Abstract
We have established that CpG oligodeoxynucleotide 5mers, of sequence type CGNNN (N = A, G, C or T), rapidly induce apoptosis/cell cycle arrest in human leukaemia lines. The 5′-CpG is obligatory for these effects. Induction of apoptosis in MOLT-4 cells did not require new protein synthesis and was insensitive to the caspase 3 inhibitor, Ac-DEVD-CHO, although the latter abrogated DNA laddering, phosphatidylserine externalization and collapse of the mitochondrial transmembrane potential. A subline of MOLT-4 cells, MOLT-4CpGR, was selected for acquired resistance to CpG 5mers. Differences in gene expression between MOLT-4 and MOLT-4CpGR cells were identified following three independent reciprocal cDNA subtractions, consensus selection and virtual cloning through targeted display. Several known genes were implicated in the action of or resistance to CpG oligodeoxynucleotide 5mers. Their protein products listed below immediately suggest cell signalling pathways/processes worthy of further investigation in elucidating the mechanism of CpG 5mer activity: caspase 2, the transcription factors Atf4, Hic, HoxB3 and Rqcd1, the splicing factors Rbmx, Sfrs5 and Sfrs7, the DNA replication factors Mcm5 and Brd4, phosphoinositide-3-kinase, annexin A1, mucosa-associated lymphoid tissue lymphoma translocation 1 and three enzymes involved in protein ubiquitylation, Siah1, Gsa7 and Nin283.
INTRODUCTION
During the course of work on inhibiting oncogene expression with antisense oligonucleotide analogues, a control, chimeric methylphosphonodiester/phosphodiester 15mer oligodeoxynucleotide of randomly selected sequence was observed to induce apoptosis rapidly in MOLT-4 and Jurkat E6 T-lymphocytic leukaemia cells following intracytoplasmic delivery. A series of further methylphosphonate substitutions, and mutations and truncations of the oligodeoxynucleotide served to establish that the phosphodiester-linked sequence CGGTA present within the 15mer was responsible for this biological activity (1). Isolated CpG oligodeoxynucleotide 5mers, end-protected against exonucleases, of sequence type CGNNN (N = A, G, C or T) exhibited a range of apoptosis-inducing potency depending on the 3′ sequence, with CGTTA being the most active. The effects were apparently quite specific, as the presence of the 5′-CpG was obligatory. In a separate study, conflicting results on the proliferation of leukaemia cells were observed with c-myc antisense oligonucleotides of different chemistries targeting different sites in c-myc mRNA/pre-mRNA. RNase H-active, chimeric methylphosphonodiester/phosphodiester antisense oligodeoxynucleotides targeting bases 1147–1166 of c-myc mRNA downregulated c-Myc protein and induced apoptosis and cell cycle arrest, respectively, in cultures of MOLT-4 and KYO1 human leukaemia cells. In contrast, an RNase H-inactive, morpholino antisense oligonucleotide analogue 28mer, simultaneously targeting the exon 2 splice acceptor site and initiation codon, reduced c-Myc protein to barely detectable levels but did not affect cell proliferation in these or other leukaemia lines (2,3). It was noted that the RNase H-active oligodeoxynucleotide 20mers contained the phosphodiester-linked motif CGTTG. An isolated 5mer of this sequence mimicked the antiproliferative effects of the 20mer, in the absence of any antisense activity against c-myc mRNA. In contrast, the c-myc antisense 20mer still reduced expression of c-myc in a subline of MOLT-4 cells that had been selected for resistance to CGTTA, but in this case the oligodeoxynucleotide failed to induce apoptosis or cell cycle arrest (3). It was concluded that the biological activity of the chimeric c-myc antisense 20mers resulted from a non-antisense mechanism related to the CGTTG motif contained within the sequence, and not through downregulation of c-myc. Although the oncogene may have been implicated in the aetiology of the original leukaemias, expression of c-myc is apparently no longer required to sustain continuous cell proliferation in these culture lines. The actual role of c-Myc may be to stimulate cell proliferation and simultaneously to select for mutant cells that are insensitive to its other activities, such as induction of apoptosis (4–9). The accompanying genetic changes could contribute to malignant progression without the further requirement for elevated c-Myc protein thereafter.
The results with c-Myc underscore a major problem in rational drug development, namely that of target selection. It is increasingly being recognized that signalling systems in living cells are webs with multiple nodes rather than separate one-dimensional arrays of sequentially interacting components (10). Microbial and animal ‘knockout’ experiments have demonstrated that deleting a gene product at a node with few inputs/outputs may have little effect on the cell or organism as a result of the ability of the web to compensate for such a change. Drastic effects may only occur through ‘hits’ at nodes with many interactions. It is apparent that CpG 5mer oligodeoxynucleotides are acting at such a node, and its biochemical characterization could provide valuable insight into part of the signalling system of human cells. The onset of apoptosis was rapid and widespread following delivery of CpG motif 5mer oligodeoxynucleotides into MOLT-4 cells. DNA laddering and redistribution of phosphatidylserine to the outer surface of the plasma membrane were marked by 160 min, and mitochondrial transmembrane potential collapsed over roughly the same time scale (1). Similar effects were observed in Jurkat E6 cells. In MOLT-4 cells, pro-caspase 8 was reduced within 130 min, and the proteolytically activated caspase 8 substrate Bid was also reduced by this time, implicating release of cytochrome c from mitochondria by the active 15 kDa fragment of Bid (1). Substantial proteolytic activation of pro-caspase 3 was relatively delayed.
In the current work reported here, we have undertaken further characterization of the induction of apoptosis by CpG 5mers in MOLT-4 cells. Induction of apoptosis in MOLT-4 cells did not require new protein synthesis and was insensitive to the caspase 3 inhibitor, Ac-DEVD-CHO, although the latter abrogated DNA laddering, phosphatidylserine externalization and collapse of the mitochondrial transmembrane potential. Consequently, it would appear that, contrary to our original conclusions, the mitochondrial mechanism is collateral damage that is not obligatory for CpG oligodeoxynucleotide-induced cell death. The determination of differential gene expression between CpG-sensitive MOLT-4 cells and a subline with acquired resistance has given some insight into the signalling pathways that may be involved in CpG oligodeoxynucleotide 5mer-induced apoptosis.
MATERIALS AND METHODS
Cell culture
The human T-cell leukaemia line MOLT-4 was supplied by the European Collection of Animal Cell Cultures (PHLS Centre for Applied Microbiology and Research, Porton Down, Wiltshire, UK). Cells were maintained in exponential growth in RPMI 1640 medium supplemented with l-glutamine (Life Technologies Ltd, Paisley, Renfrewshire, UK) and 10% heat-inactivated fetal bovine serum (Harlan Sera-Lab Ltd, Crawley Down, West Sussex, UK). Cell number per ml was determined on a Coulter Counter ZM (Coulter Electronics Ltd, Luton, Bedfordshire, UK). Viable cell number per ml was measured by flow cytometry of cells excluding propidium iodide on an Ortho Cytoron Absolute (Ortho Diagnostic Systems Ltd, High Wycombe, Buckinghamshire, UK).
Oligodeoxynucleotide synthesis and cell treatment
Oligodeoxynucleotides were synthesized on a DMT-C3-succinyl-CPG support (Peninsula Laboratories Europe Ltd, St Helens, Merseyside, UK) to provide protection against 3′-exonuclease by substitution of the 3′-OH with a 3-hydroxypropyl phosphate group. 5′-Amino-modifier C6-TFA (Glen Research, Sterling, VA; UK supplier Cambio Ltd, Cambridge) was incorporated in the final cycle of the synthesis and the oligodeoxynucleotides were labelled with fluorescein, post-synthesis, using Fluos reagent (Roche Diagnostics Ltd, Lewes, East Sussex, UK). The caspase 3 inhibitor Ac-DEVD-CHO, the caspase 6/caspase 8 inhibitor Ac-IETD-CHO and the broad spectrum caspase inhibitor Z-VAD-fmk were from BIOMOL Research Laboratories Inc. (Plymouth Meeting, PA; UK supplier Affiniti Research Products Ltd, Mamhead, Exeter). Oligodeoxynucleotides and caspase inhibitors were introduced into the cytoplasm of cells by 10 min reversible permeabilization of the plasma membrane with streptolysin O (Sigma-Aldrich Company Ltd, Poole, Dorset, UK) in the presence of external concentrations of 20 µM oligodeoxynucleotide and the indicated concentrations of inhibitors in serum-free RPMI 1640, as previously described (11). A full experimental protocol for this technique is available at http://www.liv.ac.uk/∼giles. Results presented represent cultures in which >85% of the cells had taken up the oligodeoxynucleotide and subsequently resealed to exclude propidium iodide as determined by dual parameter flow cytometry. The MOLT-4CpGR subline of MOLT-4 cells was established through selection for resistance to CpG 5mer-induced apoptosis, by repeated streptolysin O-mediated intracytoplasmic introduction of CGTTA into cells from an extracellular concentration of 20 µM. Cells that survived following extended incubation after application of the treatment were expanded in culture and treated again. This process was repeated for a total of six treatments over the course of 6 months, after which the cells selected were effectively resistant to CpG 5mer-induced apoptosis.
Apoptosis assays
Redistribution of plasma membrane phosphatidylserine to the outer cell surface and collapse of mitochondrial transmembrane potential were measured by flow cytometry on cells loaded with unlabelled oligodeoxynucleotide using fluorescein isothiocyanate (FITC)-labelled annexin V (Clontech Laboratories UK Ltd, Basingstoke, Hampshire, UK) and the DePsipher kit (R&D Systems Europe Ltd, Abingdon, Oxfordshire, UK) respectively, according to the manufacturers’ instructions. DNA laddering was monitored by electro phoresis through 0.8% agarose gels containing 1 µg/ml ethidium bromide.
Western blotting
Effects on cellular content of protein were determined by densitometry of western blots as previously described (12,13). Rabbit antibodies against human caspase 3 and pro-caspase 8 were from Upstate Biotechnology (Waltham, MA; UK supplier TCS Biologicals Ltd, Buckingham) and rabbit antibody against human Bid was from R&D Systems Europe Ltd. Anti-rabbit IgG (whole molecule) alkaline phosphatase conjugate was supplied by Sigma-Aldrich Company Ltd.
Determination of differential gene expression between MOLT-4 and MOLT-4CpGR cells by reciprocal cDNA subtraction, consensus selection and virtual cloning through targeted display
Reciprocal PCR-Select cDNA subtractive hybridizations (Clontech Laboratories UK Ltd; http://www.clontech.com/techinfo/manuals/PDF/PT1117-1.pdf) were performed, following isolation of RNA from sensitive and CpG 5mer-resistant cell lines and its conversion to SMART PCR cDNA (Clontech Laboratories UK Ltd; http://www.clontech.com/techinfo/manuals/PDF/PT3041-1.pdf). It was expected that there would be comparatively few differences in gene expression between the two cell lines and, as Clontech caution, this can lead to a very high percentage of false positives in libraries generated by PCR-Select cDNA subtraction. Therefore, as a first step, mirror orientation selection (14) was applied to subtracted cDNA populations to reduce the level of false positives somewhat. A total of three independent, reciprocal cDNA subtraction experiments were performed, and a novel procedure of consensus selection was devised and applied to these to select for and amplify true differentially expressed cDNAs. Consensus selection is based on the principle that it is unlikely that the same false-positive cDNA molecules would slip through the net and subsequently become PCR amplified in both orientations relative to the adaptors in independent PCR-Select cDNA subtraction experiments. On the other hand, true differentially expressed cDNAs should be amplified each time. Therefore, we selected for and amplified sequences that were present in all subtracted cDNA populations derived from the three independent subtraction experiments, A, B and C. The subtracted cDNA populations were first digested with RsaI, to remove the residual adaptors, and were separated from the cleaved oligodeoxynucleotides on Microcon PCR devices (Millipore Corporation, Bedford, MA). Subtraction of the cDNAs was repeated exactly as specified by Clontech, except that this time the drivers were the pooled subtracted cDNAs from three experiments, and the adaptors 1 and 2R were ligated individually to cDNAs from two different subtraction experiments to generate the tester. Therefore, the tester could only be amplified during suppression PCR if the same double-stranded sequences were present originally in the subtracted cDNA from the two experiments, to give subsequently hybrids with different adaptors at each end. Performing consensus selection across three independent reciprocal cDNA subtraction experiments produced three consensus template mixtures, A–B, B–C and A–C, from resistant cells and three from the reciprocal subtractions against cDNA from sensitive cells. Following overnight second hybridization, the primary suppression PCR step was 20 cycles and the secondary PCR with nested primers was run for 16 cycles. Consensus-subtracted cDNAs were subjected to virtual cloning by targeted display using 76 different primer combinations (15). Where three intense bands of identical size on agarose gels were observed for one cell line but not the other, they were excised, reamplified and sequenced. The corresponding genes were identified by BLAST searches with the sequence fragments against GenBank and the human genome sequence.
RESULTS AND DISCUSSION
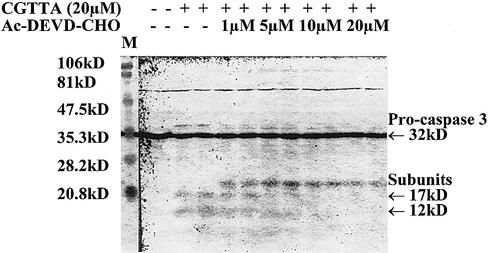
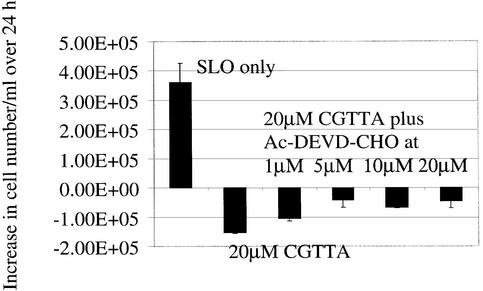
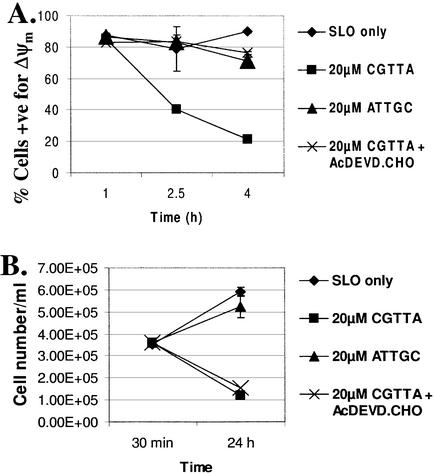
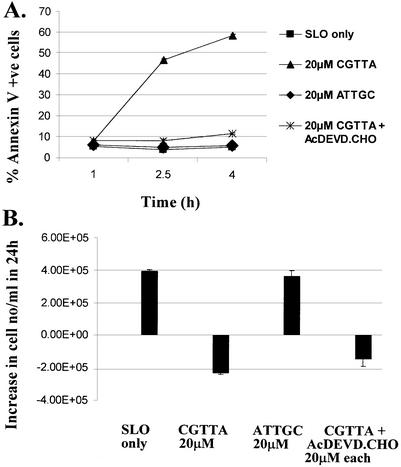
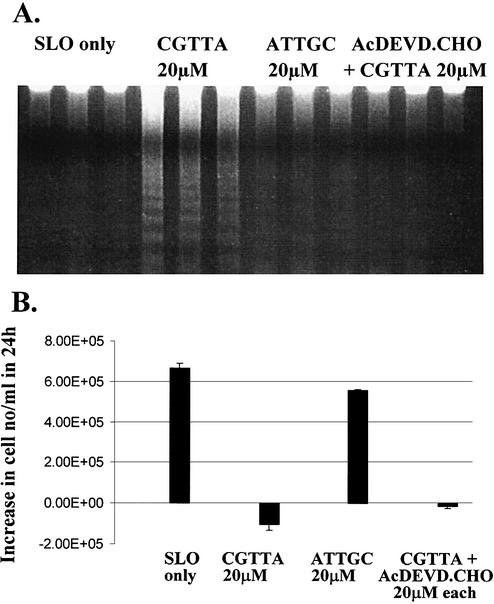
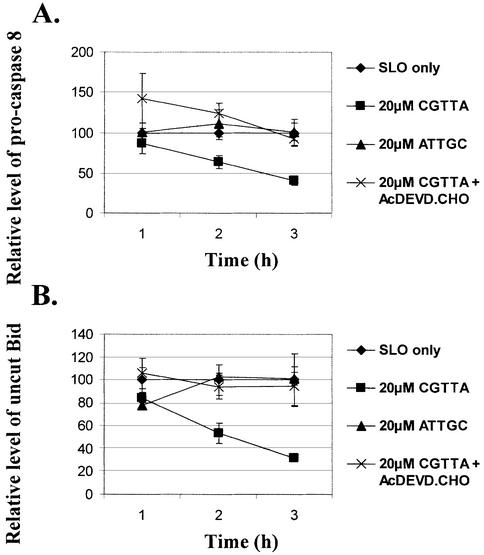
In our previous report (1), we concluded that apoptosis induced by CpG oligodeoxynucleotide 5mers in T-lymphocytic leukaemia cells exhibited some of the characteristics of that in ‘type II cells’ triggered into mitochondria-dependent programmed cell death by cross-linking of CD95 (16,17). However, the caspase 3 inhibitor Ac-DEVD-CHO failed to protect the cells. Therefore, since activation of caspase 3 is apparently central to CD95-induced apoptosis in type II cells, it was of interest to look more closely into why the inhibitor failed to afford any protection against CpG 5mers. Low levels of the active 12 and 17 kDa subunits of caspase 3 are detectable at early times after the initiation of treatment within MOLT-4 cells, but these represent only a fraction of the level of 32 kDa pro-caspase 3 present (Fig. 1). Delivery of the caspase 3 inhibitor Ac-DEVD-CHO along with the oligodeoxynucleotide abrogated the production of the active 12 and 17 kDa subunits. However, the inhibitor did not prevent upstream proteolytic processing of pro-caspase 3, as evidenced by the accumulation of the initial cleavage product of ∼20 kDa (Fig. 1). This fragment is seen on western blots of type I cells undergoing apoptosis, but does not normally accumulate in type II cells (16). As shown in Figure 2, inhibition of caspase 3 failed to protect the cells against induction of apoptosis by CGTTA. The caspase inhibitor alone was completely non-toxic to MOLT-4 cells at up to 40 µM, the highest concentration tested (data not shown). The caspase 6/caspase 8 inhibitor Ac-IETD-CHO and the broad spectrum caspase inhibitor Z-VAD-fmk also failed to protect cells against the CpG 5mer (data not shown). However, the caspase 3 inhibitor did abrogate collapse of the mitochondrial transmembrane potential, Δψm (Fig. 3), externalization of phosphatidylserine as measured by annexin V binding (Fig. 4), and DNA laddering (Fig. 5). All these classic characteristic features of cells undergoing programmed cell death were nullified through application of caspase 3 inhibition, but the cells were still dying at essentially the same rate following intracytoplasmic delivery of CGTTA. The caspase 3 inhibitor also prevented proteolytic processing of pro-caspase 8 and Bid (Fig. 6), suggesting that these events are not upstream of caspase 3 activation, as had been inferred previously (1). It would appear that, contrary to our original conclusions (1), the mitochondrial mechanism is collateral damage that is not obligatory for CpG oligodeoxynucleotide-induced cell death.
Figure 1.
Effect of the caspase 3 inhibitor, Ac-DEVD-CHO on CpG 5mer-mediated induction of caspase 3 active subunits at 4 h in MOLT-4 cells. The oligodeoxynucleotide CGTTA and caspase 3 inhibitor were delivered into cells by streptolysin O permeabilization at 0 h from the indicated extracellular concentrations.
Figure 2.
Lack of effect of the caspase 3 inhibitor Ac-DEVD-CHO on CpG 5mer-mediated induction of apoptosis in MOLT-4 cells. The oligodeoxynucleotide CGTTA was delivered into cells by streptolysin O permeabilization from an extracellular concentration of 20 µM alone or in combination with the indicated concentrations of caspase 3 inhibitor. The increase in the number of cells per ml excluding propidium iodide over 24 h was determined by flow cytometry. The starting cell density was 4 × 105 cells/ml. SLO only: cells were permeabilized with streptolysin O in serum-free medium alone. Plotted are the mean and standard deviation of two replicates.
Figure 3.
Inhibition of CpG 5mer-induced collapse of Δψm in MOLT-4 cells by Ac-DEVD-CHO without increase in cell survival. (A) The percentage of cells exhibiting red fluorescence from the dye JC-1, typical of normal mitochondrial transmembrane potential, was determined by flow cytometry. The oligodeoxynucleotide CGTTA, inverse control ATTGC, and caspase 3 inhibitor were delivered into cells by streptolysin O permeabilization at 0 h all from an extracellular concentration of 20 µM. SLO only: cells were permeabilized with streptolysin O in serum-free medium alone. (B) Cell number per ml excluding propidium iodide was determined by flow cytometry. Plotted are the mean and standard deviation of two replicates.
Figure 4.
Inhibition of CpG 5mer-induced phosphatidylserine externalization on MOLT-4 cells by Ac-DEVD-CHO. (A) The percentage of cells exhibiting externalization of phosphatidylserine through binding of fluorescein-labelled annexin V was determined by flow cytometry. The oligodeoxynucleotide CGTTA, inverse control ATTGC and caspase 3 inhibitor were delivered into cells by streptolysin O permeabilization at 0 h, all from an extracellular concentration of 20 µM. SLO only: cells were permeabilized with streptolysin O in serum-free medium alone. (B) The increase in the number of cells per ml excluding propidium iodide over 24 h was determined by flow cytometry. The starting cell density was 4 × 105 cells/ml. Plotted are the mean and standard deviation of two replicates.
Figure 5.
Inhibition by Ac-DEVD-CHO of CpG 5mer-induced MOLT-4 DNA laddering at 2.5 h. (A) The oligodeoxynucleotide CGTTA, inverse control ATTGC, and caspase 3 inhibitor were delivered into cells by streptolysin O permeabilization at 0 h, all from an extracellular concentration of 20 µM. SLO only: cells were permeabilized with streptolysin O in serum-free medium alone. DNA was isolated and separated by electrophoresis through a 0.8% agarose gel containing 1 µg/ml ethidium bromide. The three lanes corresponding to each treatment are three replicates of the treatment. (B) The increase in the number of cells per ml excluding propidium iodide over 24 h was determined by flow cytometry. The starting cell density was 4 × 105 cells/ml. Plotted are the mean and standard deviation of three replicates.
Figure 6.
Inhibition of CpG 5mer-induced proteolytic processing of pro- caspase 8 and Bid in MOLT-4 cells by Ac-DEVD-CHO. The oligodeoxynucleotide CGTTA, inverse control ATTGC and caspase 3 inhibitor were delivered into cells by streptolysin O permeabilization at 0 h, all from an extracellular concentration of 20 µM. SLO only: cells were permeabilized with streptolysin O in serum-free medium alone. Protein extracts were prepared from 7.5 × 105 cell samples taken at the indicated times and 25 µg of total protein extract per lane submitted to western blotting analysis. Levels of the proteins were determined in arbitrary units by densitometry of western blots and are normalized to the values for the SLO only controls taken as 100% at each time point. Plotted are the mean and standard deviation of three replicates.
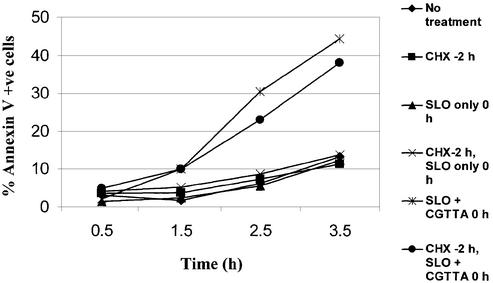
One hypothesis that was entertained for the potential mechanism of action of CpG 5mer oligodeoxynucleotides was that they might be disrupting synthesis of anti-apoptotic proteins or relieving suppression of pro-apoptotic proteins through interactions with transcription factors. CpG dinucleotides are found commonly in enhancer elements and transcription factor-binding sites. Although these are necessarily double stranded, it is not inconceivable that a large molar excess of single-stranded CpG 5mer could block transcription factor binding. The CpG could interact reversibly with those residues of the protein normally contacting these bases, and inhibit binding in a manner reminiscent of the mechanisms of classical competitive inhibitors of enzyme activity. The resulting imbalance between pro-apoptotic and anti-apoptotic proteins could trigger an irreversible commitment to programmed cell death. However, this scenario would require new protein synthesis. The application of a concentration of cycloheximide that completely inhibited protein synthesis (12) for 2 h prior to introduction of CGTTA did not significantly affect the onset of apoptosis in MOLT-4 cells, as demonstrated by the externalization of phosphatidylserine (Fig. 7). Consequently, it was apparent that investigation of changes in gene expression following delivery of oligodeoxynucleotide, in the absence of cycloheximide, would be uninformative in terms of the mechanism of action of CpG 5mers. On the other hand, alteration in cell constitution to counter the activity of CpG oligodeoxynucleotides would be highly informative in defining the processes affected by the oligodeoxynucleotides, and would help to pinpoint the actual target. Development of resistance to CpG 5mers would involve significant changes at several neighbouring nodes in the intracellular signalling web to isolate the target site and thereby to compensate for the vulnerability of a principal signalling node upon application of the oligomers.
Figure 7.
Total inhibition of de novo protein synthesis with cycloheximide fails to significantly affect CpG 5mer-induced phosphatidylserine externalization on MOLT-4 cells. The oligodeoxynucleotide CGTTA was delivered into cells by streptolysin O permeabilization at 0 h from an extracellular concentration of 20 µM. Where indicated by ‘CHX at –2 h’, cells from no treatment control, streptolysin permeabilization only control and streptolysin permeabilization with CGTTA were pre-treated for 2 h with 20 µg/ml cycloheximide to achieve prior inhibition of protein synthesis. Following resuspension in fresh medium at 0 h, cycloheximide at 20 µg/ml was added to all treatments, both to maintain protein synthesis inhibition and to control for any posssible apoptosis-inducing activity of the inhibitor itself. The percentage of cells exhibiting externalization of phosphatidylserine through binding of fluorescein-labelled annexin V was determined by flow cytometry.
A subline of MOLT-4 cells, MOLT-4CpGR, was established through selection for resistance to CpG 5mer-induced apoptosis, by repeated streptolysin O-mediated intracytoplasmic introduction of CGTTA into cells that had survived and were expanded in culture following the previous treatment. An investigation was undertaken to identify differences in gene expression between the parent sensitive MOLT-4 cells and the resistant subline. It was anticipated that differences in gene expression between the two cell lines would be relatively few, and indeed several rounds of targeted display (15) with total cDNA prepared from the cells failed to show any detectable differences in banding patterns on 1.5% agarose– ethidium bromide gels. Clontech’s PCR-Select reciprocal cDNA subtraction procedure was used to enrich for differentially expressed cDNAs. Mirror orientation selection (14) was applied to the subtracted cDNA populations to reduce the percentage of false positives, which can be very high in PCR-Select-subtracted cDNA libraries when differences in gene expression between the cells being compared are few. The PCR-Select cDNA subtraction procedure is based upon the principle that homohybrids of the tester (the cDNA population being subtracted with the driver cDNA without end adaptors) formed during the first subtractive hybridization step, with the same adaptor ligated at each end, will not be amplified during the subsequent round of suppression PCR (18). Only hybrids with different adaptors at each end, formed during the second hybridization step, will be amplified in the suppression PCR step. It was apparent that the same technique could be exploited to achieve consensus selection across independent subtraction experiments. Consensus selection is based upon the principle that it is unlikely that the same false-positive cDNA molecules would fall through the net and subsequently become PCR amplified in both orientations with respect to the adaptors in independent PCR-Select cDNA subtraction experiments. On the other hand, true differentially expressed cDNAs should be amplified each time. If the two different adaptors 1 and 2R are ligated individually to subtracted cDNAs from two independent subtractions, only cDNA sequences and their complements that are present in both cDNA populations will form hybrids with different adaptors at each end upon mixing, dissociation and reannealing. These, which are likely to be true differentially expressed cDNAs, may then be amplified selectively by suppression PCR. Consensus selection as described in Materials and Methods was applied across three independent reciprocal cDNA subtraction experiments, A, B and C, between CpG 5mer-sensitive and resistant MOLT-4 cells. This generated three template mixtures, A–B, B–C and A–C, for the resistant cells and three for the reciprocally subtracted parent MOLT-4 cells. Following suppression PCR, and then PCR with nested primers, clear differences between the cell lines in banding patterns extending across all three lanes were observed for total amplified products on 1.5% agarose–ethidium bromide gels. Differences were also readily detected by targeted display (15). The three consensus-selected, subtracted cDNA populations from resistant cells and the three from sensitive cells were subjected to virtual cloning by targeted display using 76 different primer combinations. Where intense bands of identical size were observed on agarose gels in all three lanes for one cell line but not the other, the bands were excised, reamplified and sequenced. The corresponding genes were identified by BLAST searches against GenBank and the human genome sequence. Genes that were downregulated in CpG 5mer-resistant cells are listed in Table 1, while Table 2 presents upregulated genes.
Table 1. Genes downregulated in CpG oligodeoxynucleotide 5mer-resistant MOLT-4CpGR cells.
| Caspase 2, apoptosis-related cysteine protease (neural precursor cell expressed, developmentally downregulated 2) (CASP2) |
| APMCF1 protein (apoptotic-related gene in MCF-7 cell line 1) (APMCF1) |
| MCM5 minichromosome maintenance-deficient 5, cell division cycle 46 (S.cerevisiae) (MCM5) |
| Bromodomain-containing 4 (BRD4) |
| Activating transcription factor 4 (tax-responsive enhancer element B67) (ATF4) |
| Inositol 1,4,5-trisphosphate 3-kinase B (ITPKB) |
| Putative chemokine receptor; GTP-binding protein (HM74) |
| I-mfa domain-containing protein (HIC) |
| Ribosomal protein L28 (RPL28) |
| LOC144100, Model mRNA XM_084732, Model Protein XP_084732 |
| UniGene Cluster Hs.85573. Hypothetical protein MGC10911 (MGC10911) |
| UniGene Cluster Hs.43145. Similar to sp:P51814-ZN41_HUMAN Zinc finger protein 41 |
| UniGene Cluster Hs.180428. KIAA1181 protein (KIAA1181) |
NCBI LocusLink locus name in parentheses. http://www.ncbi.nlm.nih.gov/LocusLink/
Table 2. Genes upregulated in CpG oligodeoxynucleotide 5mer-resistant MOLT-4CpGR cells.
| Phosphoinositide-3-kinase, catalytic, γ polypeptide (PIK3CG) |
| Annexin A1 (ANXA1) [synonyms: annexin A1, lipocortin, calpactin; proteins that inhibit phospholipase A2 (PLA2) activity] |
| Transmembrane 4 superfamily member 2 (TM4SF2) (synonym: T-cell acute lymphoblastic leukaemia-associated antigen 1, TALLA-1) |
| RNA-binding motif protein, X chromosome (RBMX) |
| Splicing factor, arginine/serine-rich 5 (SFRS5) (alternative symbols: HRS, SRP40) |
| Splicing factor, arginine/serine-rich 7, 35 kDa (SFRS7) (alternative symbols: 9G8, HSSG1. Alias: heat-shock suppressed protein 1) |
| Homeobox B3 (HOXB3) |
| Required for cell differentiation1 homologue (S.pombe) (RQCD1) |
| Seven in absentia homologue 1 (Drosophila) (SIAH1) (E3 ubiquitin ligase) |
| Ubiquitin-activating enzyme E1-like protein (human glucose-induced selective autophagy 7) (GSA7) |
| RUN and FYVE domain-containing 2 (RUFY2) |
| Nerve injury gene 283 (NIN283) (member of a novel family of E3 ubiquitin ligases) |
| Mucosa-associated lymphoid tissue lymphoma translocation gene 1 (MALT1/MLT1) |
| Histidyl-tRNA synthetase 2 (HARS2) |
| Adenylate kinase 2, mitochondrial (ATP–AMP transphosphorylase) (AK2). (AK2 is localized in the mitochondrial intermembrane space and may play a role in apoptosis) |
| Mitochondrial ribosomal protein L48 (MRPL48) |
| Ribosomal protein S11 (RPS11) |
| UniGene Cluster Hs.313342. Homo sapiens mRNA; cDNA DKFZp313F0317 (from clone DKFZp313F0317) |
| UniGene Cluster Hs.57932. H.sapiens cDNA FLJ14134 fis, clone MAMMA1002708 (similar to 2004399A. Chromosomal protein gi: 740170) |
| UniGene Cluster Hs.200595. KIAA0562 gene product [similar in part to Rattus norvegicus glycine-, glutamate-, thienylcyclohexyl- piperidine-binding protein (LOC246295)] |
| UniGene Cluster Hs.16229. KIAA1373 protein. Hit in putative intron of the gene |
| UniGene Cluster Hs.23850. Unknown gene defined by expressed sequence tags |
| UniGene Cluster Hs.179112. Expressed sequence tags, highly similar to T43444 hypothetical protein DKFZp434C1715.1–human (fragment) |
| UniGene Cluster Hs.6451. PRO0659 protein product, function unknown (PRO0659) |
| UniGene Cluster Hs.58217. H.sapiens, clone MGC: 10002 IMAGE: 3882800, mRNA, complete cds |
| UniGene Cluster Hs.289455. Expressed sequence tags |
| UniGene Cluster Hs.16063. Hypothetical protein FLJ21877. (Alias: death receptor-interacting protein) |
| SPTF-associated factor 65γ [STAF65(γ)] |
NCBI LocusLink locus name in parentheses. http://www.ncbi.nlm.nih.gov/LocusLink/
A picture is starting to emerge. Caspase 2 has been implicated as mediating the DNA damage response (19–21), and the CpG 5mers could well be triggering this pathway. It is noteworthy that several nucleic acid-interacting proteins are also involved. In addition to the transcription factors Atf4, Hic, Hoxb3 and Rqcd1, and splicing factors Rbmx, Sfrs5 and Sfrs7, for example, Mcm5 is one member of a hexameric complex that is a key component of the pre-replication complex that assembles at DNA replication origins during early G1 phase (22). Brd4 carries two bromodomains implicated in binding to chromatin, and the protein also interacts with DNA replication factor C (23). However, the possible involvement of p53 may be excluded from consideration for the mechanism of induction of apoptosis by CpG oligodeoxynucleotides, since the p53 pathway is inoperative in the MOLT-4 cell line used in our experiments. These cells were found to be heterozygous for the p53 gene, with one allele carrying a mutation at the mutation hot spot, codon 248, where CGG was mutated to CAG, converting an arginine to glutamine in the protein product of the mutant allele (24). This mutation has been shown previously to have a dominant-negative effect on the activity of the product of the wild-type allele (25).
The apoptosis-resistant cells have enhanced protein ubiquitylation capacity with upregulated Siah1, Gsa7 and Nin283, and this could be contributing to the protection against programmed cell death through enhanced proteolysis of apoptosis-inducing factors (26–28). Phosphoinositide-3-kinase has been recognized as playing a role in cell survival in the PTEN/PIK3/AKT pathway (29–31), and increased annexin A1 may protect against apoptosis through inhibition of phospholipase A2 (32,33). Enhanced levels of Malt1 in the resistant cells may result in the activation of NF-κB (34). Adenylate kinase 2 is localized in the mitochondrial intermembrane space and is released into the cytosol in apoptotic cells, but its role during apoptosis is not established (35,36).
In conclusion, it would appear that apoptosis induced by CpG oligodeoxynucleotide 5mers may entrain elements of the classical mitochondrial and cell surface receptor-mediated pathways (16,17), but that these are not essential for induction of cell death. CpG 5mer-induced apoptosis may proceed in the absence of caspase 3 activity and without the characteristic DNA laddering, phosphatidylserine externalization and collapse of the mitochondrial transmembrane normally associated with apoptosis triggered by other natural and artificial inducers. Further work on elucidating the mandatory upstream initiator and downstream effector apoptotic pathways activated by these oligomers will proceed from the leads provided by the cDNA subtraction experiments with sensitive and resistant MOLT-4 cells.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by the Leukaemia Research Fund of Great Britain.
REFERENCES
- 1.Tidd D.M., Spiller,D.G., Broughton,C.M., Norbury,L.C., Clark,R.E. and Giles,R.V. (2000) Oligodeoxynucleotide 5mers containing a 5′-CpG induce apoptosis through a mitochondrial mechanism in T lymphocytic leukaemia cells. Nucleic Acids Res., 28, 2242–2250. [DOI] [PMC free article] [PubMed]
- 2.Giles R.V., Spiller,D.G., Clark,R.E. and Tidd,D.M. (1999) Antisense morpholino oligonucleotide analog induces missplicing of c-myc mRNA. Antisense Nucleic Acid Drug Dev., 9, 213–220. [DOI] [PubMed]
- 3.Tidd D.M., Giles,R.V., Broughton,C.M. and Clark,R.E. (2001) Expression of c-myc is not critical for cell proliferation in established human leukemia lines. BMC Mol. Biol., 2, 13–00. (http://www.biomedcentral.com/1471-2199/2/13) [DOI] [PMC free article] [PubMed]
- 4.Thompson E.B. (1998) The many roles of c-Myc in apoptosis. Annu. Rev. Physiol., 60, 575–600. [DOI] [PubMed]
- 5.Lowe S.W. and Lin,A.W. (2000) Apoptosis in cancer. Carcinogenesis, 21, 485–495. [DOI] [PubMed]
- 6.Tsuneoka M. and Mekada,E. (2000) Ras/MEK signalling suppresses Myc-dependent apoptosis in cells transformed by c-myc and activated ras. Oncogene, 19, 115–123. [DOI] [PubMed]
- 7.Jamerson M.H., Johnson,M.D. and Dickson,R.B. (2000) Dual regulation of proliferation and apoptosis: c-myc in bitransgenic murine mammary tumor models. Oncogene, 19: 1065–1071. [DOI] [PubMed]
- 8.Henriksson M., Selivanova,G., Lindstrom,M. and Wiman,K.G. (2001) Inactivation of Myc-induced p53-dependent apoptosis in human tumors. Apoptosis, 6, 133–137. [DOI] [PubMed]
- 9.Pelengaris S., Khan,M. and Evan,G.I. (2002) Suppression of Myc-induced apoptosis in β cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell, 109, 321–334. [DOI] [PubMed]
- 10.Jeong H., Mason,S.P., Barabasi,A.-L. and Oltvai,Z.N. (2001) Lethality and centrality in protein networks. The most highly connected proteins in the cell are the most important for its survival. Nature, 411, 41–42. [DOI] [PubMed]
- 11.Giles R.V., Grzybowski,J., Spiller,D.G. and Tidd,D.M. (1997) Enhanced antisense effects resulting from an improved streptolysin-O protocol for oligodeoxynucleotide delivery into human leukaemia cells. Nucl. Nucl., 16, 1155–1163.
- 12.Spiller D.G., Giles,R.V., Broughton,C.M., Grzybowski,J., Ruddell,C.J., Tidd,D.M. and Clark,R.E. (1998) The influence of target protein half-life on the effectiveness of antisense oligonucleotide analog-mediated biologic responses. Antisense Nucleic Acid Drug Dev., 8, 281–293. [DOI] [PubMed]
- 13.Giles R.V., Spiller,D.G., Grzybowski,J., Clark,R.E., Nicklin,P. and Tidd,D.M. (1998) Selecting optimal oligonucleotide composition for maximal antisense effect following streptolysin O-mediated delivery into human leukaemia cells. Nucleic Acids Res., 26, 1567–1575. [DOI] [PMC free article] [PubMed]
- 14.Rebrikov D.V., Britanova,O.V., Gurskaya,N.G., Lukyanov,K.A., Tarabykin V.S. and Lukyanov,S.A. (2000) Mirror orientation selection (MOS): a method for eliminating false positive clones from libraries generated by suppression subtractive hybridization. Nucleic Acids Res., 28, e90. [DOI] [PMC free article] [PubMed]
- 15.Brown A.J.H., Hutchings,C., Burke,J.F. and Mayne,L.V. (1999) Application of a rapid method (targeted display) for the identification of differentially expressed mRNAs following NGF-induced neuronal differentiation in PC12 cells. Mol. Cell. Neurosci., 13, 119–130. [DOI] [PubMed]
- 16.Scaffidi C., Fulda,S., Srinivasan,A., Friesen,C., Li,F., Tomaselli,K.J., Debatin,K.-M., Krammer,P.H. and Peter,M.E. (1998) Two CD95 (APO-1/Fas) signaling pathways. EMBO J., 17, 1675–1687. [DOI] [PMC free article] [PubMed]
- 17.Scaffidi C., Schmitz,I., Zha,J., Korsmeyer,S.J., Krammer,P.H. and Peter,M.E. (1999) Differential modulation of apoptosis sensitivity in CD95 type I and type II cells. J. Biol. Chem., 274, 22532–22538. [DOI] [PubMed]
- 18.Gurskaya N.G., Diatchenko,L., Chenchik,A., Siebert,P.D., Khaspekof,G.L., Lukyanov,K.A., Vagner,L.L., Ermolaeva,O.D., Lukyanov,S.A. and Sverdlov,E.D. (1996) Equalizing cDNA subtraction based on selective suppression of polymerase chain reaction: cloning of Jurkat cell transcripts induced by phytohemaglutinin and phorbol 12-myristate 13-acetate. Anal. Biochem., 240, 90–97. [DOI] [PubMed]
- 19.Zhu J.H., Nozell,S., Wang,J., Jiang,J.Y., Zhou,W.J. and Chen,X.B. (2001) p73 cooperates with DNA damage agents to induce apoptosis in MCF7 cells in a p53-dependent manner. Oncogene, 20, 4050–4057. [DOI] [PubMed]
- 20.Robertson J.D., Enoksson,M., Suomela,M., Zhivotovsky,B. and Orrenius,S. (2002) Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J. Biol. Chem., 277, 29803–29809. [DOI] [PubMed]
- 21.Lassus P., Opitz-Araya,X. and Lazebnik,Y. (2002) Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science, 297, 1352–1354. [DOI] [PubMed]
- 22.Tye B.K. (1999) MCM proteins in DNA replication. Annu. Rev. Biochem., 68, 649–686. [DOI] [PubMed]
- 23.Maruyama T., Farina,A., Dey,A., Cheong,J., Bermudez,V.P., Tamura,T., Sciortino,S., Shuman,J., Hurwitz,J. and Ozato,K. (2002) A mammalian bromodomain protein, Brd4, interacts with replication factor C and inhibits progression to S phase. Mol. Cell. Biol., 22, 6509–6520. [DOI] [PMC free article] [PubMed]
- 24.Ruddell C.J. (2000) Antisense oligonucleotide-mediated inhibition of mutant p53 expression in cultured human cells. PhD thesis, The University of Liverpool, UK. [DOI] [PubMed]
- 25.O’Connor P.M., Jackman,J., Jondle,D., Bhatia,K., Magrath,I. and Kohn,K.W. (1993) Role of the p53 tumor-suppressor gene in cell-cycle arrest and radiosensitivity of Burkitt’s-lymphoma cell-lines. Cancer Res., 53, 4776–4780. [PubMed]
- 26.Johnsen S.A., Subramaniam,M., Monroe,D.G., Janknecht,R. and Spelsberg T.C. (2002) Modulation of transforming growth factor beta (TGF beta)/Smad transcriptional responses through targeted degradation of TGF beta-inducible early gene-1 by human seven in absentia homologue. J. Biol. Chem., 277, 30754–30759. [DOI] [PubMed]
- 27.Wilson R., Goyal,L., Ditzel,M., Zachariou,A., Baker,D.A., Agapite,J., Steller,H. and Meier,P. (2002) The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nature Cell Biol., 4, 445–450. [DOI] [PubMed]
- 28.Jesenberger V. and Jentsch,S. (2002) Deadly encounter: ubiquitin meets apoptosis. Nature Rev. Mol. Cell. Biol., 3, 112–121. [DOI] [PubMed]
- 29.Maehama T. and Dixon,J.E. (1998) The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem., 273, 13375–13378. [DOI] [PubMed]
- 30.Cantley L.C. and Neel,B.G. (1999) New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc. Natl Acad. Sci. USA, 96, 4240–4245. [DOI] [PMC free article] [PubMed]
- 31.Viglietto G., Motti,M.L., Bruni,P., Melillo,R.M., D’Alessio,A., Califano,D., Vinci,F., Chiappetta,G., Tsichlis,P., Bellacosa,A., Fusco,A. and Santoro,M. (2002) Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27Kip1 by PKB/Akt-mediated phosphorylation in breast cancer. Nature Med., 8, 1136–1144. [DOI] [PubMed]
- 32.Wu Y.L., Jiang,X.R., Lillington,D.M., Newland,A.C. and Kelsey,S.M. (2000) Upregulation of lipocortin 1 inhibits tumour necrosis factor-induced apoptosis in human leukaemic cells: a possible mechanism of resistance to immune surveillance. Br. J. Haematol., 111, 807–816. [PubMed]
- 33.Zhao M., Brunk,U.T. and Eaton,J.W. (2001) Delayed oxidant-induced cell death involves activation of phospholipase A2. FEBS Lett., 509, 399–404. [DOI] [PubMed]
- 34.Lucas P.C., Yonezumi,M., Inohara,N., McAllister-Lucas,L.M., Abazeed,M.E., Chen,F.F. and Yamaoka,S. (2001) Bcl10 and MALT1, independent targets of chromosomal translocation in MALT lymphoma, cooperate in a novel NF-kappa B signaling pathway. J. Biol. Chem., 276, 19012–19019. [DOI] [PubMed]
- 35.Kohler C., Gahm,A., Noma,T., Nakazawa,A., Orrenius,S. and Zhivotovsky,B. (1999) Release of adenylate kinase 2 from the mitochondrial intermembrane space during apoptosis. FEBS Lett., 447, 10–12. [DOI] [PubMed]
- 36.Lai Q.A., Hu,J.J. and Sun,J.R. (2001) Adenylate kinase and cellular apoptosis. Prog. Biochem. Biophys., 28, 444–446.