Abstract
Human thymine-DNA glycosylase (TDG) is well known to excise thymine and uracil from G·T and G·U mismatches, respectively, and was therefore proposed to play a central role in the cellular defense against genetic mutation through spontaneous deamination of 5-methylcytosine and cytosine. In this study, we characterized two newly discovered orthologs of TDG, the Drosophila melanogaster Thd1p and the Schizosaccharomyces pombe Thp1p proteins, with an objective to address the function of this subfamily of uracil-DNA glycosylases from an evolutionary perspective. A systematic biochemical comparison of both enzymes with human TDG revealed a number of biologically significant facts. (i) All eukaryotic TDG orthologs have broad and species-specific substrate spectra that include a variety of damaged pyrimidine and purine bases; (ii) the common most efficiently processed substrates of all are uracil and 3,N4- ethenocytosine opposite guanine and 5-fluorouracil in any double-stranded DNA context; (iii) 5-methylcytosine and thymine derivatives are processed with an appreciable efficiency only by the human and the Drosophila enzymes; (iv) none of the proteins is able to hydrolyze a non-damaged 5′-methylcytosine opposite G; and (v) the double strand and mismatch dependency of the enzymes varies with the substrate and is not a stringent feature of this subfamily of DNA glycosylases. These findings advance our current view on the role of TDG proteins and document that they have evolved with high structural flexibility to counter a broad range of DNA base damage in accordance with the specific needs of individual species.
INTRODUCTION
The human thymine-DNA glycosylase (TDG) was first discovered as an enzymatic activity in extracts of HeLa cells that excised thymine from G·T mismatched oligonucleotide DNA duplexes (reviewed in 1). Today, we know that this DNA repair enzyme belongs to a large family of conserved and omnipresent proteins with a common α/β-fold structure and a general ability to excise uracil from DNA (2). All these uracil-DNA glycosylases (UDGs) have a monofunctional mode of action. They all employ a common base-flipping, DNA intercalation strategy for substrate recognition and binding, and catalyze the hydrolysis of the N-glycosidic bond of the flipped-out substrate base, thereby generating an abasic site (AP-site) in the DNA backbone (reviewed in 3). However, minor variations in the active site architecture and the catalytic mechanism define a number of subfamilies of UDGs with distinct properties. The eukaryotic UDGs belong to the UNG, MUG and SMUG subfamilies (2), with TDG representing the founding member of the MUG branch. Most, if not all, organisms possess at least one DNA uracil-processing activity, but none of the UDG subfamilies is represented ubiquitously. The UNG and MUG proteins show the broadest distribution and are found in species ranging from bacteria to human, whereas SMUG proteins appear to occur only in multicellular eukaryotes excluding plants and nematodes (2). Strikingly, Drosophila melanogaster lacks an UNG but has a MUG and a SMUG, whereas the budding yeast Saccharomyces cerevisiae has an UNG but neither a MUG nor a SMUG, and the fission yeast Schizosaccharomyces pombe has both an UNG and a MUG, but no SMUG. The enzymatic differences between the UDG protein subfamilies are best illustrated by a comparison of UNG with TDG. Unlike the very potent UNG-type UDGs that remove uracil from single-stranded (ss) and double-stranded DNA (dsDNA) with a high catalytic rate and selectivity (4), TDG processes both uracil and thymine with an appreciable rate but only when they are in a mispairing configuration with guanine (5,6). Since both the G·U and the G·T mismatches arise in DNA mainly as a consequence of spontaneous hydrolytic deamination of cytosine or 5-methylcytosine, respectively, the G mismatch dependence was taken as evidence for TDG being responsible for the initiation of base excision repair (BER) at sites of cytosine or 5-methylcytosine deamination in DNA.
The first TDG homologs to be described were proteins of Escherichia coli and Serratia marcescens that share ∼37% amino acid sequence identity with the human TDG and align with its catalytic core domain (7). The in vitro translated E.coli enzyme showed UDG activity on a G·U substrate but failed to process a G·T mismatch under physiological conditions. Like human TDG, the bacterial protein did not excise uracil from A·U base pairs or ssDNA efficiently and was therefore named Mug (for mismatch-specific uracil-DNA glycosylase). Mutational analysis of the human enzyme confirmed that its conserved central domain (residues 112–360) is indeed sufficient for processing of a G·U substrate, while efficient hydrolysis of thymine from a G·T mismatch requires additional N‐terminal sequence (6,7). Further insight into the substrate spectra and requirements of the MUG subfamily of UDGs came with the observation that both the human and the E.coli enzymes were capable of excising the 3,N4-ethenocytosine (εC) from DNA (8). In fact, Mug was reported to be the major activity in E.coli to eliminate this highly mutagenic lesion from DNA (9). Other substrates described for the human TDG were the cytotoxic anti-cancer agent 5-fluorouracil (6) and 5-methylcytosine (10).
These findings raise the question of whether MUG proteins, rather than dealing specifically with the deamination products of cytosine or 5-methylcytosine, are more generally involved in the repair of DNA base damage. The crystal structure available for E.coli Mug shows that, in principle, this is possible. Unlike the restricting active site geometry of the highly uracil-specific UNG proteins, the active site pocket of Mug would easily accommodate a wide spectrum of pyrimidine derivatives (11,12) and, owing to the high degree of structural conservation, this is likely to be the case also for other members of the protein subfamily. Thus, a systematic examination of the substrate spectra of MUG proteins of different species seems imperative for a comprehensive understanding of their conserved functions and, ultimately, their biological roles. This study introduces two novel members of the MUG protein family, which we identified in the partially sequenced genomes of D.melanogaster and S.pombe (1). We report their molecular cloning and biochemical characterization, and show a systematic comparison of the substrate spectra and requirements of the fission yeast and insect enzymes with those of the human TDG. The data establish that eukaryotic MUG proteins have divergent substrate spectra which include a wide range of damaged cytosine, 5-methylcytosine, thymine and even adenine bases. The G·U mismatch is the common most efficiently processed substrate, whereas 5-methylcytosine and thymine derivatives are processed at a physiologically relevant rate only by the human enzyme. Also, the characteristic double strand and mismatch dependency of the MUG proteins varies with the substrate and is not strictly conserved between species. These findings suggest that MUG proteins have evolved with a high degree of flexibility to counter particular forms of DNA base damage according to the specific needs of individual species.
MATERIALS AND METHODS
Reagents and oligonucleotides
All standard oligonucleotides were synthesized by Microsynth (Switzerland). The oligonucleotides containing εC and HmU were supplied by Gemini Biotech (USA). All substrate oligonucleotides were purified by PAGE after synthesis. All enzymes were supplied by New England Biolabs (USA). All other chemicals and reagents were purchased from Sigma (Switzerland).
Plasmids and expression vectors
pPRS270. An NheI–SalI PCR fragment of the full-length cDNA coding for S.pombe Thp1p was cloned into the respective restriction sites of the plasmid pET28c from Novagen (PCR primers were Thp1-NheATG, 5′-CGATCGGCTAGCATGAACGACATTGAGACGA; and Thp1-TAASal, 5′-TCCGTGTCGACTTAGACTGCATGTTTCAC).
pPRS250. A 1242 bp segment of the Drosophila Thd1 cDNA encoding amino acids 650M–1063N of Thd1p was cloned as a BglII–SalI PCR fragment into the BamHI–SalI restriction sites of plasmid pQE30 (Qiagen) (PCR primers were Thd1-BglATG, 5′-GTACAGATCTATGCATTCACCATCACATCG; and Thd1-TAGSal, 5′-CCGACGTCGACCTAATTATGATTAGACACTGGGAG).
pPRS125. pET28c (Novagen) containing an NheI–SalI PCR fragment of full-length cDNA encoding the human AP endonulease (APE1) cloned into the respective restriction sites (PCR primers were APE1-NheATG, 5′-CGATCGGCTAGCATGCCGAAGCGTGGGAAAAAG-3′; and APE1-TGASal, 5′-TCCGTGTCGACTCACAGTGCTAGGTATAGG-3′).
These cloning strategies generated fusion open reading frames (ORFs) encoding His6-Thp1p, His6-Thd1p (residues 650M–1063N) and His6-APE1, respectively, downstream of a promoter–operator element consisting of phage T7 promoter and a lac operator sequence.
Purification of recombinant proteins
pPRS270 expressing S.pombe Thp1p was transformed into E.coli BL21 (DE3) cells by electroporation, and transformants were selected on LB plates containing 50 µg/ml kanamycin and 2% glucose. Expression cultures of 2 l of LB medium containing 50 µg/ml kanamycin were inoculated with 50 ml of an overnight culture and incubated at 30°C to an OD600 of 1.2. Following induction of protein expression by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG; to 500 µM) and further incubation at 30°C for 3 h, the cells were harvested by centrifugation (Sorvall SLA-3000, 6000 r.p.m., 4°C, 30 min) and subsequently stored at –80°C. For protein extraction, thawed cell pellets were resuspended in 3 ml/g sonication buffer (sb) [50 mM Na-phosphate pH 8.0, 300 mM NaCl, 10% glycerol, 1 mM imidazole, 10 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF)] and lysed by sonication on ice (25 × 10 s bursts with intermittent chilling for 10 s). After removal of cell debris by centrifugation (Sorvall SS34, 15 000 r.p.m., 4°C, 30 min), 1 ml of sb-equilibrated Ni-NTA–agarose (Qiagen) was added to the crude lysate and incubated for 1 h at 4°C with gentle shaking. The suspension was then packed into a disposable column from which unbound protein was washed out with 2 × 15 column volumes (cv) of sb and 2 × 5 cv of sb containing 20 mM imidazole. Finally, bound His6-tagged Thp1p was eluted with 5 × 1 cv of sb containing 300 mM imidazole. The 300 mM imidazole fractions were pooled and diluted 1:3 with dilution buffer (50 mM Tris–HCl pH 8.0, 10% glycerol, 5 mM β-mercaptoethanol) resulting in an NaCl concentration of 100 mM. After loading the diluted fraction onto a binding buffer-equilibrated (50 mM Tris–HCl pH 8.0, 10% glycerol, 5 mM β-mercaptoethanol, 100 mM NaCl) 1.3 ml UNO S1 FPLC column (Bio-Rad) and washing with 10 ml of binding buffer, bound protein was eluted with a linear gradient of 100–500 mM NaCl in 20 ml. The nearly homogeneous S.pombe Thp1p (>98% pure) eluted as a major protein peak in fractions containing ∼300 mM NaCl. After a last dialysis step against storage buffer (50 mM Tris–HCl pH 8.0, 50 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol), the pure Thp1p was stored in aliquots at –80°C.
pPRS250 expressing Drosophila His6-Thd1p was co- transformed with the lacI-repressor encoding pREP4 plasmid (Qiagen) into E.coli BL21 (DE3) cells by electroporation. Transformants were selected on LB plates containing 100 µg/ml ampicillin, 50 µg/ml kanamycin and 2% glucose after incubation at 30°C. Protein expression was done using the induction and growth regimen described for human TDG (6). Cell lysis and purification on Ni-NTA–agarose were performed as described above for Thp1p. All 300 mM imidazole elution fractions were pooled, dialyzed extensively against binding buffer (50 mM Tris–HCl pH 7.5, 10% glycerol, 5 mM β-mercaptoethanol) and loaded onto an equilibrated 1.3 ml UNO S1 FPLC column (Bio-Rad). After washing with 10 ml of binding buffer, bound protein was eluted with a linear gradient of 0–500 mM NaCl in 25 ml. Thd1p eluted as a major protein peak in fractions containing ∼300 mM NaCl, and the nearly homogeneous (>90% pure) protein was dialyzed against storage buffer and stored in aliquots at –80°C.
Recombinant human His6-TDG was expressed in E.coli and purified as described before (6).
The expression construct for APE1 was transformed into E.coli BL21 (DE3) cells and expression was achieved under the same conditions as described above for Thp1p. The only differences were the use of 1 l of expression culture and induction at OD600 of 0.8. Cell lysis and purification on Ni-NTA–agarose were performed essentially as described for Thp1p, with slightly different wash conditions using sb with the following imidazole concentrations: 1 × 15 cv, 1 mM imidazole; 5 × 5 cv, 20 mM imidazole; 1 × 5 cv, 60 mM imidazole. Bound histidine-tagged APE1 protein was eluted with 5 × 1 cv of sb containing 300 mM imidazole. All elution fractions were pooled, dialyzed extensively against binding buffer (50 mM Tris–HCl pH 8.0, 10% glycerol, 5 mM β-mercaptoethanol) and loaded onto an equilibrated 1.3 ml UNO S1 FPLC column (Bio-Rad). After washing with 10 ml of binding buffer, bound protein was eluted with a linear gradient of 0–500 mM NaCl in 15 ml. APE1 eluted as a major protein peak in fractions containing ∼350 mM NaCl, and the nearly homogeneous (>97% pure) protein was dialyzed against storage buffer and stored in aliquots at –80°C.
Glycosylase activity assay
The enzymatic activities of the recombinant S.pombe Thp1p, D.melanogaster Thd1p and human TDG proteins were measured by means of a standardized nicking assay essentially as described before (6). The oligonucleotide substrates used are illustrated in Figure 2 and described in Hardeland et al. (6). The nicking reactions were carried out in a total volume of 20 µl of 1× nicking buffer [50 mM Tris–HCl pH 8.0, 1 mM dithiothreitol (DTT), 0.1 mg/ml bovine serum albumin (BSA), 1 mM EDTA, 0.5 U of uracil-DNA glycosylase inhibitor (Ugi)] containing 1 pmol of substrate DNA and 1 pmol of either of the recombinant proteins. Unless indicated otherwise, the reactions were incubated for 15 min at 37°C and then stopped by addition of 1 M NaOH to a final concentration of 90 mM, heating to 99°C for 10 min and subsequent ethanol precipitation. APE1-mediated cleavage of AP-sites was assayed by the addition of 5 pmol of human APE1 and 5 mM MgCl2, and incubation for 5 min at 37°C, followed by ethanol precipitation. After ethanol precipitation, the dried pellets were resuspended in 10 µl of formamide gel loading buffer (90% formamide, 1× TBE) containing 200 mM NaBH4 to limit spontaneous cleavage of the AP-site. Finally, the reaction products were separated on 15% denaturing polyacrylamide gels after short heat denaturation for 1 min at 60°C.
Figure 2.
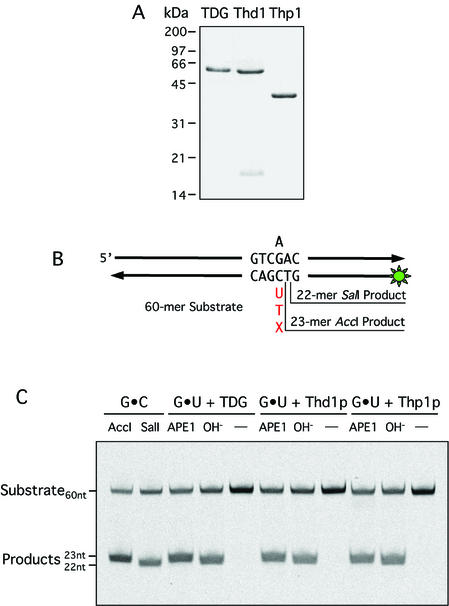
Glycosylase activity of TDG, Thd1p and Thp1p. (A) Fractions of purified TDG, Thd1p and Thp1p analyzed by SDS–PAGE. A 2 µg aliquot of each protein was loaded onto a 12.5% gel. The proteins were visualized by Coomassie brilliant blue staining. (B) The 60mer oligonucleotide duplex DNA used as substrate. An unlabeled upper strand oligonucleotide contains guanine or adenine at the position indicated. The complementary 5′-fluorescein-labeled (green asterisk) lower strand positions either a cytosine, a thymine, a uracil or any other target base of interest opposite the guanine as indicated. AccI and SalI digestions result in 22mer and 23mer product formation, respectively. (C) Thd1p and Thp1p generate APE1 and alkaline-sensitive AP-sites in a G·U substrate. Standard base release reactions were done in 20 µl volumes in the presence of 1 pmol of substrate DNA and 1 pmol of protein. The reaction products shown were analyzed on 15% denaturing polyacrylamide gels. The positions of the 60mer substrate DNA, the respective product fragments, and the 22mer and 23mer AccI and SalI restriction fragments are indicated.
In kinetic assays, the reactions were set up in a 200 µl volume with the substrate and enzyme concentrations described above. At the indicated time points, 20 µl samples were withdrawn, stopped by NaOH and treated further as described above. Due to the instability of εC under alkaline conditions and heat treatment, the reaction products of these substrates were cleaved by APE1 as follows. Ethanol-precipitated DNA was dissolved in 10 µl of 10 mM Tris–HCl pH 8.0 and the reaction performed in a total volume of 20 µl of 1× APE1 buffer (50 mM Tris–HCl pH 8.0, 1 mM DTT, 5 mM MgCl2) containing 4 pmol of APE1. After incubation for 10 min at 37°C, the reactions were stopped by ethanol precipitation and analyzed on 15% denaturing polyacrylamide gels as described above.
Electrophoretic mobility shift assays (EMSAs)
EMSAs were performed essentially as described before for human TDG (6). In standard EMSA, 5 pmol of Thp1p, or 4 pmol of Thd1p and TDG were incubated in 10 µl reaction mixtures containing 1 pmol of labeled oligonucleotide substrate, 10 pmol of unlabeled homoduplex competitor DNA, 50 mM Tris–HCl pH 8.0, 1 mM DTT, 5% glycerol and 1 mM EDTA. After 15 min at 37°C, the reactions were loaded immediately onto 6% non-denaturing polyacrylamide gels (Bio-Rad, Mini Protean II cell) in 0.5× TBE, and electrophoresis was carried out for 50 min at 100 V at room temperature.
RESULTS
TDG homologs of Drosophila melanogaster and Schizosaccharomyces pombe
The genome sequences of D.melanogaster and S.pombe revealed ORFs encoding homologs of human TDG. We cloned the cDNAs and named the genes thd1 and thp1 for TDG homolog of D.melanogaster and S.pombe, respectively. Drosophila thd1 is located on chromosome IV and has four introns, one of them located in the 5′-untranslated region of the mRNA. Strikingly, this gene encodes a large protein of 1738 amino acids with a calculated molecular mass of 191.5 kDa. The S.pombe thp1 is an intron-less gene of chromosome III and produces a protein of 325 amino acids with a molecular mass of 36.5 kDa. mRNA analyses by northern blotting revealed transcripts of the thd1 and the thp1 genes in proliferating Drosophila (Schneider S3) and in vegetatively growing as well as meiotically differentiating fission yeast cells, respectively. Three thd1-specific signals were detectable, the predominant one being the shortest of ∼6100 nucleotides (nt) length, matching the size of the cloned cDNA, and the longer species most probably representing differently spliced precursors. thp1-specific mRNA appeared as a single band corresponding to a length of 1200 nt, which is predicted from its genomic sequence (data not shown). Hence, thd1 and thp1 are both expressed genes but they have never been described genetically.
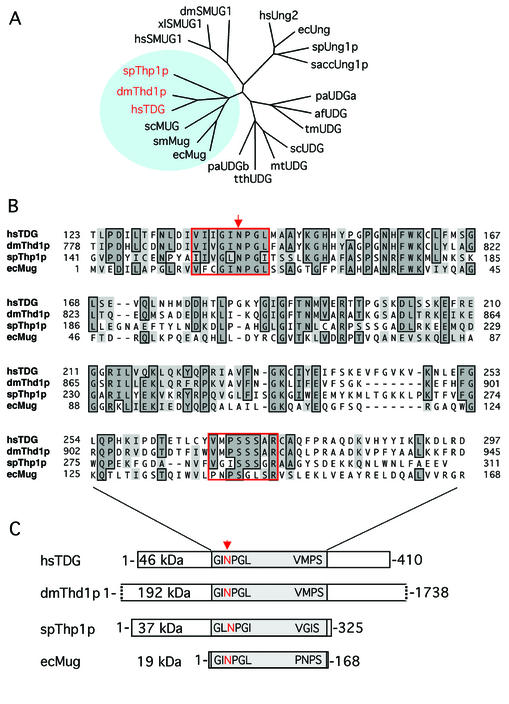
Phylogenetic analyses of Thd1p and Thp1p clearly associated both proteins with the MUG branch of UDGs (Fig. 1A), and an amino acid sequence alignment of both with human TDG and E.coli Mug (7) revealed a moderate degree of conservation that is confined to the catalytic core domains of the proteins (Fig. 1B). The core domains show the typical signature of α/β-fold UDGs of the MUG type (2), including the highly conserved active site motif, as inferred from structural and biochemical analyses of the E.coli Mug (11) and the human TDG proteins (6) (Fig. 1B). This GINPGL motif has a minor variation in Thp1p where the isoleucine and the leucine residues appear to be swapped to give a GLNPGI sequence. The invariant asparagine within this motif (Asn140 in TDG; Asn18 in Mug) is critical for catalysis. It is proposed to mediate hydrolysis of the N-glycosidic bond linking the DNA base with the sugar moiety of the nucleoside by positioning and activating a water molecule for a hydrophilic attack (6,11). Asn795 and Asn158 occupy structurally equivalent positions in Thd1p and Thp1p, respectively, and are thus considered the putative active site residues of these enzymes. A second, less conserved C-terminal sequence motif (VMPSSSAR in TDG) also contributes to the active site in MUG proteins. Residues of this motif were shown to establish specific contacts with the DNA substrate during the base hydrolysis process, ensuring both stability of the DNA helical structure and substrate specificity (6,11). Closely related sequence elements are present at equivalent positions in Thd1p (VMPSSSAR) and in Thp1p (VGISSSGR). Interest ingly, the extra N- and C-terminal domains of the eukaryotic MUG proteins appear to be species specific and vary in sequence composition and size (Fig. 1C). However, the overall architecture and the specific structural elements of the conserved domains imply that Thd1p and Thp1p are MUG-type UDGs.
Figure 1.
Conservation of MUG-type UDGs. (A) Shown are the phylogenetic relationships between representative UDGs of the α/β-fold superfamily. Included are Homo sapiens UNG2 (hsUNG2; accession no. P22674), TDG (hsTDG; Q13569) and SMUG1 (hsSMUG1; O95862); Drosophila melanogaster Thd1p (dmThd1p; Q9V4D8) and SMUG1 (dmSMUG1; Swiss-Prot, Q9VEM1); Xenopus laevis SMUG1 (xlSMUG1; Q9YGN6) Schizosaccharomyces pombe Ung1p (spUng1p; O74834) and Thp1p (spThp1p: O59825); Saccharomyces cerevisiae Ung1p (saccUng1p; P12887); Serratia marcescens Mug (smMug; P43343); Escherichia coli Ung (ecUng; P12295) and Mug (ecMug; P43342); Streptomyces coelicolor UDGb (scUDGb; NP_626251) and MUG (scMug; NP_625542); Pyrobaculum aerophilum UDGa (paUDGa; NP_558739) and UDGb (paUDGb: NP_559226); Thermus thermophilus UDG (tthUDG; CAD29337); Mycobacterium tuberculosis UDG (mtUDG; NP_335742); Thermotoga maritima UDG (tmUDG; NP_228321); and Archaeoglobus fulgidus UDG (afUDG; NP_071102). (B) Amino acid sequence alignment of the catalytic core domains of spThp1p, dmThd1p with hsTDG and ecMug. The conserved MUG characteristic active site motif G(I/L)NPG(L/I) in the N‐terminal part of the catalytic domain and the less conserved C-terminal residues that are critical for a specific interaction with the substrate DNA are framed in red; other conserved residues are framed and shaded. The arrow indicates the position of the critical catalytic residue (N140) of TDG (6). (C) Schematic representation of the overall domain organization of the same members of the MUG subfamily of UDGs. The conserved core domains of the proteins are shaded, and the relative positions of the active site motifs indicated. All sequence analyses were done with the ClustalW routine (Blosum30 matrix; Gap penalty, 10; Gap extension penalty, 0.1) of the MacVector Sequence analysis software (Version 7.1.1; Accelrys, Burlington, USA).
Recombinant Thd1p and Thp1p possess DNA glycosylase activity
To facilitate enzymatic studies, we produced the Drosophila and S.pombe TDG homologs in E.coli and purified the recombinant proteins to near homogeneity. In the case of the large Thd1p, we engineered a vector expressing a segment of 415 amino acids (650M–1063N) including the putative catalytic domain (see Fig. 1) plus an extra His6 tag at the N-terminus. Thp1p was expressed as a full-length protein with an N‐terminal His6 tag. Both proteins were expressed in soluble form in E.coli and, following a two-step purification scheme including Ni-NTA and FPLC-UNO S1 chromatography, we were able to recover 2–4 mg of highly pure protein per liter of bacterial culture. Like human TDG (46 kDa), the 46 kDa Thd1p polypeptide showed a retarded mobility in SDS–PAGE (∼60 kDa), whereas migration of the 37 kDa Thp1p was as expected (Fig. 2A).
To monitor the catalytic activities of Thd1p and Thp1p, we employed a standard base release assay previously developed for human TDG (6). The assay measures the excision of mispaired (e.g. G·U) and/or damaged (e.g. G·εC) DNA bases from synthetic 60mer oligonucleotide duplex substrates (Fig. 2B). Incubation of Thd1p and Thp1p alone with a G·U substrate did not yield any incised product (Fig. 2C). Only when the samples were incubated further with the human AP-endonuclease APE1 or heat treated in the presence of 90 mM NaOH did product bands with the expected length of 23 nt appear on the gel. The slightly higher electrophoretic mobility of the chemical cleavage products is accounted for by the negatively charged 3′ phosphate generated by the β,δ- elimination reaction involved. Neither APE1 nor NaOH treatment alone generated any detectable incision products (data not shown). These data illustrate that recombinant Thd1p and Thp1p act as monofunctional DNA-glycosylases with properties similar to the human TDG.
We further investigated the effects of temperature, pH and possible cofactors on G·U processing by Thd1p and Thp1p. Both enzymes were most active at 37°C with a pH between 7 and 9. Testing the requirement for divalent metal ions and ATP revealed that neither of the combinations of Mg2+, Mn2+ or Fe2+ with ATP had any effect on the nicking activities of the two enzymes. However, Zn2+ at concentrations >1 mM inhibited the G·U processing activity of both enzymes as well as of human TDG. Therefore, we chose a metal ion- and ATP-free buffer at pH 8.0 and an incubation temperature of 37°C for all subsequent comparisons.
TDG homologs all process uracil in DNA but act differently on G·T mispairs
The human TDG excises thymine from G·T and uracil from G·U mismatched substrates (13,14), whereas the E.coli Mug protein processes G·U substrate but fails to act on G·T mismatches with an appreciable efficiency (7). To compare the substrate preferences of the eukaryotic MUG proteins, we examined their abilities to process the ‘classical’ G·T, G·U and A·U substrates as well as U in ssDNA. While the G·U mispair was efficiently converted to product G·AP-sites by all enzymes, the G·T mismatch was only a poor substrate for Thd1p and totally resisted processing by Thp1p (Fig. 3). Even in the presence of high concentrations of Thp1p and after extended incubation, the G·T substrate remained fully intact (data not shown). Strikingly, unlike the human and Drosophila enzymes, Thp1p showed little preference for mismatched substrate and processed uracil opposite A or in an ssDNA context with remarkable efficiency.
Figure 3.
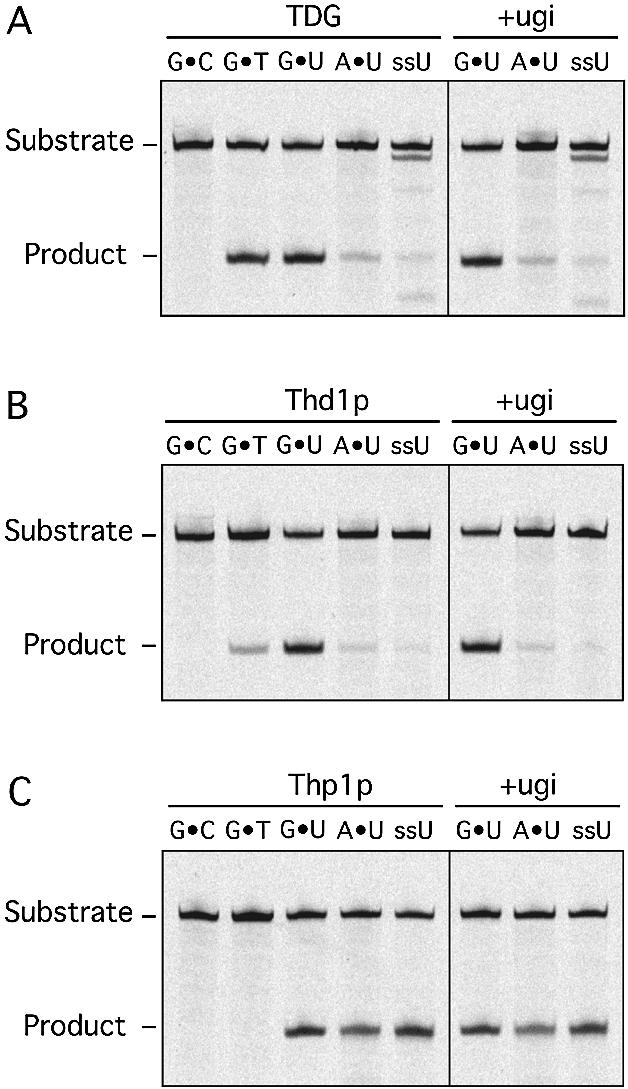
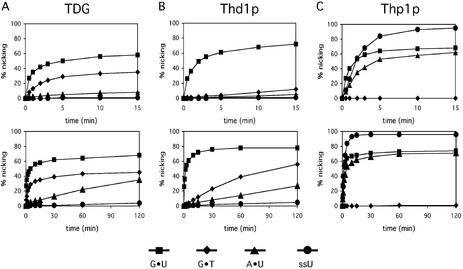
Differential G·T processing by TDG, Thd1p and Thp1p. The ability to generate alkaline-sensitive sites in standard substrates was assayed for TDG (A), Thd1p (B) and Thp1p (C) in the absence or presence of 1 U of Ugi. Shown are the results obtained with 60mer dsDNA substrates containing either G·C, G·T, G·U or A·U base pairs or a single uracil in ssDNA at identical positions. All reactions were done in a 20 µl volume containing equimolar amounts (50 nM) of substrate DNA and enzyme, and the products were separated on 15% denaturing polyacrylamide gels. The positions of the 60mer substrate DNA and 23mer product fragment are indicated.
All UNG-type UDGs are inhibited by the small polypeptide Ugi encoded by the bacteriophage PBS2 (15). Although UNG and MUG proteins appear to have similar active site architectures (11), E.coli Mug and human TDG are not inhibited by Ugi (7), and this is also true for Thd1p and Thp1p (Fig. 3). All substrates tested were processed to the same extent by both enzymes in the absence or presence of an excess of Ugi peptide (Fig. 3). Identical reaction conditions fully inhibited an equimolar amount of the E.coli Ung enzyme (data not shown).
The differences in substrate spectra and mismatch dependencies prompted us to investigate the base release reaction in more detail. Because TDG is fully product inhibited owing to its tight binding to the product AP-site (6,16,17), we used a previously standardized method to measure and compare single turnover kinetics of AP-site formation (6). Time course experiments with TDG, Thd1p and Thp1p and G·U substrate revealed that all reactions proceeded relatively quickly in an initial phase, and then leveled off to a plateau where no more substrate was processed (Fig. 4). We confirmed that neither of the enzymes lost activity under assay conditions and that the product:enzyme ratios never exceeded 1:1 even when a vast excess of substrate over enzyme was provided (data not shown). Thus, the plateaus reflect product inhibition of Thd1p and Thp1p.
Figure 4.
Kinetic properties of Thd1p and Thp1p in comparison with TDG. The time-dependent generation of alkaline-sensitive AP-sites was measured by incubation of TDG (A), Thd1p (B) and Thp1p (C) with double-stranded 60mer substrates containing either a single G·U, G·T or A·U mismatch, or ssDNA containing U as indicated. The substrate and enzyme concentrations were 50 nM as described for the standard TDG assay (6). All reactions were performed at 37°C and stopped after the indicated times by the addition of NaOH. Product formation was monitored and quantified after denaturing gel electrophoresis and fluorescent scanning. Shown are nicking efficiencies averaged from at least three independent experiments over short (upper panels) and longer (lower panels) time courses; standard deviations are listed in Table 1.
As expected, the kinetic time courses with the substrates G·T, A·U and U in ssDNA showed interesting qualitative differences. G·T was processed with an intermediate efficiency by TDG, with low efficiency by Thd1p but not detectably by Thp1p. The comparably slow initial rate of Thd1p most probably reflects a poor affinity of the protein for the G·T mismatch so that initial mismatch binding would be the rate-limiting step in this reaction. A poor affinity for the G·T mismatch was shown previously for human TDG (6) and could also help to explain the lack of G·T processing by Thp1p and E.coli Mug. On the other hand, A·U and U in ssDNA (ssU) were processed with an appreciable rate by Thp1p only. The reaction with A·U was initially slower but plateaued at a level comparable with that obtained with G·U, whereas single-stranded uracil substrate was processed with an intermediate rate and to a higher product yield. Since Thp1p also failed to turn over on these substrates (data not shown), we conclude that the substrate-dependent uracil excision kinetics reflect differences in enzyme–substrate complex formation rather than in catalysis. The kinetic properties further show that catalysis by Thp1p is less dependent on a specific interaction with the base opposite uracil, implicating a catalytic mechanism which differs from that proposed for the human and bacterial proteins (6,11). Taken together, G·U appears to be a common efficiently processed substrate for all MUG-type UDGs but neither the G·T processing activity nor the mismatch dependency of the proteins appear to be strictly conserved.
Comparison of DNA-binding properties of Thd1p, Thp1p and TDG
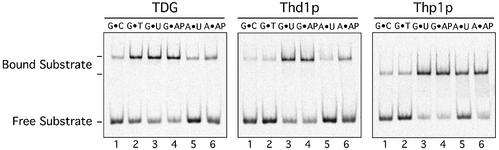
The lack of turnover in base release assays with dsDNA substrates indicated that Thd1p and Thp1p are strongly product inhibited. We therefore examined the ability of the two enzymes to bind different DNA substrates and product AP-sites in the presence or the absence of homoduplex competitor DNA. EMSAs in the presence of a 10-fold excess of competitor DNA revealed that both enzymes bound G·C homoduplex and G·T heteroduplex DNA with significantly lower affinity than the G·U substrate (Fig. 5, lanes 1–3). This resembled the substrate-binding preferences of a catalytically inactive variant of TDG (6). The apparently higher affinity of TDG for the G·T substrate reflects the ability of this enzyme to process the mismatch under EMSA conditions (6, and unpublished data), while Thd1p and Thp1p are unable to do so. Hence, the assay measures product AP-site binding for TDG but G·T binding for Thd1p and Thp1p. The same caveat of substrate conversion also applies when binding of Thd1p and Thp1p to the well processed G·U substrate is examined. The finding that both enzymes bound to G·U duplexes nearly as robustly as to G·AP-site substrates (Fig. 5, lanes 3 and 4) must therefore be interpreted to reflect their high compound efficiencies of uracil excision and G·AP-site interaction. Of all the enzymes, Thp1p was the only one to bind an A·U substrate with an appreciable efficiency and an A·AP-site duplex with a high affinity (Fig. 5, lanes 5 and 6). This is consistent with its ability to release uracil from A·U base pairs in a reaction that is product AP-site inhibited (Fig. 4). In contrast, Thd1p and TDG bound efficiently to G·AP substrates whereas A·AP binding was hardly above homoduplex background (Fig. 5, lanes 4 and 6).
Figure 5.
Substrate and product DNA-binding properties. Comparative EMSAs were performed with TDG, Thd1p and Thp1p as indicated. The DNA-binding reactions were performed in a 10 µl volume with 1 pmol of labeled homoduplex (G·C), substrate (G·T, G·U, A·U) or product DNA (G·AP, A·AP) in the presence of 10 pmol of unlabeled homoduplex competitor DNA and either 4 pmol of TDG, 4 pmol of Drosophila Thd1p or 5 pmol of S.pombe Thp1p. The AP-site-containing DNAs were generated by incubation of uracil substrate DNA with E.coli UDG and subsequent purification as described (6). Bound fluorescein-labeled DNA was separated from free substrate DNA in 6% native polyacrylamide gels. Shown are fluorograms of representative gels.
These data document that the complementary base facing the AP-site rather than the AP-site itself determines the DNA-binding affinities of MUG enzymes, and that this mode of opposite base interaction may differ slightly between Thp1p and Thd1p or TDG. For the reasons discussed above, a more precise assessment of substrate affinities is difficult with active DNA glycosylases and will require the use of either non-hydrolyzable DNA substrates (18), or of catalysis-deficient but substrate interaction-proficient mutant proteins (6).
Comparison of the substrate spectra of Thd1p, Thp1p and human TDG
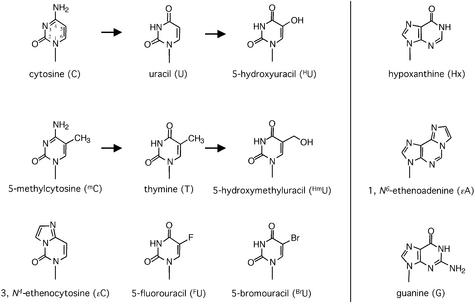
Unlike UNG proteins, which have a restricting active site geometry designed for accommodation of and specific interaction with uracil (19), MUG proteins appear to form a comparably loose binding pocket, where no specific contacts with the substrate base are established (12). This observation led us to investigate the substrate spectrum of the eukaryotic MUG proteins in greater detail. We thus examined the abilities of Thd1p, Thp1p and TDG to excise various DNA base lesions including oxidation, alkylation and deamination products of cytosine, 5-methylcytosine, thymine and adenine (Fig. 6). Because these enzymes are fully product inhibited, we based our comparisons on measurements of single turnover kinetics under conditions that were standardized for TDG (6). For each substrate, we determined the parameters Pmax, maximum of substrate processing within an unlimited period of time; and T50, the time required to reach 50% of the product plateau level (Pmax) (6). To facilitate comparison, we calculated a relative processing efficiency (EffRel) for each enzyme and substrate by dividing Pmax by T50. The values obtained from at least three independent measurements are listed in Table 1, and the relative processing efficiencies for all double-stranded substrates are plotted in Figure 7. Because the substrates 5-hydroxyuracil (HU) and 5-hydroxycytosine (HC) were provided in a slightly different sequence context and required an autoradiography-based method of evaluation, these results are not directly comparable with all others and therefore are only mentioned in the text.
Figure 6.
Substrate DNA bases examined. Shown are the chemical structures of purine and pyrimidine DNA bases with modifications relevant to this study.
Table 1. Relative substrate processing efficiencies of TDG, Thd1p and Thp1p.
| Substrate | TDG | Thd1p | Thp1p | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Pmax | T50 | Erel | Pmax | T50 | Erel | Pmax | T50 | Erel | |
| G·U | 68 ± 2 | 0.9 | 76 | 78 ± 3 | 1.2 | 65 | 73 ± 3 | 0.8 | 91 |
| A·U | 35 ± 1 | 40 | 0.9 | 27 ± 1 | 60 | 0.5 | 70 ± 4 | 2.0 | 35 |
| ssU | <5 | >60 | <0.08 | <5 | >60 | <0.08 | 95 ± 2 | 1.8 | 53 |
| G·FU | 90 ± 2 | 0.6 | 150 | 83 ± 2 | 0.9 | 92 | 80 ± 3 | 0.5 | 160 |
| A·FU | 75 ± 3 | 4.2 | 18 | 81 ± 2 | 15 | 5.4 | 72 ± 3 | 1.0 | 72 |
| ssFU | 95 ± 2 | 9.2 | 10 | 93 ± 1 | 20 | 4.7 | 96 ± 1 | 0.7 | 137 |
| G·BrU | 68 ± 2 | 1.5 | 45 | 66 ± 2 | 2.0 | 33 | 73 ± 1 | 13 | 5.6 |
| A·BrU | 17 ± 1 | 55 | 0.3 | <5 | >60 | <0.08 | 29 ± 1 | 30 | 1.0 |
| ssBrU | <5 | >60 | <0.08 | <2 | >60 | <0.02 | 73 ± 1 | 14 | 5.2 |
| G·T | 45 ± 2 | 2.2 | 20 | 56 ± 4 | 40 | 1.4 | <2 | >60 | <0.03 |
| G·HmU | 49 ± 1 | 0.8 | 61 | 68 ± 3 | 5.5 | 12 | <2 | >60 | <0.03 |
| G·εC | 72 ± 5 | 2.5 | 29 | 86 ± 3 | 4.6 | 19 | 86 ± 3 | 0.7 | 123 |
| A·εC | 55 ± 2 | 9.1 | 6.0 | 72 ± 3 | 15 | 4.8 | 83 ± 5 | 0.9 | 82 |
| ssεC | <5 | >60 | <0.08 | <5 | >60 | <0.08 | 87 ± 7 | 1.8 | 48 |
| G·Hx | 42 ± 3 | 42 | 1.0 | 47 ± 1 | 45 | 1.0 | 70 ± 2 | 0.5 | 140 |
| T·Hx | <5 | >60 | <0.08 | <5 | >60 | <0.08 | 71 ± 4 | 0.5 | 142 |
| ssHx | <2 | >60 | <0.03 | <2 | >60 | <0.03 | 92 ± 2 | 0.3 | 307 |
| G·εA | <2 | >60 | <0.03 | <2 | >60 | <0.03 | 65 ± 2 | 3.8 | 17 |
| T·εA | <2 | >60 | <0.03 | <2 | >60 | <0.03 | 68 ± 1 | 3.6 | 19 |
| ssεA | <2 | >60 | <0.03 | <2 | >60 | <0.03 | 60 ± 2 | 12 | 5.0 |
| G·G | <2 | >60 | <0.03 | <2 | >60 | <0.03 | 54 ± 1 | 42 | 1.3 |
| G·mC | <1 | >60 | <0.02 | <1 | >60 | <0.03 | <1 | >60 | <0.02 |
The generation of alkaline-sensitive sites was assessed by measurement of single turnover parameters under standardized reaction conditions. The substrate and enzyme concentrations were 50 nM each and the reaction products were quantified after denaturing gel electrophoresis and fluorescence scanning. Shown are the plateau levels of substrate nicking (Pmax) and the time required for processing of 50% of Pmax (T50) for all enzymes with the respective substrates, as indicated. Relative processing efficiency was calculated as: Erel = Pmax/T50.
Figure 7.
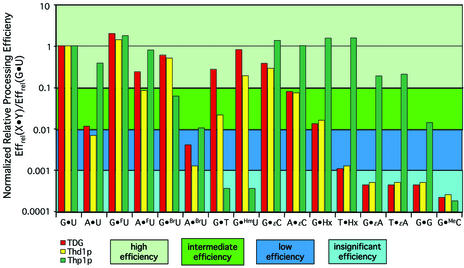
Substrate processing ability of eukaryotic MUG proteins. The substrate processing efficiencies of TDG, Thd1p and Thp1p were determined in kinetic assays as described in the text. Each experiment was repeated at least three times, and the resulting Pmax, T50 and Effrel (= Pmax/T50) values are listed in Table 1. Graphically illustrated are the relative processing efficiencies (Effrel) obtained with dsDNA substrates showing comparable kinetic properties. For ease of comparison, the Effrel values are normalized to the relative efficiency of G·U processing for each individual enzyme [= Effrel(X·Y)/Effrel(G·U)].
With the exception of HC (not shown), human TDG removed the substrate pyrimidines 5-fluorouracil (FU), 5-hydroxymethyluracil (HmU), 5-bromouracil (BrU), εC and thymine paired with guanine with high relative efficiencies (Table 1, Fig. 7). HU mispaired with guanine was also excised from DNA with a catalytic efficiency in the range of that obtained for G·HmU (data not shown). In addition, TDG efficiently excised εC opposite adenine and FU from any dsDNA or ssDNA substrate as observed previously (6,8). Remarkably, we also found a low but significant level of hypoxanthine (Hx) processing when the base was located opposite guanine. Since Hx is the deamination product of adenine, it would arise most frequently in DNA opposite thymine, but TDG processed a Hx·T substrate with an insignificant efficiency only. All data together give the following order of substrate preference for TDG: high efficiency, G·FU > G·U > G·HmU > G·HU > G·BrU > G·εC > G·T = A·FU; intermediate efficiency, A·εC > G·Hx > A·U; low efficiency, A·BrU = T·Hx; insignificant efficiency, G·εA = T·εA = G·G > G·mC = G·HC (Table 1, Fig. 7).
Thd1p showed a substrate preference very similar to that of TDG except that it was significantly less effective (5–10-fold) on the derivatives of 5-methylcytosine (G·HmU, G·T). Like TDG, Thd1p clearly preferred guanine as an opposite base and, with the notable exception of FU, did not process damaged bases in ssDNA efficiently (Table 1, Fig. 7). Thd1p thus processes dsDNA substrates with the following preference: high efficiency, G·FU > G·U > G·BrU > G·εC > G·HmU; intermediate efficiency, A·FU > A·εC > G·T = G·Hx; low efficiency, A·U > A·BrU = T·Hx; insignificant efficiency, G·εA = T·εA = G·G > G·mC.
Thp1p was a surprise. This enzyme processed all substrates containing U, FU, εC and HU irrespective of whether they were in a dsDNA or a ssDNA context. Thp1p also excised the purine derivatives Hx, 1,N6-ethenoadenine (εA) and even a regular guanine in a G·G mismatch. However, the enzyme showed no activity towards T, HmU or HC. In general, Thp1p hydrolyzed the target base fastest when it was opposite a guanine, but for most substrates the Pmax values reached similar levels independent of the complementary base. Thp1p thus prefers dsDNA substrates in the order: high efficiency, G·FU > T·Hx = G·Hx > G·εC > A·εC > A·FU > A·U > T·εA = G·εA; intermediate efficiency, G·HU > G·BrU > G·G > A·BrU; insignificant efficiency, G·T = G·HmU > G·mC (Table 1, Fig. 7).
The most impressive difference between the substrate spectra of the three MUG proteins was the inability of Thp1p to process the 5-methylcytosine-derived G·T and G·HmU mispairs on the one hand, and its broader range of efficiently processed substrates, including the adenine derivatives Hx and εA and the G·G mismatch, on the other hand. Another obvious difference was the lack of opposite base and/or dsDNA dependency in the action of Thp1p on virtually all substrates tested. The common, most efficiently processed substrates for all enzymes were the cytosine-derived G·U, G·HU, G·εC and A·εC mispairs as well as the non-physiological but therapeutically interesting FU. Recent reports documented the excision of 5-methylcytosine in a hemimethylated CpG sequence context by bacterially expressed human and chicken TDG. This activity was 30-fold lower than that of thymine excision from G·T mismatches (10). Using different hemi- and symmetrically methylated DNA substrates (mC·G/G·C, mC·G/G·mC) and applying similar assay conditions, we were unable to detect a release of mC by TDG or by either the Drospophila or the S.pombe proteins. Finally, the DNA sequence context only marginally affected the relative efficiencies of uracil excision by all three enzymes (data not shown).
DISCUSSION
In this study, we introduce two novel members of the MUG family of DNA glycosylases, Thd1p of D.melanogaster and Thp1p of S.pombe. A comparative enzymatic characterization of the human, Drosophila and fission yeast proteins justifies the following general conclusions: (i) eukaryotic MUG proteins have divergent and species-specific substrate spectra that include a wide range of damaged cytosine, 5-methylcytosine, thymine and even adenine bases; (ii) the common, most efficiently processed substrate is a G·U mismatch, whereas 5-methylcytosine and thymine derivatives are processed with a physiologically relevant rate only by the human enzyme; and (iii) the double strand and mismatch dependency of the MUG enzymes varies with the substrate and is not strictly conserved between the species. These findings open up novel perspectives with regard to the possible biological roles of MUG proteins and provide a solid basis for functional studies in prominent genetic model organisms.
Like the bacterial and human orthologs, the Drosophila and fission yeast MUG enzymes are monofunctional, they hydrolyze the N-glycosidic bond of a target base without incising the DNA strand at the resulting AP-site. The substrate spectra of Thd1p and TDG appear to be similar, with the caveat that the preferences of the full-length Thd1p may deviate slightly from those of the extended glycosylase domain studied here. Thp1p, however, shows some interesting and distinctive features. It fails to process G·T and G·HmU but efficiently excises damaged and/or mispaired purine bases, and lacks dsDNA or opposite base dependency.
A lack of G·T and G·HmU processing is understandable from the standpoint that S.pombe apparently does not methylate cytosine in DNA (20). Obviously, this obviates the need for an enzyme dealing with 5-methylcytosine damage. In organisms that have cytosine methylation, however, evolution appears to have selected for MUG proteins with an acquired function to excise the respective deamination and oxidation products. In this context, it is noteworthy that the domain architecture of the MUG proteins documents a modular evolution that most probably started from a simple DNA glycosylase fold to which activities modulating N- and C-terminal sequences were added in the process of adaptation to specialized functions. For instance, the ability to process G·T or G·HmU efficiently appears to be related to the structure of the non-conserved N-termini of these proteins rather than to the geometry of their active sites. Deletion of the N-terminus of TDG was shown to virtually abolish G·T but not G·U processing activity, and E.coli Mug that lacks a comparable N-terminus does not process G·T mismatches with a physiologically significant rate (7). Also, the ability of MUG proteins to hydrolyze purine-derived substrate bases (e.g. Hx and εA) implicates that bases far bulkier than thymine can fit into their active site pockets. This suggests that the initial formation of a productive enzyme–substrate complex and/or catalysis rather than a steric exclusion of thymine from the binding pocket is rate limiting for G·T processing by Thp1p and, by inference, by E.coli Mug and the N‐terminally truncated human TDG protein. However, the resolution of exactly how the N-terminus of TDG provides G·T processing ability will require more detailed structure–function studies.
TDG and E.coli Mug show relatively low catalytic efficiency as compared with the functionally related UNG enzymes, and both need to establish a stable interaction with the DNA strand opposite the mismatched substrate base whereby specific opposite base contacts play an important role (6,11,12). Interestingly, Thp1p shows little double strand or complementary base dependency; similar to UNG enzymes, but unlike TDG and Thd1p, it excised uracil from both dsDNA and ssDNA substrates with comparable efficiencies. This is consistent with the DNA-binding properties of the glycosylases; Thp1p could stably interact with a guanine and an adenine opposite an AP-site, whereas the human and Drosophila enzymes showed a strong preference for the G·AP substrate and A·AP binding was only insignificantly above homoduplex background. Thus, an increased affinity for a complementary adenine would give Thp1p the power to interact stably with and process uracil in any DNA substrate that has the potential to establish a secondary structure involving the substrate base. Obviously, precisely this must be avoided for thymine-processing MUG enzymes such as TDG to ensure the discrimination between a regular thymine opposite an adenine and a mutagenic thymine mispaired with guanine, which prevents processing of normal A·T base pairs.
Besides uracil and thymine, several other DNA lesions are processed by MUG proteins. To date, we can distinguish three classes of substrates: those that represent damaged cytosine bases, including U, HU or εC; those that arise from 5-methylcytosine, namely T and HmU; and the purine derivatives Hx and εA. However, uracil in a G·U mismatch and εC in a G·εC context appear to be the only substrates processed efficiently by all MUG enzymes tested to date. We would therefore conclude that the conserved function of MUG DNA glycosylases is the repair of cytosine damage in DNA.
In the absence of genetic evidence, the biological significance of the divergent substrate spectra of the MUG proteins remains uncertain. The range of possible substrates predicts that the MUG proteins are functionally redundant with other DNA glycosylases including UDGs and enzymes that process oxidized pyrimidines (e.g. NTH1) and deamination or alkylation products of purines (e.g. MAG1). Considering only the repair of cytosine or 5-methylcytosine damage in mammalian cells, there are at least three enzymes in addition to TDG that are capable of excising the respective deamination and/or oxidation products: UNG is a very potent enzyme known to excise uracil from DNA with a high turnover rate (4); SMUG1 was first described as a single-strand-selective UDG (21), but was then shown to process G·U, A·U, A·HmU and G·HmU substrates too (22,23); MBD4, which is related to bacterial EndoIII-type DNA glycosylases and contains a methyl-CpG-binding domain, excises thymine and uracil from G·T and G·U mismatches, respectively, but also FU and εC when paired with guanine (24,25). It is difficult to assess the significance of these apparent functional overlaps at this point, but it is likely that each of these enzymes evolved to fulfill specialized functions that are temporally and/or spatially separated from each other in the cellular context. For instance, the nuclear form of human UNG (UNG2) was shown to co-localize with proliferating cell nuclear antigen (PCNA) and replication protein A (RPA) in replication foci (26). No such localization was found for SMUG1 (23) and TDG (our unpublished data). ung–/– knockout mice do not exhibit an increased spontaneous mutation frequency, most probably because some uracil-processing activity remains in these cells (27). In tissue extracts of ung–/– mice, this activity processes G·U and less efficiently U opposite A or in ssDNA and can be partly attributed to the SMUG1 enzyme (22). Thus, UNG2 may have evolved as a uracil-processing enzyme with a specialized role in the elimination of dUMP misincorporated opposite adenine during DNA synthesis, whereas the mutagenic G·U mispairs resulting from cytosine deamination may be repaired preferentially by one of the other uracil-excising activities in the cell (SMUG1, TDG or MBD4). Considering G·T repair in mammalian cells, TDG may associate with and be active in transcribed regions of the genome through its interaction with transcription factors (28–30), whereas MBD4 could be targeted to transcriptionally silent genomic regions with dense CpG methylation through its methylated DNA-binding domain (24,31). Finally, the MUG homologs in general may play an additional role in repairing other pyrimidine damage including εC, HU and HmU, and possibly purine damage such as εA or Hx.
We conclude that MUG proteins form part of a DNA repair network consisting of base damage recognition functions with overlapping substrate specificities. Our data suggest that the catalytic properties of individual members of the MUG subfamily of UDGs have evolved in a species-specific manner. TDG, Thp1p and Thd1p are thus illustrative examples of how nature can take advantage of a simple but versatile catalytic polypeptide to develop specialized enzymes by the addition of activity-modulating extra domains. It is our hope that the study of these enzymes in organisms easily amenable to genetic manipulation will allow us to discover their true biological role.
Acknowledgments
ACKNOWLEDGEMENTS
We express our gratitude to Orlando Schärer, Christophe Kunz and Yusuke Saito for stimulating discussions and critical reading of the manuscript. We also thank Margaret Fäsi for excellent technical assistance. This study was supported by grants from the Sassella Stiftung (U.H. and P.S.) and the Swiss National Science Foundation (M.B. and P.S.).
REFERENCES
- 1.Hardeland U., Bentele,M., Lettieri,T., Steinacher,R., Jiricny,J. and Schär,P. (2001) Prog. Nucleic Acids Res. Mol. Biol., 68, 235–252. [DOI] [PubMed]
- 2.Aravind L. and Koonin,E.V. (2000) The alpha/beta fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol., 1, 1–8. [DOI] [PMC free article] [PubMed]
- 3.Schärer O.D. and Jiricny,J. (2001) Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays, 23, 270–281. [DOI] [PubMed]
- 4.Slupphaug G., Eftedal,I., Kavli,B., Bharati,S., Helle,N.M., Haug,T., Levine,D.W. and Krokan,H.E. (1995) Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry, 34, 128–138. [DOI] [PubMed]
- 5.Waters T.R. and Swann,P.F. (1998) Kinetics of the action of thymine DNA glycosylase. J. Biol. Chem., 273, 20007–20014. [DOI] [PubMed]
- 6.Hardeland U., Bentele,M., Jiricny,J. and Schär,P. (2000) Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. J. Biol. Chem., 275, 33449–33456. [DOI] [PubMed]
- 7.Gallinari P. and Jiricny,J. (1996) A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature, 383, 735–738. [DOI] [PubMed]
- 8.Saparbaev M. and Laval,J. (1998) 3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc. Natl Acad. Sci. USA, 95, 8508–8513. [DOI] [PMC free article] [PubMed]
- 9.Lutsenko E. and Bhagwat,A.S. (1999) The role of the Escherichia coli mug protein in the removal of uracil and 3,N(4)-ethenocytosine from DNA. J. Biol. Chem., 274, 31034–31038. [DOI] [PubMed]
- 10.Zhu B., Zheng,Y., Hess,D., Angliker,H., Schwarz,S., Siegmann,M., Thiry,S. and Jost,J.P. (2000) 5-Methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc. Natl Acad. Sci. USA, 97, 5135–5139. [DOI] [PMC free article] [PubMed]
- 11.Barrett T.B., Savva,R., Panayotou,G., Barlow,T., Brown,T., Jiricny,J. and Pearl,L.H. (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell, 92, 117–129. [DOI] [PubMed]
- 12.Barrett T.E., Schärer,O.D., Savva,R., Brown,T., Jiricny,J., Verdine,G.L. and Pearl,L.H. (1999) Crystal structure of a thwarted mismatch glycosylase DNA repair complex. EMBO J., 18, 6599–6609. [DOI] [PMC free article] [PubMed]
- 13.Neddermann P. and Jiricny,J. (1993) The purification of a mismatch-specific thymine DNA glycosylase from HeLa cells. J. Biol. Chem., 268, 21218–21224. [PubMed]
- 14.Neddermann P., Gallinary,P., Lettieri,T., Schmid,D., Truong,O., Hsuan,J.J., Wiebauer,K. and Jiricny,J. (1996) Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem., 271, 12767–12774. [DOI] [PubMed]
- 15.Wang Z. and Mosbaugh,D.W. (1989) Uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 encodes a binding protein specific for uracil-DNA glycosylase. J. Biol. Chem., 264, 1163–1171. [PubMed]
- 16.Waters T.R., Gallinari,P., Jiricny,J. and Swann,P.F. (1999) Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem., 274, 67–74. [DOI] [PubMed]
- 17.Hardeland U., Steinacher,R., Jiricny,J. and Schar,P. (2002) Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J., 21, 1456–1464. [DOI] [PMC free article] [PubMed]
- 18.Schärer O.D., Kawate,T., Gallinari,P., Jiricny,J. and Verdine,G.L. (1997) Investigation of the mechanisms of DNA binding of the human G/T glycosylase using designed inhibitors. Proc. Natl Acad. Sci. USA, 94, 4878–4883. [DOI] [PMC free article] [PubMed]
- 19.Mol C.D., Arvai,A.S., Sanderson,R.J., Slupphaug,G., Kavli,B., Krokan,H.E., Mosbaugh,D.W. and Tainer,J.A. (1995) Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell, 82, 701–708. [DOI] [PubMed]
- 20.Antequera F., Tamame,M., Villanueva,J.R. and Santos,T. (1984) DNA methylation in the fungi. J. Biol. Chem., 259, 8033–8036. [PubMed]
- 21.Haushalter K.A., Todd Stukenberg,M.W., Kirschner,M.W. and Verdine,G.L. (1999) Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol., 9, 174–185. [DOI] [PubMed]
- 22.Nilsen H., Haushalter,K.A., Robins,P., Barnes,D.E., Verdine,G.L. and Lindahl,T. (2001) Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J., 20, 4278–4286. [DOI] [PMC free article] [PubMed]
- 23.Kavli B., Sundheim,O., Akbari,M., Otterlei,M., Nilsen,H., Skorpen,F., Aas,P.A., Hagen,L., Krokan,H.E. and Slupphaug,G. (2002) hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem., 277, 39926–39936. [DOI] [PubMed]
- 24.Hendrich B., Hardeland,U., Ng,H.H., Jiricny,J. and Bird,A. (1999) The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature, 401, 301–304. [DOI] [PubMed]
- 25.Bellacosa A. (2001) Role of MED1 (MBD4) gene in DNA repair and human cancer. J. Cell. Physiol., 187, 137–144. [DOI] [PubMed]
- 26.Otterlei M., Warbrick,E., Nagelhus,T.A., Haug,T., Slupphaug,G., Akbari,M., Aas,P.A., Steinsbekk,K., Bakke,O. and Krokan,H.E. (1999) Post-replicative base excision repair in replication foci. EMBO J., 18, 3834–3844. [DOI] [PMC free article] [PubMed]
- 27.Nilsen H., Rosewell,I., Robins,P., Skjelbred,C., Andersen,S., Slupphaug,G., Daly,G., Krokan,H.E., Lindahl,T. and Barnes,D.E. (2000) Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell, 5, 1059–1065. [DOI] [PubMed]
- 28.Um S., Harbers,M., Benecke,A., Pierrat,B., Losson,R. and Chambon,P. (1998) Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J. Biol. Chem., 273, 20728–20736. [DOI] [PubMed]
- 29.Missero C., Pirro,M.T., Simeone,S., Pischetola,M. and Di Lauro,R. (2001) The DNA glycosylase T:G mismatch-specific thymine DNA glycosylase represses thyroid transcription factor-1-activated transcription. J. Biol. Chem., 276, 33569–33575. [DOI] [PubMed]
- 30.Tini M., Benecke,A., Um,S.J., Torchia,J., Evans,R.M. and Chambon,P. (2002) Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell, 9, 265–277. [DOI] [PubMed]
- 31.Hendrich B. and Bird,A. (1998) Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol., 18, 6538–6547. [DOI] [PMC free article] [PubMed]