Abstract
We specifically sought genes within the yeast genome controlled by a non-conventional translation mechanism involving the stop codon. For this reason, we designed a computer program using the yeast database genomic regions, and seeking two adjacent open reading frames separated only by a unique stop codon (called SORFs). Among the 58 SORFs identified, eight displayed a stop codon bypass level ranging from 3 to 25%. For each of the eight sequences, we demonstrated the presence of a poly(A) mRNA. Using isogenic [PSI+] and [psi–] yeast strains, we showed that for two of the sequences the mechanism used is a bona fide readthrough. However, the six remaining sequences were not sensitive to the PSI state, indicating either a translation termination process independent of eRF3 or a new stop codon bypass mechanism. Our results demonstrate that the presence of a stop codon in a large ORF may not always correspond to a sequencing error, or a pseudogene, but can be a recoding signal in a functional gene. This emphasizes that genome annotation should take into account the fact that recoding signals could be more frequently used than previously expected.
INTRODUCTION
It has been known since the 1980s that several mRNAs can be obtained from the same gene, by post-transcriptional modifications like editing and splicing (1–5). Alternatively, recoding events which act at the translational level can occur by subverting the normal decoding rules allowing the synthesis of two related proteins from the same mRNA (6). All of these events enhance the coding potential of complex genomes and may fill the gap between the number of genes and the number of polypeptides present in a given organism (7). In most cases, recoding events are used to graft a new biological function onto a protein. This is spectacularly illustrated in retroviruses, in which recoding is almost always necessary to produce the Gag-Pol polyprotein which bears, among others, the reverse transcriptase activity (8,9). In the vast majority of reported recoding events, either frameshifting or readthrough of stop codons is involved (6,10–12). Although the mechanisms are different, the end result is very similar, since both finish in the skipping of the natural stop codon and the synthesis of an extended protein.
Most genes controlled by recoding have thus far been observed in small autonomous genetic elements, such as RNA viruses and transposable elements, but very few in chromosomal genomes (13–19) with the exception of euplotes genome (20). This bias may be the result of a selective pressure favoring translational control in compact genomes. However, an easy explanation could be that these events are more easily identifiable in small genomes in which a large amount of information is available. In particular, the pattern of protein expression is often known and can be related to the nucleic acid sequence. This is obviously not the case for even the smallest chromosomal genomes, where there is a large knowledge gap between the nucleotide sequence and the pattern of protein expression. One may thus suppose that recoding events might be more frequent in large genomes than can be extrapolated from current data (10,21).
We have previously reported a computational analysis to identify readthrough controlled genes in the yeast Saccharomyces cerevisiae, using a stop codon nucleotide context corresponding to a consensus readthrough motif (15). This allowed us to identify eight sequences, among which we have demonstrated that translational readthrough on the PDE2 gene induces a high instability of the Pde2p protein, which in turn affects the cAMP level in the cell. This mechanism could explain some physiological modifications associated with presence of the yeast prion [PSI+] (22). Although this approach has been fruitful, one cannot exclude that we had missed other readthrough events. Termination is still one of the least understood aspects of translation, especially in eukaryotes. In particular, the precise biochemical mechanisms involved remain to be elucidated, as recently in prokaryotes (23). In addition, other mechanisms like ribosome hopping/sliding can also lead, as does readthrough, to stop codon bypass (24,25).
In this study, we developed a strategy to identify genes in the yeast S.cerevisiae, whose expression is controlled by a stop codon bypass, without a priori knowledge of the mechanism involved. We used a computer program to determine the genomic regions where two adjacent open reading frames are separated simply by a unique stop codon, termed ‘semi-open reading frames’ (SORFs). A subset of 60 candidate regions was found. We then quantified the stop codon bypass efficiency of each. Eight regions showed a bypass efficiency 10-fold higher than background; of these, two corresponded to classical readthrough events and six to other mechanisms. These SORFs, showing a high level of stop codon bypass efficiency, were named ‘bypass of stop codon’ (BSC). Several of these SORFs are genes with known functions, which will allow further analysis of the physiological role of the recoding event. Overall, these results demonstrate that bypassing stop codons may be a more frequent event in eukaryotes than was thought.
MATERIALS AND METHODS
Yeast strains and media
The strains Y349 (MATa lys2Δ201 leu2-3,112 his3Δ200 ura3-52) and 74-D694 (MATa ade1-14 trp1-289 his3Δ200 leu2-3,112 ura3-52 [psi–] or [PSI+]) were used in this study. YNB (0.67% yeast nitrogen base; 2% glucose) missing the appropriate amino acids was used to prepare media for standard growth conditions. The 74-D694 [psi–] and [PSI+] strains carry the ade1-14 allele which corresponds to a nonsense UGA mutation. This allows direct visualization of the PSI state of the strain through the accumulation of a red pigment in the [psi–] and not in the [PSI+] strain. The PSI status of the strains was verified by the red or white color of the colonies they give and by routine determinations of readthrough efficiency directed by sensitive stop codons (26).
Plasmids
The pAC99 reporter plasmid has been previously described (15). Constructs were obtained by inserting a PCR fragment containing the stop codon into the cloning site, between the lacZ and luc genes in the plasmid pAC99. For readthrough measurements, an in-frame control was used (pAC-TQ) which allowed the production of 100% fusion protein (β-galactosidase-luciferase). The region including the inserted fragment was sequenced in the newly constructed plasmids.
Enzymatic activities and readthrough frequency
The yeast strains were transformed with the reporter plasmids using the lithium acetate method according to Ito et al. (27). In each case, at least three transformants, cultured in the same conditions, were assayed. Cells were broken using acid-washed glass beads; luciferase and β-galactosidase activities were assayed in the same crude extract, as previously described (28). Readthrough frequency is defined as the ratio of luciferase activity to β-galactosidase activity. To establish the relative activities of β-galactosidase and luciferase when expressed in equimolar amounts, the ratio of luciferase activity to β-galactosidase from an in-frame control plasmid was taken as a reference. Readthrough frequency, expressed as percentage, was calculated by dividing the luciferase/β-galactosidase ratio obtained from each test construct by the same ratio obtained with the in-frame control construct (29).
Molecular biology procedures and RT–PCR
Each SORF fragment was amplified from Y349 genomic DNA by PCR, using Pfu polymerase (Stratagene), and cloned into the pAC99 vector.
Total RNA was extracted, as described by Schmitt et al., from 5 ml of exponential yeast culture (30). Each RNA extraction was subjected to a DNA digestion with 10 U of RNAse-free DNAse I (Boerhinger) at 37°C for 1 h. DNAse I was inactivated by heating at 90°C for 5 min as recommended by the manufacturer. RNA was reverse-transcribed with oligo(dT) primer by AMV reverse transcriptase (Stratagene) for PCR amplification with Taq polymerase® (Amersham) in a GeneAmp 2600 thermocycler (Perkin Elmer). PCR fragments were visualized in a 1.5% agarose gel. The sequences of the primers used either in SORF amplification or RT–PCR experiments are shown in Tables S1 and S2, which are published as Supplementary Material.
Computer program
The SORFS have been automatically identified in the nucleic sequence of the entire genome of S.cerevisiae, with a simple computer program dedicated to this search witten in Pascal and available upon request. The nucleic sequence and the coordinates (start and stop codons location) of all the ORFs were retrieved from the Stanford Genome Database (March 1999).
For each SORF, the program retrieves the following characteristics: (i) the length of the 3′ extension (ORF2); (ii) the position of the first codon ATG (if any) being present in the ORF2; (iii) the sequence surrounding (3 nt before and 6 nt after) the termination codon of ORF1; (iv) if the ORF2 is overlapping an already annotated ORF (existing in the SGD database).
This was done for each strand separately, and resulted in a list of all the SORFs being present in the S.cerevisiae genome. The maximal length observed for ORF2 was 3375 nt, the minimal length being 3 nt (when the stop codon of ORF1 is immediately followed by another stop codon). We observed that ∼90% of the annotated ORFs exhibit an extension (ORF2) which is smaller or equal to 150 nt, and that 2% of the annotated ORFs are followed by an ORF longer than 200 nt. We pursued our analysis with the latter sequences in order to be able to perform experiments on all of them.
RESULTS
Identification of SORFs
The goal of this study was to identify ORFs exhibiting a genomic organization compatible with a translational readthrough-dependent mode of expression. We chose to restrict our search to ORFs already annotated, in order to investigate those followed by a sequence without a second immediate termination codon, in the same reading frame (Fig. 1). This 3′ extension was named ‘ORF2’, ORF1 being the annotated ORF. Those two ORFs were referred to as SORFs. We initially retrieved the entire S.cerevisiae DNA sequence and the coordinates of all the ORFs from the Stanford Genome Database. We then constructed a computer program to identify automatically all possible SORFs (see Materials and Methods). For each, we measured the ORF2 length and pursued our analysis only with SORFs containing an ORF2 with a minimal length of 200 nt (this limitation is discussed in Discussion).
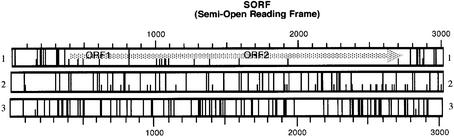
Figure 1.
Schematic representation of a SORF (YER109c). The three possible reading frames of the mRNA are represented in lines 1, 2 and 3. A plain vertical bar corresponds to a stop codon, a semi bar is an ATG codon. Numbers represent the length in nucleotides from the beginning of the sequence.
Preliminary experiments have shown that fragments containing an in-frame ATG codon within the 50 nt after the stop codon sometimes promoted translation, possibly through translational reinitiation (data not shown). We thus excluded all fragments containing an in-frame ATG codon in the first 50 nt after the stop codon. Sixty SORFs corresponding to these criteria are listed in Table 1.
Table 1. List of SORFs.
| SORF name | ORF1a length | ORF2b length |
|---|---|---|
| YAL019W | 570 | 3495 |
| YAL021C | 2514 | 228 |
| YAL024C | 4308 | 216 |
| YAR066W | 612 | 1284 |
| YCL006C | 330 | 246 |
| YCL014W | 4104 | 807 |
| YCL022C | 515 | 570 |
| YCL074W | 927 | 291 |
| YDL037C | 987 | 2271 |
| YDL050C | 372 | 378 |
| YDR082W | 1485 | 1299 |
| YDR275W | 708 | 216 |
| YDR509W | 348 | 348 |
| YER039C-a | 237 | 807 |
| YER109C | 426 | 1974 |
| YFL052W | 1398 | 246 |
| YGL241W | 3015 | 906 |
| YGR037C | 264 | 225 |
| YGR046W | 1158 | 351 |
| YGR096W | 945 | 414 |
| YHL006C | 480 | 207 |
| YHR058C | 888 | 771 |
| YHR148W | 552 | 240 |
| YHR155W | 3687 | 228 |
| YHR214W | 612 | 1284 |
| YIL023C | 1041 | 243 |
| YIR044C | 186 | 951 |
| YJL028W | 336 | 414 |
| YJL075C | 417 | 402 |
| YJL162C | 1449 | 303 |
| YJL169W | 369 | 456 |
| YJL170C | 552 | 246 |
| YJL217W | 597 | 225 |
| YJR116W | 840 | 228 |
| YKL020C | 3249 | 1488 |
| YKL031W | 414 | 618 |
| YKR089C | 2733 | 267 |
| YKR103W | 3657 | 1020 |
| YLR465C | 309 | 780 |
| YMR057C | 372 | 1045 |
| YMR273C | 2748 | 387 |
| YNL091W | 3723 | 315 |
| YNL235C | 432 | 423 |
| YNL269W | 396 | 321 |
| YNL305C | 894 | 309 |
| YNR014W | 639 | 534 |
| YNR069C | 1470 | 1056 |
| YOL137W | 1494 | 201 |
| YOR024W | 222 | 1392 |
| YOR030W | 1860 | 372 |
| YOR051C | 1239 | 576 |
| YOR158W | 957 | 492 |
| YOR178C | 2382 | 252 |
| YOR268C | 399 | 222 |
| YOR324C | 1809 | 204 |
| YOR332W | 702 | 327 |
| YPL076W | 843 | 408 |
| YPR003C | 2265 | 225 |
| YPR143W | 753 | 240 |
| YPR158W | 759 | 210 |
aORF1 length is given in nucleotides between the AUG codon and the first stop codon found.
bORF2 length is given in nucleotides between the first stop codon and the second one.
Quantification of stop codon bypass efficiency
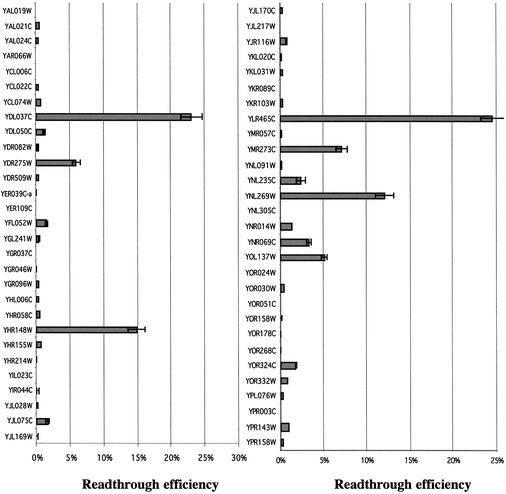
These 60 SORFs were identified without any criteria on stop codon nucleotide context. Therefore, it cannot be predicted if ribosomes can actually bypass the unique stop codon present between ORF1 and ORF2. To quantify stop codon bypass frequency, each fragment (about ±50 nt either side of the stop codon) was amplified by PCR (see Supplementary Material Table S1 for the list of oligonucleotides used) from genomic DNA of a wild-type yeast strain (Y349) and cloned into the pAC99 dual reporter vector (15). Each construct was then sequenced to verify the presence of the stop codon and to verify that no error occurred during PCR amplification. Among the 60 SORFs analyzed, two (YCL014w and YJL162c) did not show the presence of the expected stop codon. Since the strain used here (Y349) is not the SC288C strain that has been used for the yeast sequencing project, either a sequencing/annotation error or a gene polymorphism could explain this discrepancy. Among the 58 remaining candidates, eight fragments displayed a stop codon bypass efficiency >10-fold of the background (≥3%) and six displayed a bypass efficiency between 1 and 3% (Fig. 2). The eight sequences with a bypass efficiency >3% have been called BSC and were retained for further study.
Figure 2.
Quantification of the stop codon bypass efficiency. Each sequence surrounding the stop codon (±50 nt) has been amplified by PCR and cloned in the pAC99 reporter plasmid. For each construct at least three clones in two independent experiments have been used. The value is the average of the results. Standard deviation is <15% in all cases. Only sequences allowing >3% of stop codon bypass efficiency were retained for further analysis.
Expression of BSC genes
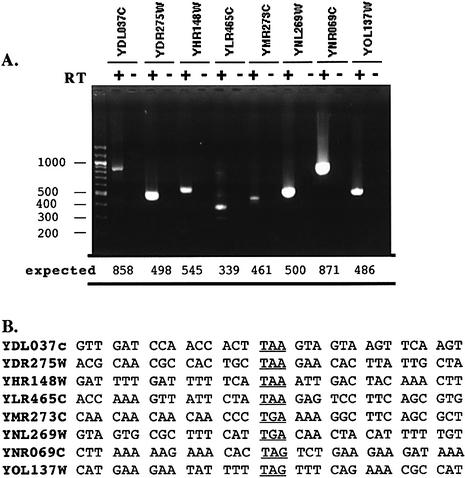
To confirm the presence of an mRNA encompassing the leaky stop codon, we examined each of the eight BSC genes identified above by RT–PCR, using first a reverse transcriptase step with an oligo dT primer, and secondly a PCR step with an upper primer located in ORF1 and a lower primer located far beyond the stop codon in ORF2 (see Supplementary Material Table S2 for the list of oligonucleotides used). A unique specific amplification was obtained in each case only in the presence of reverse transcriptase (Fig. 3A). All of them have been sequenced (Fig. 3B)
Figure 3.
RT–PCRs. (A) Total RNA was extracted as described in Materials and Methods, and treated by DNAse I. RT–PCR was carried out in two steps. First, a reverse transcription was done using a oligo(dT) primer, allowing only the reverse transcription of poly(A) mRNA. Then a standard PCR was performed on the mRNA before (lane –) and after (lane +) the reverse transcription. PCR products were visualized in a 2% agarose gel stained with ethidium bromide. All bands in the ladder are spaced by 100 nt. A single amplification product was seen in all lanes, the expected size is indicated (in nucleotides) for each product at the bottom of the gel. However, several faint bands were present in lane YLR465c (+). This could be due to the reiterated parts of the SORF YLR465C in the telomere regions of the chromosomes. (B) Sequences surrounding the stop codon (±15 nt). The stop codon is underlined.
These results demonstrate that the same molecule of mRNA carries both the ORFs for each BSC tested. Moreover, the amplification indicated that these mRNAs are polyadenylated. In these conditions, a readthrough product could in fact be produced from these BSC. An analysis of the polyadenylation signal in yeast has indicated that the mean length of 3′ non-coding sequence, before the poly(A) tail, is ∼60 nt (31). Our analysis confirmed these data except for BSC genes which possess a long 3′ extension between the stop codon and the polyadenylation signal (Fig. 3 and data not shown).
Similarities or motifs existing in identified BSC genes
We analyzed databases in order to determine if these BSC genes exhibit any similarity with known genes, or carried any known motifs. The results are shown in Table 2.
Table 2. Homology of Bsc proteins found in the databases.
| ORF or gene name | ORF1 | ORF2 | Motif found |
|---|---|---|---|
| YDL037c (BSC1) | 55% identity with Muc1p (3e–21)a | PRM7/YDL039c | 12 TSSST in ORF1 and 4 TTSVDPTTS motifs |
| YDR275w (BSC2) | – | – | – |
| YHR148w (IMP3) | IMP3 | – | RNA binding motif in ORF1 |
| YLR465c (BSC3) | – | 88% identity with Yel076w-cp (1.6e–45)a | – |
| YMR273c (ZDS1) | 36% with Zds2p (1.1e–26)a | – | 8 CAA (gln) repeat just before stop codon |
| YNL269w (BSC4) | – | – | – |
| YNR069c (BSC5) | 35% identity with Bul1p (1.6e–45)a | – | |
| YOL137w (BSC6) | 30% identity with B15I20.50 of Neurospora crassa (2.4e–25)a | – | 8 or 10 putative transmembrane segments |
aE-value of the corresponding alignment (% of identity) obtained with FASTA against Swissprot, Pfam and Prosite databases.
BSC5 (YNR069c) is referenced as a pseudogene in the database. However, we have demonstrated that a unique poly(A) mRNA exists, strongly suggesting that it can in fact be expressed.
BSC6 (YOL137w) has eight putative transmembrane segments in ORF1, and two in ORF2. The protein resulting from the fusion of the two ORFs is expected to have a structure different to that synthesized from ORF1 only. This novel structure would probably modify the specificity or the activity of the protein.
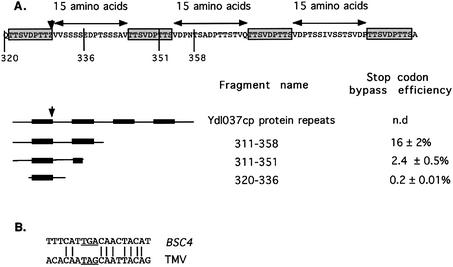
Finally, BSC1 (YDL037c) displays significant homology with Muc1p (cell surface flocculin) on both ORF1 and ORF2. It also bears two large nucleotide repetitions, one of 94 nt in ORF1 and one of 111 nt in ORF2. Furthermore, there are several imperfect nucleotide motif repeats around the stop codon between ORF1/ORF2. Strikingly, these imperfect nucleotide repeats correspond to two perfect repeats of nine amino acids, exactly spaced by 15 amino acids (Fig. 4A).
Figure 4.
(A) Schematic representation of the amino acids repeats found in the BSC1 protein sequence. Deletion of repeat No. 2 abolished totally the stop codon bypass. The position of the stop codon is represented by a ‘Z’ and a downward arrow. (B) Comparison of the BSC4 stop codon context, with the well known TMV context.
To determine either these BSC genes are limited to S.cerevisiae, or are found in other yeast genomes, a BLAST comparison has been done with the five other yeast genomes almost completed. Preliminary data suggest that homologous extensions can be found at least for some BSC genes (data not shown). However, the data available on the other yeast genomes are still too preliminary to draw any definitive conclusion.
Analysis of stop codon nucleotide context
Numerous experiments have demonstrated that the proximate nucleotide context, surrounding the stop codon, is a key determinant for readthrough efficiency (26,32–35). We have previously proposed a 3′ consensus pattern which yields high readthrough efficiency in yeast (26). This motif, STOP-CAA N(-A)A, very similar to the sequence first identified in the TMV virus, is found in BSC4 (Fig. 4B), but not in the other BSC. We postulated that the 3′ motif of the BSC4 is actually responsible for the high readthrough level obtained, and to test this we cloned an 18 bp fragment from BSC4 (Fig. 4B) into the pAC99 vector. This fragment gave a readthrough efficiency of 9%, similar to that directed by the original 123 bp fragment. This result clearly indicates that the immediate nucleotide context of the BSC4 stop codon is sufficient to obtain a high readthrough level.
As quoted above, BSC1 displays an unusual nucleotide structure around the stop codon with imperfect nucleotide repeats and perfect amino acid repeats (Fig. 4A). We have investigated the role of these repetitions in the stop codon bypass mechanism, by quantifying the stop codon bypass efficiency of several deletion mutants. Our results, shown in Figure 4A, indicate that this region plays an important role in stop codon bypass efficiency.
Effect of [PSI+] on bypassing efficiency
How the stop codon bypass occurs in the BSC is clearly understood only for BSC4, where we suspect a suppression by a natural tRNA, promoted by the stop codon proximal nucleotide context. For the other sequences, the mechanism remains unknown, since several events could lead to the same result. Among these are translational readthrough, splicing, editing, ribosome hopping, etc. As a first test to discriminate between these possibilities, the RT–PCR products obtained in the experiment shown in Figure 3 were sequenced. We confirmed the presence of in-frame stop codons, but in no case did we observe RNA post-transcriptional modification (data not shown).
We then tested the selected stop codon regions in isogenic [psi–] and [PSI+] strains. [PSI+] is an epigenetic element corresponding to intracellular aggregates of the Sup35p translation termination factor (36). In a [PSI+] strain, there is an overall decrease of translation termination efficiency, due to the depletion of soluble eRF3 termination factor which favors natural suppressor tRNAs (37,38).
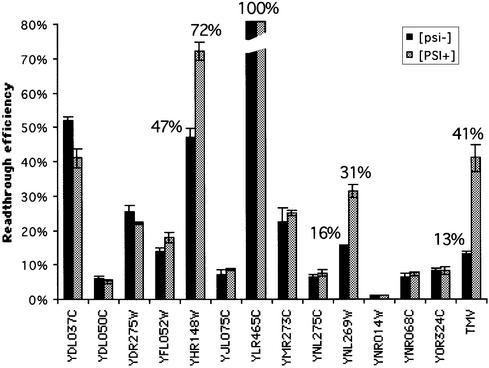
The same constructs were used to transform isogenic [psi–] and [PSI+] strains. As shown in Figure 5, the basal level of readthrough was increased at least 3-fold in the 74-D694 [psi–] strain as compared to the Y349 strain used in the previous experiment. This increase is, however, moderate since the [psi–] 74-D694 strain ([ade–]), contrary to its [PSI+] derivative, gives rise to red colonies, characteristic of a low level of readthrough. Such limited variability in basal readthrough efficiency has already been observed (15,29) and is probably due to different genetic backgrounds. The most striking observation is that, for most of the BSC, the bypass frequency is unaffected by the [PSI+] context. Only IMP3 and BSC4 showed a significant increase of stop codon readthrough level when associated with the depletion of Sup35p (Fig. 5). We concluded from these data that only the stop codons from both these genes are bypassed by a mechanism depending on eRF3.
Figure 5.
Effect of the [PSI+] factor on the stop codon bypass efficiency. See Figure 2 legend for details. The TMV readthrough sequence (CAA TAG CAA TTA) has been quantified in [psi–] and [PSI+] strains to check the ability of the [PSI+] strain to stimulate the readthrough efficiency.
DISCUSSION
This work describes a comprehensive analysis of the S.cerevisiae genome which attempts to identify cellular recoding events occurring during translational termination. We have developed a genomic approach, seeking genes with an extended coding potential, through the stop codon, without prior bias from existing ideas on termination codon suppression mechanism. The candidate genes are composed of two ORFs separated by a unique stop codon named SORF. We fixed the minimal length of the second ORF (ORF2) at 200 nt, and only SORFs without an ATG codon in the first 50 nt after the stop codon were retained for analysis. This allowed us to analyze a reasonable number of candidates, and excluded from the analysis genes possibly expressed through internal ribosome entry or translational reinitiation. Our preliminary data suggest that such genes, controlled through translation initiation, could also be retrieved by this kind of approach (O.Namy, I.Hatin and J.P.Rousset, unpublished results).
Recently, Harrison et al. have published an analysis of the yeast genome seeking ‘disabled ORF’, calling these dORF or mORF (39). Their results overlap only slightly with ours, because we did not base our analysis on homologies with known genes, and limit our research of 3′ extensions to official ORFs. They identified 11 of the SORFs characterized here (YDR082W, YIR044C, YER039C-a, YHR058C, YKL031W, YKL020C, YLR465C, YMR057C, YNR069C, YOR024W, YOR051C), two of which (YLR465C and YNR069C) display a significant stop codon bypass efficiency.
Our approach identified 58 SORFs in the yeast genome. Eight sequences displayed a stop codon bypass efficiency 10-fold higher than background. For each of these candidates, a unique mRNA covering both ORFs is present in the cell. Although it is only for those SORFs showing the highest bypass levels that one could expect to detect an mRNA editing mechanism, we did sequence the RT–PCR products for each SORF. No RNA post-transcriptional modification was identified. Moreover, from the amplification of the mRNA using a poly(dT) primer at the reverse transcription step, we concluded that these mRNA are polyadenylated and not rapidly degraded.
We quantified the stop codon bypass efficiency for each of the eight BSC genes in two isogenic [PSI+] and [psi–] yeast strains. In the [PSI+] strain, we expected that if the bypass mechanism is bona fide readthrough, the stop codon efficiency should increase. In fact, we observed a stop codon bypass increase only for the genes BSC4 and IMP3. For the other sequences, no such difference between the [psi–] and the [PSI+] strains was observed; thus for these sequences, the termination process is less dependent on the concentration of the eRF3 release factor, or the stop codon is bypassed by another translational mechanism independent of eRF3. The discrepancy between the results obtained with both wild-type [psi–] strains (Y349 and 74-D694) is not clear. In any case, these results indicate that the genetic background influences these unknown stop codon bypass mechanisms.
Recoding signals are usually composed of two elements, the sequence where the recoding takes place, and a stimulatory sequence. In a few examples, the stimulatory sequence is simply the immediate 3′ stop codon nucleotide context (26,34). However, most frequently the stimulatory sequence is a pseudoknot (or a stem–loop), and may be as far as 4 kb from the recoding site (40–44). A computational analysis of the stop codon 3′ nucleotide context of the BSC genes did not reveal any significant secondary structure in the vicinity of the stop codon. However, the BSC1 and BSC4 sequences displayed an unusual sequence pattern. The BSC1 sequence included four repeats of a nine amino acid motif that correspond to imperfect nucleotide repeats. This suggests that the role of these motifs is at the level of the protein and not of the mRNA. The exact role of these motifs in protein function is still unknown; however, our results indicate that this region is essential to obtain a high level of stop codon bypass, which suggests that the nucleotide motif is involved in the bypass mechanism. It is interesting to note that another recoding event, hopping, uses a codon repetition to promote stop codon bypassing (25,45). One can thus speculate that the mechanism active in BSC1 would not be readthrough, but hopping. This could explain the lack of effect of the PSI factor on the stop codon bypass efficiency. More experiments are necessary to elucidate the precise mechanism.
The study of BSC4 indicated that the stop codon is present in a typical readthrough context. This observation is coherent with the increase of the stop codon bypass efficiency observed in the [PSI+] strain. Our results demonstrate that the immediate stop codon context is in fact sufficient to promote a high level readthrough of the BSC4 stop codon.
Overall, our results emphasize that recoding events take place at the stop codon more often than expected. These events should be carefully sought during genome annotation as shown recently in euplotes (20). They also suggest that mechanisms other than readthrough are possibly used by cells to allow ribosomes to bypass the stop codon.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are especially indebted to Maryse Godon for genetics and molecular biology experiments. We thank Ian Stansfield, Laure Bidou and Guillaume Stahl for fruitful discussions. We are grateful to Ian Brierley and Sawsan Naphtine for their crucial help and comments on the manuscript. This work was supported in part by the Association pour la Recherche contre le Cancer (contract 4699 to J.P.R.) by the Groupe d’Intérêt Scientifique: Infections à prions (project A74) and by a CNES grant on the Preparatory Program Mars Sample Analysis.
REFERENCES
- 1.Lewin R. (1983) Editing mRNA precursors. Science, 222, 493. [DOI] [PubMed]
- 2.Shaw J.M., Feagin,J.E., Stuart,K. and Simpson,L. (1988) Editing of kinetoplastid mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell, 53, 401–411. [DOI] [PubMed]
- 3.Benne R., Van den Burg,J., Brakenhoff,J.P., Sloof,P., Van Boom,J.H. and Tromp,M.C. (1986) Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell, 46, 819–826. [DOI] [PubMed]
- 4.Amara S.G., Jonas,V., Rosenfeld,M.G., Ong,E.S. and Evans,R.M. (1982) Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature, 298, 240–244. [DOI] [PubMed]
- 5.King C.R. and Piatigorsky,J. (1983) Alternative RNA splicing of the murine alpha A-crystallin gene: protein-coding information within an intron. Cell, 32, 707–712. [DOI] [PubMed]
- 6.Gesteland R.F. and Atkins,J.F. (1996) Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem., 65, 741–768. [DOI] [PubMed]
- 7.Roberts G.C. and Smith,C.W. (2002) Alternative splicing: combinatorial output from the genome. Curr. Opin. Chem. Biol., 6, 375–383. [DOI] [PubMed]
- 8.Jacks T., Madhani,H.D., Masiarz,F.R. and Varmus,H.E. (1988) Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell, 55, 447–458. [DOI] [PMC free article] [PubMed]
- 9.Farabaugh P.J., Qian,Q. and Stahl,G. (2000) Programmed translational frameshifting, hopping and readthrough of termination codons. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. CSHL Press, Cold Spring Harbor, New York, NY, pp. 741–761.
- 10.Baranov P.V., Gesteland,R.F. and Atkins,J.F. (2002) Recoding: translational bifurcations in gene expression. Gene, 286, 187–201. [DOI] [PubMed]
- 11.Atkins J.F., Weiss,R.B. and Gesteland,R.F. (1990) Ribosome gymnastics—degree of difficulty 9.5, style 10.0. Cell, 62, 413–423. [DOI] [PMC free article] [PubMed]
- 12.Bertram G., Innes,S., Minella,O., Richardson,J. and Stansfield,I. (2001) Endless possibilities: translation termination and stop codon recognition. Microbiology, 147, 255–269. [DOI] [PubMed]
- 13.Craigen W.J. and Caskey,C.T. (1987) The function, structure and regulation of E. coli peptide chain release factors. Biochimie, 69, 1031–1041. [DOI] [PubMed]
- 14.Morris D.K. and Lundblad,V. (1997) Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol., 7, 969–976. [DOI] [PubMed]
- 15.Namy O., Duchateau-Nguyen,G. and Rousset,J.P. (2002) Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae. Mol. Microbiol., 43, 641–652. [DOI] [PubMed]
- 16.Shigemoto K., Brennan,J., Walls,E., Watson,C.J., Stott,D., Rigby,P.W. and Reith,A.D. (2001) Identification and characterisation of a developmentally regulated mammalian gene that utilises –1 programmed ribosomal frameshifting. Nucleic Acids Res., 29, 4079–4088. [DOI] [PMC free article] [PubMed]
- 17.Rom E. and Kahana,C. (1994) Polyamines regulate the expression of ornithine decarboxylase antizyme in vitro by inducing ribosomal frame-shifting. Proc. Natl Acad. Sci. USA, 91, 3959–3963. [DOI] [PMC free article] [PubMed]
- 18.Asakura T., Sasaki,T., Nagano,F., Satoh,A., Obaishi,H., Nishioka,H., Imamura,H., Hotta,K., Tanaka,K., Nakanishi,H. et al. (1998) Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene, 16, 121–130. [DOI] [PubMed]
- 19.Shah A.A., Giddings,M.C., Parvaz,J.B., Gesteland,R.F., Atkins,J.F. and Ivanov,I.P. (2002) Computational identification of putative programmed translational frameshift sites. Bioinformatics, 18, 1046–1053. [DOI] [PubMed]
- 20.Klobutcher L.A. and Farabaugh,P.J. (2002) Shifty ciliates. Frequent programmed translational frameshifting in euplotids. Cell, 111, 763–766. [DOI] [PubMed]
- 21.Hammell A.B., Taylor,R.C., Peltz,S.W. and Dinman,J.D. (1999) Identification of putative programmed –1 ribosomal frameshift signals in large DNA databases. Genome Res., 9, 417–427. [PMC free article] [PubMed]
- 22.True H.L. and Lindquist,S.L. (2000) A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature, 407, 477–483. [DOI] [PubMed]
- 23.Zavialov A.V., Buckingham,R.H. and Ehrenberg,M. (2001) A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell, 107, 115–124. [DOI] [PubMed]
- 24.Herr A.J., Nelson,C.C., Wills,N.M., Gesteland,R.F. and Atkins,J.F. (2001) Analysis of the roles of tRNA structure, ribosomal protein L9 and the bacteriophage T4 gene 60 bypassing signals during ribosome slippage on mRNA. J. Mol. Biol., 309, 1029–1048. [DOI] [PubMed]
- 25.Huang W.M., Ao,S.Z., Casjens,S., Orlandi,R., Zeikus,R., Weiss,R., Winge,D. and Fang,M. (1988) A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science, 239, 1005–1012. [DOI] [PubMed]
- 26.Namy O., Hatin,I. and Rousset,J.P. (2001) Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep., 2, 787–793. [DOI] [PMC free article] [PubMed]
- 27.Ito H., Fukuda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed]
- 28.Stahl G., Bidou,L., Rousset,J.P. and Cassan,M. (1995) Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res., 23, 1557–1560. [DOI] [PMC free article] [PubMed]
- 29.Bidou L., Stahl,G., Hatin,I., Namy,O., Rousset,J.P. and Farabaugh,P.J. (2000) Nonsense-mediated decay mutants do not affect programmed –1 frameshifting. RNA, 6, 952–961. [DOI] [PMC free article] [PubMed]
- 30.Schmitt M.E., Brown,T.A. and Trumpower,B.L. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091–3092. [DOI] [PMC free article] [PubMed]
- 31.van Helden J., del Olmo,M. and Perez-Ortin,J.E. (2000) Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res., 28, 1000–1010. [DOI] [PMC free article] [PubMed]
- 32.Bonetti B., Fu,L.W., Moon,J. and Bedwell,D.M. (1995) The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol., 251, 334–345. [DOI] [PubMed]
- 33.Buckingham R.H. (1994) Codon context and protein synthesis: enhancements of the genetic code. Biochimie, 76, 351–354. [DOI] [PubMed]
- 34.Mottagui-Tabar S., Tuite,M.F. and Isaksson,L.A. (1998) The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem., 257, 249–254. [DOI] [PubMed]
- 35.Cassan M. and Rousset,J.P. (2001) UAG readthrough in mammalian cells: effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol. Biol., 2, 3. [DOI] [PMC free article] [PubMed]
- 36.Paushkin S.V., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J., 15, 3127–3134. [PMC free article] [PubMed]
- 37.Wickner R.B., Masison,D.C. and Edskes,H.K. (1995) [PSI] and [URE3] as yeast prions. Yeast, 11, 1671–1685. [DOI] [PubMed]
- 38.Wickner R.B. (1996) Prions and RNA viruses of Saccharomyces cerevisiae. Annu. Rev. Genet., 30, 109–139. [DOI] [PubMed]
- 39.Harrison P., Kumar,A., Lan,N., Echols,N., Snyder,M. and Gerstein,M. (2002) A small reservoir of disabled ORFs in the yeast genome and its implications for the dynamics of proteome evolution. J. Mol. Biol., 316, 409–419. [DOI] [PubMed]
- 40.Brierley I., Rolley,N.J., Jenner,A.J. and Inglis,S.C. (1991) Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol., 220, 889–902. [DOI] [PMC free article] [PubMed]
- 41.Bidou L., Stahl,G., Grima,B., Liu,H., Cassan,M. and Rousset,J.P. (1997) In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem–loop stimulatory signal. RNA, 3, 1153–1158. [PMC free article] [PubMed]
- 42.Paul C.P., Barry,J.K., Dinesh-Kumar,S.P., Brault,V. and Miller,W.A. (2001) A sequence required for –1 ribosomal frameshifting located four kilobases downstream of the frameshift site. J. Mol. Biol., 310, 987–999. [DOI] [PubMed]
- 43.Le S.Y., Shapiro,B.A., Chen,J.H., Nussinov,R. and Maizel,J.V. (1991) RNA pseudoknots downstream of the frameshift sites of retroviruses. Genet. Anal. Tech. Appl., 8, 191–205. [DOI] [PMC free article] [PubMed]
- 44.Parkin N.T., Chamorro,M. and Varmus,H.E. (1992) Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol., 66, 5147–5151. [DOI] [PMC free article] [PubMed]
- 45.Herr A.J., Gesteland,R.F. and Atkins,J.F. (2000) One protein from two open reading frames: mechanism of a 50 nt translational bypass. EMBO J., 19, 2671–2680. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.