Abstract
Polyhydroxybutyrate (PHB) is a member of a class of thermoelastic polymers called polyhydroxyalkanoates that serve many bacteria as intracellular storage molecules for carbon and energy. Transgenic plants provide a potential means of producing this polymer cost-effectively. To date, however, few reports of the successful production of this polymer have been published, with the exception of work with transgenic Arabidopsis. Using a variety of chimeric constructs, we have determined that the constitutive, chloroplast-localized expression of one of the genes involved in PHB production—the β-ketothiolase (phbA) gene—is detrimental to the efficient production of transgenic PHB. The alternate use of either inducible or somatically activated promoters allowed the construction of transgenic PHB-producing potato (Solanum tuberosum) and tobacco (Nicotiana tabacum) plants, although the amount of PHB formed was still rather low. Taking advantage of an inducible promoter, the maximal amount of PHB produced in transgenic potato was 0.09 mg g−1 dry weight. In transgenic tobacco using a somatically activated promoter, up to 3.2 mg g−1 dry weight was accumulated. In Arabidopsis, the formation of high levels of PHB had previously been shown to be accompanied by severe negative effects on growth and development of the plant. Phasins are proteins known from PHB-producing bacteria speculated to serve as protectants against the highly hydrophobic surface of the PHB granules in the bacterial intracellular milieu. Co-expression of the phasin gene in parallel with the PHB synthesis genes, however, did not lead to reduced symptom development.
The use of transgenic plants for the production of novel compounds represents one important aspect of plant biotechnology. Whereas multiple examples exist with respect to the production of novel compounds to be used in food and feed, much less work has been done with respect to the use of transgenic plants for production of compounds for non-food purposes. Polyhydroxyalkanoates (PHAs) are polymers that serve many bacteria as intracellular storage molecules for carbon and energy. These molecules occur alone or together with triacylglycerols (Alvarez et al., 2000). The homopolymer poly(3-hydroxybutyrate) (PHB) is the most common type of PHA, although many other PHAs are known (Steinbüchel and Valentin, 1995). Starting from acetyl-CoA, PHB is synthesized in bacteria by the consecutive action of three genes: β-ketothiolase (phbA), acetoacetyl CoA reductase (phbB), and PHB synthase (phbC). Thermoelastic polymers such as PHB have a number of advantages over polymers derived from oil: They are biodegradable, their production is essentially neutral with respect to carbon dioxide balance, and they are biologically renewable and are, therefore, in line with the goal of sustainable development (Steinbüchel and Füchtenbusch, 1998).
The use of recombinant micro-organisms to efficiently produce PHB and PHAs has been described extensively. The main disadvantage of this production route is its high cost (Choi and Lee, 1999). Plants obviously represent an attractive alternative for PHB production. Pioneering work performed by Poirier et al. (1992) in Arabidopsis showed that PHB synthesis is feasible in plants. A dramatic increase in the amount of PHB produced by Arabidopsis plants was achieved when PHB synthesis was engineered in chloroplasts (Nawrath et al., 1994). However, Arabidopsis obviously will not be the crop used for PHB production. Surprisingly, few reports of PHB production in crop plants have appeared, and those that have reported only very low amounts of PHB being produced (compare with e.g. John and Keller, 1996; Nakashita et al., 1999). The only exception is the work resulting in transgenic Brassica napus plants with seeds expressing up to 7% (w/v) PHB by weight (Valentin et al., 1999; Houmiel et al., 1999).
We recently reported the efficient production of PHB in Arabidopsis leaf chloroplasts where leaf levels up to 40% of dry weight were reached, thus, confirming and extending earlier observations by Nawrath et al. (1994). These high amounts were, however, accompanied by severe growth reduction of the plant and the development of chlorotic leaves.
We, therefore, started a series of experiments with essentially two goals: analysis of plants other than Arabidopsis (notably potato [Solanum tuberosum] and tobacco [Nicotiana tabacum]) for their suitability as hosts for efficient PHB production; and analysis of strategies to prevent the negative effects of PHB synthesis on growth and development.
Evidence will be described suggesting that the constitutive expression of one of the PHB synthesis genes (phbA) is detrimental to plant growth as early as during the transformation step. When put either under an inducible or a somatically activated promoter, transgenic tobacco and potato plants were obtained that expressed all three PHB genes. However, the amount of PHB formed was nevertheless low, suggesting that, in this case, what is true for Arabidopsis is not necessarily true for potato and tobacco. Results obtained following two strategies trying to circumvent the negative effects of PHB accumulation on growth and development in Arabidopsis will also be described.
RESULTS
Transformation of Tobacco and Potato with Chimeric Gene Constructs Containing the Three PHB Genes under a Constitutive Promoter Leads to a Low Number of Transgenic Plants, Only a Few of Which Produce Minor Amounts of PHB
The construct pBI ABC, which contains the three PHB-synthesis genes fused to a plastid-targeting signal under the control of the constitutive cauliflower mosaic virus 35S (CaMV 35S) promoter, was used for Agrobacterium tumefaciens-mediated transformation experiments using leaf discs of tobacco and potato. In tobacco, a much reduced transformation frequency was observed yielding only 47 lines in total from several transformation batches, which represents about one-tenth of the number of lines expected when compared with other transformations. In potato, the situation was even worse. Of five transformation experiments, which normally would have yielded several hundred transformants, no transgenic lines were obtained at all. The transgenic tobacco lines grown in tissue culture were directly screened for their 3-hydroxybutyrate content via gas chromatography-mass spectroscopy (GC-MS) analysis. PHB accumulation was observed in only three lines, the maximal amount being 91 μg 3-hydroxybutyrate g−1 fresh weight (Table I). After transfer to the greenhouse, this already low level fell even further (< 5 μg 3-hydroxybutyrate g−1 fresh weight). These results were most surprising given the fact that the identical construct resulted in transgenic Arabidopsis plants containing up to 40% of their dry weight as PHB (Bohmert et al., 2000).
Table I.
PHB accumulation in mature leaves of transgenic lines of different plant species
| Species | Name of Transgenic Line | No. of Lines Screened for PHB Production | Maximal Amount of 3HB | |
|---|---|---|---|---|
| mg g−1 fresh wt | mg g−1 dry wt | |||
| Tobacco | ABC | 47 | 0.09a | 0.8 |
| prpABC | 66 | 0.05 | 0.4 | |
| acABC | 80 | 0.38 | 3.2 | |
| Potato | ABC | No transformants | – | – |
| prpABC | 24 | 0.01 | 0.09 | |
| acABC | 13 | 0.002 | 0.02 | |
| Arabidopsis | ABC | 86 | 42b | |
| prpABC | 151 | 11.6 | 132 | |
| acABC | 80 | 1.4 | 17 | |
| ABC P | 166 | 4.9 | 58 | |
The amount of 3HB per gram fresh weight was measured in mature leaves by GC-MS analysis as described in “Materials and Methods”; the dry weight values were calculated from observed fresh weight to dry weight ratios. Samples from all prpABC lines were taken after induction with salicylic acid.
Value was detected from plants grown in tissue culture and was not reproducible under greenhouse conditions.
Expressing Chimeric Genes Bearing the β-Ketothiolase Gene (phbA) under the Control of a Constitutive Promoter Leads to a Reduced Transformation Efficiency
One possible explanation for the low efficiency of transformation described above is that one of the three chimeric genes directing PHB synthesis is specifically detrimental for plant growth and development during the transformation procedure. To investigate this hypothesis and possibly to identify the gene responsible for this effect, we performed a series of transformation experiments where, in each case, only one of the three genes (constructs pBI-TP-Thio, pBI-TP-Red, and pBI-TP-Syn; Nawrath et al., 1994) was used. Whereas transformation of either tobacco or potato using the construct pBI-TP-Syn led to normal transformation efficiencies and the use of the construct encoding the acetoacetyl-CoA reductase led to only a slightly decreased transformation efficiency, the use of the construct pBI-TP-Thio encoding the β-ketothiolase led to a drastically reduced transformation efficiency in case of both tobacco and potato (Fig. 1).
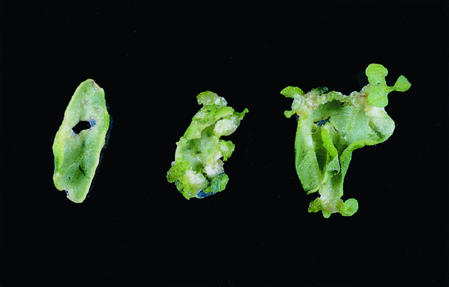
Figure 1.
Callus initiation in tobacco leaves 4 weeks after A. tumefaciens-mediated transformation using the constructs pBI-TP-Thio (left), pBI-TP-Red (center), and pBI-TP-Syn (right), which lead to the expression of the genes phbA (β-ketothiolase), phbB (acetoacetyl-CoA reductase), and phbC (PHB synthase), respectively. Callus development and number of regenerated plants increases from left to right.
To determine whether this negative influence of the acetoacetyl-CoA reductase gene on transformation frequency was also true in Arabidopsis, we analyzed the efficiency of transformation using all three genes singly and in various combinations. The single constructs pBI-TP-Red and pBI-TP-Syn and a chimeric construct containing both the PHB synthase and the acetoacetyl-CoA reductase gene led to normal transformation efficiencies (between 1% and 3% of the seeds being transformed). In strong contrast to this, the rate dropped down to 0.01% whenever the β-ketothiolase gene was contained in the construct either alone (pBI-TP-Thio) or in combination with the acetoacetyl-CoA reductase gene or the acetoacetyl-CoA reductase plus the PHB synthase gene (data not shown).
These experiments, thus, strongly suggest that in all three plant species analyzed, i.e. tobacco, potato, and Arabidopsis, the expression of the β-ketothiolase gene has detrimental effects on transformation efficiency, thus, preventing the analysis of these plant species with respect to their suitability to serve as hosts for PHB production.
Construction of Chimeric Genes Allowing the Inducible or Somatically Activated Expression of the Thiolase Gene
The data described above suggest the expression of the phbA gene during the transformation procedure is responsible for the reduced transformation efficiency. We, therefore, followed two strategies to alter the expression of phbA. In the first, we used the prp-1 promoter from potato. The prp-1 promoter was originally identified as a pathogen-inducible promoter (Martini et al., 1993), but it can also be induced via external application of salicylic acid. Inducing the expression of phbA after transgenic plant regeneration should prevent any potentially deleterious effects of phbA expression during the transformation and regeneration. In the second approach, we took advantage of the maize transposable element Ac. The Ac element was inserted between the CaMV 35S promoter and the coding region of the β-ketothiolase gene, thus, allowing expression of the β-ketothiolase gene only after the mobile element has been excised due to the establishment of the link between the promoter and the coding sequence. Frequent somatic excision of Ac has been reported in tobacco, potato, and Arabidopsis (Van Sluys et al., 1987; Knapp et al., 1988; Taylor et al., 1989; Keller et al., 1993; Lawson et al., 1994).
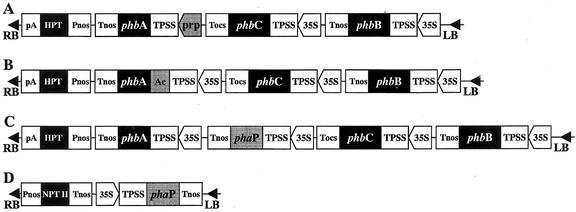
Both constructs (pBinARHygprpABC for inducible expression of phbA and pBinARHygacABC bearing the transposable element; see Fig. 2) also contain the acetoacetyl-CoA reductase gene (phbB) and the PHB synthase gene (phbC), both of which are constitutively expressed under the control of the CaMV 35S promoter. All proteins are targeted to the plastid.
Figure 2.
T-DNA of the binary vectors used for plant transformation. A, pBinARHygprpABC; B, pBinARHygAcABC; C, pBinARHygABC P; and D, pBI P. LB and RB, Left and right T-DNA border, respectively; Pnos and Tnos, promoter region and terminator region of nopalin synthase gene of A. tumefaciens, respectively; Tocs, terminator region of octopin synthase gene of A. tumefaciens; pA, polyadenylation signal of gene 7 of A. tumefaciens; HPT and NPT II, hygromycin phosphotransferase gene and neomycin phosphotransferase gene; 35S, CaMV 35S promoter; prp, prp-1 promoter of pathogen-related protein of potato (Martini et al., 1993); Ac, transposable Ac-element of maize (Zea mays); TPSS, transit peptide of the small subunit of ribulose-bisphosphate carboxylase of pea (Pisum sativum); phbA, -B, and -C, β-ketothiolase, acetoacetyl-CoA reductase, and PHB synthase gene of Ralstonia eutropha, respectively; and phaP, phasin gene of R. eutropha. The backbone of pBI P is pBI 121 (CLONTECH), the backbone for the other three vectors is pBinARHyg (Becker, 1990).
Alternative Constructs Lead to a Significant Increase in Transformation Efficiency for Both Tobacco and Potato
A. tumefaciens-mediated transformation of tobacco with the construct pBinARHygprpABC led to 66 transgenic tobacco prpABC lines. All lines were grown in the greenhouse and sprayed with 5 mm salicylic acid once a day for a period of 15 weeks. Leaf samples were taken from the edge of the top one-third of mature leaves, which is the oldest part of the leaf (analysis of the distribution of PHB throughout the whole leaf revealed that this part accumulates the highest amount of PHB; data not shown). Samples were taken 10 d before the beginning of the induction with salicylic acid and 4, 22, 44, and 88 d after the 1st d of induction. PHB accumulation was shown to take place in 23 of the 66 transgenic tobacco prpABC lines screened, the maximal amount of PHB observed being 50 μg 3-hydroxybutyrate g−1 fresh weight (corresponding to 420 μg 3-hydroxybutyrate g−1 dry weight [Table I]), detected 32 d after onset of induction. Further analysis showed that the promoter was leaky in some of the transgenics. No significant changes in phenotype were observed.
Using the construct pBinARHygacABC, 80 transgenic tobacco acABC lines were obtained. PHB accumulation was detected in 21 of the 80 plants analyzed with a maximal production of 375 μg 3-hydroxybutyrate g−1 fresh weight, corresponding to 3.2 mg 3-hydroxybutyrate g−1 dry weight (Table I). Some of the lines displayed phenotypic changes such as chlorosis, changes in leaf morphology or increased number of trichomes. These changes could not be correlated, however, with the amounts of PHB produced.
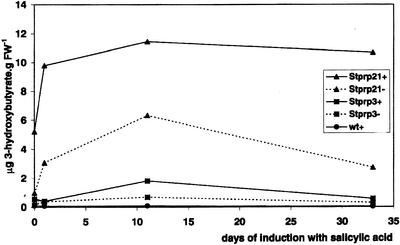
Both constructs were also used for A. tumefaciens-mediated transformation of potato. Transformation using the construct pBinARHygprpABC led to 24 transgenic potato prpABC lines. Ten transgenic potato prpABC lines accumulated some PHB, the maximal level after induction being 11 μg 3-hydroxybutyrate g−1 fresh weight (corresponding to 87 μg 3-hydroxybutyrate g−1 dry weight; Table I). Analysis of negative controls, which were not sprayed, showed that the promoter was leaky in potato, too (Fig. 3).
Figure 3.
PHB accumulation in transgenic potato prpABC lines. The amount of 3-HB per gram fresh weight was measured in mature leaves by GC-MS analysis as described in “Material and Methods.” Stprp21+, Stprp3+, and wt+ refer to lines sprayed with 1 mm salicylic acid once a day. Stprp21− and Stprp3− are nontreated controls.
Using the construct pBinARHygacABC for potato transformation, 13 transformants were generated, of which two lines produced some PHB (maximal 2 μg 3-hydroxybutyrate g−1 fresh weight; Table I). In addition to leaf extracts, tubers of all transgenic potato lines were also screened for PHB accumulation. They contained detectable but very low amounts of PHB (< 1 μg 3-hydroxybutyrate g−1 fresh weight). Neither the prpABC transgenics nor the acABC lines displayed outward phenotypic changes.
Creation and PHB Analysis of Transgenic acABC and prpABC Lines in Arabidopsis
Both constructs, pBinARHygprpABC and pBinARHygacABC, were also used to transform Arabidopsis. This was done to check the functionality of the constructs in a known system on one hand, and on the other hand, to see whether the use of these different expression systems may allow the negative effects on growth observed earlier to be overcome (Bohmert et al., 2000).
In the pBinARHygprpABC construct, 151 transgenic prpABC lines were obtained. After 3 weeks, the Arabidopsis plants had developed a rosette and started to bolt. Leaf samples were taken from all plants before onset of induction with salicylic acid and 18 d after induction.
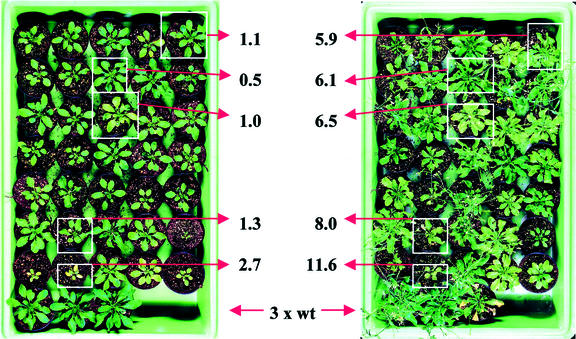
Most of the transgenic Arabidopsis prpABC lines produced PHB before the first treatment with salicylic acid. The plant that accumulated the highest amount of PHB before spraying (2.7 mg 3-hydroxybutyrate g−1 fresh weight) showed severe growth reduction (Fig. 4). The same plant produced the maximal amount of PHB measured among all Arabidopsis prpABC lines after 18 d of spraying (11 mg 3-hydroxybutyrate g−1 fresh weight; Table I). Other plants accumulated up to 6 mg 3-hydroxybutyrate g−1 fresh weight after the induction period and displayed no negative effects on their growth.
Figure 4.
PHB accumulation in transgenic Arabidopsis prpABC lines. The figure shows the same set of plants before induction with salicylic acid (left side) and after 18 d of daily spraying with 1 mm salicylic acid (right side). The three plants at the bottom are wild type. The given values for the accumulation of 3-hydroxybutyrate were determined by GC-MS. They are presented as milligrams per gram fresh weight.
Transformation of Arabidopsis with the construct pBinARHygacABC generated 80 transgenic acABC lines. The maximal amount of PHB observed was 1.4 mg 3-hydroxybutyrate g−1 fresh weight (Table I).
Co-Expression of the Phasin Gene in PHB-Producing Arabidopsis Plants Does Not Alleviate the Negative Effects on Growth and Development
Phasins are proteins expressed in PHA-producing bacteria that are associated with the surface of the PHA granules. Bacteria devoid of phasins produce less PHB and, furthermore, display a slightly reduced growth rate. Phasin production has, therefore, been suggested to be a protective mechanism against the highly hydrophobic PHB granule (Steinbüchel et al., 1995). To test whether phasins may exert such a protective effect in plants, we decided to express phasins in transgenic Arabidopsis plants in the presence or absence of PHB synthesis genes.
To test the effects of phasin proteins alone, Arabidopsis was transformed with the construct pBI P encoding a phasin protein fused to a chloroplast transit peptide and driven by the constitutive CaMV 35S promoter (Fig. 2). Seventy-two transgenic lines were analyzed, none of which showed any alteration in outward phenotype. RNA and protein-blot analysis substantiated the expression of the phasin gene (data not shown).
In a next step, the construct pBinARHygABC P, containing the three genes needed for PHB synthesis and the phasin gene (all under the control of the CaMV 35S promoter and all with a chloroplast-targeting sequence), was used (Fig. 2). In total, 166 transgenic Arabidopsis lines were generated and analyzed for PHB production via GC-MS analysis. The maximal amount of PHB measured in leaf samples was 4.9 mg 3-hydroxybutyrate g−1 fresh weight (Table I). High PHB-producing lines showed reduced growth and a chlorotic pattern in their leaves. The highest amount of PHB produced in transgenic lines without phenotypic changes was 1.5 mg 3-hydroxybutyrate g−1 fresh weight.
Transmission electron microscopy of leaf samples of PHB-producing ABC P lines revealed that the polymer accumulated in granules located in the stroma of the plastids. In comparison with former results from transgenic lines with constitutive expression of the three phb genes without the phasin gene (Bohmert et al., 2000), no difference in granule size was observed. Preliminary results from immunolocalization of the phasin protein suggest predominant localization of the phasin protein at the surface of the PHB granules (data not shown). The signal density obtained for the transgenic lines was comparable with those in R. eutropha controls (data not shown).
DISCUSSION
Despite the impressive, large amounts of PHB produced using Arabidopsis as host plant for PHB expression and despite the obvious economic potential of plants as factories for PHB production, there is only one report in the literature of a crop plant producing significant levels of PHB (in seeds of B. napus; Valentin et al., 1999; Houmiel et al., 1999). The data described here enabled us to identify one critical parameter with respect to PHB production in plants: the expression of the β-ketothiolase gene, phbA. Results obtained with a variety of chimeric constructs used for transformation of tobacco, potato, and Arabidopsis indicate that the constitutive expression of the β-ketothiolase gene is responsible for the drastically reduced transformation efficiency observed. To test this hypothesis, two chimeric genes were used where either the expression of phbA was put under the control of an inducible promoter or the coding sequence of phbA was separated from the promoter by the presence of a transposable element, thus, leading only to somatic activation of expression. In both cases, generation of transgenic lines was possible in all three plant species.
These data suggest that preventing the expression of phbA during the transformation/regeneration procedure is sufficient to allow generation of transgenic plants. The toxic effects exerted by phbA expression could result from PHB biosynthesis intermediates (acetoacetyl-CoA and 3-hydroxybutyryl-CoA) or derivatives of these intermediates, the depletion of the acetyl CoA pool, or interactions of the β-ketothiolase with other proteins or substrates.
One candidate interfering substance, acetoacetyl-CoA, is present in the cytosol, where it is an intermediate of the mevalonate pathway leading to isopentenyl diphosphate, the central precursor for all isoprenoids. In the plastid, isopentenyl diphosphate proceeds via the dioxyxylulose 5-phosphate pathway (Arigoni et al., 1997; Lichtenthaler et al., 1997; Lange et al., 2000). Therefore, acetoacetyl-CoA is not expected to be present in the plastid. Another observation supports this assumption: Measurements of transgenic Arabidopsis, tobacco, and potato lines with constitutive expression of phbB and phbC targeted to the plastids did not reveal any PHB accumulation (data not shown).
Because acetoacetyl-CoA is quite similar to β-ketoacyl ACP, one may speculate about an interference of acetoacetyl-CoA with the reaction of β-ketoacyl ACP synthase (KASIII) in fatty acid biosynthesis. To further analyze this possibility, fatty acids were determined in PHB-producing representatives of the lines ABC, prpABC, and ABC P. None of the analyzed lines displayed changes in total fatty acid levels or in fatty acid composition (data not shown), which is in agreement with similar data obtained for PHB-producing Brassica napus seeds (Houmiel et al., 1999). Therefore, these data do not support an interference of acetoacetyl-CoA with the reaction catalyzed by KASIII.
The second aim of this work was to test possible strategies to prevent the severe impairment in growth and development observed in Arabidopsis plants accumulating high amounts of PHB (≥3 mg g−1 fresh weight; Bohmert et al., 2000). Essentially two strategies were followed. First, using an inducible promoter system transgenic prpABC, Arabidopsis lines were grown until they developed a fully expanded rosette and were subsequently treated with salicylic acid to induce phbA gene expression. This procedure led to transgenic lines accumulating 6 mg 3-hydroxybutyrate g−1 fresh weight without any alteration in phenotype. This is twice the amount observed in phenotypically comparable Arabidopsis lines constitutively expressing the phbA gene (Bohmert et al., 2000). Transgenic lines bearing the inducible promoter produced up to 11 mg PHB g−1 fresh weight. This is a fairly high value considering that full phbA gene expression was present for only 18 d. This line, however, displayed growth retardation.
The second strategy for overcoming growth reduction in PHB-accumulating Arabidopsis lines was directed toward possible negative effects caused by the PHB itself. One could imagine that PHB granules may interfere with plastidial structures and, thereby, hinder their function. In PHB-accumulating bacteria, PHB granules are associated with proteins called phasins. One could speculate that the phasins prevent interference of PHA granules with other structures such as thylakoid membranes. To test possible positive effects of phasins on PHB accumulation in transgenic plants, the phasin gene phaP of R. eutropha was constitutively expressed in Arabidopsis. Expression of the phaP gene alone did not cause any outward phenotypic changes. Expression of the three phb genes in conjunction with phaP led to PHB production in transgenic lines; however, higher PHB concentrations were again accompanied by growth reduction. Therefore, phaP gene expression does not mask or prevent the deleterious effects of PHB expression.
In conclusion, the data presented suggest that constitutive expression of β-ketothiolase leads to problems during transformation of different plant species using an A. tumefaciens-mediated in planta transformation procedure (Arabidopsis) and tissue culture transformation procedures (tobacco and potato). Strategies that overcome this problem include the use of an inducible promoter system or a somatically activated expression system. Unfortunately, the association of growth impairment with high PHB production could not be overcome using these strategies.
MATERIALS AND METHODS
Plasmid Constructions
A 439-bp BamHI fragment of the plasmid pBS42-1 (Martini et al., 1993) was ligated in sense orientation into the BamHI restriction site of plasmid pBS-SK (Stratagene, La Jolla, CA). The resulting plasmid was digested with the restriction enzymes SmaI and NotI, and the overhanging ends were filled by T4 DNA polymerase. This 462-bp fragment containing the prp1-1 promoter of potato (Solanum tuberosum; Martini et al., 1993) was ligated into the plasmid pBI-TP-Thio (Nawrath et al., 1994), which had been digested with the restriction enzymes HindIII and XbaI and which had the 5′ ends filled-in, resulting in plasmid pBIprpA.
Plasmid pBI-TP-Syn (Nawrath et al., 1994) was digested with the restriction enzymes EcoRI and XbaI and had the 5′ ends filled-in. This 2.1-kb fragment was ligated in sense orientation into the SmaI site of plasmid pBinARHyg (Becker, 1990). The resulting plasmid, pBinARHygC, was linearized with EcoRI, and the 5′ ends were filled-in. A HindIII/EcoRI digestion of plasmid pBI-TP-Red (Nawrath et al., 1994) generated a 2.1-kb fragment ligated in sense orientation into the linearized plasmid pBinARHygC. The resulting plasmid, pBinARHygBC, was linearized with HindIII, and the 5′ ends were filled-in. An XbaI/EcoRI digestion of the plasmid pBIprpA generated a 2.3-kb fragment that was ligated in sense orientation into the linearized plasmid pBinARHygBC to generate the construct pBinARHygprpABC. This construct contained three cassettes, each of them bearing one of the three phb genes with a plastidial-targeting sequence, a poly(A) adenylation sequence, and either the CaMV 35S promoter (phbB and phbC) or the inducible prp1-1 promoter (phbA).
The constitutive CaMV 35S promoter of plasmid pBI121 (CLONTECH, Palo Alto, CA) was amplified by PCR using the 37-mer oligonucleotide 5′-ggT CTA gAA CTA gTg TCC CCC gTg TTC TCT CCA AAT g-3′ complementary to the promoter's 5′ end in combination with the 37-mer oligonucleotide 5′-gCg CAA gCT TgC ATg CCT gCA ggT CCC CAg ATT AgC C-3′ (PHB-2) to generate a SpeI site upstream of the XbaI site at the promoter's 3′ end. After digestion of the 800-bp PCR fragment with the restriction enzymes XbaI and HindIII, the promoter was ligated together with a 1.8-kb XbaI/EcoRI fragment of plasmid pBI-TP-Thio (Nawrath et al., 1994) into the HindIII/EcoRI site of plasmid pBS-SK (Stratagene), resulting in plasmid pSKASpe. Plasmid pBinARHygBC was digested with the restriction enzyme HindIII, and the 5′ ends were filled-in. A HindIII/EcoRI digestion of plasmid pSKASpe led to a 2.6-kb fragment that was ligated in sense orientation into the linearized plasmid pBinARHygBC to generate the construct pBinARHygSpeABC.
The oligonucleotides 5′-ggA CTA gTC Agg gAT gAA AgT Agg ATg-3′ (PHB-3) 27-mer and 5′-ggA CTA gTA ggg ATg AAA Acg gTC gg-3′ (PHB-4) were used to amplify the transposable Ac element from maize from the plasmid pT12 (T. Altmann, unpublished data; Müller-Neumann et al., 1984; Pohlmann et al., 1984) adding SpeI restriction sites at both ends. The 4.5-kb PCR-product was digested with SpeI and ligated in sense orientation into the vector pBinARHygSpeABC linearized with SpeI, resulting in plasmid pBinARHygacABC. This plasmid contains three cassettes, each of them bearing one of the three phb genes with a plastidial-targeting sequence, a poly(A) adenylation sequence, and the CaMV 35S promoter. The transposable Ac element of maize was located upstream of the phbA gene and its plastidial-targeting sequence.
The plasmid pSKSaE17 bearing a 1,692-bp subfragment of E5000 (Wieczorek et al., 1995) was used as a template to amplify the 579-bp coding region of the phaP gene of R. eutropha (Hanley et al., 1999). Using the 30-mer oligonucleotides 5′-AAT CCC ggg TgA TCC TCA CCC Cgg AAC AAg-3′ (PHB-7) and 5′-ggg gAg CTC TTC AAC gCA ggC AgT TAT CAg-3′ (PHB-8) led to the introduction of a SmaI restriction site at the 5′ end and a SacI restriction site at the 3′ end. The resulting PCR product was cut with SmaI and SacI and was ligated into the SmaI/SalI site of the vector pBI-TP-Red (Nawrath et al., 1994). The resulting construct, pBI P, harbored phaP under control of the CaMV 35S promoter and targeted to the plastid.
The constitutive CaMV 35S promoter of the plasmid pBinARHyg (Becker, 1990) was amplified by PCR using the 40-mer oligonucleotide 5′-gCg CAA gCT TAC TAg Tgg ATC CgC Atg CCT gCA ggT CCC C-3′ (PHB-10) and the 39-mer oligonucleotide 5′-ggT CTA gAg TCC CCC gTg TTC TCT CCA AAT gAA ATg AAC-3′ (PHB-9). After digestion of the 800-bp PCR fragment with the restriction enzymes HindIII and XbaI, the promoter was ligated to the HindIII/XbaI site of the plasmid pBI-TP-Thio (Nawrath et al., 1994). The 2.4-kb HindIII/EcoRI fragment of the resulting plasmid was introduced into the HindIII site of the plasmid pBinARHygBC to create the plasmid pBinARHyg2000SpeABC. The 1.9-kb HindIII/EcoRI fragment of plasmid pBI P was blunt-end-ligated to the SpeI site of pBinARHyg2000SpeABC. The resulting plasmid, pBinARHygABC P, contained four cassettes, each of them bearing one of the three phb genes or the phasin gene. Every gene contained a plastidial-targeting sequence, the CaMV 35S promoter, and a poly(A) adenylation sequence.
Construction of the plasmids pBI AB, pBI ABC, pBI-TP-Thio, pBI-TP-Red, and pBI-TP-Syn has been described previously (Nawrath et al., 1994; Bohmert et al., 2000).
Plant Material and Transformation
The plasmids pBI-TP-Thio, pBI-TP-Red, pBI-TP-Syn, pBinARHygprpABC, pBinARHygacABC, pBI P, and pBinARHygABC P were electroporated into Agrobacterium tumefaciens C58C1 harboring plasmid pMP90 and used to transform Arabidopsis C24 plants via dipping (Clough and Bent, 1998). Transformed seeds were selected on Murashige and Skoog medium (Murashige and Skoog, 1962; DUCHEFA, Haarlem, The Netherlands) containing 1% (w/v) Suc and 100 μg mL−1 kanamycin or 20 μg mL−1 hygromycin. Plants were grown at 20°C with a 16-h-light, 8-h-dark regime with approximately 250 μmol photons m−2 s−1.
Potato cv Desiree was obtained from Saatzucht Lange AG (Bad Schwartau, Germany). Transformation of potato was performed using the protocol of Rocha-Sosa et al. (1989). Depending on the construct used, transgenic plants were selected on kanamycin- or hygromycin-containing medium (Dietze et al., 1995). Plants were maintained in tissue culture with a 16-h-light, 8-h-dark regime on Murashige and Skoog medium (Murashige and Skoog, 1962; DUCHEFA) containing 2% (w/v) Suc. In the greenhouse, plants were grown under the same light regime with a minimum of 250 μmol photons m−2 s−1 at 22°C.
Seeds of tobacco (Nicotiana tabacum L. cv Samsun NN) were obtained from Vereinigte Saatzuchten (Ebstorf, Germany). Tobacco transformation using A. tumefaciens-mediated gene transfer was carried out as described previously (Rosahl et al., 1987; Komari, 1989). Plants in tissue culture were grown with a 16-h-light, 8-h-dark regime on Murashige and Skoog medium (Murashige and Skoog, 1962; DUCHEFA) containing 2% (w/v) Suc. In the greenhouse, plants were grown under the same light regime with a minimum of 250 μmol photons m−2 s−1 at 25°C during the light period and 20°C during the dark period.
Treatment of Transgenic prpABC Lines with Salicylic Acid
All experiments with transgenic prpABC lines were performed in the greenhouse. The plants were sprayed once a day. Samples were taken from mature leaves. Transgenic prpABC lines of tobacco were sprayed with 5 mm salicylic acid for a period of 15 weeks. Leaf samples were taken from the edge of the top one-third of mature leaves. Sampling was performed 10 d before induction with salicylic acid and 4, 22, 44, and 88 d after the 1st d of induction.
Transgenic prpABC lines of potato were sprayed with 1 mm salicylic acid for a period of 5 weeks. Leaf samples were taken the day before the beginning of the induction with salicylic acid, the day after the first spraying, and 11 and 33 d after the first induction.
First spraying of transgenic prpABC lines of Arabidopsis with 1 mm salicylic acid was performed on plants beginning to bolt (4 weeks after germination) and was continued for a period of 18 d. Leaf samples were taken before the first salicylic acid induction and at the end of the spraying period.
Transgenic prpABC lines of B. napus were sprayed with 1 mm salicylic acid during 6 weeks. Leaf samples were taken from the edge of the top one-third of mature leaves. The first samples were taken before induction with salicylic acid. The next were taken the next day, after which sampling was continued weekly.
Analysis of PHB by GC-MS
Extraction and derivatization of 20 to 250 mg of leaf material or 100 mg of tuber material was performed as described previously (Bohmert et al., 2000). An aliquot (1 μL) of the derivative was injected into a GC-MS system (AS 2000 autosampler, GC 8000 gas chromatograph and a MD 800 quadrupole mass spectrometer; all ThermoQuest, Manchester, UK) using a split ratio of 25:1. Single-ion monitoring was used to detect the m/z 189, which corresponds to the major fragment of 3-trimethylsilyl oxybutanoic acid ethyl ester after 3,4 cleavage. Chromatography was carried out using a 30-m × 250-μm DB 5MS column (J&W Scientific, Folsom, CA).
ACKNOWLEDGMENT
We thank Megan McKenzie (Max-Planck-Institut für Molekulare Pflanzenphysiologie, Golm/Potsdam, Germany) for critical reading of the manuscript.
Footnotes
This work was supported by the Bundesministerium für Landwirtschaft (to K.B. and I.B.) and by the German Agricultural Ministry.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010615.
LITERATURE CITED
- Alvarez HM, Kalscheuer R, Steinbüchel A. Accumulation and mobilization of storage lipids by Rhodococcus ruber NCIMB 40126. Appl Microbiol Biotechnol. 2000;54:218–223. doi: 10.1007/s002530000395. [DOI] [PubMed] [Google Scholar]
- Arigoni D, Sagner S, Latzel C, Eisenreich W, Bacher A, Zenk MH. Terpenoid biosynthesis from 1-deoxy-d-xylulose in higher plants by intramolecular skeletal rearrangement. Proc Natl Acad Sci USA. 1997;94:10600–10605. doi: 10.1073/pnas.94.20.10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D. Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 1990;18:203–204. doi: 10.1093/nar/18.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert K, Balbo I, Kopka J, Mittendorf V, Nawrath C, Poirier Y, Tischendorf G, Trethewey R, Willmitzer L. Transgenic Arabidopsis plants can accumulate polyhydroxybutyrate to up to 4% of their fresh weight. Planta. 2000;211:841–845. doi: 10.1007/s004250000350. [DOI] [PubMed] [Google Scholar]
- Choi J, Lee SY. Factors affecting the economics of polyhydroxyalkanoate production by bacterial fermentation. Appl Microbiol Biotechnol. 1999;51:13–21. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dietze J, Blau A, Willmitzer L. Agrobacterium-mediated transformation of potato (Solanum tuberosum) In: Potrykus I, Spangenberg G, editors. Gene Transfer to Plants XXII. Berlin: Springer Verlag; 1995. pp. 24–29. [Google Scholar]
- Hanley SZ, Pappin DJC, Rahman D, White AJ, Elborough KM, Slabas AR. Re-evaluation of the primary structure of Ralstonia eutropha phasin and implications for polyhydroxyalkanoic acid granule binding. FEBS Lett. 1999;447:99–105. doi: 10.1016/s0014-5793(99)00235-5. [DOI] [PubMed] [Google Scholar]
- Houmiel KL, Slater S, Broyles D, Casagrande L, Colburn S, Gonzalez K, Mitsky TA, Reiser SE, Shah D, Taylor NB et al. Poly(β-hydroxybutyrate) production in oilseed leukoplasts of Brassica napus. Planta. 1999;209:547–550. doi: 10.1007/s004250050760. [DOI] [PubMed] [Google Scholar]
- John ME, Keller G. Metabolic pathway engineering in cotton: biosynthesis of polyhydroxybutyrate in fiber cells. Proc Natl Acad Sci USA. 1996;93:12768–12773. doi: 10.1073/pnas.93.23.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller J, Jones JDG, Harper E, Lim E, Carland F, Ralston EJ, Dooner HK. Effects of gene dosage and sequence modification on the frequency and timing of transposition of the maize element Activator(Ac) in tobacco. Plant Mol Biol. 1993;21:157–170. doi: 10.1007/BF00039626. [DOI] [PubMed] [Google Scholar]
- Knapp S, Coupland G, Uhrig H, Starlinger P, Salamini F. Transposition of the maize transposable element Ac in Solanum tuberosum. Mol Gen Genet. 1988;213:285–290. [Google Scholar]
- Komari T. Transformation of callus cultures of nine species mediated by Agrobacterium tumefaciens. Plant Sci. 1989;60:223–229. [Google Scholar]
- Lange M, Rujan T, Martin W, Croteau R. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathway across genomes. Proc Natl Acad Sci USA. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson EJR, Scofield SR, Sjodin C, Jones JDG, Dean C. Modification of the 5′ untranslated region of the maize Activator element leads to increased activity in Arabidopsis. Mol Gen Genet. 1994;245:608–615. doi: 10.1007/BF00282223. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Schwender J, Disch A, Rohmer M. Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Lett. 1997;400:271–274. doi: 10.1016/s0014-5793(96)01404-4. [DOI] [PubMed] [Google Scholar]
- Martini N, Egen M, Rüntz I, Strittmatter G. Promoter sequences of a potato pathogenesis-related gene mediate transcriptional activation selectively upon fungal infection. Mol Gen Genet. 1993;236:179–186. doi: 10.1007/BF00277110. [DOI] [PubMed] [Google Scholar]
- Müller-Neumann M, Yoder JI, Starlinger P. The DNA sequence of the transposable element Ac of Zea mays L. Mol Gen Genet. 1984;198:19–24. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nakashita H, Arai Y, Yoshioka K, Fukui T, Doi Y, Usami R, Horikoshi K, Yamaguchi I. Production of biodegradable polyester by transgenic tobacco. Biosci Biotechnol Biochem. 1999;63:870–874. doi: 10.1271/bbb.63.870. [DOI] [PubMed] [Google Scholar]
- Nawrath C, Poirier Y, Somerville C. Targeting of the polyhydroxybutyrate biosynthetic pathway to the plastids of Arabidopsis thaliana results in high levels of polymer accumulation. Proc Natl Acad Sci USA. 1994;91:12760–12764. doi: 10.1073/pnas.91.26.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann RF, Fedoroff NV, Messing J. The nucleotide sequence of the maize controlling element Activator. Cell. 1984;37:635–643. doi: 10.1016/0092-8674(84)90395-7. [DOI] [PubMed] [Google Scholar]
- Poirier Y, Dennis DE, Klomparens K, Somerville C. Polyhydroxybutyrate, a biodegradable thermoplastic, produced in transgenic plants. Science. 1992;256:520–523. doi: 10.1126/science.256.5056.520. [DOI] [PubMed] [Google Scholar]
- Rocha-Sosa M, Sonnewald U, Frommer W, Stratmann M, Schell J, Willmitzer L. Both developmental and metabolic signals activate the promoter of the class I patatin gene. EMBO J. 1989;8:23–29. doi: 10.1002/j.1460-2075.1989.tb03344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosahl S, Schell J, Willmitzer L. Expression of a tuber-specific storage protein in transgenic tobacco plants: demonstration of an esterase activity. EMBO J. 1987;6:1155–1159. doi: 10.1002/j.1460-2075.1987.tb02348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbüchel A, Aerts K, Babel W, Föllner C, Liebergesell M, Madkour MH, Mayer F, Pieper-Fürst U, Pries A, Valentin HE et al. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can J Microbiol Suppl. 1995;41:94–105. doi: 10.1139/m95-175. [DOI] [PubMed] [Google Scholar]
- Steinbüchel A, Füchtenbusch B. Bacterial and other biological systems for polyester production. Trends Biotechnol. 1998;16:419–427. doi: 10.1016/s0167-7799(98)01194-9. [DOI] [PubMed] [Google Scholar]
- Steinbüchel A, Valentin HE. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol Lett. 1995;128:219–228. [Google Scholar]
- Taylor BH, Finnegan EJ, Dennis ES, Peacock WJ. The maize transposable element Ac excises in progeny of transformed tobacco. Plant Mol Biol. 1989;13:109–118. doi: 10.1007/BF00027339. [DOI] [PubMed] [Google Scholar]
- Valentin HE, Broyles DL, Casagrande LA, Colburn SM, Creely WL, DeLaquil PA, Felton HM, Gonzalez KA, Houmiel KL, Lutke K et al. PHA production, from bacteria to plants. Int J Biol Macromol. 1999;25:303–306. doi: 10.1016/s0141-8130(99)00045-8. [DOI] [PubMed] [Google Scholar]
- Van Sluys MA, Tempe J, Federoff N. Studies on the introduction and mobility of the maize Activator element in Arabidopsis thaliana and Daucus carota. EMBO J. 1987;6:3881–3889. doi: 10.1002/j.1460-2075.1987.tb02728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek R, Pries A, Steinbüchel A, Mayer F. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J Bacteriol. 1995;177:2425–2435. doi: 10.1128/jb.177.9.2425-2435.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]