Abstract
RNA interference (RNAi), mediated by either long double-stranded RNA (dsRNA) or short interfering RNA (siRNA), has become a routine tool for transient knockdown of gene expression in a wide range of organisms. The antisense strand of the siRNA duplex (antisense siRNA) was recently shown to have substantial mRNA depleting activity of its own. Here, targeting human Tissue Factor mRNA in HaCaT cells, we perform a systematic comparison of the activity of antisense siRNA and double-strand siRNA, and find almost identical target position effects, appearance of mRNA cleavage fragments and tolerance for mutational and chemical backbone modifications. These observations, together with the demonstration that excess inactive double-strand siRNA blocks antisense siRNA activity, i.e. shows sequence-independent competition, indicate that the two types of effector molecules share the same RNAi pathway. Interest ingly, both FITC-tagged and 3′-deoxy antisense siRNA display severely limited activity, despite having practically wild-type activity in a siRNA duplex. Finally, we find that maximum depletion of target mRNA expression occurs significantly faster with antisense siRNA than with double-strand siRNA, suggesting that the former enters the RNAi pathway at a later stage than double-strand siRNA, thereby requiring less time to exert its activity.
INTRODUCTION
In Caenorhabditis elegans specific depletion of mRNA was found both with the antisense and, surprisingly, the sense strand of RNA (1), leading to the discovery of RNA interference (RNAi) by Andrew Fire and co-workers (2). The potent RNAi process, whereby double-stranded (ds)RNA causes specific interference with the expression of the corresponding endogenous genes, appears to function in the defence against virus and transposons. This defence has subsequently been shown to exist in a wide range of species (3–5). With the demonstration of the efficacy of short interfering RNAs (siRNA) in human cells (6–8), a valuable tool for both research and therapeutics was created. Now the development has come full circle with the recent reports that ∼21 nt single-stranded antisense siRNA is almost as potent as the siRNA duplex (9–11).
The double-strand siRNA appears to be incorporated into an inactive RISC complex, requiring unwinding of the duplex with concomitant loss of its sense strand for conversion into an active complex (RISC*) (12). The antisense siRNA is also incorporated into RISC in HeLa cell extracts and supports RISC-specific target RNA cleavage, although at lower efficiency than the siRNA duplex (9,10). The highly diverging estimates of the size of RISC (10,12,13), together with the reports of additional RISC-like complexes (14–17) associated with both siRNA and the related microRNAs (18), suggest the existence of various distinct complexes with possible involvement in different RNAi pathways.
Here we compare antisense and double-strand siRNA using a range of different oligos, demonstrating that the two types of effector molecules behave very similarly with regard to mRNA target position effects, cleavage fragment production and tolerance for chemical and mutational modifications. Further more, antisense siRNA can be blocked by competition with excess inactive double-strand siRNA. Interesting exceptions are antisense siRNAs modified in the 3′-terminus with either a 3′-deoxyribose instead of ribose or a FITC group in the 3′-OH position. These display severely limited activity, despite having practically wild-type activity in a siRNA duplex. Time-course experiments also showed a difference. They demonstrated that antisense siRNA depleted target mRNA faster than siRNA, possibly due to the direct incorporation of the former into RISC. Overall, our data suggest that antisense and double-strand siRNA work through the same RNAi pathway, although the 3′ end of the antisense strand may be treated differently in double-strand siRNA and antisense siRNA.
MATERIALS AND METHODS
RNAi antisense preparation
The 21 nt RNAs were synthesized as described (11,19). Antisense RNA were designated as-N, with N being the corresponding siRNA and mRNA target position (19). The various mutated and chemically modified versions of as-167 were designated as-X, where X is the corresponding siRNA (11). A previously synthesized version of as-167 (as-3d) had a 3′-deoxyribose as the 3′-terminal nucleotide. The siRNAs mentioned in this paper are summarised in Table 1.
Table 1. List of RNA molecules synthesized.
| Unmodified double-stranded and antisense siRNA | Strands/target position in human Tissue Factor |
| hTF167i | double-strand siRNA at hTF167 |
| as-77 | antisense siRNA at hTF77 |
| as-158 | antisense siRNA at hTF158 |
| as-161 | antisense siRNA at hTF161 |
| as-164 | antisense siRNA at hTF164 |
| as-167 | antisense siRNA at hTF167 |
| as-170 | antisense siRNA at hTF170 |
| as-173 | antisense siRNA at hTF173 |
| as-176 | antisense siRNA at hTF176 |
| as-256 | antisense siRNA at hTF256 |
| as-372 | antisense siRNA at hTF372 |
| as-478 | antisense siRNA at hTF478 |
| as-562 | antisense siRNA at hTF562 |
| as-929 | antisense siRNA at hTF929 |
| Modified versions of antisense siRNA as-167 | Modification in antisense strand (nt, nucleotide, numbering from 5′ end) |
| as-s3 | mutated (C→G) at nt 17 |
| as-s7 | mutated (G→C) at nt 13 |
| as-s10 | mutated (C→G) at nt 10 |
| as-s13 | mutated (G→C) at nt 7 |
| as-s16 | mutated (G→C) at nt 4 |
| as-M0+2 | 2-O-methylated 2 nt on 3′ |
| as-M1+1 | 2-O-methylated 1 nt on 5′ and 3′ |
| as-M2+2 | 2-O-methylated 2 nt on 5′ and 3′ |
| as-M2+4 | 2-O-methylated 2 nt on 5′ and 4 nt on 3′ |
| as-M4+4 | 2-O-methylated 4 nt on 5′ and 3′ |
| as-M4+6 | 2-O-methylated 4 nt on 5′ and 6 nt on 3′ |
| as-M6+6 | 2-O-methylated 6 nt on 5′ and 3′ |
| as-M6+8 | 2-O-methylated 6 nt on 5′ and 8 nt on 3′ |
| as-FITC | FITC-group on 3′ position on 3′ nt |
| as-3d | 3′-terminal ribose replaced with 3′-deoxyribose |
| Modified versions of double-strand siRNA hTF167i | Modifications in both strands (except hTF167-3d) |
| M0+2 | 2-O-methylated 2 nt on 3′ |
| M1+1 | 2-O-methylated 1 nt on 5′ and 3′ |
| M2+2 | 2-O-methylated 2 nt on 5′ and 3′ |
| M2+4 | 2-O-methylated 2 nt on 5′ and 4 nt on 3′ |
| M4+4 | 2-O-methylated 4 nt on 5′ and 3′ |
| M4+6 | 2-O-methylated 4 nt on 5′ and 6 nt on 3′ |
| M6+6 | 2-O-methylated 6 nt on 5′ and 3′ |
| M6+8 | 2-O-methylated 6 nt on 5′ and 8 nt on 3′ |
| hTF167-3d | Antisense strand 3′-terminal ribose replaced with 3′-deoxyribose |
Cell culture and transfections
The human keratinocyte cell line HaCaT was cultured in serum-free keratinocyte medium (Gibco BRL) supplemented with 2.5 ng/ml epidermal growth factor and 25 µg/ml bovine pituitary extract. The cell line was regularly passaged at sub-confluence and plated 1 or 2 days before transfection. HaCaT cells in 6-well plates were transfected at low confluency (<40%) with 1.0 ml 100 nM double-strand siRNA in serum-free medium, using Lipofectamine 2000. For complexation, a 10 µM stock solution of double-strand siRNA or 20 µM stock solution of antisense siRNA was diluted with a 10× volume of serum-free medium and mixed with an equal volume of medium-diluted Lipofectamine 2000, at a ratio of liposome to siRNA of 5:2 v/w. Batch dilutions of liposomes were performed for each 6-well plate and preincubated at room temperature for 5–7 min before addition to the medium-diluted siRNA. Complexes were replaced with full medium 5 h after initiation of transfection. For standard assays of activity, cells were harvested the day after transfection. For longer incubations and time-course experiments, medium was replaced every second day after transfection. In time-courses with actinomycin D (Sigma), 10 µg/ml was added 2 h into the transfection period.
Northern analysis
Polyadenylated mRNA was isolated using Dynabeads oligo(dT)25 (Dynal). Isolated mRNA was fractionated by electrophoresis for 16–18 h on 1.3% agarose/formaldehyde (0.8 M) gels and blotted onto nylon membranes (MagnaCharge; Micron Separations). Membranes were hybridised with random primed Tissue Factor (position 61–1217 in cDNA) and GAPDH (1.2 kb) cDNA probes in PerfectHyb hybridisation buffer (Sigma).
RESULTS
Antisense siRNA shows lower activity than double-strand siRNA
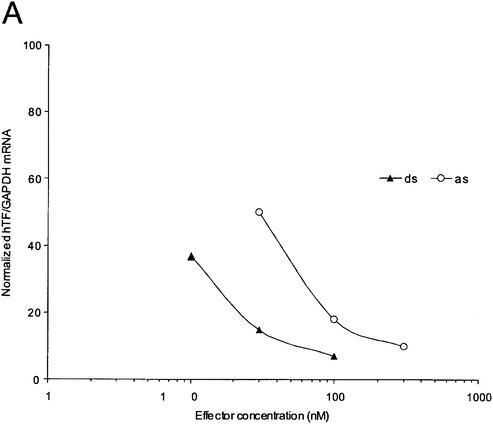
We have previously shown that the maximum depletion of Tissue Factor mRNA by antisense siRNA, at 200 nM concentration, approaches that achieved by the corresponding siRNA at 100 nM (19). As double-strand siRNA-mediated inhibition reaches saturation at substantially lower concentrations than that used for antisense siRNA, the latter might still be substantially less active than double-strand siRNA at lower concentrations. Dose dependence experiments demonstrated that double-strand siRNA was active at ∼5–6-fold lower concentrations than antisense siRNA (Fig. 1A). Their IC50 values were estimated at 5 and 30 nM, respectively.
Figure 1.
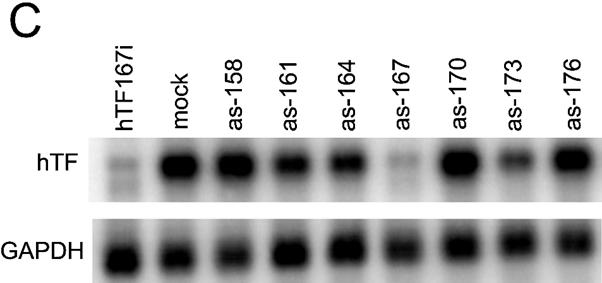
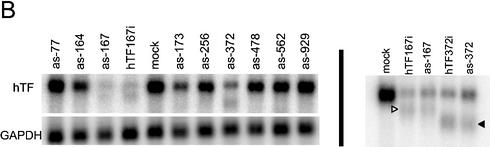
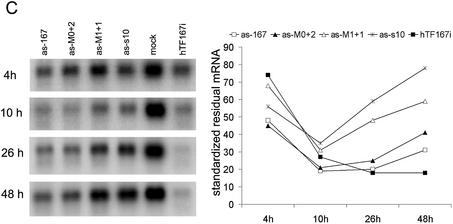
Antisense and double-strand siRNA show identical positional effects against the same target sites on Tissue Factor mRNA. (A) Dose dependence of double-strand (ds) and antisense (as) siRNA. Complexation with Lipofectamine 2000 was performed in one batch for all samples and complexes were diluted in medium immediately before addition to cells. (B) Global and (C) local target position effect. (B and C) Northern analysis of Tissue Factor mRNA after transfection of HaCaT cells with various antisense siRNA (200 nM) or double-strand (100 nM) siRNA. GAPDH was used as a control. Arrowheads (B, right panel) indicate cleavage products for antisense and double-strand siRNA.
Antisense siRNA has target position dependence
A comparison between antisense and double-strand siRNA cannot, of course, conclusively prove a shared RNAi pathway, but major deviations between the two could very well decisively disprove our hypothesis of a shared RNAi pathway. We therefore decided to investigate whether antisense siRNA would have the same position effects demonstrated previously for double-strand siRNA (19,20). The efficacies of antisense RNAs targeting sites in the Tissue Factor mRNA coding region from the start codon (as-77) to the stop codon (as-929) were evaluated in a quantitative northern assay (Fig. 1B). The results confirmed the position dependence of antisense siRNA efficacy, with the rank order of effectivity correlating strongly with inhibitory data for the corresponding double-strand siRNAs (hTF167 > hTF372 > hTF173, hTF164 > hTF256 > hTF161, hTF562) (Pearson’s coefficient for bivariate correlation of inhibition 0.967, P < 0.001, n = 7).
Going from global position effects to investigate the local area on the mRNA close to the best antisense siRNA, as-167, using overlapping oligos differing only by 3 nt from each neighbour, we found that the activity differed sharply with each nucleotide triplet shift (Fig. 1C). The order of effectivity also correlated here with the double-strand siRNA efficacy order.
Antisense siRNA produces mRNA cleavage fragments
Evidence for RNAi-induced endonucleolytic cleavage of mRNA in mammalian cells was obtained in our previous work (19). Similar cleavage fragments were generated by the antisense siRNAs as-167 and as-372, targeting the two most accessible sites (Fig. 1B), with as-167 producing a larger cleavage fragment than as-372, as the as-372 target site is closer to the 3′ end on the target mRNA. This shows that antisense and double-strand siRNA both mediate endonucleolytic cleavage of target mRNA in a manner resulting in an apparent partial protection of a part of the degraded mRNA, possibly caused by kinetic or structural conditions. This strengthens the case for a common RNAi pathway.
Antisense siRNA is gradually affected by mutations
We have earlier demonstrated a relatively high tolerance for mutations in a double-strand siRNA with high activity (11). These mutated siRNAs, which display a wide range of activities, provided a further test of the correspondence between antisense and double-strand siRNA activity. Screen ing of a set of mutated antisense siRNAs demonstrated the same rank order of activity for the antisense candidates as for the corresponding siRNA duplexes (Fig. 2).
Figure 2.
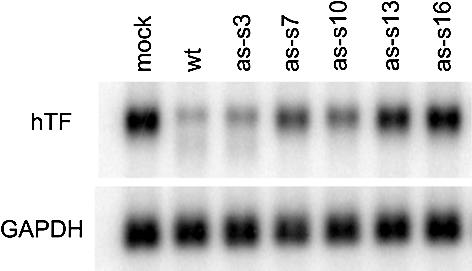
Mutational inactivation of the antisense siRNA as-167. Northern analysis of Tissue Factor mRNA after transfection of HaCaT cells with 200 nM as-167 (wild-type, wt) or single mutated (as-s3, as-s7, as-s10, as-s13 or as-s16; the numerals refer to the position of the mutation, counted from the 5′ end of the siRNA sense strand) versions of antisense siRNA as-167. GAPDH was used as a loading control.
Antisense siRNA is gradually affected by nucleotide methylation
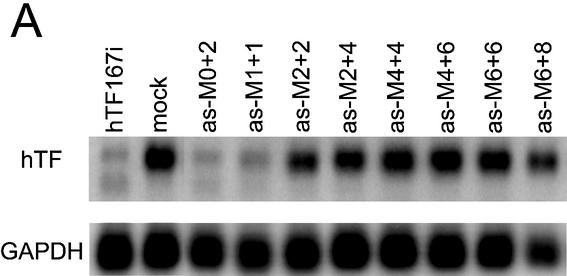
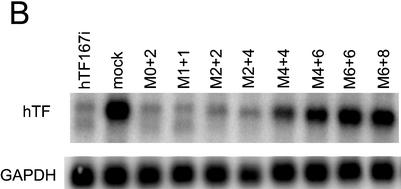
In a further characterisation of the functional anatomy of antisense siRNA, we investigated the effects on activity of gradually increasing degrees of 2′-O-methylation, as previously reported for double-strand siRNA (11). The mRNA depletion achieved with chemically modified antisense siRNA (Fig. 3A) mirrored the previously observed gradual attenuation of double-strand siRNA activity by chemical modification (11) (Fig. 3B). The antisense siRNA with very few modifications (as-M0+2 and as-M1+1) had almost the same activity as the wild-type as-167, while more extensive methylation resulted in a gradual decline in activity, more pronounced with antisense siRNA. Exceptions here are M2+2 and M2+4, which lost disproportionally more activity.
Figure 3.
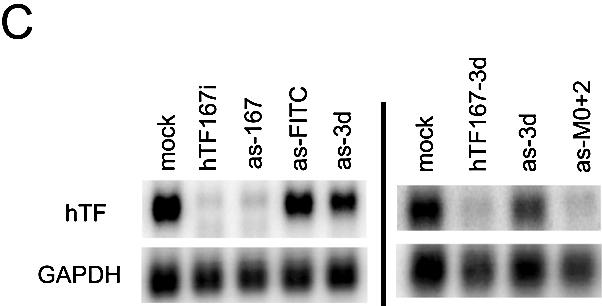
Comparison of the influence of chemical modification on antisense and double-strand siRNA. (A) Inactivation by methylation of antisense siRNA as-167. (B) Inactivation by methylation of double-strand siRNA hTF167i. (C) Inactivation of antisense siRNA by modification of the 3′-OH of the 3′-terminal nucleotide. as-FITC contains a FITC group in the 3′ position whereas in as-3d, 3′-deoxyribose is substituted for the terminal ribose. hTF167-3d is a double-strand siRNA in which as-3d is paired with a wild-type sense strand. (A–C) Northern analysis of Tissue Factor mRNA after transfection of HaCaT cells with 200 nM antisense siRNA or 100 nM double-strand siRNA. GAPDH was used as a loading control.
Antisense siRNA modified in the 3′-terminal 3′-OH position displays much lower activity than corresponding double-stranded siRNA
Another interesting difference in the effect of antisense and double-strand siRNA was seen with antisense siRNA tagged at the 3′ end with a fluorescent FITC group. This RNA oligo, when paired with the wild-type sense strand of the double-strand siRNA hTF167i, has previously been shown to have near wild-type activity (19). It was thus somewhat surprising that even at the highest achievable concentrations possible in our assays, the FITC-tagged antisense siRNA (as-FITC) demonstrated only very limited activity (data not shown). We first reasoned that the lack of activity was due to faulty incorporation into the RISC complex due to the bulky 3′ end fluorescent group. However, we find that an antisense siRNA with a 3′-deoxyribose modification (as-3d) of its 3′ end, although somewhat more active than the 3′-FITC version, has a substantially impaired activity compared with the wild- type antisense. In the corresponding siRNA duplex it has comparable activity with wild-type siRNA (Fig. 3C).
An excess of double-strand siRNA can competitively block antisense siRNA
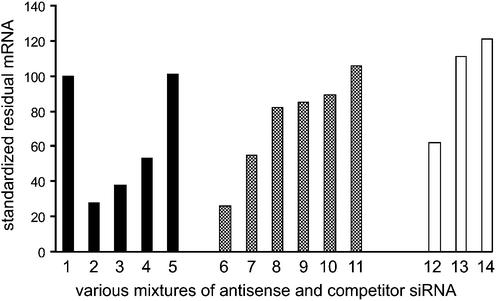
The close similarity of antisense and double-strand siRNA mRNA depletion behaviour with regard to position effects, cleavage fragment formation, methylation and mutation tolerance suggest at least in part a common pathway for antisense and double-strand siRNA. If this is the case, the effect of antisense siRNA should be susceptible to competition by an inactive double-strand siRNA, in a manner previously shown for inactive and active siRNA (19,21). Competition experiments were therefore performed in order to test this hypothesis. These experiments are not straightforward to do, as discussed below. Nevertheless, a surplus of various inactive competitor double-strand siRNAs seemed to block the effect of the active antisense siRNA as-167 under various transfection conditions (Fig. 4).
Figure 4.
Competition between double-strand and antisense siRNA. Results in the black, grey and white columns are from three separate experiments. Experiments were performed in HaCaT cells as previously described, unless otherwise stated, and relative Tissue Factor/GAPDH expression normalised to levels in mock-transfected cells in each experiment. Column 1, mock; column 2, 50 nM antisense siRNA as-167; columns 3–5, 50 nM as-167 + 75 nM various inactive (column 3, hTF167-ds10/16) and irrelevant (column 4, PSK229i; column 5, BCR-ABL-1i) double-strand siRNA; column 6, 200 nM as-167; column 7, 50 nM as-167; columns 8–11, 50 nM as-167 + increasing concentrations (75, 100, 150 and 200 nM) of irrelevant double-strand siRNA (PSK208i); column 12, 200 nM as-167; column 13, 200 nM as-167 + 2000 nM PSK208i; column 14, 2000 nM PSK208i. In columns 12–14, cells were transfected with the less toxic agent Oligofectamine (Gibco BRL), enabling the use of higher total concentrations of RNA and excess of competitor, at the expense of reduced transfection efficiency.
Antisense siRNA reaches maximum activity faster than double-strand siRNA
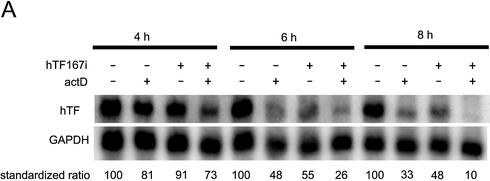
Standard double-strand siRNA, in our Tissue Factor system, reaches maximum effect only after ∼24 h, after which the silencing fades out over 3–5 days (19). The causes of the slow onset of silencing are not known. In our earlier work we proposed that RNAi depletion might be relatively slow and in a near equilibrium kinetic balance between mRNA depletion and mRNA transcription (19). Testing this concept by blocking mRNA transcription with actinomycin D showed that the natural turnover of mRNA, over a short period of 6 h, was faster than RNAi (Fig. 5A). This is consistent with the reported 1–2 h half-life of Tissue Factor mRNA (22,23). GAPDH has a reported half-life of >24 h (24,25), thus the 6 h exposure to actinomycin D should only reduce the GAPDH signal by <16%. Even when not considering decay of GAPDH, combining double-strand siRNA and actinomycin D gives an additive effect at all measured time points (Fig. 5A, lanes 4, 8 and 12). This might be interpreted as supporting the notion that RNAi has independent exonucleases for mRNA degradation (19,26,27).
Figure 5.
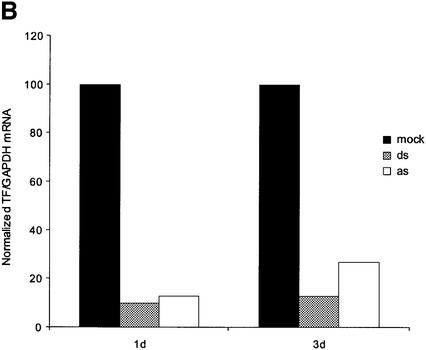
Time-courses of the efficacy of double-strand and antisense siRNA. (A) Actinomycin D time-course. Cells were transfected with double-strand siRNA (100 nM) hTF167i or mock control as indicated above each lane. Actinomycin D (10 µg/ml) was added to the medium 2 h later and cells harvested at 4, 6 and 8 h post-transfection as indicated. The Tissue Factor signal was standardised to GAPDH and normalized to levels in the control at each time point. The results are representative for two independent experiments. (B) Long-term effect of antisense (as) and double-strand (ds) siRNA targeting hTF167. Tissue Factor mRNA was measured at days 1 and 3 post-transfection and standardized to a GAPDH control. Data are from one of two independent experiments. (C) Short-term time-course of antisense and double-strand siRNA mRNA depletion. Cells were transfected as described and harvested at the indicated time points. Tissue Factor/GAPDH expression was normalised to levels in mock-transfected cells. The results are representative of two independent experiments.
Time-course experiments with antisense and double-strand siRNA demonstrated that the effect of the former was more transient, although effective silencing was still evident 3 days post-transfection (Fig. 5B). In a further comparison of double-strand and antisense siRNA function, we have investigated the time dependence of the onset of mRNA depletion. Surprisingly, we found that the antisense siRNA effect was somewhat stronger than that of double-strand siRNA at 10 h post-transfection. The maximum effect of antisense siRNA seemed to be achieved already at this time point, as mRNA levels increased at later time points (Fig. 5C). This behaviour extended to the double-strand siRNA/antisense siRNA pair targeting hTF372 (data not shown). The faster onset of antisense siRNA-mediated silencing may be due to direct incorporation of single-stranded RNA into the active nuclease complex (RISC*) (12).
DISCUSSION
Antisense siRNA provides another efficient tool for use in transient knockdown of gene expression, but questions remain regarding its mechanism of function. This work has compared a range of different antisense siRNA oligos to their corresponding double-strand siRNA, seeking to disprove the hypothesis of a shared RNAi effector pathway by finding major differences between them in our different assays.
Antisense siRNA is somewhat less efficient than the corresponding double-strand siRNA, displaying an ∼5-fold higher IC50. The lower specific activity of antisense siRNA compared to double-strand siRNA may be due to a lower intracellular stability of the former. We expect antisense oligos to be protected by the transfecting agent until released from the lipid miscelles, a process that can take some time (19); following release from lipid miscelles and prior to incorporation into RISC, the antisense siRNA should be more vulnerable to degradation than double-strand siRNA. The above-mentioned drawbacks combined may result in a more transient nature of silencing, although our data suggest that antisense siRNA is able to maintain partial silencing for at least 3 days.
A standard set-up with 100 nM double-strand siRNA compared with 200 nM antisense siRNA was chosen to ensure equal amounts of both nucleic acids and the transfection agent. We found that the position dependence of antisense siRNA correlated with that of the corresponding double-strand siRNA. This observation has one important practical implication: it reduces the cost of screening for optimal target sites for siRNA. Despite its lower specific activity, antisense siRNA efficacy still approached that of the corresponding double-strand siRNA at moderate and non-toxic concentrations (200–300 nM).
When comparing antisense and double-strand with regard to both global and local mRNA target position effects, the appearance of cleavage fragments and the tolerance to mutational and chemical backbone modifications, a striking similarity of behaviour is evident. One would expect different effector complexes to have different target or oligo preferences, as previously reported for the different position dependencies of RNase H-competent ribozymes and siRNA (19,21). In the absence of significant differences between antisense and double-strand siRNA for a series of oligos, we propose that antisense and double-strand siRNA mediate RNAi through the same pathway, although we are not able to provide conclusive evidence for an identity of mechanism. An exception to the similarity of behaviour is as-FITC, a version of as-167 tagged with a bulky fluorescent FITC group at the 3′-OH position of the 3′-terminal nucleotide, which displayed very poor activity. We first interpreted these results to indicate the impairment of incorporation of the antisense strand into the active RISC* due to the presence of a bulky functional group. However, a version with 3′-deoxyribose instead of ribose at the 3′-terminus of the antisense siRNA, as-3d, showed similar behaviour. Efforts to investigate this phenomenon further are currently under way.
Direct proof of a shared RNAi pathway in vivo would be the demonstration of inhibition by competition. Unfortunately, this proof is hard to come by as lipid toxicity sets an upper limit for oligo concentrations and antisense siRNA requires higher doses than double-strand siRNA. Furthermore, difficulties arise from the surplus capacity of the RNAi system under optimal conditions, where blocking part of the excess capacity, for example by lowering of the oligo concentration or partial inactivation of oligos by chemical modification or mutation, does not affect the final mRNA depletion activity (11,19). Finally, the entry of antisense siRNA at a supposedly later point in the pathway leaves antisense siRNA alone in the RISC* for a limited period, setting further limitations on a competition assay. This last point has been emphasised by the recent in vitro demonstration that competition for formation of a target-specific RISC required the simultaneous addition of both active and competitor siRNA to the lysate; competitor added 15 min after initiation of incubation could no longer compete with the active siRNA (10). The relevance of this observation for a biologically necessary turnover in vivo over hours and days is doubtful. Still, with the caveats noted, it has proved possible to block antisense siRNA with inactive or irrelevant double-strand siRNA, thus strengthening the hypothesis of a shared pathway.
Finally, we demonstrate that natural turnover of Tissue Factor mRNA is faster than RNAi, and that the two effects are additive. This can be interpreted as support for the existence of independent RNAi exonucleases (19,26,27). Interestingly, the slow onset of double-strand siRNA-mediated was contrasted by the early onset of antisense siRNA-mediated mRNA depletion. This argues that the antisense siRNA enters the RNAi pathway at a stage closer to the effector nuclease than double-strand siRNA. The unwinding of the RNA strands, perhaps mediated by one of the putative helicases recently found to be associated with RISC and RISC-like complexes (14,17), could thus constitute a rate-limiting step in the pathway, partially explaining the slow onset of double-strand siRNA-mediated silencing (19). Furthermore, this discovery of differential depletion speed could give rise to two different classes of therapeutic drugs if RNAi finds clinical application.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Norwegian Cancer Society, Health and Rehabilitation, and the Research Council of Norway (RCN) to H.P. T.H. is a fellow of RCN.
REFERENCES
- 1.Guo S. and Kemphues,K. (1995) par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell, 81, 611–620. [DOI] [PubMed]
- 2.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed]
- 3.Zamore P.D. (2002) Ancient pathways programmed by small RNAs. Science, 296, 1265–1269. [DOI] [PubMed]
- 4.Ahlquist A. (2002) RNA-dependent RNA polymerases, viruses, and RNA silencing. Science, 296, 1270–1273. [DOI] [PubMed]
- 5.Plasterk R.H.A. (2002) RNA silencing: the genome’s immune system. Science, 296, 1263–1265. [DOI] [PubMed]
- 6.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed]
- 7.Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21 and 22 nt RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed]
- 8.Caplen N.J., Parrish,S., Imani,F., Fire,A. and Morgan,R.A. (2001) Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl Acad. Sci. USA, 98, 9742–9747. [DOI] [PMC free article] [PubMed]
- 9.Schwarz D.S., Hutvágner,G., Haley,B. and Zamore,P.D. (2002) Evidence that siRNA function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell, 10, 537–548. [DOI] [PubMed]
- 10.Martinez J., Patkaniowska,A., Urlaub,H., Lührmann,R. and Tuschl,T. (2002) Single-stranded antisense siRNA guide target RNA cleavage in RNAi. Cell, 110, 563–574. [DOI] [PubMed]
- 11.Amarzguioui M., Holen,T., Babaie,E. and Prydz,H. (2003) Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res., 31, 589–595. [DOI] [PMC free article] [PubMed]
- 12.Nykänen A., Haley,B. and Zamore,P.D. (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell, 107, 309–321. [DOI] [PubMed]
- 13.Hammond S.M., Boettcher,S., Caudy,A.A., Kobayashi,R. and Hannon,G.J. (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293, 1146–1150. [DOI] [PubMed]
- 14.Mourelatos Z., Dostie,J., Paushkin,S., Sharma,A., Charrouz,B., Abel,L., Rappsilber,J., Mann,M. and Dreyfuss,G. (2002) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev., 16, 720–728. [DOI] [PMC free article] [PubMed]
- 15.Hutvagner G. and Zamore,P.D. (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297, 2056–2060. [DOI] [PubMed]
- 16.Caudy A.A., Myers,M., Hannon,G.J. and Hammond,S.M. (2002) Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev., 19, 2491–2496. [DOI] [PMC free article] [PubMed]
- 17.Ishizuka A., Siomi,M.C. and Siomi,H. (2002) A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev., 19, 2497–2508. [DOI] [PMC free article] [PubMed]
- 18.Ambros V. (2001) MicroRNAs: tiny regulator with great potential. Cell, 107, 823–826. [DOI] [PubMed]
- 19.Holen T., Amarzguioui,M., Wiiger,M.T., Babaie,E. and Prydz,H. (2002) Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res., 30, 1757–1766. [DOI] [PMC free article] [PubMed]
- 20.Yang D., Buchholz,F., Huang,Z., Goga,A., Chen,C.-Y., Brodsky,F.M. and Bishop,J.M. (2002) Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 9942–9947. [DOI] [PMC free article] [PubMed]
- 21.McManus M.T., Haines,B.B., Dillon,C.P., Whitehurst,C.E., Van Parijs,L., Chen,J. and Sharp,P.A. (2002) Small interfering RNA-mediated gene silencing in T lymphocytes. J. Immunol., 169, 5754–5760. [DOI] [PubMed]
- 22.Amarzguioui M., Brede,G., Babaie,E., Grotli,M., Sproat,B. and Prydz,H. (2000) Secondary structure prediction and in vitro accessibility of mRNA as tools in the selection of target sites for ribozymes. Nucleic Acids Res., 28, 4113–4124. [DOI] [PMC free article] [PubMed]
- 23.Ahern S.M., Miyata,T. and Sadler,J.E. (1993) Regulation of human tissue factor expression by mRNA turnover. J. Biol. Chem., 268, 2154–2159. [PubMed]
- 24.Lekas P., Tin,K.L., Lee,C. and Prokipcak,R.D. (2000) The human cytochrome P450 1A1 mRNA is rapidly degraded in HepG2 cells. Arch. Biochem. Biophys., 384, 311–318. [DOI] [PubMed]
- 25.Waterboer T., Rahaus,M. and Wolff,M.H. (2002) Varicella-zoster virus (VZV) mediates a delayed host shutoff independent of open reading frame (ORF) 17 expression. Virus Genes, 24, 49–56. [DOI] [PubMed]
- 26.Ketting R.F., Haverkamp,T.H., van Luenen,H.G. and Plasterk,R.H. (1999) Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell, 99, 133–141. [DOI] [PubMed]
- 27.Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed]