Abstract
Members of the Tcf/Lef family of the HMG box transcription factors are nuclear effectors of the Wnt signal transduction pathway. Upon Wnt signaling, TCF/LEF proteins interact with β-catenin and activate transcription of target genes, while, in the absence of the Wnt signal, TCFs function as transcriptional repressors. All vertebrate Tcf/Lef transcription factors associate with TLE/Groucho-related co-repressors, and here we provide evidence for an interaction between the C-terminus of the TCF-4 HMG box protein and the C-terminal binding protein 1 (CtBP1) transcriptional co-repressor. Using Wnt-1-stimulated human embryonic kidney 293 cells, we show that CtBP1 represses the transcriptional activity of a Tcf/β-catenin-dependent synthetic promoter and, furthermore, decreases the expression of the endogenous Wnt target, Axin2/Conductin. The CtBP1-mediated repression was alleviated by trichostatin A treatment, indicating that the CtBP inhibitory mechanism is dependent on the activity of histone deacetylases.
INTRODUCTION
Members of the Wnt family of growth factors control numerous developmental processes and aberrant activation of Wnt genes, and Wnt signaling is also implicated in deregulated cell growth and cancer (reviewed in 1,2). Insights into the mechanisms of the Wnt signaling pathway have emerged from genetic studies in Drosophila, biochemical experiments in cell culture, ectopic gene expression in early Xenopus embryos and from target gene inactivation in mice (3). A striking characteristic of Wnt signaling is its conservation during evolution. Components of the Wnt signaling pathway have been found in such evolutionary distant species as slime mould Dictyostelium, diploblastic metazoan Hydra, Caenorhabditis elegans and in higher vertebrates, including mammals (4–6). This indicates that the Wnt signaling cascade is one of the primary regulatory mechanisms controlling cell fate decisions in multicellular organisms.
A brief outline of the canonical Wnt pathway is the following (for more detailed information, refer to the Wnt homepage at http://www.stanford.edu/∼rnusse/wntwindow.html). The interaction of extracellular Wnt ligands with the Frizzled/LRP receptor complex results in increased intracellular levels of β-catenin in the target cell (7–9). In unstimulated cells, free cytoplasmic β-catenin is marked on N-terminal serine and threonine residues by phosphorylation, and then ubiquitinated and rapidly degraded by the proteasome pathway (10–13). The β-catenin degradation machinery includes casein kinase Iα (CKIα) and a multiprotein cytoplasmic complex containing serine–threonine kinase GSK-3, Axin1 (or its homolog Axin2/Conductin) and the APC tumor suppressor (14–20). When cells are stimulated by Wnt proteins, the cytoplasmic phosphoprotein Dishevelled is recruited to the plasma membrane and, by a so far unknown mechanism, inhibits the function of the GSK-3/Axin/APC complex (21,22). The unphosporylated stable β-catenin molecules accumulate in the cytoplasm and also translocate into the nucleus. Nuclear β-catenin forms heterocomplexes with TCF/LEF proteins (23–25). The TCF/β-catenin heterodimers act as bipartite transcription factors and activate the expression of the specific Wnt responsive genes (26,27). Approximately 50 Wnt targets have been identified to date, and several of these genes encode proteins related to cell cycle regulation, e.g. c-myc, Cyclin D1 and Pitx2 (28–31).
Relay and final processing of the Wnt signal in the target cell is controlled at several levels. In the extracellular space, proteins of the Dkk, WIF and SFRP families interact directly with Wnt ligands or their receptors and inhibit transmission of the signal into the cell (32–37). In the cytoplasm, LIT-1/Nemo-like kinase phosphorylates TCFs and regulates the DNA binding and subcellular distribution of TCF/LEF factors (38–40). β-Catenin activity in the nucleus may be promoted by interaction with a number of proteins including Pontin52, Brg-1, Pygopus, p300 and Lines (41–45). β-Catenin-mediated transcription is repressed by Reptin52 (a Pontin52 homolog), ICAT, XSox17α/β and XSox-3 protein (46,47). Other interacting partners modulate the function of TCF/LEF proteins.
In mammals, the Tcf/Lef family consists of four genes: Tcf-1, Lef-1, Tcf-3 and Tcf-4. All TCF/LEF proteins display several common structural features (48,49). They contain a nearly identical DNA-binding domain, the HMG box, recognizing the consensus sequence A/T A/T CAAA. The extreme N-terminus harbors the β-catenin interacting domain. Although capable of binding to DNA, TCF/LEF proteins possess only limited ability to activate transcription. They might rather be viewed as nuclear vehicles targeting other auxiliary proteins to a specific set of promoters. β-Catenin possesses a strong transcription activation domain, so it is an example of an activating interacting partner. In contrast, all mammalian TCFs also associate with TLE/Groucho co-repressors and, in the absence of the Wnt signal, repress the transcription of the Tcf-dependent synthetic reporter or endogenous Tcf targets (50). TLE/Groucho proteins interact with the hypoacetylated N-terminal tail of histone H3 and also with human histone deacetylase 1 (HDAC 1) and its Drosophila homolog Rpd3 (51,52). This suggests that the function of TLE/Groucho is to form a specialized repressive chromatin structure that prevents the inappropriate activation of β-catenin/TCF target genes.
The participation of Wnt signaling in multiple developmental programs leads to the question of how β-catenin/TCF complexes discriminate between different subsets of all potential Wnt target genes at a given time and cellular background. Functional differences between individual TCF/LEF proteins could be one possible explanation. Although Tcf/Lef mRNAs are expressed in complex and often overlapping patterns during embryogenesis and in adult tissues, gene targeting experiments in mice revealed that Tcf-1, Lef-1 and Tcf-4 execute different developmental programs and their functions are only partially redundant (53–55). Additionally, several observations indicated that TCFs interact with distinct partners and show individual context-dependent DNA-binding and transactivation properties. For example, it has been reported that LEF-1 cooperates with microphthalmia- associated transcription factor (MITF) to up-regulate the dopachrome tautomerase gene promoter in melanocytes (56), and that activation of the T-cell receptor alpha enhancer by LEF-1 depends on ALY, a nuclear protein that specifically associates with LEF-1 and AML-1 (57). The activity of LEF-1 is suppressed by its association with PIASy, a nuclear matrix-associated SUMO E3 ligase, which sequesters LEF-1 into nuclear bodies (58) and, furthermore, direct interaction of LEF-1 with HDAC 1 mediates a repressive effect of LEF-1 in the absence of Wnt signaling (59). Another Tcf family member, TCF-3, interacts with casein kinase 1ε (CK1ε), and phosphorylation of TCF-3 by CK1ε stimulates its binding to β-catenin (60). In Xenopus laevis, XTCF-3 requires XCtBP to function as a transcriptional repressor (61). Recently, Hecht and Stemmler showed that TCF-4 and LEF-1 exhibit significant differences in regulating the promoter of two Wnt-responsive genes, Cdx-1 and Siamois, in human embryonic kidney cells (62). Cdx-1 activation strictly depended on the promoter-specific transactivation domain located at the TCF-4 C-terminus. In addition, multiple isoforms are generated from Tcf/Lef mRNAs by way of alternative splicing or by use of dual promoters, and these isoforms display diverse functional properties (63–65). Thus, selective interaction of members of the Tcf/Lef family with various partners and their intrinsic DNA-binding properties can support the execution of different developmental programs.
To study the regulatory mechanisms of Wnt signaling, we screened for proteins interacting with human TCF-4. In this report, we show that the C-terminal binding protein 1 (CtBP1) associates with the TCF-4 C-terminus and that CtBP1 overexpression in Wnt-1-stimulated 293 cells represses transcription from the Wnt-responsive reporter and also decreases the activity of the endogenous Axin2 promoter. The CtBP1-mediated repression is alleviated by trichostatin A treatment, indicating that the CtBP inhibitory mechanism is dependent on the action of histone deacetylases.
MATERIALS AND METHODS
Plasmids
Constructs encoding proteins containing N-terminal EGFP were generated in pEGFP/C vector (BD Clontech). Myc-tagged cDNAs were prepared in pK-Myc vector. This vector was created by replacing the EGFP region in pEGFP/C with a T7 promoter and Myc tag coding sequence. Full-length cDNAs without N-terminal tags contained either natural initiator or a PCR-introduced consensus Kozak sequence. The expression plasmids for Tcf-4 were derived from human Tcf-4E cDNA (GenBank accession No. NM_030756) using naturally occurring restriction enzyme sites: SmaI (position 236 bp from ATG), NsiI (995) and NdeI (1159). Plasmids encoding full-length or truncated forms of mCtBP1 were prepared in an analogous way using BglII, PvuII, KpnI and PstI internal restriction enzyme sites. Detailed information about the constructs is available upon request. The mouse Lef-1 cDNA was obtained from a mouse testis cDNA library (a gift from Stoil Dimitrov, IMG AS CR, Prague, Czech Republic). The TCF4–LEF-1 chimeric construct was generated by an exchange of the 3′ coding region in Tcf-4 with the C-terminal coding sequence of mouse Lef-1 using the ApaLI restriction enzyme site conserved in both genes. Human β-catenin cDNA was obtained from a bone marrow MATCHMAKER cDNA library (BD Clontech). Human CtBP1 cDNA was amplified from the cDNA of 293 cells, mouse CtBP2 was cloned from day-19 mouse embryo cDNA. Mouse Wnt-1 cDNA was a gift from Marc van Dijk (University Hospital Utrecht, Utrecht, The Netherlands). For retroviral transduction, Wnt-1 or EGFP-tagged mCtBP1 were ligated into pLNIT (generously provided by Fred H. Gage, The Salk Institute, La Jolla, CA) or pLNCX vector (BD Clontech). Platinum Pfx DNA Polymerase (Invitrogen) was used for all PCR amplifications. PCR-derived constructs were verified by sequencing.
Yeast two-hybrid screen
A pre-transformed mouse 17-day embryo MATCHMAKER cDNA library (Clontech) was screened with the last 185 amino acids of the human TCF-4 protein fused to the GAL-4 DNA-binding domain (DBD) in pGBKT7 as bait. Positive clones were subjected to the specificity test using the GAL-4 DBD or GAL-4 DBD–lamin fusion protein as bait. Those clones that interacted specifically with the GAL-4 DBD–TCF-4 bait were sequenced.
Cell culture, transfections and retrovirus infection
Human embryonic kidney (HEK) cell lines 293, monkey kidney cells COS-7, rat fibroblast cell line Rat2, EcoPack-293 and AmphoPack-293 (Clontech) cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Hyclone). Transfections were performed using the lipofectamine reagent (Invitrogen), as described by the manufacturer. For retrovirus production, ecotropic EcoPack-293 or amphotropic AmphoPack-293 packaging cells were seeded in 6-well plates 12 h prior to transfection and then transfected with 3 µg of a particular retroviral expression construct. After 24 h, the growth medium was replaced with 2 ml of fresh medium. After a further 24 h incubation, the supernatant containing non-replicating forms of the virus was harvested. Target cell lines were infected in 6-well plates with the virus supernatant in the presence of 5 µg of polybrene (Sigma) per ml. Twenty-four hours later, the medium was replaced by complete DMEM supplemented with either G418 (concentration 1 mg per ml) or hygromycin B (0.5 mg per ml; both antibiotics were purchased from Invitrogen) and doxycycline (Sigma), concentration 1 µg per ml [expression of the transduced cDNAs in pLNIT is regulated by doxycycline (66)]. After a 10 day selection, the growing (resistant) cells were washed extensively with DMEM and subsequently cultivated in fresh selective medium without doxycycline. The expression of the transferred genes was confirmed by western blot analysis.
Reporter gene assays, trichostatin A treatment and Wnt-1 stimulation experiments
For reporter gene assays, 293 cells were seeded into 24-well plates (approximately 105 cells per well) and transfected 2 h later with a lipofectamine mixture containing 50 ng Renilla pRL-SV40 plasmid (Promega) as an internal control, 400 ng luciferase reporter plasmid and up to 1 µg of the particular expression vector. The total amount of DNA was kept constant by adding empty expression vector where necessary. The firefly luciferase reporter constructs pTOPFLASH and pFOPFLASH, containing either three copies of the optimal Tcf motif GATCAAAGG or three copies of the mutant motif GGCCAAAGG, respectively, were described previously (27). The lipofectamine–DNA mixtures for reporter gene assays in 293-EGFP-CtBP1/Dox contained 50 ng Renilla pRL-SV40 internal control plasmid and 400 ng luciferase reporter plasmid only, i.e. no empty vector was added. COS-7 cells were seeded in 12-well plates and transfected by lipofection with 50 ng Renilla pRL-SV40 plasmid, 400 ng luciferase reporter plasmid and various combinations of the following plasmids (see Fig. 5A): TCF-4 expression vector (400 ng), β-catenin expression vector (800 ng) and CtBP1 expression vector (0.5–2 µg). Additional empty pK-Myc plasmid was added when necessary to make the total amount of DNA equivalent. Firefly and Renilla luciferase activities in cell lysates were determined 15 h post-transfection using the dual luciferase system (Promega), according to the protocol supplied by the manufacturer, and a single tube luminometer Sirius (Berthold). All reporter gene assays were done in triplicate. The reporter gene activities shown are average values, along with the standard deviations from at least three independent experiments after normalization against the Renilla luciferase activities. Treatments with the deacetylase inhibitor trichostatin A (Sigma) were performed using the drug at 300 nM concentration for 12 h. Wnt-1 stimulation experiments were performed as follows. 3 × 105 293 cells were seeded into 6-well plates and transfected by lipofectamine–DNA mixtures. The amounts of individual DNAs in the mixtures were proportionally increased by a factor of three as compared to the experiments in 24-well plates. Four hours post-transfection, the cells were washed extensively with DMEM and then 1 × 105 Rat2-Wnt-1/Const cells expressing stable levels of Wnt-1 protein were plated over the target cells. Parental Rat2 cells were used as a negative control. After co-cultivation for 3–30 h, the cells were harvested together and further processed for the reporter gene assay, RNA isolation, electrophoretic-mobility shift assay (EMSA) or immunoblotting. When feeders with inducible Wnt-1 expression (Rat2-Wnt-1/Dox) were used, the experimental procedure was modified as follows. We regularly grew the feeder cells in a complete medium supplemented with 2 ng of doxycycline per ml. This low concentration of the antibiotic was sufficient to suppress Wnt-1 production, and, additionally, Wnt-1 protein started to appear almost instantly upon the removal of doxy cycline. Transfected 293 cells were covered with Rat2-Wnt-1/ Dox cells and co-cultivated in medium without doxycycline. Cell cultures growing in the presence of doxycycline (5 ng per ml) were used as negative controls. The regulation of CtBP1 expression in 293-EGFP–CtBP/Dox cells was rather ‘leaky’, thus we cultivated these cells in higher concentrations of Dox (1 µg/ml). The full induction of the CtBP1 transgene was then achieved during 15 h growth in the absence of doxycycline.
Figure 5.
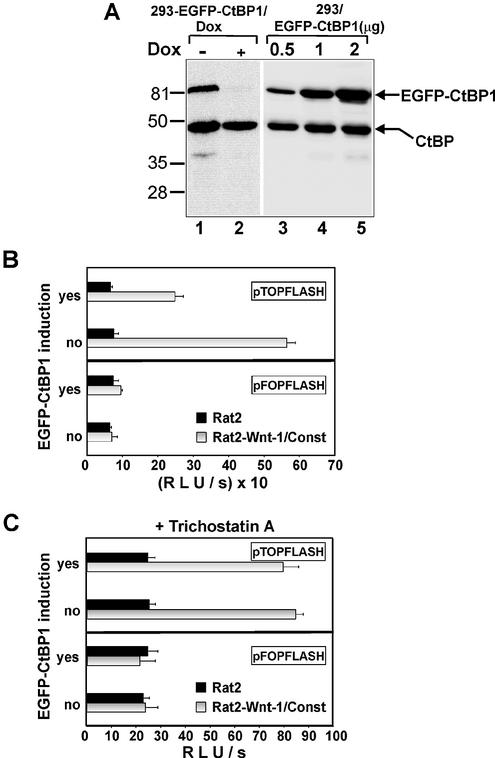
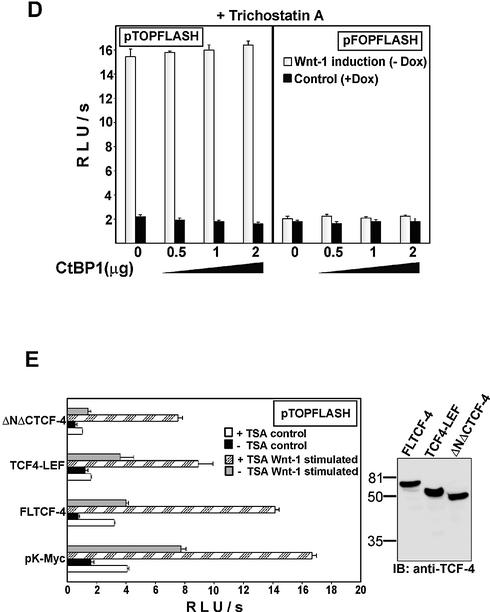
Histone deacetylases inhibitor trichostatin A alleviates the repressive effect of CtBP1 on the Wnt-responsive promoter. (A) 293-EGFP–CtBP1/Dox cells produce EGFP-tagged mCtBP1 in quantities that represent one-half of the amount of endogenous CtBPs. Total cell lysates from 293 cells expressing EGFP–CtBP1 from a doxycycline-repressive promoter growing in the absence (EGFP–CtBP1 induced, lane 1) or presence (EGFP–CtBP1 repressed, lane 2) of doxycycline (1 µg/ml) and from the parental 293 cell line transiently transfected with different amounts of plasmid encoding EGFP-tagged CtBP1 (lanes 3–5, micrograms of transfected construct are indicated on the top) were analyzed by western blot analysis. The anti-CtBP monoclonal antibody used to visualize proteins on the blots recognizes both human CtBP1 and CtBP2 proteins. The positions of molecular weight markers in kDa are indicated at the left. (B) Expression of the EGFP–CtBP1 transgene down-regulates activity of the Wnt-resposive promoter in 293-EGFP–CtBP1/Dox cells. 293-EGFP–CtBP1/Dox cell line was transfected with the indicated Tcf reporter constructs. Four hours post-transfection, DNA mixtures were removed, the cells washed extensively and Rat2-Wnt-1/Const fibroblasts (steadily producing the Wnt-1 protein) or parental Rat2 cells were subsequently plated over the 293 transfectants. Cultures were further grown for 15 h either in the absence (EGFP–CtBP induction: yes) or presence (EGFP–CtBP induction: no) of doxycycline (1 µg/ml). (C) Trichostatin A treatment releases the repressive function of EGFP–CtBP1. The experiment was performed as described above in (B) except that trichostatin A (final concentration 300 nM) was added simultaneously with the feeder cells. (D) Trichostatin A treatment alleviates the CtBP1-mediated repression in transient transfection assay. Human 293 embryonic kidney cells were co-transfected with the indicated amounts of CtBP1 expression plasmid and the Tcf reporter constructs using the lipofectamine reagent. Four hours post-transfection, DNA–lipofectamine mixtures were removed and 293 cells were covered with Rat2-Wnt-1/Dox fibroblasts. The cultures were further grown in the presence (control) or absence of doxycycline (Wnt-1 stimulated). Trichostatin A (300 mM final) was added simultaneously with the feeder cells. Following co-cultivation for 15 h, the cells were harvested and luciferase and Renilla luciferase activities were determined in cell lysates. (E) Tcf-mediated repression is partially insensitive to trichostatin A. Human 293 embryonic kidney cells were co-transfected with empty expression vector pK-Myc or TCF-4 deletion constructs indicated on the y-axis and the Tcf reporter plasmid pTOPFLASH. TCF-4 proteins were produced at comparable levels in the transfected cells as shown by western blot analysis (right panel). Four hours post-transfection, cells were covered with Rat2 cells (control) or Rat2-Wnt-1/Const cells (Wnt-1 stimulated), and half of the samples were further treated with trichostatin A (TSA, final concentration 300 nM). Following an additional 12 h, the cells were harvested, and luciferase and Renilla luciferase activities were determined in cell lysates. Transfections were done in triplicate. Average luciferase light units per second (RLU/s) corrected to Renilla luciferase activities and their standard deviations are shown.
GST interaction assays
Full-length GST–mCtBP1 and GST–mCtBP2 fusion proteins were expressed in the BL21 (DE3) strain of Escherichia coli using the pET-42b vector (Novagen). Full-length or truncated TCF-4 and full-length mCtBP1 protein were produced in vitro using the TNT Coupled Reticulocyte System (Promega) and corresponding pK-Myc constructs. All coupled transcription–translations were performed in a total volume of 50 µl using 10 µl of [35S]methionine (ICN Biomedicals) per reaction. Twenty microliters of radiolabeled TCFs or mCtBP1 were incubated with GST–mCtBP1 or GST–mCtBP2 proteins bound to glutathione–Sepharose 4B beads (Amersham Pharmacia Biotech) in GST binding buffer [phosphate-buffered saline, pH 7.4, 0.5% (v/v) Nonidet P-40, 20 µM NADH (Sigma) and Protease Inhibitor Cocktail (Sigma)] for 1 h at 4°C. Beads were collected by centrifugation and washed five times in GST binding buffer. Bound proteins were separated by SDS–PAGE and analyzed by autoradiography.
Antibodies and immunoblotting
The following commercially available antibodies were used: β-catenin, CtBP, mouse monoclonal (Santa Cruz); EGFP, mouse monoclonal (BD Clontech); Myc tag, mouse monoclonal 9E10 (Roche Molecular Biochemicals); TCF-4, mouse monoclonal (Sigma); Wnt-1, rabbit polyclonal (Santa Cruz); and LEF-1, goat polyclonal (Santa Cruz). To monitor protein expression, 2 × 105 293 cells, seeded into 6-well plates, were transfected with 3 µg of expression vector. Twenty-four hours later cells were harvested and disrupted directly in SDS–PAGE sample buffer. The cell lysates were cleared of cell debris and chromosomal DNA by ultracentrifugation at 200 000 g, then loaded onto SDS–PAGE gels, transferred to polyvinylidene fluoride membranes (Pall Gelman Laboratory) and immunoblotted with the appropriate antibodies. Proteins were visualized with an enhanced chemiluminiscence system (Pierce).
EMSA
The nuclear extracts were prepared according to Dignam et al. (67), snap frozen in liquid nitrogen and stored at –70°C. The final buffer composition was 20 mM HEPES, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF and 20% glycerol. Extracts were prepared from intact nuclei that were washed three times to avoid contamination with cytoplasmic β-catenin. As the optimal Tcf/Lef probe, we used a double-stranded 19-nucleotide oligomer AGAACCCTTTGATCTTAGG; the control probe was AGAACCCTTTGGCCTTAGG. The oligonucleotides were end-labeled using T4 polynucleotide kinase (Fermentas) and [γ-32P]ATP (3000 Ci/mmol; ICN Biomedicals). A typical binding reaction contained 8 µg of nuclear protein, 1 ng of radiolabeled probe and 300 ng of poly-(deoxyinosic–deoxycytidylic) acid (poly [dIdC]; Sigma) in 25 µl of binding buffer (60 mM KCl, 1 mM EDTA, 1 mM DTT, 10 mM HEPES, pH 6.9 and 10% glycerol). Samples were incubated for 30 min at room temperature, an antibody was added and the samples were incubated for a further 30 min. The binding reactions were loaded onto 5% polyacrylamide gels with 0.5× Tris–borate–EDTA running buffer. Electrophoresis was carried out at 15°C at 220 V constant voltage. Gels were transferred onto Whatman paper, dried and exposed to the BAS-phosphoimager screen (Fuji).
Real-time quantitative RT–PCR
One hundred micrograms of total RNA from Wnt-1- stimulated 293 or control cells isolated by the guanidine thiocyanate (Fluka) method (68) were further purified using the Micro-to-Midi total RNA isolation system (Invitrogen). Random or oligo dT-primed cDNA was prepared in a 20 µl reaction from 2.5 µg of total RNA using Superscript II RNAseH– reverse transcriptase (Invitrogen). One percent of the resulting cDNA was used for one quantitative PCR reaction. The 10 µl reaction mixtures also contained 1× Platinum Quantitative PCR SuperMix-UDG (Invitrogen), acetylated BSA (final concentration 3 µg per ml; New England Biolabs), Sybr Green I (final dilution 1:40 000; Amresco) and 5 pmol of each specific primer. All primers were calculated using Primer 3 computer services at http://www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi/primer3_www.cgi. The primer pairs were designed and tested to be specific for human genes, i.e. the primers did not prime on rat or mouse cDNA. We used primers for the following human genes (the primers are written in the 5′ to 3′ direction, the first primer is derived from the plus and the second primer from the minus DNA strand): Axin1, CCTGTGGTCTACCCGTGTCT, GCTATGAGGAGTGGTCCAGG; Axin2, CTGGCTTTGGTGAACTGTTG, AGTTGCTCACAGCCAAGACA; GAPDH, CACCACACTGAATCTCCCCT, CCCCTCTTCAAGGGGTCTAC; CyclinD1, CCATCCAGTGGAGGTTTGTC, AGCGTATCGTAGGAGTGGGA; SDHA, AGATTGGCACCT AGTGGCTG, ACAAAGGTAAGTGCCACGCT; CtBP1, CCTTCGCGTTCCTCGTTA, AAGAACGTTCATGGGAGAATAA; Tcf-1, CCTCTGCCTCCCTAGCTTTT, ATGGGGGAGATGGGTAGAGA; Lef-1, CTGCTAGAGACGCT GATCCA, TGGCTCTTGCAGTAGACGAA. cDNAs were produced from at least two independent RNA isolations, and the PCR reactions were performed in triplicate for each primer set. The specificity of the PCR products was verified by sequencing. Two housekeeping genes, Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Succinate dehydrogenase complex, subunit A (SDHA), were used as internal control genes to standardize the quality of different cDNA preparations (69). The cycling was performed in a LightCycler instrument (Roche Molecular Biochemicals). The results were analyzed using the LightCycler 3 software package. The relative abundance of Axin1 and Axin2 mRNA in 293 cells stimulated by Rat-2-Wnt-1/Const cells versus control 293 cells (co-cultivated with the parental Rat2 cell line) was derived from the average CT values of each triplicate after normalizing against the levels of SDHA cDNA. Results of a representative experiment are shown.
RESULTS
TCF-4 interacts with the transcriptional co-repressors CtBP1 and CtBP2
To gain insight into the molecular function of TCF-4 protein, we started a search for novel TCF-4-interacting proteins using a yeast two-hybrid system. We selected a 180 amino acid region distal to the HMG box covering the C-terminus of TCF-4 as bait. This region has a unique primary structure within the Tcf/Lef family and displays only limited homology to the C-terminus of TCF-3, the closest relative of TCF-4 (Fig. 1). One strong positive obtained from a day-17 mouse embryo cDNA library corresponded to CtBP1, a widely expressed transcriptional co-repressor. The cDNA encoded the entire 430 amino acid CtBP1 polypeptide. The CtBP1– Gal4 activation domain fusion protein interacted specifically with the TCF-4 bait and not with the Gal-4 DBD alone or with the Gal-4 DBD–lamin fusion protein, respectively (see Materials and Methods).
Figure 1.
Amino acid comparison of the C-terminal regions of human TCF-4 and TCF-3. Amino acid identities are indicated by corresponding letters; amino acid similarities are indicated by double dots (closely related residues) or single dots (distantly related residues). Sequences were aligned with CLUSTALW. Overlining indicates highly conserved HMG box sequences; two putative CtBP binding sites are boxed. The arrows depict the amino acid sequence used in a yeast two-hybrid screen.
We further attempted to delineate the minimal region of CtBP1 required for association with TCF-4. We generated a series of N- and C-terminal deletions of CtBP1 fused to the Gal4 activating domain and tested their interaction with Gal4 DBD–TCF-4 in a yeast two-hybrid system. Each truncation of CtBP1 displayed a dramatically reduced ability to bind TCF-4 (data not shown). This indicates the complex multidomain mode of interaction between CtBP1 and TCF-4.
Human CtBP1 was originally discovered during a screen for cellular proteins that complex with the C-terminal region of the adenoviral E1A protein (70). Subsequently, a highly homologous human polypeptide termed CtBP2 was identified by analysis of EST database sequences (71). Other vertebrates such as rodents and Xenopus also contain two CtBP homologs, while invertebrates have a single CtBP gene. The CtBP proteins bind to a short sequence motif PLDLS conserved among the E1A proteins of all human and primate adenoviruses. Different variants of this motif are also present in other CtBP-interacting partners that function mainly as negative regulators of transcription (reviewed in 72). Two putative CtBP-binding sequences are also present in the C-termini of TCF-3 and TCF-4 proteins (Fig. 1); Brannon and others showed that simultaneous mutations in both these sites abolished the association of XTCF-3 with XCtBP, a Xenopus homolog of CtBP1, in vitro (61).
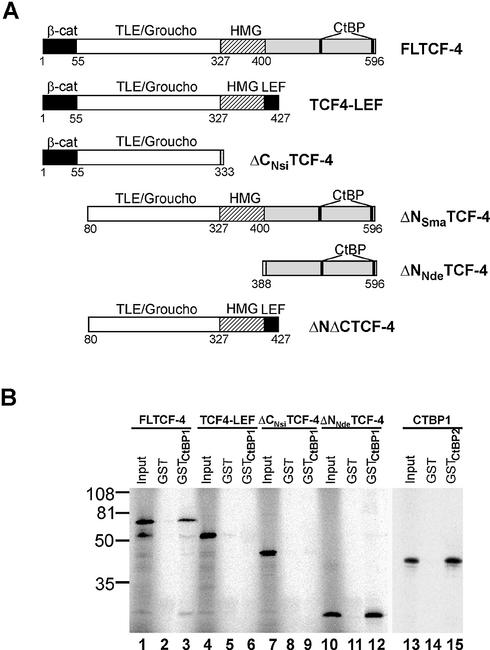
Given the putative CtBP-binding motifs in the C-terminus of TCF-4, we tested the interaction using bacterially purified GST–CtBP1 or GST–CtBP2 and various forms of TCF-4 translated in vitro (Fig. 2A). As shown in Figure 2B, a GST fusion protein containing complete CTBP1 interacted with [35S]methionine-labeled full-length TCF-4 (FLTCF-4) or with its C-terminal fragment (ΔNNdeTCF-4). Truncated proteins that lacked either sequences distal to the HMG box (TCF-4– LEF-1 chimera) or the entire C-terminal region (ΔCNsiTCF-4) failed to interact with mCtBP1. GST-tagged CtBP2 showed an identical mode of interaction (not shown), and, additionally, we observed efficient heterodimerization of CtBP1 and CtBP2 in vitro (Fig. 2B). Taken together, these data indicate that TCF-4 associates with CtBP1 and CtBP2 proteins and that the TCF-4 C-terminus is indispensable for the interaction.
Figure 2.
The C-terminus of TCF-4 interacts with CtBP1. (A) A schematic representation of the human TCF-4 deletion constructs used in this study. All constructs contained the N-terminal Myc-tag (not depicted). β-cat, β-catenin interaction domain; TLE/Groucho, TLE/Groucho binding domain; CtBP, CtBP-binding sites; HMG, DNA-binding domain; LEF, LEF-1 C-terminus. (B) In vitro interaction of TCF-4 with CtBP1. GST–CtBP1 and GST–CtBP2 fusion proteins were conjugated to glutathione–Sepharose beads and incubated with the indicated 35S-labeled TCF-4 proteins translated in vitro. After washing and recovery of the beads, associated proteins were resolved by SDS–PAGE and analyzed by autoradiography. In lanes 1, 4, 7, 10 and 13, 10% of the input labeled proteins was applied directly onto the gel. Full-length TCF-4 (lane 3) and the TCF-4 C-terminal fragment (lane 12) bind to GST–CtBP1. TCF4–LEF-1 chimera (line 6) or the N-terminal fragment of TCF-4 lacking CtBP-binding sites (lane 9) do not interact with GST–CtBP1. In vitro labeled CtBP1 associates with GST–CtBP2 (lane 15). None of the proteins bind to GST-bound Sepharose beads (lanes 2, 5, 8, 11 and 14). The positions of molecular weight markers in kDa are indicated at the left.
Wnt signaling is activated in 293 cells co-cultivated with Wnt-1 protein-producing feeder cells
By retrovirus infection we generated several polyclonal rat fibroblast cell lines with constitutive (Rat2-Wnt-1/Const) or doxycycline-repressible (Rat2-Wnt-1/Dox) expression of the mouse Wnt-1 gene (Fig. 3A). Because Wnt molecules possess only limited solubility and mostly stay attached to the cell surface of the producing cell, we activated Wnt signaling in 293 cells by co-cultivation with Wnt-1-expressing feeder cells. The human embryonic kidney 293 cells selected for the study activate the Tcf/β-catenin-dependent reporter if transiently transfected with Wnt-1 expression plasmid, and, additionally, express high levels of Tcf-4 mRNA and produce predominantly a longer variant, i.e. the TCF-4E form, of TCF-4 protein, which contains both CtBP-binding motifs [(27); Fig. 3B].
Figure 3.
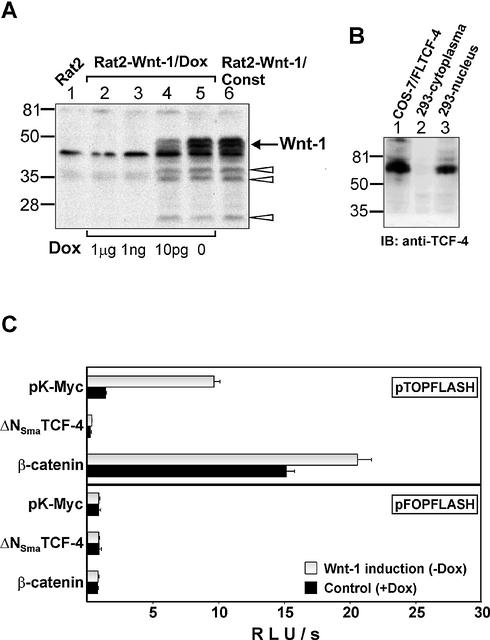
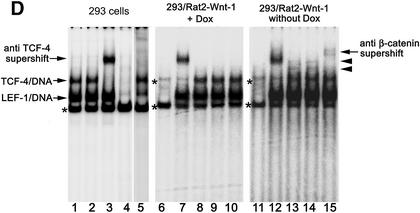
Wnt-1 protein-producing Rat2 feeder cells activate Wnt signaling in 293 cells. (A) Wnt-1 expression is strictly regulated in Rat2 fibroblasts by doxycycline. Western blot analysis of total cell lysates from parental Rat2 cells (lane 1), Rat2 cells with doxycycline-regulated Wnt-1 expression (Rat2-Wnt-1/Dox, lanes 2–5) and Rat2 cells with constitutive production of Wnt-1 (Rat2-Wnt-1/Const, lane 6). The Rat2-Wnt-1/Dox cells were grown at different concentrations (1 µg, 1 ng and 10 pg per ml) or in the absence of doxycycline, as indicated at the bottom. SDS–PAGE and immunoblotting with Wnt-1 antibody resolved cell lysates. The arrow shows the position of the putative Wnt-1 protein; the positions of shorter degradation products are indicated by open arrowheads. Molecular weight markers in kDa are at the left. (B) 293 cells express a longer form of TCF-4 protein. A total cell lysate from COS-7 cells transfected with expression vector encoding full-length TCF-4E protein (lane 1), and from cytoplasmic and nuclear fractions prepared from 293 cells (lanes 2 and 3, respectively) were resolved by SDS–PAGE and immunoblotted with an anti-TCF-4 antibody recognizing an epitope proximal to the HMG box. Molecular weight markers in kDa are at the left. (C) Wnt-1 expressing Rat2 cells induce the activation of the Tcf reporter in 293 cells. The human embryonic kidney 293 cell line was co-transfected with the expression plasmids indicated on the y-axis and the Tcf reporter construct pTOPFLASH (containing wild-type Tcf-binding sites) or pFOPFLASH (with mutated Tcf motifs) as a negative control. Four hours post-transfection, DNA mixtures were removed and Rat2-Wnt-1/Dox fibroblasts were subsequently plated over the 293 cells. Cultures were further grown either in the absence (Wnt-1 induction) or presence of doxycycline (control) for 15 h, then the cells were harvested together and processed to assay the reporter gene activities. Luciferase activities [shown on the x-axis as relative light units per second (RLU/s)] were corrected for the efficiency of transfection using the internal control Renilla pRL-SV40 expression plasmid. Average values and their standard deviations from three independent experiments are shown. (D) Wnt-1 induces nuclear TCF–β-catenin complexes in 293 cells. A gel retardation assay performed with nuclear extracts from parental 293 cells (left) and from 293 cells co-cultivated with Rat2-Wnt-1/Dox cells growing in the presence (control, middle) or absence (Wnt-1 induction, right) of doxycycline. Samples in lanes 1, 8 and 13 were incubated under standard conditions. Anti-β-catenin antibody was added to the samples in lanes 2, 10 and 15. Anti-TCF-4 antibody was added to the samples in lanes 3, 7 and 12. Anti-LEF-1 antibody was added to the sample in lane 5. Filled arrowheads indicate the positions of the TCF/β-catenin complexes. A control antibody (anti-HA tag) was added to the samples in lanes 9 and 14. Asterisks indicate non-specific bands also observed with a probe mutated in the Tcf-binding site (lanes 4, 6 and 11).
As shown in Figure 3C, Wnt-1-producing feeders stimulated the Tcf-reporter pTOPFLASH 6–7-fold as compared to control feeders. Co-transfection of an expression construct encoding dominant negative TCF-4, which lacks the N-terminal β-catenin interacting domain (ΔNSmaTCF-4), completely abrogated the reporter gene activity, whereas a β-catenin expression plasmid strongly enhanced transcription from the pTOPFLASH reporter. The activity of the negative control reporter pFOPFLASH did not display any significant changes. We also employed a gel retardation supershift assay to determine whether Wnt signaling induces a nuclear TCF-4/β-catenin complex in stimulated 293 cells. We did not detect TCF–DNA complexes in nuclear extracts prepared from parental Rat2 fibroblast cells or Rat2-Wnt-1-transfected cells (not shown), therefore we performed gel retardations with compound nuclear extracts isolated from 293 and feeder cells. Using an optimal Tcf-motif as probe, we readily obtained two types of TCF–DNA complexes from parental and from both unstimulated and stimulated 293 cells; one of these complexes could be supershifted with a monoclonal antibody recognizing human TCF-4 (Fig. 3D). Although 293 cells express all four Tcf/Lef genes, previous studies showed that TCF-1 and TCF-3 remain associated with insoluble nuclear components and are not eluted from nuclei by conventional extraction procedures (73,74). Thus, we concluded that the second gel-retarded complex contained endogenous LEF-1 protein and we attempted to supershift this complex with an antibody recognizing LEF-1. Addition of the LEF-1-specific antibody to the binding reaction led to almost complete diminishing of the original protein–DNA complex while the TCF-4–DNA complex stayed intact (Fig. 3D). We assumed that the second TCF–DNA complex indeed contains LEF-1 protein, and that the interaction between LEF-1 and the DNA probe was destabilized by the antibody added. Stimulation of 293 cells by Rat2-Wnt-1 cells resulted in the appearance of additional bands that could be further supershifted with a β-catenin antibody (Fig. 3D). Taken together, these data provide clear proof that the exogenous Wnt signal delivered by the Wnt-1-producing feeder cells, is effectively transmitted into human 293 cells, stimulates the formation of TCF/β-catenin complexes and subsequently activates the transcription of Wnt-responsive promoters. We further used this cellular system to study the effects of CtBP1 on Wnt-induced transcription.
CtBP1 represses TCF/β-catenin-dependent transcription by a trichostatin A-sensitive mechanism
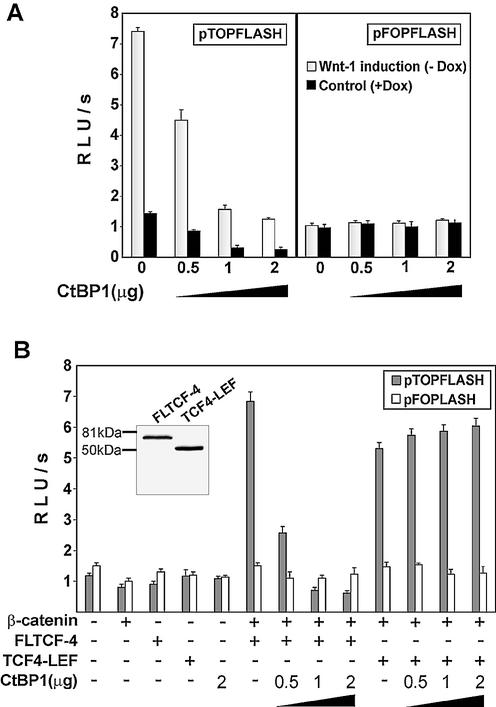
CtBP proteins function as short-range transcriptional co-repressors, therefore we examined the functional relevance of the TCF-4–CtBP1 interaction in reporter assays. It has been reported previously that N-terminal modification of the CtBP1 protein may change its association with histone deacetylases (75); for that reason, we tested the activity of three different CtBP1 constructs encoding mCtBP1 with an N-terminal Myc-tag, EGFP-tagged mCtBP1 and, finally, full-length human CtBP1 without N-terminal modification. All three constructs repressed transcription from the Tcf-dependent pTOPFLASH reporter in a concentration-dependent manner in both unstimulated and stimulated 293 cells, whereas the activity of the control reporter pFOPFLASH was not affected by CtBP1 (Fig. 4A, only data for human CtBP1 are shown). CtBP1 was originally described as a protein interacting with the adenoviral oncogene E1A. Since 293 cells express E1A, we used an alternative cell system to avoid the possibility that the observed effects are dependent on E1A expression. We utilized the previously established transient β-catenin–Tcf reporter assay. In this assay, co-transfection of TCF-4 and β-catenin into COS-7 cells results in transactivation of the Tcf-dependent luciferase reporter gene (Fig. 4B). Upon addition of increasing amounts of CtBP1, the luciferase activity decreases proportionally only when the construct encoding full-length TCF-4 (FLTCF-4) capable of binding to CtBP was co-transfected. Transactivation mediated by β-catenin and C-terminally truncated TCF-4 (TCF4–LEF) was not affected by CtBP1. The reporter gene assays indicated that Tcf/β-catenin transcriptional activity is down-regulated by TCF-4–CtBP1 interaction. To study the mechanisms of CtBP1 action in greater detail, we generated 293 cells with inducible expression of EGFP-tagged mCtBP1. We obtained four polyclonal cell lines (293-EGFP–CtBP1/Dox) producing EGFP–mCtBP1 protein in the quantities representing approximately one-half of the amount of endogenous CtBPs (Fig. 5A). For further analysis we utilized a mixture of these cell lines. We further compared EGFP–CtBP1 production from the CtBP1 transgene with the EGFP–CtBP1 levels obtained in transient transfections. Results of a western blot analysis showed that transfection of 0.5 µg of the CtBP1 expression construct generated cellular amounts of EGFP–CtBP1 protein similar to the levels obtained in 293-EGFP–CtBP1/Dox cells upon removal of doxycycline. Transfection of a larger quantity of the construct produced even more EGFP–CtBP1 protein than the amount of endogenous CtBPs (Fig. 5A). Nevertheless, the relatively moderate expression of the EGFP–CtBP1 transgene significantly decreased pTOPFLASH transcription in Wnt-1- stimulated cells, whereas the activities of this reporter in 293-EGFP–CtBP1/Dox cells growing in the presence of doxycycline, i.e. with repressed CtBP1, were comparable to values obtained in the parental cell line (Fig. 5B). We readily obtained almost identical amounts of TCF/β-catenin complexes in Wnt-1-stimulated 293-EGFP–CtBP1/Dox cells independently of the expression status of the CtBP1 transgene (not shown). This indicates that CtBP levels do not interfere directly with the formation of such complexes or with the upstream steps of Wnt signaling. We further determined whether the mechanism of CtBP1 repression relies on histone deacetylases. As shown in Figure 5C, treatment with trichostatin A, a histone deacetylase inhibitor, alleviated the repressive effect of CtBP1 in 293-EGFP–CtBP1/Dox cells. The trichostatin A treatment also completely neutralized the repressive effect of CtBP1 in transient transfection assays (Fig. 5D). We conclude that CtBP1 acts as a repressor of Wnt-mediated transcription via recruitment of histone deacetylases to the target promoter.
Figure 4.
CtBP1 represses TCF/β-catenin transcription. (A) CtBP1 represses TCF/β-catenin transcription in 293 cells. Human 293 embryonic kidney cells were co-transfected with the indicated amounts of CtBP1 expression plasmid and the Tcf reporter construct pTOPFLASH or the negative control reporter pFOPFLASH using the lipofectamine reagent. Four hours post-transfection, DNA–lipofectamine mixtures were removed and 293 cells were covered with Rat2-Wnt-1/Dox fibroblasts containing the Wnt-1 gene driven by the doxycycline-repressed promoter. The cultures were further grown in the presence (control) or absence of doxycycline (Wnt-1 stimulated). Following co-cultivation for 15 h, the cells were harvested, and luciferase (firefly) and Renilla luciferase activities were determined in cell lysates. (B) CtBP1 can repress transactivation mediated by TCF-4 and β-catenin in COS-7 cells. COS-7 cell line was co-transfected with the Tcf reporter constructs and a specific TCF-4 construct, β-catenin and with the indicated amount of CtBP1 plasmid. Luciferase and Renilla luciferase activities were determined in cell lysates 15 h following transfection. Whole cell extracts were analyzed by western blotting with TCF-4 monoclonal antibody (inset). All transfections were done in triplicate. Relative luciferase light units per second (RLU/s) are average values corrected for the efficiency of transfection by determining the luciferase/Renilla ratio.
Besides CtBP, 293 cells express other TCF-4-interacting proteins, the TLE/Groucho co-repressors (52). Therefore, we asked next whether both types of negative regulators utilize the same inhibitory mechanisms. We examined the impact of different TCF-4 deletions on the activity of the pTOPFLASH reporter in unstimulated and stimulated 293 cells (see Fig. 2A for details about the constructs used). TCF-4 constructs, when overexpressed in 293 cells, decreased the basal and the Wnt-1-primed activity of pTOPFLASH but, strikingly, TCF4-LEF, which interacts only with TLE/Groucho and lacks the C-terminal CtBP-binding domain, appeared to be more potent repressor than full-length TCF-4 (Fig. 5E). In addition, only repression mediated by full-length TCF-4, i.e. by a protein containing both TLE/Groucho and CtBP interaction regions, has been almost completely alleviated by the trichostatin A treatment. These results indicate that the TLE/Groucho proteins may utilize different mechanisms than do CtBPs to repress the Tcf-responsive promoter and/or that the action of these repressors might be mutually exclusive.
CtBP1 down-regulates the expression of Axin2, a Tcf/β-catenin target in 293 cells
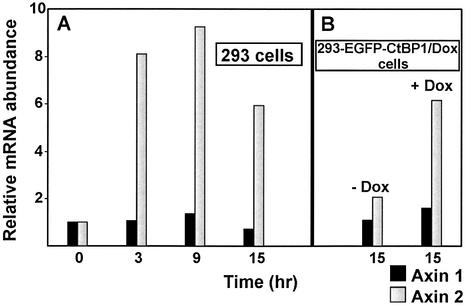
We examined the expression levels of several human genes related to Wnt signaling in Wnt-1-stimulated 293 cells. We designed sets of human-specific primers that selectively amplified cDNA produced from 293 mRNA and did not function with rat or mouse cDNAs, respectively (see Materials and Methods). This allowed us to assess the level of expression of the selected human genes even in 293/Rat2 mixed RNA samples. Upon Wnt-1 stimulation, we noticed significant changes in expression of several genes: (i) we detected a moderate up-regulation of presumptive Wnt targets Cyclin D1, Tcf-1 and Lef-1 (see Supplementary Material) and (ii) we also detected strong transcriptional activation of Axin2/Conductin. Axin2 was recently identified by DNA-array technology as a gene positively regulated by TCF/β-catenin complexes in colorectal and liver tumors and in rat RK3E epithelial cells (76,77). We tested the time course of induction of the Axin2 transcript in stimulated cells (Fig. 6A). We detected an 8-fold increase in Axin2 mRNA as early as 3 h after stimulation (a 3 h interval was a minimal time period necessary for attachment of feeder cells onto the 293 cells). The maximal quantity of Axin2 mRNA was reached after 9 h of co-cultivation. Following this time point, Axin2 levels started to decrease but, nevertheless, after 15 h still remained approximately six times higher than in control 293 cells. Expression of the Axin1 gene, an Axin2 homolog, did not change significantly at any time point tested. We further examined the influence of CtBP1 overexpression on the activation of the Axin2 gene. Axin2 mRNA in 293-EGFP–CtBP1/Dox cells stimulated by Rat2-Wnt-1/Const feeder cells and with the repressed EGFP–CTBP1 transgene was up-regulated 6-fold (compared to the same cells co-cultivated with parental Rat2 fibroblasts). However, the induction of CtBP1 in Wnt-1-activated cells significantly down-regulated Axin2 expression (Fig. 6B). These results indicate that a cellular dose of CtBPs modulates the activity of the Axin-2 promoter and, by implication, might selectively regulate the expression of other Wnt target genes.
Figure 6.
CtBP1 down-regulates the expression of Axin2 in 293 cells. (A) Wnt-1 activates Axin2 mRNA in 293 cells. The results of quantitative real-time PCR performed with cDNA generated from 293 cells stimulated by Wnt-1-producing feeder cells (Rat2-Wnt-1/Const) or by control Rat2 cells are shown. The 293 and feeder cells were co-cultivated for the indicated period of time, then harvested and random primed cDNA was prepared from total RNA. The PCR reactions were performed for each primer set in triplicate using cDNAs produced from at least two independent RNA isolates. (B) EGFP–CtBP1 decreases responsiveness of the Axin2 gene to the Wnt-1 stimulation but has no effect on the expression of the Axin1 gene. 293-EGFP–CtBP1/Dox cells were co-cultivated with feeder cells stably producing Wnt-1 protein (Rat2-Wnt-1/Const) or control Rat2 fibroblasts as a negative control. Cells were co-cultivated for 15 h in the presence (EGFP– CtBP1 repressed) or absence (EGFP–CtBP1 overexpressed) of doxycycline (1 µg/ml). The random primed cDNAs generated from the relevant RNAs were analyzed. The PCR reactions were performed for each primer set in triplicate using cDNAs produced from at least two independent RNA isolates. The results of a representative experiment are shown. The results were analyzed using the LightCycler 5.1 software package, and the values of a representative experiment are shown. The relative abundance of Axin1 and Axin2 mRNA in Wnt-1-stimulated versus control cells was derived from the average CT values of each triplicate after normalizing to the levels of SDHA cDNA.
DISCUSSION
In this report we provide evidence for an interaction between the TCF-4 protein, a member of the Tcf/Lef family of nuclear mediators of Wnt signaling, and the transcriptional co-repressor CtBP1. Using Wnt-1-stimulated human embryonic kidney 293 cells, we demonstrate that CtBP1 represses the transcriptional activity of a Tcf/β-catenin-dependent synthetic promoter and, furthermore, decreases the expression of the endogenous Wnt target Axin2. We further show that the inhibitory effect of CtBP is neutralized by the histone deacetylase inhibitor trichostatin A.
TCF/LEF proteins represent a specific type of transcription factor. They harbor β-catenin and TLE/Groucho interaction domains that determine whether, under given circumstances, the protein will act as a transcriptional activator or repressor. TCFs display essentially identical DNA-binding specificities, thus they have been considered as equivalent nuclear components of the Wnt signaling pathway. However, recent studies revealed functional differences among TCF family members. The most obvious feature of individual TCFs is their ability to associate with various cellular proteins. From that point of view, LEF-1 and TCF-3 proteins have been the most thoroughly studied Tcf/Lef family members (see Introduction). The main focus of this study is TCF-4 protein.
The limited solubility of Wnt proteins is a critical obstacle to studying the biological function of these molecules in vitro. We developed a dual cell system for Wnt pathway activation in which the target cells are stimulated by cells producing Wnt factors and growing in close proximity. This type of stimulation presumably corresponds well to the situation occurring in vivo in solid tissues and organs. At first, we generated rat fibroblast cell lines with constitutive or inducible Wnt-1 expression, and, using a Tcf/β-catenin-dependent reporter assay and a β-catenin supershift assay, we showed that these cell lines evidently activate Wnt signaling in neighboring 293 cells. β-Catenin overexpression from a transiently transfected construct resulted in a robust activation of the Tcf/β- catenin-dependent reporter, which was further augmented if the cells were simultaneously stimulated by Wnt-1-producing feeders (Fig. 3C). This indicates that β-catenin is indeed a key and also a limiting molecule in Wnt signaling. We further utilized this cell system to test the function of CtBP1 in the context of a Wnt-responsive promoter. Since 293 cells express E1A protein and CtBP1 was originally described as a protein interacting with this adenoviral oncoprotein, we used the COS-7 cell system in parallel to avoid the possibility that the observed effects are dependent on E1A expression. The repressive effect of CtBP1 depended directly on the expression levels obtained from either transiently or steadily transfected CtBP1 plasmids. This dosage-dependent repression corresponds well to the phenotype of mice harboring various combinations of CtBP1 and CtBP2 mutant alleles. CtBP1-null mice are small but viable, whereas CtBP2 mutants die by E10.5 due to aberrant extraembryonic development. Different CtBP1/CtBP2 compound mutant mice display additional dose-sensitive defects in a wide range of developmental processes (78). It is surprising that although 293 cells produce an abundance of LEF-1 (79), which is a protein that does not interact with CtBP, higher amounts of CtBP1 efficiently suppressed transcription from the Wnt-responsive reporter to basal levels (Fig. 4A). This fact might be explained by the greater ability of TCF-4 to form a ternary complex with DNA and β-catenin (80).
TCF-4 (and TCF-3) resembles two other repressors, Hairy and Brinker, which also interact with both TLE/Groucho and CtBP proteins (81,82). The precise mechanisms of how TLE/Groucho and CtBP contribute to gene repression remain to be elucidated. In Drosophila, Groucho functionally interacts with the histone deacetylase Rapd3 (51). Recruitment of Rapd3 to target promoters would result in the formation of a more compact chromatin structure followed by a transcriptionally repressed state. The recruitment of histone deacetylases is likely to be the main function of the TLE/Groucho proteins, although only the partial release of TLE/Groucho-Rapd3 repression by the histone deacetylase inhibitor trichostatin A indicates that additional mechanisms of repression may exist. CtBP also induces transcriptional silencing in a histone deacetylase-dependent or -independent manner. CtBP1 associates with class I and class II histone deacetylases, and CtBP-related down-regulation of certain promoters has been reported to be sensitive to trichostatin A (83). In contrast, repression of several other promoters was insensitive to trichostatin A treatment (75). We detected a complete release of the CtBP-mediated repression by trichostatin A. This clearly indicates the involvement of the histone deacetylases in the repression mechanism. Since 293 cells express both TLE/Groucho and CtBP proteins (52 and this manuscript), we further evaluated the contribution of the TLE/Groucho and CtBP co-repressors to the repression of the activity of the pTOPFLASH reporter. Overexpression of TCF-4–LEF deletion containing only the TLE/Groucho interaction domain significantly inhibited the basal and Wnt-1-dependent activity of the Tcf reporter. This is in agreement with the observation that the C-terminally truncated forms of XTCF-3 keep their repressive potential in Xenopus embryos, even if they lack the CtBP-interacting domains (61). Surprisingly, the putative repressive action of TLE/Groucho was only partly suppressed by trichostatin A, and a greater sensitivity to this chemical was only revealed by the full-length TCF-4-mediated repression (Fig. 5E). This implies the possibility of parallel repressive action of CtBP and TLE/Groucho, which might contribute to the regulation of different Wnt targets. Since the TLE/Groucho and CtBP-binding sites are quite far apart in TCF-4 and TCF-3 proteins, we assume that there is no steric competition for these sites as in Hairy protein (82).
We took advantage of the different origin of human 293 and rat feeder cells and used real-time quantitative RT–PCR to analyze the changes in expression of selected genes upon Wnt stimulation. The strongest up-regulation was observed for Axin2 mRNA. This finding was rather expected because Axin2 was recently identified as a gene activated by β-catenin overexpression in the related rat RKE3 cells (76). We further evaluated the impact of CtBP1 overexpression on Axin2 activation and detected a strong reduction in the levels of Axin2 mRNA. Such a robust effect was surprising because the quantities of the ectopically expressed CtBP1 protein represented approximately one-half of the amount of endogenous CtBPs. 293 cells express all four TCF/LEF proteins, although at different levels, TLE/Groucho, and both CtBP1 and CtBP2 co-repressors (V. Korinek, unpublished data). TCF-4 associates with TLE/Groucho proteins and also with CtBPs, and these proteins also form homo- or heterodimers. The strength of the interaction between CtBPs and their partners can be further potentiated by the levels of nuclear NAD(H) (84,85). We propose that the composition of various Wnt signaling pathway components and their expression levels dictate the primary sensitivity of a given cell to the Wnt signal. The balance in such a system can be disturbed by changing the levels of only one component. Likewise the defects in other negative regulators of Wnt signaling (e.g. in the tumor suppressor APC), mutations that inactivate CtBP proteins could also lead to inappropriate activation of the Wnt pathway. On the other hand, non-physiological increases in the expression levels of CtBPs might by implication result in a significant reduction in Wnt signaling output in the stimulated cell.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank F. H. Gage for the pLNIT vector, M. van Dijk for the Wnt-1 construct and S. Dimitrov for the mouse testis cDNA library. We are grateful to L. Cermak for excellent help with the real-time PCR experiments, to O. Horvath for expert computer work, to L. Andera for helpful discussion and to J. Dutt for critically reading the manuscript. We wish to express our gratitude to V. Horejsi for continuous and generous support. This work was supported from the projects Center of Molecular and Cellular Immunology (LN00A026), by the Grant Agency of the Czech Republic grant 312/99/0348 (to V.K.) and by the Grant Agency of Academy of Sciences of the Czech Republic grant A5052905 (to V.K.).
REFERENCES
- 1.Cadigan K.M. and Nusse,R. (1997) Wnt signaling: a common theme in animal development. Genes Dev., 11, 3286–3305. [DOI] [PubMed]
- 2.Bienz M. and Clevers,H. (2000) Linking colorectal cancer to Wnt signaling. Cell, 103, 311–320. [DOI] [PubMed]
- 3.Miller J.R. and Moon,R.T. (1996) Signal transduction through beta-catenin and specification of cell fate during embryogenesis. Genes Dev., 10, 2527–2539. [DOI] [PubMed]
- 4.Coates J.C., Grimson,M.J., Williams,R.S., Bergman,W., Blanton,R.L. and Harwood,A.J. (2002) Loss of the beta-catenin homologue aardvark causes ectopic stalk formation in Dictyostelium. Mech. Dev., 116, 117–127. [DOI] [PubMed]
- 5.Hobmayer B., Rentzsch,F., Kuhn,K., Happel,C.M., von Laue,C.C., Snyder,P., Rothbacher,U. and Holstein,T.W. (2000) WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature, 407, 186–189. [DOI] [PubMed]
- 6.Grimson M.J., Coates,J.C., Reynolds,J.P., Shipman,M., Blanton,R.L. and Harwood,A.J. (2000) Adherens junctions and beta-catenin-mediated cell signalling in a non-metazoan organism. Nature, 408, 727–731. [DOI] [PubMed]
- 7.Pinson K.I., Brennan,J., Monkley,S., Avery,B.J. and Skarnes,W.C. (2000) An LDL-receptor-related protein mediates Wnt signalling in mice. Nature, 407, 535–538. [DOI] [PubMed]
- 8.Tamai K., Semenov,M., Kato,Y., Spokony,R., Liu,C., Katsuyama,Y., Hess,F., Saint-Jeannet,J.P. and He,X. (2000) LDL-receptor-related proteins in Wnt signal transduction. Nature, 407, 530–535. [DOI] [PubMed]
- 9.Wehrli M., Dougan,S.T., Caldwell,K., O’Keefe,L., Schwartz,S., Vaizel-Ohayon,D., Schejter,E., Tomlinson,A. and DiNardo,S. (2000) arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature, 407, 527–530. [DOI] [PubMed]
- 10.Aberle H., Bauer,A., Stappert,J., Kispert,A. and Kemler,R. (1997) beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J., 16, 3797–3804. [DOI] [PMC free article] [PubMed]
- 11.Jiang J. and Struhl,G. (1998) Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature, 391, 493–496. [DOI] [PubMed]
- 12.Marikawa Y. and Elinson,R.P. (1998) beta-TrCP is a negative regulator of Wnt/beta-catenin signaling pathway and dorsal axis formation in Xenopus embryos. Mech. Dev., 77, 75–80. [DOI] [PubMed]
- 13.Hart M., Concordet,J.P., Lassot,I., Albert,I., del los Santos,R., Durand,H., Perret,C., Rubinfeld,B., Margottin,F., Benarous,R. et al. (1999) The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr. Biol., 9, 207–210. [DOI] [PubMed]
- 14.Hart M.J., de los Santos,R., Albert,I.N., Rubinfeld,B. and Polakis,P. (1998) Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr. Biol., 8, 573–581. [DOI] [PubMed]
- 15.Fagotto F., Jho,E., Zeng,L., Kurth,T., Joos,T., Kaufmann,C. and Costantini,F. (1999) Domains of axin involved in protein–protein interactions, Wnt pathway inhibition, and intracellular localization. J. Cell Biol., 145, 741–756. [DOI] [PMC free article] [PubMed]
- 16.Liu C., Li,Y., Semenov,M., Han,C., Baeg,G.H., Tan,Y., Zhang,Z., Lin,X. and He,X. (2002) Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell, 108, 837–847. [DOI] [PubMed]
- 17.Yanagawa S., Matsuda,Y., Lee,J.S., Matsubayashi,H., Sese,S., Kadowaki,T. and Ishimoto,A. (2002) Casein kinase I phosphorylates the Armadillo protein and induces its degradation in Drosophila. EMBO J., 21, 1733–1742. [DOI] [PMC free article] [PubMed]
- 18.Behrens J., Jerchow,B.A., Wurtele,M., Grimm,J., Asbrand,C., Wirtz,R., Kuhl,M., Wedlich,D. and Birchmeier,W. (1998) Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science, 280, 596–599. [DOI] [PubMed]
- 19.Kishida M., Koyama,S., Kishida,S., Matsubara,K., Nakashima,S., Higano,K., Takada,R., Takada,S. and Kikuchi,A. (1999) Axin prevents Wnt-3a-induced accumulation of beta-catenin. Oncogene, 18, 979–985. [DOI] [PubMed]
- 20.Zeng L., Fagotto,F., Zhang,T., Hsu,W., Vasicek,T.J., Perry,W.L.,3rd, Lee,J.J., Tilghman,S.M., Gumbiner,B.M. and Costantini,F. (1997) The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell, 90, 181–192. [DOI] [PubMed]
- 21.Boutros M., Mihaly,J., Bouwmeester,T. and Mlodzik,M. (2000) Signaling specificity by Frizzled receptors in Drosophila. Science, 288, 1825–1828. [DOI] [PubMed]
- 22.Axelrod J.D., Miller,J.R., Shulman,J.M., Moon,R.T. and Perrimon,N. (1998) Differential recruitment of Dishevelled provides signaling specificity in the planar cell polarity and Wingless signaling pathways. Genes Dev., 12, 2610–2622. [DOI] [PMC free article] [PubMed]
- 23.Huber O., Korn,R., McLaughlin,J., Ohsugi,M., Herrmann,B.G. and Kemler,R. (1996) Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech. Dev., 59, 3–10. [DOI] [PubMed]
- 24.Behrens J., von Kries,J.P., Kuhl,M., Bruhn,L., Wedlich,D., Grosschedl,R. and Birchmeier,W. (1996) Functional interaction of beta-catenin with the transcription factor LEF-1. Nature, 382, 638–642. [DOI] [PubMed]
- 25.Molenaar M., Roose,J., Peterson,J., Venanzi,S., Clevers,H. and Destree,O. (1998) Differential expression of the HMG box transcription factors XTcf-3 and XLef-1 during early Xenopus development. Mech. Dev., 75, 151–154. [DOI] [PubMed]
- 26.Morin P.J., Sparks,A.B., Korinek,V., Barker,N., Clevers,H., Vogelstein,B. and Kinzler,K.W. (1997) Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science, 275, 1787–1790. [DOI] [PubMed]
- 27.Korinek V., Barker,N., Morin,P.J., van Wichen,D., de Weger,R., Kinzler,K.W., Vogelstein,B. and Clevers,H. (1997) Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC–/– colon carcinoma. Science, 275, 1784–1787. [DOI] [PubMed]
- 28.He T.C., Sparks,A.B., Rago,C., Hermeking,H., Zawel,L., da Costa,L.T., Morin,P.J., Vogelstein,B. and Kinzler,K.W. (1998) Identification of c-MYC as a target of the APC pathway. Science, 281, 1509–1512. [DOI] [PubMed]
- 29.Kioussi C., Briata,P., Baek,S.H., Rose,D.W., Hamblet,N.S., Herman,T., Ohgi,K.A., Lin,C., Gleiberman,A., Wang,J. et al. (2002) Identification of a Wnt/Dvl/beta-catenin → Pitx2 pathway mediating cell-type-specific proliferation during development. Cell, 111, 673–685. [DOI] [PubMed]
- 30.Shtutman M., Zhurinsky,J., Simcha,I., Albanese,C., D’Amico,M., Pestell,R. and Ben-Ze’ev,A. (1999) The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl Acad. Sci. USA, 96, 5522–5527. [DOI] [PMC free article] [PubMed]
- 31.Tetsu O. and McCormick,F. (1999) Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature, 398, 422–426. [DOI] [PubMed]
- 32.Fedi P., Bafico,A., Nieto Soria,A., Burgess,W.H., Miki,T., Bottaro,D.P., Kraus,M.H. and Aaronson,S.A. (1999) Isolation and biochemical characterization of the human Dkk-1 homologue, a novel inhibitor of mammalian Wnt signaling. J. Biol. Chem., 274, 19465–19472. [DOI] [PubMed]
- 33.Glinka A., Wu,W., Delius,H., Monaghan,A.P., Blumenstock,C. and Niehrs,C. (1998) Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature, 391, 357–362. [DOI] [PubMed]
- 34.Hsieh J.C., Kodjabachian,L., Rebbert,M.L., Rattner,A., Smallwood,P.M., Samos,C.H., Nusse,R., Dawid,I.B. and Nathans,J. (1999) A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature, 398, 431–436. [DOI] [PubMed]
- 35.Rattner A., Hsieh,J.C., Smallwood,P.M., Gilbert,D.J., Copeland,N.G., Jenkins,N.A. and Nathans,J. (1997) A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc. Natl Acad. Sci. USA, 94, 2859–2863. [DOI] [PMC free article] [PubMed]
- 36.Leyns L., Bouwmeester,T., Kim,S.H., Piccolo,S. and De Robertis,E.M. (1997) Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell, 88, 747–756. [DOI] [PMC free article] [PubMed]
- 37.Melkonyan H.S., Chang,W.C., Shapiro,J.P., Mahadevappa,M., Fitzpatrick,P.A., Kiefer,M.C., Tomei,L.D. and Umansky,S.R. (1997) SARPs: a family of secreted apoptosis-related proteins. Proc. Natl Acad. Sci. USA, 94, 13636–13641. [DOI] [PMC free article] [PubMed]
- 38.Rocheleau C.E., Yasuda,J., Shin,T.H., Lin,R., Sawa,H., Okano,H., Priess,J.R., Davis,R.J. and Mello,C.C. (1999) WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C.elegans. Cell, 97, 717–726. [DOI] [PubMed]
- 39.Ishitani T., Ninomiya-Tsuji,J., Nagai,S., Nishita,M., Meneghini,M., Barker,N., Waterman,M., Bowerman,B., Clevers,H., Shibuya,H. et al. (1999) The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature, 399, 798–802. [DOI] [PubMed]
- 40.Meneghini M.D., Ishitani,T., Carter,J.C., Hisamoto,N., Ninomiya-Tsuji,J., Thorpe,C.J., Hamill,D.R., Matsumoto,K. and Bowerman,B. (1999) MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature, 399, 793–797. [DOI] [PubMed]
- 41.Bauer A., Chauvet,S., Huber,O., Usseglio,F., Rothbacher,U., Aragnol,D., Kemler,R. and Pradel,J. (2000) Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J., 19, 6121–6130. [DOI] [PMC free article] [PubMed]
- 42.Barker N., Hurlstone,A., Musisi,H., Miles,A., Bienz,M. and Clevers,H. (2001) The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J., 20, 4935–4943. [DOI] [PMC free article] [PubMed]
- 43.Hecht A., Vleminckx,K., Stemmler,M.P., van Roy,F. and Kemler,R. (2000) The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J., 19, 1839–1850. [DOI] [PMC free article] [PubMed]
- 44.Kramps T., Peter,O., Brunner,E., Nellen,D., Froesch,B., Chatterjee,S., Murone,M., Zullig,S. and Basler,K. (2002) Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell, 109, 47–60. [DOI] [PubMed]
- 45.Parker D.S., Jemison,J. and Cadigan,K.M. (2002) Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development, 129, 2565–2576. [DOI] [PubMed]
- 46.Zorn A.M., Barish,G.D., Williams,B.O., Lavender,P., Klymkowsky,M.W. and Varmus,H.E. (1999) Regulation of Wnt signaling by Sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol. Cell, 4, 487–498. [DOI] [PubMed]
- 47.Tago K., Nakamura,T., Nishita,M., Hyodo,J., Nagai,S., Murata,Y., Adachi,S., Ohwada,S., Morishita,Y., Shibuya,H. et al. (2000) Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev., 14, 1741–1749. [PMC free article] [PubMed]
- 48.Clevers H. and van de Wetering,M. (1997) TCF/LEF factor earn their wings. Trends Genet., 13, 485–489. [DOI] [PubMed]
- 49.Eastman Q. and Grosschedl,R. (1999) Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr. Opin. Cell Biol., 11, 233–240. [DOI] [PubMed]
- 50.Roose J., Molenaar,M., Peterson,J., Hurenkamp,J., Brantjes,H., Moerer,P., van de Wetering,M., Destree,O. and Clevers,H. (1998) The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature, 395, 608–612. [DOI] [PubMed]
- 51.Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999) A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed]
- 52.Brantjes H., Roose,J., van De Wetering,M. and Clevers,H. (2001) All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res., 29, 1410–1419. [DOI] [PMC free article] [PubMed]
- 53.van Genderen C., Okamura,R.M., Farinas,I., Quo,R.G., Parslow,T.G., Bruhn,L. and Grosschedl,R. (1994) Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev., 8, 2691–2703. [DOI] [PubMed]
- 54.Verbeek S., Izon,D., Hofhuis,F., Robanus-Maandag,E., te Riele,H., van de Wetering,M., Oosterwegel,M., Wilson,A., MacDonald,H.R. and Clevers,H. (1995) An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature, 374, 70–74. [DOI] [PubMed]
- 55.Korinek V., Barker,N., Moerer,P., van Donselaar,E., Huls,G., Peters,P.J. and Clevers,H. (1998) Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nature Genet., 19, 379–383. [DOI] [PubMed]
- 56.Yasumoto K., Takeda,K., Saito,H., Watanabe,K., Takahashi,K. and Shibahara,S. (2002) Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling. EMBO J., 21, 2703–2714. [DOI] [PMC free article] [PubMed]
- 57.Bruhn L., Munnerlyn,A. and Grosschedl,R. (1997) ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev., 11, 640–653. [DOI] [PubMed]
- 58.Sachdev S., Bruhn,L., Sieber,H., Pichler,A., Melchior,F. and Grosschedl,R. (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev., 15, 3088–3103. [DOI] [PMC free article] [PubMed]
- 59.Billin A.N., Thirlwell,H. and Ayer,D.E. (2000) Beta-catenin-histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol. Cell. Biol., 20, 6882–6890. [DOI] [PMC free article] [PubMed]
- 60.Lee E., Salic,A. and Kirschner,M.W. (2001) Physiological regulation of [beta]-catenin stability by Tcf3 and CK1epsilon. J. Cell Biol., 154, 983–993. [DOI] [PMC free article] [PubMed]
- 61.Brannon M., Brown,J.D., Bates,R., Kimelman,D. and Moon,R.T. (1999) XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development, 126, 3159–3170. [DOI] [PubMed]
- 62.Hecht A. and Stemmler,M.P. (2002) Identification of a promoter-specific transcriptional activation domain at the C-terminus of the Wnt-effector protein TCF4. J. Biol. Chem., 22, 22. [DOI] [PubMed]
- 63.Hovanes K., Li,T.W., Munguia,J.E., Truong,T., Milovanovic,T., Lawrence Marsh,J., Holcombe,R.F. and Waterman,M.L. (2001) Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nature Genet., 28, 53–57. [DOI] [PubMed]
- 64.Roose J., Huls,G., van Beest,M., Moerer,P., van der Horn,K., Goldschmeding,R., Logtenberg,T. and Clevers,H. (1999) Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science, 285, 1923–1926. [DOI] [PubMed]
- 65.Gradl D., Konig,A. and Wedlich,D. (2002) Functional diversity of Xenopus lymphoid enhancer factor/T-cell factor transcription factors relies on combinations of activating and repressing elements. J. Biol. Chem., 277, 14159–14171. [DOI] [PubMed]
- 66.Hoshimaru M., Ray,J., Sah,D.W. and Gage,F.H. (1996) Differentiation of the immortalized adult neuronal progenitor cell line HC2S2 into neurons by regulatable suppression of the v-myc oncogene. Proc. Natl Acad. Sci. USA, 93, 1518–1523. [DOI] [PMC free article] [PubMed]
- 67.Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed]
- 68.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed]
- 69.Vandesompele J., De Preter,K., Pattyn,F., Poppe,B., Van Roy,N., De Paepe,A. and Speleman,F. (2002) Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol., 3, RESEARCH0034. [DOI] [PMC free article] [PubMed]
- 70.Boyd J.M., Subramanian,T., Schaeper,U., La Regina,M., Bayley,S. and Chinnadurai,G. (1993) A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J., 12, 469–478. [DOI] [PMC free article] [PubMed]
- 71.Katsanis N. and Fisher,E.M. (1998) A novel C-terminal binding protein (CTBP2) is closely related to CTBP1, an adenovirus E1A-binding protein, and maps to human chromosome 21q21.3. Genomics, 47, 294–299. [DOI] [PubMed]
- 72.Chinnadurai G. (2002) CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell, 9, 213–224. [DOI] [PubMed]
- 73.Korinek V., Barker,N., Willert,K., Molenaar,M., Roose,J., Wagenaar,G., Markman,M., Lamers,W., Destree,O. and Clevers,H. (1998) Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol. Cell. Biol., 18, 1248–1256. [DOI] [PMC free article] [PubMed]
- 74.Van de Wetering M., Castrop,J., Korinek,V. and Clevers,H. (1996) Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol. Cell. Biol., 16, 745–752. [DOI] [PMC free article] [PubMed]
- 75.Koipally J. and Georgopoulos,K. (2000) Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem., 275, 19594–19602. [DOI] [PubMed]
- 76.Leung J.Y., Kolligs,F.T., Wu,R., Zhai,Y., Kuick,R., Hanash,S., Cho,K.R. and Fearon,E.R. (2002) Activation of AXIN2 expression by beta-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J. Biol. Chem., 277, 21657–21665. [DOI] [PubMed]
- 77.Lustig B., Jerchow,B., Sachs,M., Weiler,S., Pietsch,T., Karsten,U., van de Wetering,M., Clevers,H., Schlag,P.M., Birchmeier,W. et al. (2002) Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol., 22, 1184–1193. [DOI] [PMC free article] [PubMed]
- 78.Hildebrand J.D. and Soriano,P. (2002) Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol., 22, 5296–5307. [DOI] [PMC free article] [PubMed]
- 79.Porfiri E., Rubinfeld,B., Albert,I., Hovanes,K., Waterman,M. and Polakis,P. (1997) Induction of a beta-catenin-LEF-1 complex by wnt-1 and transforming mutants of beta-catenin. Oncogene, 15, 2833–2839. [DOI] [PubMed]
- 80.Pukrop T., Gradl,D., Henningfeld,K.A., Knochel,W., Wedlich,D. and Kuhl,M. (2001) Identification of two regulatory elements within the high mobility group box transcription factor XTCF-4. J. Biol. Chem., 276, 8968–8978. [DOI] [PubMed]
- 81.Zhang H., Levine,M. and Ashe,H.L. (2001) Brinker is a sequence-specific transcriptional repressor in the Drosophila embryo. Genes Dev., 15, 261–266. [DOI] [PMC free article] [PubMed]
- 82.Poortinga G., Watanabe,M. and Parkhurst,S.M. (1998) Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J., 17, 2067–2078. [DOI] [PMC free article] [PubMed]
- 83.Sundqvist A., Sollerbrant,K. and Svensson,C. (1998) The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein-histone deacetylase complex. FEBS Lett., 429, 183–188. [DOI] [PubMed]
- 84.Kumar V., Carlson,J.E., Ohgi,K.A., Edwards,T.A., Rose,D.W., Escalante,C.R., Rosenfeld,M.G. and Aggarwal,A.K. (2002) Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol. Cell, 10, 857–869. [DOI] [PubMed]
- 85.Zhang Q., Piston,D.W. and Goodman,R.H. (2002) Regulation of corepressor function by nuclear NADH. Science, 295, 1895–1897. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.