Abstract
The genes encoding the 1,3-propanediol (1,3-PD) operon of Clostridium butyricum VPI1718 were characterized from a molecular and a biochemical point of view. This operon is composed of three genes, dhaB1, dhaB2, and dhaT. When grown in a vitamin B12-free mineral medium with glycerol as carbon source, Escherichia coli expressing dhaB1, dhaB2, and dhaT produces 1,3-PD and high glycerol dehydratase and 1,3-PD dehydrogenase activities. dhaB1 and dhaB2 encode, respectively, a new type of glycerol dehydratase and its activator protein. The deduced proteins DhaB1 and DhaB2, with calculated molecular masses of 88,074 and 34,149 Da, respectively, showed no homology with the known glycerol dehydratases that are all B12 dependent but significant similarity with the pyruvate formate lyases and pyruvate formate lyases activating enzymes and their homologues. The 1,158-bp dhaT gene codes for a 1,3-PD dehydrogenase with a calculated molecular mass of 41,558 Da, revealing a high level of identity with other DhaT proteins from natural 1,3-PD producers. The expression of the 1,3-PD operon in C. butyricum is regulated at the transcriptional level, and this regulation seems to involve a two-component signal transduction system DhaAS/DhaA, which may have a similar function to DhaR, a transcriptional regulator found in other natural 1,3-PD producers. The discovery of a glycerol dehydratase, coenzyme B12 independent, should significantly influence the development of an economical vitamin B12-free biological process for the production of 1,3-PD from renewable resources.
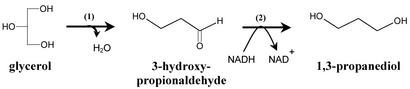
For a long time, 1,3-propanediol (1,3-PD) has been considered a specialty chemical. However, the recent development of a new polyester called poly(propylene terephtalate), with unique properties for the fiber industry (1, 2), necessitates a drastic increase in the production of this chemical. There are currently two processes for chemical synthesis of 1,3-PD. Both of these processes produce toxic intermediates and require a reduction step under high hydrogen pressure (3). Recently, several patents (see ref. 4 for the most recent) were filed describing an environmentally friendly biological process for the conversion of renewable resources, such as glucose, to 1,3-PD. This process uses recombinant microorganisms expressing Saccharomyces cerevisiae genes encoding a glycerol-3-phosphate dehydrogenase and a glycerol-3-phosphate phosphatase for the conversion of glucose derived dihydroxyacetone-phosphate to glycerol and Klebsiella pneumoniae genes encoding a coenzyme B12-dependent glycerol dehydratase and a 1,3-PD dehydrogenase for the reduction of glycerol to 1,3-PD (Fig. 1). One of the key limitations of this biological process is that it uses a coenzyme B12-dependent glycerol dehydratase, requiring the addition of large amounts of a high-cost molecule, vitamin B12, to the culture medium (4).
Figure 1.
Bacterial conversion of glycerol to 1,3-PD: (1) glycerol dehydratase, (2) 1,3-PD dehydrogenase.
In all of the natural 1,3-PD producers characterized to date, i.e., K. pneumoniae, Citrobacter freundii, and Clostridium pasteurianum, glycerol conversion to 1,3-PD involves a coenzyme B12-dependent glycerol dehydratase (5–10). We recently reported that the glycerol dehydratase of Clostridium butyricum VPI1718, extracted from 1,3-PD-producing cells, was not stimulated by coenzyme B12 and was extremely oxygen sensitive, which suggested it might be a coenzyme B12-independent enzyme (11). A B12-independent diol dehydratase, inactive on glycerol, was previously purified from Clostridium glycolycum and was proposed to be a diiron-tyrosyl radical protein (12).
In this paper, we report on the cloning, sequencing, and characterization of the genes encoding the glycerol dehydratase and the 1,3-PD dehydrogenase of C. butyricum VPI 1718. By both heterologous expression and biochemical characterization, we have demonstrated that this glycerol dehydratase does not require coenzyme B12 for activity. To our knowledge, this report represents the first description of such an enzyme and suggests that it belongs to a new family of coenzyme B12-independent glycerol dehydratases. This discovery should allow the development of an economical vitamin B12-free biological process for the production of 1,3-PD from renewable resources.
Materials and Methods
Bacterial Strains and Plasmids.
C. butyricum VPI 1718 was obtained from the Virginia Polytechnic Institute (Blacksburg, VA). E. coli DH5α [supE44Δlac U169 (φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used as a host for all cloning steps and was obtained from New England Biolabs.
E. coli MG1655 ΔadhE (I. Mortier and P.S., unpublished results) was used for expression of the 1,3-PD operon to avoid possible inactivation of activated DhaB1 by AdhE as demonstrated for activated PflB (13). The plasmid pUC18 (New England Biolabs) was used for construction of the genomic libraries and the pGEM-T (Promega) for PCR products cloning. The E. coli–Clostridium acetobutylicum shuttle vector pSOS95 was used for gene expression (GenBank accession no. AY187686), whereas the pIMP1 plasmid (14) was used as a control.
Growth Conditions.
Batch cultures of C. butyricum VPI 1718 for genomic DNA preparation were grown anaerobically at 37°C in 2× YT medium (yeast extract, 10 g/liter; bactotryptone, 16 g/liter; NaCl, 4 g/liter) supplemented with 2% glucose. E. coli was routinely grown aerobically at 37°C in LB (15) supplemented when needed with ampicillin (100 μg⋅ml−1), isopropyl-β-d-thiogalactopyranoside (50 μg⋅ml−1), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (40 μg⋅ml−1).
Continuous cultures of C. butyricum for total RNA extraction were grown in phosphate-limited synthetic medium containing (per liter): KH2PO4, 0.1 g; KCl, 0.65 g; MgSO4, 7H2O, 0.2 g; FeSO4, 7H2O, 0.028 g; NH4Cl, 1.5 g; CoCl2, 6H2O, 0.01 g; para-aminobenzoic acid, 8 mg; biotin, 0.04 mg; struktol J633 antifoam, 0.1 g, with either glucose (30 g) or glycerol (27 g) plus glucose (3 g) as the carbon and energy source. The dilution rate was 0.05 h−1, the temperature was controlled at 37°C and the pH automatically maintained at 6.0 by addition of 6 M NH4OH.
For enzyme assays and heterologous gene expression, recombinant E. coli (pSPD5) and E. coli (pIMP1) were grown anaerobically in LB in the presence of nitrilotriacetic acid (200 mg⋅liter−1), FeSO4 (50 mg⋅liter−1), K2HPO4 (0.5 g⋅liter−1), Na selenate (30 μg⋅liter−1), supplemented with 40 g⋅liter−1 glycerol, ampicillin (100 μg⋅ml−1), and erythromycin (200 μg⋅ml−1) as the selection pressure.
Preparation of Cell Extracts and Enzyme Assays.
E. coli ΔadhE (pSPD5) cell extracts were prepared by sonication by using the anaerobic procedure of Vasconcelos et al. (16). 1,3-PD dehydrogenase activity was determined spectrophotometrically by the procedure of Boenigk et al. (7). The assay mixture contained in a 1-ml final volume, 30 mM ammonium sulfate, 100 mM potassium carbonate (pH 9.0), 2 mM DTT, 2 mM NAD+, and 100 mM 1,3-PD. Glycerol dehydratase activity was assayed by a method derived from that initially described by Toraya et al. (17), based on NADH consumption when the aldehydes formed by the dehydratase are reduced to the corresponding alcohol by an excess of yeast alcohol dehydrogenase. The assay mixture contained in a 1-ml final volume: 0.03 M (NH4)2SO4, 0.1 M 1,2-propanediol, 0.1 M potassium carbonate buffer pH 7.0, 2 mM DTT, and 10 mM NADH. Coenzyme B12 (10 μM) or S-adenosylmethionine (4 mM) was added or omitted from this reaction mixture. NADH consumption was followed continuously at 340 nm.
All enzyme assays were performed under anaerobic conditions at 30°C. One unit of enzyme activity is defined as the amount of enzyme that catalyses the conversion of 1 μmol of substrate per min at 30°C.
All assays were performed in duplicate on two different cell extracts, and the reported values are the average of four assays with the calculated standard deviation.
The protein content of the extracts was determined by the method of Bradford (18) with BSA as the standard.
Analysis of Fermentation Products.
The concentrations of substrates (glucose or glycerol) and fermentation products (1,3-PD, acetate, butyrate, lactate, succinate) were measured by HLPC (model 5810 HPLC pump from Waters) equipped with an automatic sampler (SP 8775, Spectra Physic France, Les Ulis, France) and an integrator (Intersmat ICR 1B, Shimadzu). The separation was obtained with an Aminex HPX-87H (Bio-Rad) column (300 × 7.8 mm) and the following operating conditions: temperature, 25°C; mobile phase, 0.031 mM H2SO4; flow rate, 0.7 ml⋅min−1.
Computer Programs.
DNA and amino acid analyses used the pc-gene program (IntelliGenetics). Sequence comparison and homology searches were done by using blast (19) and prodom (20).
Nucleotide Sequence Accession No.
The sequence data reported here have been submitted to the GenBank database and assigned accession no. AY112989.
Results
Nucleotide Sequence Analysis of the C. butyricum Glycerol Dehydratase and 1,3-PD Dehydrogenase-Encoding Genes.
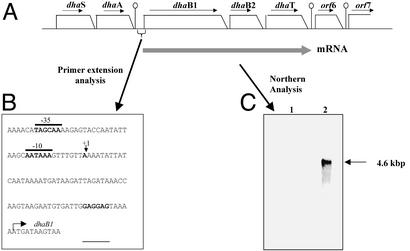
The strategy used to clone the glycerol dehydratase and 1,3-PD dehydrogenase-encoding genes and surrounding regions (9,071 bp all together) is published as supporting information on the PNAS web site, www.pnas.org. Nucleotide sequence analysis revealed six complete ORFs, dhaS, dhaA, dhaB1, dhaB2, dhaT, ORF6, and one truncated ORF7, all transcribed in the same orientation (Fig. 2).
Figure 2.
(A) Genomic arrangement of the genes encoding C. butyricum glycerol dehydratase (dhaB1, dhaB2) and 1,3-PD dehydrogenase (dhaT) and surrounding regions. (B) Promoter region sequence (from nucleotides 2580 to 2700) of the dhaB1-dhaB2-dhaT operon. The transcription start site is indicated with a vertical arrow. The putative −35 and −10 regions are overlined. The putative ribosome-binding site is underlined, and the putative ATG start codon of dhaB1 gene is shown by a bent arrow. (C) Northern blot analysis. Total RNA was isolated from C. butyricum cells grown in phosphate-limited continuous cultures containing either glucose (line 1) or glycerol (line 2) as the substrate.
Data retrieval from protein and DNA databases allowed identification of the fifth ORF (1,158 bp, from 6055 to 7212) as a putative 1,3-PD dehydrogenase gene (dhaT). A putative ribosome-binding site GAGAAG is located 7 bp upstream of the dhaT ATG initiation codon. Its sequence and location matched well those found for other clostridial genes (21). Immediately downstream of the dhaT TAA stop codon, a 22-bp inverted repeat sequence, able to form a stable stem–loop structure (ΔG = −25 kcal/mol), was identified. This structure, followed by a run of Ts, likely functions as a rho-independent transcriptional terminator.
Two complete ORFs, dhaB1 and dhaB2, were identified upstream of the dhaT start codon. The dhaB1 gene (2,364 bp, from 2690 to 5053) is immediately followed by the dhaB2 gene (915 bp, base pairs 5081 to 5995), the dhaB1 stop codon being separated from the dhaB2 start codon by 28 base pairs.
The dhaB2 ORF immediately precedes dhaT, with 58 bp separating these two genes. Two putative ribosome-binding site (GAGGAG and AAGGGGA) were found 5 and 7 bp upstream of the dhaB1 and dhaB2 start codons, respectively.
The genomic arrangement of dhaB1, dhaB2, and dhaT strongly suggests an operon organization, because a putative transcriptional terminator could be identified only downstream of dhaT.
Upstream of dhaB1, two complete ORFs were identified, dhaS (1,215 bp, from 101 to 1315) immediately followed by dhaA (1,053 bp, from 1328 to 2380), both being preceded by a putative ribosome-binding site. The genomic arrangement of dhaS and dhaA strongly suggests an operon organization, because the dhaS stop codon is separated from the dhaA start codon by only 9 bp and a putative transcriptional terminator (a 19-bp inverted repeat sequence able to form a stable stem–loop structure, ΔG = −25.2 kcal/mol) could be identified only immediately downstream of the dhaA stop codon.
The dhaT gene is followed by the complete ORF6 (564 bp, from 7463 to 8026) and the truncated ORF7 (880 bp, from 8192 to 9071), both being preceded by a putative ribosome-binding site. A 16-bp inverted-repeat sequence (ΔG = −18 kcal/mol) resembling a rho-independent terminator was found between the two genes.
Amino Acid Sequence Analysis.
The dhaT gene encodes a polypeptide of 385-aa residues with a calculated molecular mass of 41,558 Da, which is in good agreement with the molecular mass of 1,3-PD dehydrogenases. The 1,3-PD dehydrogenase from C. butyricum has a high level of identity (from 76 to 85% identity) with 1,3-PD dehydrogenases from C. pasteurianum (22), C. freundii (23), and K. pneumoniae (24). The highest level of homology was found with the C. pasteurianum enzyme (85% identity, 92% similarity), both proteins contain 385-aa residues (vs. 387-aa residues for the two enteric bacteria enzymes). A significant level of identity was also found with other type III alcohol dehydrogenases such as the methanol dehydrogenase from Bacillus methanolicus (25) (51% identity) and the Adh2 alcohol dehydrogenases from E. coli (50% identity) and Zymomonas mobilis (49% identity) (26). The putative iron-binding motif (GxxHxxAHxxGxxxxxPHG), proposed as a typical feature in this class of enzymes, is conserved in the C. butyricum dhaT gene product (amino acids 262 to 280) (27).
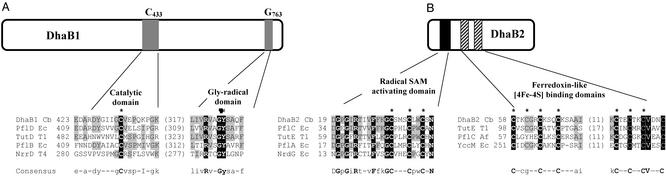
The dhaB1 gene encodes a protein of 787 amino acids with a calculated molecular mass of 88,074 Da. Homology searches in the databases revealed similarity with glycyl radical enzymes like pyruvate formate lyase (PFL) (EC 2.3.1.54) and PFL homologues. The lowest level of identity (around 25%) was found with the well characterized PFLs PflB from E. coli (28) and C. pasteurianum (29). The highest degree of identity was obtained with PflD and f810 (41% and 37%, respectively), two Pfl homologues from E. coli with unknown functions (30), and the recently characterized enzyme involved in anaerobic toluene metabolism, benzylsuccinate synthase from Thauera aromatica (32% identity) (31) and TutD from Thauera sp. T1 (30% identity) (32). Although similarity with these Pfl homologues extended over almost the entire sequence of DhaB1 (data not shown), the highest degree of identity was found between positions 595 and 785 (55% identical to PflD) and more specifically around the predicted glycyl radical site Gly-763 (Fig. 3A). In E. coli PflB, this conserved glycine (Gly-734) was shown to be essential for enzymatic function, because it was identified as being the site of formation of a free radical (Gly-radical domain, Fig. 3A) by the Pfl activase (28). The 8-aa residues surrounding the Gly-763 in DhaB1 are identical to those of the equivalent region in PflD. They also match well with the corresponding residues in PflB and NrdD, which are part of the Gly-radical domain (28, 33). Another site of strong homology is located around Cys-433 corresponding to the active Cys-419 in PflB, involved in the formation of the transient acetyl enzyme (34). Nevertheless, whereas two adjacent cysteine residues were found at this site in all of the characterized Pfl (for example, E. coli PflB), DhaB1, PflD, and TutD contain only a single cysteine residue (Fig. 3A).
Figure 3.
Schematic structure showing the domain organization and special sequence features of the C. butyricum dhaB1 and dhaB2 gene products. (A) Partial alignments and homology analysis of DhaB1 with other known and predicted glycyl radical enzymes, around the presumed active site cysteine (Catalytic domain), indicated by an asterisk (position 433 in DhaB1) (Left), and the conserved glycine residue (Gly-radical domain), indicated by a black dot (position 763 in DhaB1) (Right). (B) Partial alignments and homology analysis of dhaB2 gene product with known and predicted activating components of glycyl radical enzymes (Left), and with two predicted activases and ferredoxin (YccM) (Right). The conserved cysteines are marked by asterisks (note that one cysteine is missing in the PflC Af protein). The number (Left) indicates the location in the amino acid sequence. The last two characters of the abbreviated names refer to the organism: Cb, C. butyricum; Ec, E. coli; T1, Thauera sp. T1; Af, Archaeoglobus fulgidus, T4, bacteriophage T4. GenBank accession nos.: PflD Ec (P32674), TutD T1 (BAC5501), PflB Ec (P09373), NrrD T4 (P07071), PflC Ec (P32675), TutE T1 (AAC38452), PflA Ec (P09374), NrdG Ec (P39329), PflC Af (P070279), and YccM Ec (F64840).
The dhaB2 gene product is 304 amino acid residues long with a predicted molecular mass of 34,149 Da. Homology searches revealed that the predicted gene product shared extensive similarity with proteins activating glycyl radical enzymes. The highest degree of identity was found with Pfl homologues activating enzymes like E. coli PflC (33% identical to DhaB2) and Thauera sp. T1 TutE (36% identity). Prodom analysis revealed a well conserved N-terminal region (Fig. 3B; amino acids 19–40), containing an unusual conserved cysteine motif (CXXXCXXC) characteristic of all radical S-adenosylmethionine enzymes (35) and involved in the interaction with S-adenosylmethionine through a [4Fe-4S] cluster (36, 37). DhaB2 also contains two cysteine-rich domains CXXCXXCXXXC (Fig. 3B, amino acids 58–68 and 86–96) typically found in ferredoxins as [4Fe-4S] cluster-binding domains. The eight cysteine residues of these iron–sulfur clusters are also perfectly conserved in YccM, a putative ferredoxin from E. coli, and in TutE, the TutD-activating enzyme, whereas the activating enzymes of PFL (PlfA) and anaerobic ribonucleotide reductase (NrdG) do not contain this motif.
dhaS and dhaA encode proteins of 405 and 351 amino acid residues (molecular masses of 46,440 and 40,438 Da), respectively, with significant homology to a number of sensor histidine kinases and response regulators, respectively, which compose the archetypal prokaryotic two-component signal transduction system (38, 39).
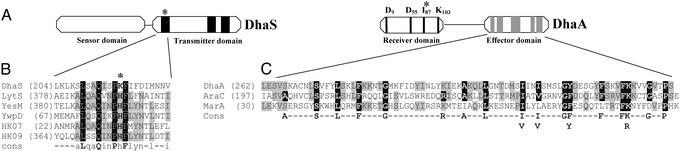
The DhaA protein shows the typical two-domain organization of the response regulators (Fig. 4A). The C-terminal part (≈90 aa), the output or effector domain, displays sequence similarities to the AraC/XylS-type DNA-binding proteins, with two HTH DNA-binding domains (40, 41) (Fig. 4C). The N-terminal part (≈125 first amino acids) represents the highly conserved receiver domain, displaying among others the invariant residue Asp-55 corresponding to Asp-57 (phosphorylation site) of CheY (42). However, the hydroxyl-containing residue (Ser/Thr-87) found in all CheY-proteins and shown to be crucial for proper function of CheY (42) is replaced by isoleucine (Ile-83) in DhaA.
Figure 4.
DhaS/DhaA two-component signal transduction system (TCS) organization. (A) Schematic structure showing the domain organization and special sequences features of the DhaS/DhaA TCS system of C. butyricum. For DhaS, the conserved sequence motifs characteristics of histidine kinases transmitter domains (H, N, and D boxes) are represented by black boxes. For DhaA, the location of the highly conserved aa residues forming the active site of the receiver domain of response regulators are indicated by vertical lines, whereas predicted α-helices of the C-terminal output or effectors domain are indicated by gray boxes. (B) Partial alignment of the putative H box (histidine phosphotransferase domain) of DhaS with the corresponding parts of selected histidine kinases from Group I. The asterisk matches the histidine residue that becomes phosphorylated, found in all histidine kinases known so far. GenBank accession nos.: LytS (A69655), YesM (D69796), YwpD (D70065), HK07 (CAB5457), and HK09 (CAB54581). (C) Comparison of the output domain amino acid sequence of DhaA with homologous regions found in the AraC/XylS-family members AraC (P03021) and MarA (P27246). Residues matching the consensus (Lower) sequence derived from more than 100 AraC/XylS-family members (31) are emphasized by black boxes, whereas additional residues identical in at least two proteins are in gray boxes.
The N-terminal domain (≈175 first amino acids) of the putative sensor protein DhaS shows significant homology (≈30% identity, 55% similarity) with the N-terminal part of the methyl-accepting chemotaxis protein from C. acetobutylicum (GenBank Accession no. AAK78412.1), and the PocR regulatory protein of Salmonella typhimurium (43). In contrast, the deduced amino acid sequence of DhaS was well conserved in the C-terminal or transmitter domain, showing the presence of a conserved ATP-binding site (HATPase-C domain, amino acids 305–405) and typical motifs, namely H, D, and N boxes (42, 44). Among the latter, the most conserved is the H box, although very surprisingly, the phosphorylable histidine residue present in all known histidine kinases is absent in DhaS, replaced by a lysine residue (Lys-216).
No obvious transmembrane segment could be predicted in DhaS. Finally, ORF6 encodes a 187-aa protein whose sequence showed homology to the alkylhydroperoxide reductase small subunit C22 (AHPC) (60% identity, 78% similarity to E. coli AHPC) involved in detoxification of hydroperoxides.
The truncated ORF7 encodes the first 293 amino acid residues of a protein homologous with the general stress protein thioredoxin reductase (TR) (50% identity, 70% similarity to Clostridium litorale TR). Indeed, ORF6 and ORF7 gene products shared homology with the large family of antioxidant enzymes described by Chae et al. (45).
Transcription Analysis of dhaB1, dhaB2, and dhaT Genes.
Northern blot analyses were conducted with total RNA isolated from C. butyricum cells grown in phosphate-limited continuous cultures containing either glucose or a glucose–glycerol mixture as substrate. The fermentation products of these two cultures were analyzed. The results were within the experimental error of our previously published data (46), and 1,3-PD was produced only by glucose–glycerol grown cells.
Total RNA was separated on a denaturing agarose gel and subjected to Northern blotting with 32a-P-labeled probes derived from different regions of the presumed (dhaB1, dhaB2, dhaT) operon. A single hybridization signal of 4.6 kbp was detected with internal fragments of either dhaB1 or dhaT genes as probes (Fig. 2). This result was in good agreement with the minimum operon size of 4.5 kbp derived from the DNA sequence analysis. Thus, it is likely that the three genes dhaB1, dhaB2, and dhaT are the components of a polycistronic operon.
The transcript was detected only with total RNA extracted from glucose–glycerol-grown cells (Fig. 2). No hybridization signal was detected with total RNA extracted from glucose-grown cells, although the ethidium bromide-stained agarose gel used for the Northern blot showed no degradation of 23S and 16S RNAs in this extract. These Northern blot results were in agreement with glycerol dehydratase and 1,3-PD dehydrogenase activities (Table 1) of cell extracts, indicating that the expression of the 1,3-PD pathway was regulated at the transcriptional level. Because dhaB1 and dhaB2 were transcribed together with dhaT, the implication of DhaB1 and DhaB2 in 1,3-PD synthesis pathway was strongly suggested, although sequence analyses of DhaB1 and DhaB2 do not by themselves provide evidence that they are responsible for the glycerol dehydratase activity. This operon was thus designed as the 1,3-PD operon.
Table 1.
Level of glycerol dehydratase and 1,3-PD dehydrogenase activities in crude extracts of E. coli ΔadhE (pSPD5) and E. coli ΔadhE (pIMP1)
| Glycerol dehydratase, units⋅mg−1 protein | 1,3-PD dehydrogenase, units⋅mg−1 protein | ||
|---|---|---|---|
| E. coli ΔadhE (pIMP1) grown on glucose | <0.01 | <0.005 | |
| E. coli ΔadhE (pSPD5) grown on glycerol | Control | 0.36 ± 0.11 | 0.3 ± 0.07 |
| + SAM | 0.92 ± 0.28 | ||
| + CoB12 | 0.35 ± 0.14 | ||
Each assay was done in duplicate on two cell extracts from the same culture. The average of the four values and the standard deviation are given. SAM, S-adenosylmethionine.
To identify the transcriptional start site of the 1,3-PD operon, primer extension was performed with total RNA extracted from C. butyricum grown on glucose–glycerol (46). A major transcriptional start site (at an A base), corresponding to position 65 upstream of dhaB1 start codon, was detected (see supporting information on the PNAS web site). The deduced promoter sequence (Fig. 2) 5′-TAGCAA-3′(−35) and 5′-AATAAA-3′ (−10) with a 19-nt spacing is close to the σA RNA polymerase recognition sequence from Gram-positive bacteria.
Characterization of the 1,3-PD Operon in E. coli.
The E. coli–C. acetobutylicum shuttle vector pSOS95 was previously constructed (GenBank accession no. AY187686) for the expression of any desirable genes by cloning PCR amplified structural genes between the constitutive promoter from C. acetobutylicum ATCC 824 thiolase gene (thl) and the rho-independent transcription terminator from C. acetobutylicum American Type Culture Collection 824 acetoacetate decarboxylase gene (adc) (see supporting information on the PNAS web site for more information on the pSOS95).
The [dhaB1, dhaB2, dhaT] operon was PCR-amplified by using a high-fidelity thermostable DNA polymerase pfu, with appropriately designed primers (see supporting information on the PNAS web site for the sequences of the primers) and cloned downstream of the pSOS95 promoter. The resulting vector was designated pSPD5. Both pSPD5 and pIMP1 (used as a control) were introduced in E. coli MG1655 ΔadhE. The recombinant E. coli ΔadhE (pSPD5) and E. coli ΔadhE (pIMP1) strains were grown anaerobically in a mineral medium that did not contain vitamin B12 and with glycerol as a carbon source. Fermentation products were analyzed by HPLC. As expected, E. coli ΔadhE (pIMP1) was unable to grow on glycerol and to produce 1,3-PD. On the other hand, E. coli ΔadhE (pSPD5) could grow on glycerol and produced 4.5 g/liter of 1,3-PD after 80 h of culture (Fig. 5).
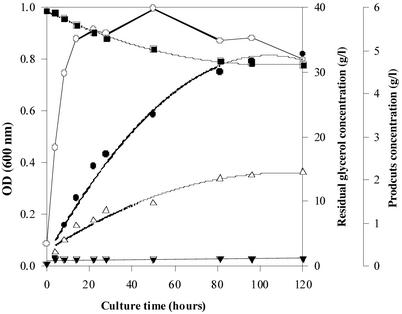
Figure 5.
Growth of E. coli ΔadhE (pSPD5) on glycerol. The batch culture was inoculated with cells grown on LB (52) under anaerobic conditions. ○ , biomass (OD 600 nm); ■, glycerol; ●, 1,3-PD; ▵ , acetate; ▾, succinate.
Crude extracts from E. coli ΔadhE harboring either pSPD5 (grown on glycerol) or pIMP1 (grown on glucose) were assayed for glycerol dehydratase and 1,3-PD dehydrogenase activities (Table 1). Specific activities of 0.36 and 0.3 units/mg for the glycerol dehydratase and the 1,3 PD dehydrogenase, respectively, were measured in an E. coli ΔadhE (pSPD5) extract, whereas no activity was detected in an E. coli ΔadhE (pIMP1) extract. Addition of coenzyme B12 did not increase glycerol dehydratase activity (Table 1), contrary to what was observed for coenzyme B12-dependent glycerol dehydratase assays in C. freundii, K. pneumoniae, and C. pasteurianum (see ref. 9 for a review). On the other hand, assay of the glycerol dehydratase activity in an E. coli ΔadhE (pSPD5) extract, in the presence of 4 mM S-adenosylmethionine, increased specific activity by more than 2.5-fold (Table 1), indicating that a reactivation of the enzyme had occurred.
Discussion
In this study, the 1,3-PD formation pathway of C. butyricum VPI 1718 has been characterized from a molecular point of view. In this microorganism, conversion of glycerol to 1,3-PD involves three genes, dhaB1, dhaB2, and dhaT. The operon arrangement of the three genes, which was suggested by DNA sequence analysis, was confirmed by Northern blot. Moreover, the transcript was detected only in cells growing on a glucose–glycerol mixture as a substrate, indicating a regulation at the transcriptional level of the expression of this operon. It seems likely that this regulation occurs through the two-component signal transduction system whose genes are located immediately upstream of the 1,3-PD operon in the C. butyricum chromosome. DhaA, a possible response regulator, falls into the new AraC-type subfamily recently described for two Streptococcus pneumoniae response regulators (42). Interestingly, this response regulator is paired with a kinase of group I according to the classification based on the sequence around the phosphorylated histidine (47), as pointed out for the few other AraC-type two-component signal transduction systems described so far for S. pneumoniae (42) or Bacillus subtilis (47). It will be very interesting to understand the exact role of the lysine residue substituting the canonical phosphorylated histidine residue present in all of the kinases of group I. Assuming that this lysine residue can be autophosphorylated on sensing the environmental signal, it is tempting to speculate that after transfer of the phosphoryl group from the sensor protein to the receiver domain of the response regulator, the latter binds to DNA and, like most members of the AraC/XylS family (40, 48), acts as a transcriptional activator. Interestingly, a DNA region showing strong homology to the consensus sequence for AraC binding (40), i.e., AGCN7TCCATA is found overlapping the −35 region of the experimentally determined promoter of the 1,3-PD operon. This would suggest direct interactions of the response regulator with C. butyricum RNA polymerase, as indicated for other AraC/XylS family members, for which the few target sequences characterized to date have been located adjacent to or overlapping the −35 region of the regulated promoters (40).
The dhaT gene encodes a 1,3-PD dehydrogenase of 385 aa with a high degree of similarity to the other sequenced DhaT proteins so far and has all of the common features of the class III alcohol dehydrogenases (27). Amino acid sequence analysis of DhaB1 and DhaB2 revealed similarity with PFL homologues and PFL activating enzyme homologues respectively. Heterologous expression of the dhaB1, dhaB2, and dhaT genes in E. coli led to the production of 1,3-PD when grown anaerobically in a glycerol mineral medium in the absence of vitamin B12. As E. coli is unable to carry out de novo synthesis of coenzyme B12, when the genes encoding DhaT and the B12-dependent glycerol dehydratase of C. freundii, K. pneumoniae, or Klebsiella oxytoca were expressed in E. coli, glycerol conversion to 1,3-PD was observed only when vitamin B12 or coenzyme B12 was added to the medium (24, 49). The E. coli recombinant strain expressing dhaB1, dhaB2, and dhaT shows high glycerol dehydratase and 1,3-PD dehydrogenase activities, whereas the level of these enzymes was below the detection limit in the same strain transformed with a control plasmid. These results demonstrate that DhaB1 and DhaB2 are responsible for the glycerol dehydratase activity, and that this enzyme is coenzyme B12 independent.
All of the glycerol and diol dehydratases characterized so far belong to class II of coenzyme B12-containing enzymes (50, 51). The radical mechanism of action of these enzymes, involving coenzyme B12 as essential cofactor, was initially proposed by Abeles and coworkers (52, 53). These enzymatic reactions are subject to suicide inactivation by the substrate glycerol (54). More recently, the crystal structure of K. oxytoca diol dehydratase–cyanocobalamin complex was solved (55).
To date, only the diol dehydratase of C. glycolycum has been shown to be independent of coenzyme B12, although this enzyme has no activity with glycerol (12). This enzyme was found to be different from the coenzyme B12-dependent dehydratases in that it (i) is extremely oxygen sensitive, (ii) is strongly associated with the cell membrane, and (iii) does not use cobalamin coenzyme as cofactor (12). Hartmanis and Stadtman (56) suggested the presence of a diiron-tyrosyl radical cofactor known to catalyze similar chemistry to ribonucleotide reductases.
The DhaB1 and DhaB2 proteins present the highest degree of homology with PFL homologues and PFL-activating enzyme homologues, whereas they did not exhibit any homology with the previously described glycerol and diol dehydratases. PFL is a glycyl radical enzyme that catalyzes the conversion of pyruvate to formate and acetyl-CoA (28). Introduction of the radical into PFL occurs anaerobically by the PFL-activating enzyme, an iron–sulfur protein that uses S-adenosylmethionine and reduced flavodoxin as cosubstrates (57). Through direct interaction of the coenzyme with the [4Fe-4S] cluster, the PFL activase carries out the reductive cleavage of S-adenosylmethionine, producing a highly reactive 5′-deoxyadenosyl radical that abstracts a hydrogen atom from the Gly-734 residue in E. coli PFLB, thus introducing a free radical into the polypeptide chain (36, 58). The radical form of PFL was shown to undergo oxygenolytic cleavage of the polypeptide chain at the site of the glycyl radical (59).
The Gly-734 residue of E. coli PFLB is conserved in DhaB1 (Gly-763). Furthermore, in a crude E. coli extract expressing dhaB1 and dhaB2, an activation of the glycerol dehydratase was observed in the presence of S-adenosylmethionine and NADH, suggesting a similar mechanism of activation as PFL. A more complete biochemical and structural characterization of dhaB1 and dhaB2 gene products should provide more insight into the activation and catalytic mechanisms of this new glycerol dehydratase.
The patented biological process for the conversion of renewable resources like glucose to 1,3-PD, relies on the use of a coenzyme-B12-dependent glycerol dehydratase (4). One of the key limitations of this current biological process is that vitamin B12, a high-cost molecule, has to be added in large amounts to the culture to sustain high productivity and titers. The discovery of a new glycerol dehydratase that is coenzyme B12-independent should significantly influence the cost of 1,3-PD production from renewable resources.
Supplementary Material
Acknowledgments
We thank Sophie Mondeil for technical assistance and Maggie Cervin, Mark Emptage, and Nicholas Lindley for critical reading of the manuscript. P.S. was the recipient of a doctoral fellowship from the Institut National de la Recherche Agronomique. This work was financially supported by the Ecotech Program (Centre National de la Recherche Scientifique–Agence de l'Environment et de la Maîtrise de l'Énergie; Grants 94N80/0168) and the European Committee Fourth and Fifth Framework Projects (FAIR-CT96-1912 and QLK5-CT1999-01364).
Abbreviations
- 1,3-PD
1,3-propanediol
- PFL
pyruvate formate-lyase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY112989).
References
- 1.Miller H. Int Fiber J. 2000;15:14–16. [Google Scholar]
- 2.Rudie R. Int Fiber J. 2000;15:8–12. [Google Scholar]
- 3.Sullivan C-J. Ullmann's Encyclopedia of Industrial Chemistry. A22. Weinheim, Germany: VCH; 1993. pp. 163–171. [Google Scholar]
- 4. Emptage, M., Haynie, S., Laffend, L., Pucci, J. & Whited, G. (2001) Patent Corporation Treaty (PCT) Int. Appl. WO 01/12833 A2.
- 5.Biebl H, Menzel K, Zeng A-P, Deckwer W D. Appl Microbiol Biotechnol. 1999;52:289–297. doi: 10.1007/s002530051523. [DOI] [PubMed] [Google Scholar]
- 6.Homann T, Tag C, Biebl H, Deckwer W-D, Schink B. Appl Microbiol Biotechnol. 1990;33:121–126. [Google Scholar]
- 7.Boenigk R, Bowien S, Gottschalk G. Appl Microbiol Biotechnol. 1993;38:453–457. [Google Scholar]
- 8.Forsberg C W. Appl Environ Microbiol. 1987;53:639–643. doi: 10.1128/aem.53.4.639-643.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel R, Bobik T-A, Gottschalk G. FEMS Microbiol Rev. 1999;22:553–566. doi: 10.1111/j.1574-6976.1998.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 10.Seyfried M, Daniel R, Gottschalk G. J Bacteriol. 1996;178:5793–5796. doi: 10.1128/jb.178.19.5793-5796.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saint-Amans S, Girbal L, Andrade J, Ahrens K, Soucaille P. J Bacteriol. 2001;183:1748–1754. doi: 10.1128/JB.183.5.1748-1754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartmanis M G N, Stadtman T C. Arch Biochem Biophys. 1986;245:144–152. doi: 10.1016/0003-9861(86)90198-0. [DOI] [PubMed] [Google Scholar]
- 13.Kessler D, Leibrecht I, Knappe J. FEBS Lett. 1991;281:59–63. doi: 10.1016/0014-5793(91)80358-a. [DOI] [PubMed] [Google Scholar]
- 14.Mermelstein L D, Papoutsakis E T. Appl Environ Microbiol. 1993;59:1077–1081. doi: 10.1128/aem.59.4.1077-1081.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Russel D W. Molecular Cloning: A Laboratory Manual. 3rd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 16.Vasconcelos I L, Girbal L, Soucaille P. J Bacteriol. 1994;176:1443–1450. doi: 10.1128/jb.176.5.1443-1450.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toraya T, Krodel E, Mildvan A-S, Abeles R-H. Biochemistry. 1979;18:417–426. doi: 10.1021/bi00570a005. [DOI] [PubMed] [Google Scholar]
- 18.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 19.Altschul S F, Madden T L, Schaffer A-A, Zhang J, Zhang Z, Lipman W M D J. Nucleic Acids Res. 1997;25:389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young M, Minton N-P, Staudenbauer W L. FEMS Microbiol Rev. 1989;63:301–326. doi: 10.1111/j.1574-6968.1989.tb03402.x. [DOI] [PubMed] [Google Scholar]
- 22.Luers F, Seyfried M, Daniel R, Gottschalk G. FEMS Microbiol Lett. 1997;154:337–345. doi: 10.1111/j.1574-6968.1997.tb12665.x. [DOI] [PubMed] [Google Scholar]
- 23.Daniel R, Boenigk R, Gottschalk G. J Bacteriol. 1995;177:2151–2156. doi: 10.1128/jb.177.8.2151-2156.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skraly F A, Lytle B L, Cameron D C. Appl Microbiol Biotechnol. 1998;64:98–105. doi: 10.1128/aem.64.1.98-105.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vries G E, Arfman N, Terpstra P, Dijkhuizen L. J Bacteriol. 1992;174:5346–5353. doi: 10.1128/jb.174.16.5346-5353.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conway T, Ingram L O. J Bacteriol. 1989;171:3754–3759. doi: 10.1128/jb.171.7.3754-3759.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bairoch A. Nucleic Acids Res. 1991;19:2241–2245. doi: 10.1093/nar/19.suppl.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner A F V, Frey M, Neugebauer F A, Schäfer W, Knappe J. Proc Natl Acad Sci USA. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weidner G, Sawers G. J Bacteriol. 1996;178:2440–2444. doi: 10.1128/jb.178.8.2440-2444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blattner F R, Plunkett G, Bloch C-A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 31.Leuthner B, Leutwien C, Schulz H, Hörth P, Haehnel W, Schiltz E, Schägger H, Heider J. Mol Microbiol. 1998;28:615–628. doi: 10.1046/j.1365-2958.1998.00826.x. [DOI] [PubMed] [Google Scholar]
- 32.Coschigano P W, Wehrman T S, Young L Y. Appl Environ Microbiol. 1998;64:1650–1656. doi: 10.1128/aem.64.5.1650-1656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Logan D T, Andersson J, Sjoberg B-M, Nordlund P. Science. 1999;283:1499–1504. doi: 10.1126/science.283.5407.1499. [DOI] [PubMed] [Google Scholar]
- 34.Plaga W, Fark R, Knappe J. Eur J Biochem. 1988;178:445–450. doi: 10.1111/j.1432-1033.1988.tb14468.x. [DOI] [PubMed] [Google Scholar]
- 35.Sofia H-J, Chen G, Hetzler B-G, Reyes-Spindola J, Miller N-E. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsby C J, Hong W, Broderick W E, Cheek J, Ortillo D, Broderick J B, Hoffman B M. J Am Chem Soc. 2002;124:3143–3151. doi: 10.1021/ja012034s. [DOI] [PubMed] [Google Scholar]
- 37.Walsby C J, Ortillo D, Broderick W E, Broderick J B, Hoffman B M. J Am Chem Soc. 2002;124:11270–11271. doi: 10.1021/ja027078v. [DOI] [PubMed] [Google Scholar]
- 38.Pao G M, Saier M., Jr J Mol Evol. 1995;40:136–154. doi: 10.1007/BF00167109. [DOI] [PubMed] [Google Scholar]
- 39.Galperin M Y, Nikolskaya A N, Koonin E V. FEMS Microbiol Lett. 2000;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 40.Gallegos M T, Schleif R, Bairoch A, Hofman K, Ramos J-L. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhee S, Martin R G, Rosner J-L, Davies D R. Proc Natl Acad Sci USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lange R, Wagner C, de Saizieu A, Flint N, Molnos J, Stieger M, Caspers P, Kamber M, Keck W, Amrein K E. Gene. 1999;237:223–234. doi: 10.1016/s0378-1119(99)00266-8. [DOI] [PubMed] [Google Scholar]
- 43.Chen P, Andersson D I, Roth J R. J Bacteriol. 1994;176:5474–5482. doi: 10.1128/jb.176.17.5474-5482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stock J B, Surette M G, Levit M, Park P. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 25–51. [Google Scholar]
- 45.Chae H Z, Robison K, Poole L B, Church G, Storz G. Proc Natl Acad Sci USA. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saint-Amans S, Soucaille P. Biotechnol Lett. 1995;17:211–216. [Google Scholar]
- 47.Fabret C, Feher V A, Hoch J A. J Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tobes R, Ramos J L. Nucleic Acids Res. 2002;30:318–321. doi: 10.1093/nar/30.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daniel R, Gottschalk G. FEMS Microbiol Lett. 1992;100:281–286. doi: 10.1111/j.1574-6968.1992.tb14053.x. [DOI] [PubMed] [Google Scholar]
- 50.Toraya T. Cell Mol Life Sci. 2000;57:106–127. doi: 10.1007/s000180050502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seifert C, Bowien S, Gottschalk G, Daniel R. Eur J Biochem. 2001;268:2369–2378. doi: 10.1046/j.1432-1327.2001.02123.x. [DOI] [PubMed] [Google Scholar]
- 52.Frey P A, Essemberg M K, Abeles R H, Kerwar S S. J Am Chem Soc. 1970;92:4488–4489. doi: 10.1021/ja00717a074. [DOI] [PubMed] [Google Scholar]
- 53.Essemberg M K, Frey P A, Abeles R H. J Am Chem Soc. 1971;93:1242–1251. doi: 10.1021/ja00734a036. [DOI] [PubMed] [Google Scholar]
- 54.Mori K, Tobimatsu T, Hara T, Toraya T. J Biol Chem. 1997;272:32034–32041. doi: 10.1074/jbc.272.51.32034. [DOI] [PubMed] [Google Scholar]
- 55.Shibata N, Masuda J, Tobimatsu T, Toraya T, Suto K, Morimoto Y, Yasuoka N. Structure (Cambridge, UK) 1999;7:997–1008. doi: 10.1016/s0969-2126(99)80126-9. [DOI] [PubMed] [Google Scholar]
- 56.Hartmanis M G N, Stadtman T C. Proc Natl Acad Sci USA. 1987;84:76–79. doi: 10.1073/pnas.84.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kulzer R, Pils T, Kappl R, Huttermann J, Knappe J. J Biol Chem. 1998;273:4897–4903. doi: 10.1074/jbc.273.9.4897. [DOI] [PubMed] [Google Scholar]
- 58.Knappe J, Neugebauer F A, Blaschkowski H P, Ganzler M. Proc Natl Acad Sci USA. 1984;81:1332–1335. doi: 10.1073/pnas.81.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sawers G, Watson G. Mol Microbiol. 1998;29:945–954. doi: 10.1046/j.1365-2958.1998.00941.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.