macke,H. and Grosse,F. (1995) Molecular cloning of the gene encoding nuclear DNA helicase II. A bovine homologue of human RNA helicase A and Drosophila Mle protein. J. Biol. Chem., 270, 16422–16427. [DOI] [PubMed]
- 5.Lee C.G., Chang,K.A., Kuroda,I.M. and Hurwitz,J. (1997) The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J., 16, 2671–2681. [DOI] [PMC free article] [PubMed]
- 6.Lee C.G., Soares,V., Newberger,C., Manova,K., Lacy,E. and Hurwitz,J. (1998) RNA helicase A is essential for normal gastrulation. Proc. Natl Acad. Sci. USA, 95, 13709–13713. [DOI] [PMC free article] [PubMed]
- 7.Nakajima T., Uchida,C., Anderson,S.F., Lee,C.G., Hurwitz,J., Parvin,J.D. and Montminy,M. (1997) RNA helicase A mediates association of CBP with RNA polymerase II. Cell, 90, 1107–1112. [DOI] [PubMed]
- 8.Anderson S.F., Schlegel,B.P., Nakajima,T., Wolpin,E.S. and Parvin,J.D. (1998) BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nature Genet., 19, 254–256. [DOI] [PubMed]
- 9.Yang J.P., Tang,H., Reddy,T.R. and Wong-Staal,F. (2001) Mapping the functional domains of HAP95, a protein that binds RNA helicase A and activates the constitutive transport element of type D retroviruses. J. Biol. Chem., 276, 30694–30700. [DOI] [PubMed]
- 10.Pellizzoni L., Charroux,B., Rappsilber,J., Mann,M. and Dreyfuss,G. (2001) A functional interaction between the survival motor neuron complex and RNA polymerase II. J. Cell Biol., 152, 75–85. [DOI] [PMC free article] [PubMed]
- 11.Kuroda M.I., Kernan,M.J., Kreber,R., Ganetzky,B. and Baker,B.S. (1991) The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell, 66, 935–947. [DOI] [PubMed]
- 12.Copps K., Richman,R., Lyman,L.M., Chang,K.A., Rampersad-Ammons,J. and Kuroda,M.I. (1998) Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. EMBO J., 17, 5409–5417. [DOI] [PMC free article] [PubMed]
- 13.Smith E.R., Pannuti,A., Gu,W., Steurnagel,A., Cook,R.G., Allis,C.D. and Lucchesi,J.C. (2000) The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol., 20, 312–318. [DOI] [PMC free article] [PubMed]
- 14.Meller V.H., Wu,K.H., Roman,G., Kuroda,M.I. and Davis,R.L. (1997) roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell, 88, 445–457. [DOI] [PubMed]
- 15.Amrein H. and Axel,R. (1997) Genes expressed in neurons of adult male Drosophila. Cell, 88, 459–469. [DOI] [PubMed]
- 16.Gu W., Wei,X., Pannuti,A. and Lucchesi,J.C. (2000) Targeting the chromatin-remodeling MSL complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. EMBO J., 19, 5202–5211. [DOI] [PMC free article] [PubMed]
- 17.Tang H., Gaietta,G.M., Fischer,W.H., Ellisman,M.H. and Wong-Staal,F. (1997) A cellular cofactor for the constitutive transport element of type D retrovirus. Science, 276, 1412–1415. [DOI] [PubMed]
- 18.Fujii R., Okamoto,M., Aratani,S., Oishi,T., Ohshima,T., Taira,K., Baba,M., Fukamizu,A. and Nakajima,T. (2001) A role of RNA helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J. Biol. Chem., 276, 5445–5451. [DOI] [PubMed]
- 19.Liao H.J., Kobayashi,R. and Mathews,M.B. (1998) Activities of adenovirus virus-associated RNAs: purification and characterization of RNA binding proteins. Proc. Natl Acad. Sci. USA, 95, 8514–8519. [DOI] [PMC free article] [PubMed]
- 20.Rzepecki R. and Fisher,P.A. (2000) During both interphase and mitosis, DNA topoisomerase II interacts with DNA as well as RNA through the protein’s C-terminal domain. J. Cell Sci., 113, 1635–1647. [DOI] [PubMed]
- 21.Hateboer G., Hijmans,E.M., Nooij,J.B., Schlenker,S., Jentsch,S. and Bernards,R. (1996) mUBC9, a novel adenovirus E1A-interacting protein that complements a yeast cell cycle defect. J. Biol. Chem., 271, 25906–25911. [DOI] [PubMed]
- 22.Desterro J.M.P., Thomson,J. and Hay,R.T. (1997) Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett., 417, 297–300. [DOI] [PubMed]
- 23.Brockdorff N., Ashworth,A., Kay,G.F., McCabe,V.M., Norris,D.P., Cooper,P.J., Swift,S. and Rastan,S. (1992) The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell, 71, 515–526. [DOI] [PubMed]
- 24.Brown C.J., Hendrich,B.D., Rupert,J.L., Lafreniere,R.G., Xing,Y., Lawrence,J. and Willard,H.F. (1992) The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell, 71, 527–542. [DOI] [PubMed]
- 25.Lanz R.B., McKenna,N.J., Onate,S.A., Albrecht,U., Wong,J., Tsai,S.Y., Tsai,M.J. and O’Malley,B.W. (1999) A steroid receptor coactivator, SRA, functions as a RNA and is present in an SRC-1 complex. Cell, 97, 17–27. [DOI] [PubMed]
- 26.Sleutels F., Zwart,R. and Barlow,D.P. (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature, 415, 810–813. [DOI] [PubMed]
- 27.Lohman T.M. (1993) Helicase-catalyzed DNA unwinding. J. Biol. Chem., 268, 2269–2272. [PubMed]
- 28.Singleton M.R., Sawaya,M.R., Ellenberger,T. and Wigley,D.B. (2000) Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell, 101, 589–600. [DOI] [PubMed]
- 29.von Hippel P.H. and Delagoutte,E. (2001) A general model for nucleic acid helicases and their ‘coupling’ within macromolecular machines. Cell, 104, 177–190. [DOI] [PubMed]
- 30.Tanner N.K. and Linder,P. (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell, 8, 251–262. [DOI] [PubMed]
- 31.Gibson T.J. and Thompson,J.D. (1994) Detection of dsRNA-binding domains in RNA helicase A and Drosophila maleless: implications for monomeric RNA helicases. Nucleic Acids Res., 22, 2552–2556. [DOI] [PMC free article] [PubMed]
- 32.Kwon H., Imbalzano,A.N., Khavari,P.A., Kingston,R.E. and Green,M.R. (1994) Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature, 370, 477–481. [DOI] [PubMed]
- 33.Imbalzano A.N., Kwon,H., Green,M.R. and Kingston,R.E. (1994) Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature, 370, 481–485. [DOI] [PubMed]
- 34.Gavin I., Horn,P.J. and Peterson,C.L. (2001) SWI/SNF chromatin remodeling requires changes in DNA topology. Mol. Cell, 7, 97–104. [DOI] [PubMed]
- 35.Mao Y., Desai,S.D. and Liu,L.F. (2000) SUMO-1 conjugation to human DNA topoisomerase II isozymes. J. Biol. Chem., 275, 26066–26073. [DOI] [PubMed]
- 36.Aratani S., Fujii,R., Oishi,T., Fujita,H., Amano,T., Ohshima,T., Hagiwara,M., Fukamizu,A. and Nakajima,T. (2001) Dual roles of RNA helicase A in CREB-dependent transcription. Mol. Cell. Biol., 21, 4460–4469. [DOI] [PMC free article] [PubMed]
- 37.Akhtar A., Zink,D. and Becker,P.B. (2000) Chromodomains are protein-RNA interaction modules. Nature, 407, 405–409. [DOI] [PubMed]
- 38.Wang Y., Knudsen,B.R., Bjergback,L., Westergaard,O. and Andersen,A.H. (1999) Stimulated activity of human topoisomerases ΙΙalpha and IIbeta on RNA-containing substrates. J. Biol. Chem., 274, 22839–22846. [DOI] [PubMed]
- 39.Spahn C.M., Kieft,J.S., Grassucci,R.A., Penczek,P.A., Zhou,K., Doudna,J.A. and Frank,J. (2001) Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science, 291, 1959–1962. [DOI] [PubMed]
- 40.Wu S. and Kaufman,R.J. (1997) A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J. Biol. Chem., 272, 1291–1296. [DOI] [PubMed]
- 41.Kobayashi M., Aita,N., Hayashi,S., Okada,K., Ohta,T. and Hirose,S. (1998) DNA supercoiling factor localizes to puffs on polytene chromosomes in Drosophila melanogaster. Mol. Cell. Biol., 18, 6737–6744. [DOI] [PMC free article] [PubMed]
- 42.Varga-Weisz P.D., Wilm,M., Bonte,E., Dumas,K., Mann,M. and Becker,P.B. (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature, 388, 598–602. [DOI] [PubMed]
- 43.Tsai S.C., Valkov,N., Yang,W.M., Gump,J., Sullivan,D. and Seto,E. (2000) Histone deacetylase interacts directly with DNA topoisomerase II. Nature Genet., 26, 349–353. [DOI] [PubMed]
- 44.Lupo R., Breiling,A., Bianchi,M.E. and Orlando,V. (2001) Drosophila chromosome condensation proteins topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol. Cell, 7, 127–136. [DOI] [PubMed]
- 45.Bhat U.G., Raychaudhuri,P. and Beck,W.T. (1999) Functional interaction between human topoisomerase IIα and retinoblastoma protein. Proc. Natl Acad. Sci. USA, 96, 7859–7864. [DOI] [PMC free article] [PubMed]
- 46.Mondal N. and Parvin,J.D. (2001) DNA topoisomerase ΙΙalpha is required for RNA polymerase II transcription on chromatin templates. Nature, 413, 435–438. [DOI] [PubMed]
- 47.Kroll D.J., Sullivan,D.M., Gutierrez-Hartmann,A. and Hoeffler,J.P. (1993) Modification of DNA topoisomerase II activity via direct interactions with the cyclic adenosine-3′,5′-monophosphate response element-binding protein and related transcription factors. Mol. Endocrinol., 7, 305–318. [DOI] [PubMed]
- 48.Kwon Y., Shin,B.S. and Chung,I.K. (2000) The p53 tumor suppressor stimulates the catalytic activity of human topoisomerase IIα by enhancing the rate of ATP hydrolysis. J. Biol. Chem., 275, 18505–18510. [DOI] [PubMed]
- 49.Harmon F.G., DiGate,R.J. and Kowalczykowski,S.C. (1999) RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol. Cell, 3, 611–620. [DOI] [PubMed]
- 50.Lebel M., Spillare,E.A., Harris,C.C. and Leder,P. (1999) The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem., 274, 37795–37799. [DOI] [PubMed]
- 51.Wu L., Davies,S.L., North,P.S., Goulaouic,H., Riou,J.F., Turley,H., Gatter,K.C. and Hickson,I.D. (2000) The Bloom’s syndrome gene product interacts with topoisomerase III. J. Biol. Chem., 275, 9636–9644. [DOI] [PubMed]




