Abstract
There is an urgent need for new drugs that can kill HIV type 1 (HIV-1)-infected cells. HIV-1 glycoprotein Env, which promotes viral membrane fusion through receptor-mediated conformational changes, is an attractive target for such agents because it is expressed on the surface of both virions and infected cells. Unfortunately, conserved binding elements on this protein frequently are buried under a canopy of flexible, glycosylated peptide loops or exposed only transiently during the fusion process. Here, we investigate the exposure of the C-terminal region of the Env ectodomain outside the context of membrane fusion. This binding element is the target of the 5-Helix protein, a designed entry inhibitor that disrupts conformational changes in Env subunit gp41, essential for the fusion process. We show that 5-Helix is capable of interacting with HIV-1 Env in a receptor-independent fashion and that a chimeric 5-Helix/Pseudomonas exotoxin protein recognizes cells expressing Env from a broad spectrum of HIV-1 strains including primary isolates from clades B, D, E, G, and H. This recombinant toxin selectively kills HIV-1-infected cells and blocks spreading infection while still maintaining potent inhibitory activity against membrane fusion. Our results demonstrate that the C-terminal region of the gp41 ectodomain is an accessible target on HIV-1-infected cells for the development of antiviral therapeutics and neutralizing antibodies.
All current therapeutic agents for HIV type 1 (HIV-1) infection are directed at the viral enzymes reverse transcriptase and protease. Despite the success of these drugs in reducing the progression of HIV-1 infection to AIDS, there are increasing problems with long-term toxicity, high cost, difficulties adhering to treatment regimens, and emergence of multiple, drug-resistant viral strains (1). Accordingly, two new types of therapeutics are needed: those that target conserved regions of proteins involved in different viral life cycle events and, thus, are likely to be active against isolates resistant to current drugs; and those that can eliminate infected cells, thereby reducing persistent and latent reservoirs of the virus.
The HIV-1 envelope (Env) glycoprotein is an attractive target for the development of antiviral agents for two reasons: (i) it is present on the surfaces of both virions and infected cells, and (ii) it mediates the initial stages of viral infection, attachment and membrane fusion, rather than the later, postentry stages of reverse transcription and proteolysis (2–4). Env is a complex of two noncovalently associated subunits, gp120 and gp41. HIV-1 gp120 is an external subunit that binds the cellular receptor CD4 and a chemokine coreceptor such as CXCR4 or CCR5. HIV-1 gp41 is a transmembrane subunit that catalyzes receptor-mediated membrane fusion. In theory, epitopes derived from either subunit are prime targets for the development of antiviral therapeutics that can either block HIV-1 entry or destroy infected cells. In practice, however, attempts to develop such compounds have been largely unsuccessful for two reasons. First, the surface of Env is poorly structured and highly variable owing to the high degree of glycosylation and the presence of many flexible, nonconserved peptide loops. Second, the HIV-1 mutation rate permits Env to escape inhibition by strain-specific antiviral agents and antibodies. These features probably enable HIV-1 to evolve in vivo to evade neutralization by the humoral immune response (2, 3, 5).
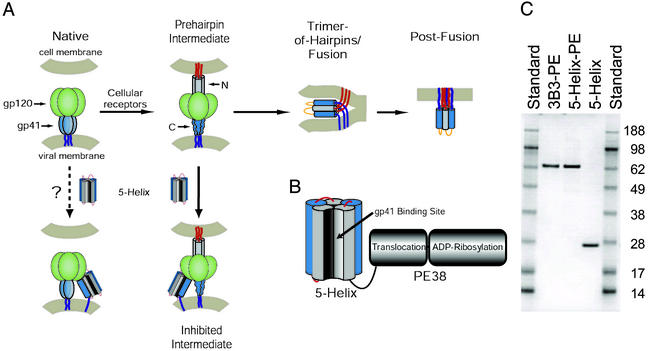
Two well conserved epitopes that are crucial to Env function are located at the N- and C-terminal regions of the gp41 ectodomain (Fig. 1A; ref. 6). In the late stages of membrane fusion, these two regions form a trimer-of-hairpins in which the C-terminal segments from three gp41 ectodomains pack as amphipathic α-helices against a central trimeric coiled coil formed by three N-terminal segments (7–9). Formation of this structure follows a cascade of conformational changes initiated by the interaction of gp120 with CD4 and the coreceptor (10). The ultimate collapse of gp41 into the trimer-of-hairpins brings the viral and cellular membranes into the close proximity necessary for membrane fusion to occur.
Figure 1.
Targeting therapeutics to the HIV-1 fusion protein. (A) Schematic of HIV-1 Env and working model of membrane fusion. The binding of gp120 to cellular receptors triggers gp41 to extend and insert its N-terminal fusion peptide into the target cell membrane (10). In this prehairpin intermediate, the N-terminal coiled coil is exposed and can be targeted by C-peptides. In the absence of inhibitor, this transient state ultimately collapses into a trimer-of-hairpins that brings the N- and C-terminal regions into close proximity, promoting membrane fusion. The entry inhibitor 5-Helix is designed to target the C-terminal region of the gp41 ectodomain and prevent the formation of the gp41 trimer-of-hairpins. The coloring scheme for gp41 is dark blue for the transmembrane sequence, light blue for the C-terminal region, gray for the N-terminal region, and red for the N-terminal fusion peptide; gp120 is colored green. (B) Design of 5-Helix-PE. The 5-Helix domain contains five of the six α-helices that constitute the gp41 trimer-of-hairpins linked together into a single polypeptide. Its C terminus is connected with a 13-aa linker to the translocation and ADP-ribosylation domains of PE38. (C) SDS-gel analysis of purified 3B3-PE, 5-Helix-PE, and 5-Helix. One microgram of each protein was analyzed by SDS/PAGE alongside SeeBlue Plus2 prestained protein standards (Invitrogen). Predicted molecular masses are 65.1 kDa for 3B3-PE, 64.6 kDa for 5-Helix-PE, and 25.4 kDa for 5-Helix.
During the fusion process, the gp41 N-terminal regions become transiently accessible to inhibitory compounds (11, 12). In this transient state, known as the prehairpin intermediate, the gp41 N terminus is inserted in the target cell membrane and the N-terminal coiled coil is exposed, but the trimer-of-hairpins has not formed yet (10). Peptides derived from the gp41 C-terminal region (denoted C-peptides) can bind to the exposed coiled coil and block the proper formation of the trimer-of-hairpins, thus preventing membrane fusion (7, 13, 14). C-peptides can be potent inhibitors of HIV-1 entry, with IC50 values as low as 1 nM in vitro. Two C-peptides, T20 and T1249, are currently in clinical trials and show antiviral activity in humans (1, 4, 15).
Recently, antiviral proteins designed to target the gp41 C-peptide region have been shown to be effective inhibitors of HIV-1 entry (16–18). Whereas the sequence of the gp41 C-peptide region is somewhat variable across HIV-1 strains, the residues on the helical face that interact with the gp41 N-terminal coiled coil are highly conserved (6, 16). The designed proteins take advantage of this specific pattern of sequence conservation by exposing all or part of a trimeric coiled coil derived from the gp41 N-terminal region. In one such protein, denoted 5-Helix, five of six helices that constitute the gp41 trimer-of-hairpins are linked covalently into a single polypeptide (16). 5-Helix lacks a third C-peptide segment, and this vacancy creates a high-affinity binding site for the C-terminal region of the gp41 ectodomain (Fig. 1A). 5-Helix is a potent and broad-spectrum inhibitor of HIV-1 membrane fusion, with IC50 values in the low nanomolar range.
Although inhibitors effectively target the gp41 ectodomain during the fusion process, it is desirable to identify critical Env epitopes that are accessible before the initiation of fusion. Agents recognizing these targets might effectively neutralize free virions or kill infected cells. The gp41 N-terminal region fails to meet this criterion because it appears to be only transiently exposed in the prehairpin intermediate state (11, 12). Therefore, we investigated whether the C-terminal region of the gp41 ectodomain might prove to be a more accessible inhibitory target. In this study, we show that 5-Helix is able to bind to cells expressing Env derived from divergent HIV-1 strains even in the absence of active fusion. Moreover, we demonstrate that this binding is sufficient to concentrate a recombinant toxin to selectively kill HIV-1-infected cells. Our results suggest that the gp41 C-peptide region is a viable target for development of antiviral therapeutics, neutralizing antibodies, and cytotoxic agents directed against infected cells.
Materials and Methods
Pull-Down Experiment.
A biotin-tagged 5-Helix protein was prepared by crosslinking PEO-maleimide activated biotin (Pierce) to a variant 5-Helix protein with a C-terminal Cys residue (5-HelixH6-GC), prepared as described (16) except that 20 mM 2-mercaptoethanol was added to all purification solutions. Cells (293T) expressing HIV-1 EnvHXB2 were exposed to biotin-tagged 5-Helix (1 μM) in the absence and presence of soluble CD4 (sCD4, 5 μg/ml; ImmunoDiagnostics, Woburn, MA) or C-peptide C37-H6 (5 μM; ref. 16) for 30 min at 37°C. After extensive washing in PBS to remove unbound 5-Helix, cells were lysed in 1% Triton X-100/50 mM Tris, pH 7.4/100 mM NaCl. After centrifugation, clarified lysates were incubated with monomeric avidin beads (Pierce) for 2 h at 4°C. The beads were washed extensively in lysis solution and eluted with lysis solution containing 10 mM biotin. The eluted fractions were separated by SDS/PAGE (NOVEX, San Diego), blotted onto nitrocellulose paper, and probed with the primary mAb Chessie 8, which recognizes an epitope in the cytoplasmic tail of gp160. The Western blots were developed by using a horseradish peroxidase-conjugated secondary antibody and the SuperSignal West Femto Maximum Sensitivity Substrate system (Pierce). The precipitated protein represented ≈1% of the total cell lysate gp41 and gp160, as expected from the stringent pull-down conditions and the intracellular localization of the bulk of HIV-1 Env (19, 20).
Protein Expression and Purification.
Overlap extension PCR was used to join the complete 5-Helix and single C34 C-peptide coding sequences to the translocation and ADP-ribosylating domains of Pseudomonas exotoxin (PE) A (PE38) in a pET-based bacterial expression vector (21). The domains were joined by a 13-aa linker sequence from fragment B of staphylococcal protein A with the amino acid sequence AKKLNDAQAPKSD (22). Constructs were verified by DNA sequencing and transformed into Escherichia coli BL21(DE3) for bacterial expression.
The 5-Helix-PE and C34-PE proteins were extracted from inclusion bodies, refolded, and purified by ion exchange and size-exclusion chromatography to >95% homogeneity as determined by SDS gel electrophoresis (23). The antibody-based immunotoxin 3B3-PE and enzymatically inactive point mutant 3B3-PEAsp-553 were prepared similarly (23). Concentrations were determined by BCA protein assay (Pierce). His-tagged 5-Helix protein was purified and quantitated as described (16). Protein binding was measured by surface plasmon resonance by using a BIAcore (Uppsala) 2000 Biosensor and CM5 chips containing C34 peptide or 5-Helix protein immobilized by amine coupling.
Cytotoxicity Assays.
The cytotoxicity of recombinant toxins was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide oxidation procedure on ENV15 cells, a stable Chinese hamster ovary (CHO) transformant that expresses HIV-1 EnvIIIB, and control CHO cells transformed with the empty expression vector (23, 24). The protein concentration at which cell viability was reduced by 50% (IC50) was calculated by fitting the data with prizm graph software.
Inhibition of Pseudovirion and Viral Infection.
Single-cycle infection of U87-CD4+-CXCR4+ cells was performed by using pseudovirions containing the pNL43-Luc-R−E− genome and encapsidated with either HIV-1 EnvHXB2 or amphotropic murine leukemia virus (MuLV) Env (25, 26). Infected cells were harvested after 48 h and assayed for luciferase activity (Promega). Under these conditions, inhibition of infection is independent of cell killing, as demonstrated by the indistinguishable activity of 3B3-PE and 3B3-PEAsp-553. For intact viral-infectivity studies, CEM cells were infected with HIV-1NL4-3 (AIDS Research and Reference Reagent Program, National Institutes of Health) for 2 h, washed extensively, and then incubated for 24–48 h at 37°C. The resulting infected cultures were washed again to remove free virus and then resuspended in medium with and without 5-Helix and 5-Helix-PE. After 5–6 days of incubation, the p24 concentrations were measured in culture supernatants by ELISA (Perkin–Elmer).
Binding to Cell-Surface Env and Inhibition of Cell Fusion.
Env-expressing HeLa effector cells were generated by infection with two vaccinia virus recombinants, one containing an HIV-1 env gene (obtained from Edward Berger, National Institute of Allergy and Infectious Diseases, or from the AIDS Research and Reference Reagent Program) and the other containing the T7 RNA polymerase gene. HeLa target cells were generated by infection with three vaccinia virus recombinants, one containing the CD4 gene, one containing the CCR5 or CXCR4 coreceptor gene, and one containing the LacZ gene linked to the T7 promoter. After 16 h at 31°C, the effector cells were incubated in the absence and presence of 12 nM 5-Helix-PE or 3B3-PE (27) at 37°C for 1 h. To test surface binding, one aliquot was incubated sequentially with mouse anti-PE mAb m40-1 and FITC-conjugated goat anti-mouse IgG and subsequently analyzed by flow cytometry (23). To test fusion inhibition, the other aliquot was incubated with the target cells for an additional 2.5 h at 37°C and subsequently assayed for β-galactosidase activity as described (28).
Results
Binding of 5-Helix Protein to Cell-Surface Env.
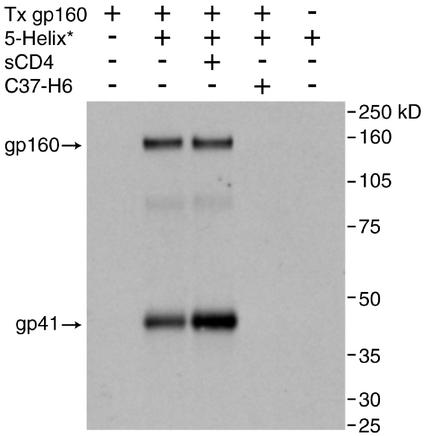
To test whether the C-terminal region of the gp41 ectodomain can interact with molecules outside the context of membrane fusion, we asked whether biotin-tagged 5-Helix is able to precipitate HIV-1 Env from cellular surfaces. Fig. 2 shows that 5-Helix precipitated both gp41 and gp160 (the uncleaved precursor of gp120 and gp41) both in the absence and presence of sCD4. Precipitation was abrogated when excess C-peptide was present, consistent with 5-Helix interacting with the C-terminal region of the gp41 ectodomain exposed on the cellular surface. The precipitation of gp41 was slightly (2- to 4-fold) but reproducibly enhanced by sCD4, suggesting that the accessibility of the gp41 C-peptide region is increased by, but not strictly dependent on, the conformational changes that accompany receptor-mediated membrane fusion. By contrast, the precipitation of the nonfusogenic precursor molecule gp160 was not increased significantly by sCD4, suggesting that its conformation is less affected by receptor binding.
Figure 2.
Interaction of 5-Helix with cell-surface gp41 and gp160. Cells (293T) expressing HIV-1 EnvHXB2 were incubated with biotin-tagged 5-Helix (1 μM) in the presence and absence of sCD4 (5 μg/ml) and C-peptide C37-H6 (2 μM). After cell washing and lysis, protein was precipitated on monomeric avidin beads and subsequently analyzed by SDS/PAGE and Western blotting. The primary detection antibody, Chessie 8, recognizes an epitope found in the cytoplasmic tail of gp41 (and, hence, gp160). The labeling above each lane refers to the initial culture-incubation conditions, and the numbers on the right refer to protein standard molecular masses. Note that no 5-Helix is added to lane 1 (leftmost) and that mock-transfected cells are used in lane 5.
Design and Cytotoxicity of a 5-Helix Recombinant Toxin.
The exposure of the gp41 C-peptide region on Env-expressing cells suggested that molecules recognizing this epitope might be used to direct therapeutic agents to HIV-1-infected cells. To test this idea, we designed a protein in which 5-Helix is fused to the translocation and ADP-ribosylating domains of a potent exotoxin from Pseudomonas (Fig. 5B). In this protein, denoted 5-Helix-PE, the 5-Helix moiety enables selective recognition of cells expressing Env, whereas the exotoxin domains promote membrane translocation and cell killing (21). 5-Helix-PE was purified to >95% homogeneity (Fig. 1C) and shown by surface plasmon resonance to bind to C-peptides in a manner similar to the starting 5-Helix protein (data not shown).
Figure 5.
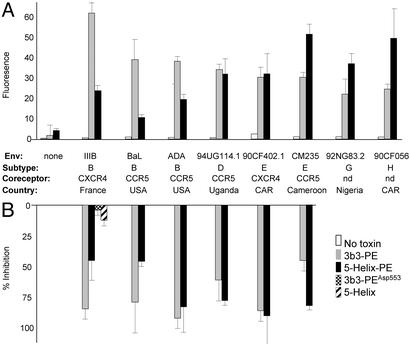
Interaction of 5-Helix-PE with diverse Env glycoproteins. HeLa effector cells expressing the indicated HIV-1 Env glycoprotein were incubated with no protein or with 12 nM 3B3-PE (27) or 5-Helix-PE for 1 h. (A) One aliquot of effector cells was incubated sequentially with a mouse anti-PE antibody and a FITC-conjugated anti-mouse antibody. The cells then were assayed for recombinant toxin binding by FACSCalibur (Becton Dickinson). Antibody binding was quantified as the median FITC fluorescence for 20,000 cellular events (cellquest software). (B) Another aliquot of effector cells was incubated for 2.5 h with target cells expressing CD4 and a coreceptor (CXCR4 or CCR5). Cell fusion was measured by assaying the activity of a β-galactosidase reporter. The data represent the percent inhibition of β-galactosidase activity relative to control wells with no added toxin. The enzymatically inactive mutant variant 3B3-PEAsp-553 and unmodified 5-Helix protein were used as controls. The results representative the mean ± SEM of two to four experiments. CAR, Central African Republic.
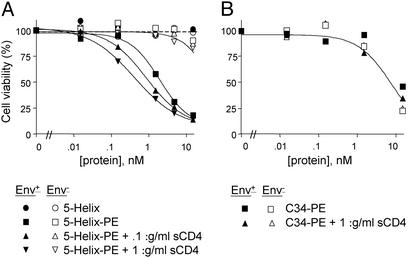
The cytotoxic activity of 5-Helix-PE was tested on CHO cells either mock-transfected or stably transfected with HIV-1 EnvIIIB. Fig. 3A shows that Env+ cells were efficiently killed by 5-Helix-PE (IC50 = 1.9 nM) whereas Env− cells were largely resistant to the recombinant toxin (IC50 > 200 nM). By contrast, neither Env− nor Env+ cells were killed by the 5-Helix protein itself, demonstrating the necessity of the bacterial toxin portion of the molecule for cytotoxicity.
Figure 3.
Cytotoxic activities of 5-Helix-PE and C34-PE. Env+ (filled symbols) and Env− (open symbols) Chinese hamster ovary cells were incubated with 5-Helix (A), 5-Helix-PE (A), or C34-PE (B) in the absence or presence of sCD4 for 48 h. Cell viability was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide oxidation procedure and normalized to 100% for no added protein. The results are representative of three independent experiments.
Consistent with the results of the precipitation studies (Fig. 2), the killing of Env+ cells by 5-Helix-PE was enhanced modestly by sCD4: The IC50 was reduced from 1.9 nM to 0.86 nM at 0.1 μg/ml sCD4 and to 0.47 nM at 1 μg/ml sCD4 (Fig. 3A). This 4-fold enhancement is specific for 5-Helix-PE because sCD4 did not have any effect on cell killing by 3B3-PE (27), an antibody-based immunotoxin that binds to the gp120 subunit of Env (data not shown).
Lack of Activity of a C-Peptide Recombinant Toxin.
In a parallel experiment, we attached PE to the C-peptide C34 (7), a potent inhibitor of HIV-1 membrane fusion. This recombinant toxin should be capable of targeting the N-terminal region of gp41 (Fig. 1A). Although surface plasmon resonance experiments showed that this molecule binds to 5-Helix protein with high affinity (data not shown), it failed to selectively kill Env+ cells either in the absence or presence of sCD4 (Fig. 3B). Instead, this protein inhibited the growth of both Env+ and Env− cells with an IC50 of ≈14 nM, more than an order of magnitude lower than the IC50 for the nonspecific killing of Env− cells by 5-Helix-PE.
Inhibition of Pseudovirion Infection by 5-Helix-PE.
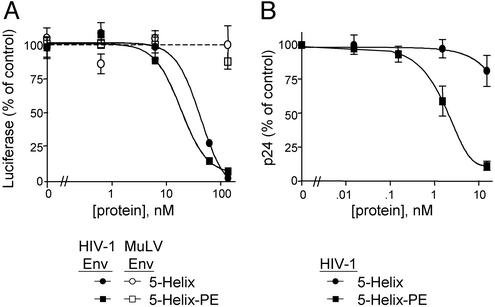
The binding of 5-Helix-PE to Env protein, implied by the observed specificity of its cytotoxic effect on Env-expressing cells, suggested that the recombinant toxin also might be an effective fusion inhibitor. To test this, we performed single-round infections with pseudovirions containing a defective env gene and a luciferase reporter construct. Cells infected with these pseudovirions do not produce any Env and, therefore, should not be killed by the recombinant toxin. Like the unmodified 5-Helix, 5-Helix-PE displayed clear inhibitory activity against viruses pseudotyped with HIV-1 EnvHXB2, but not with control virions pseudotyped with MuLV Env (Fig. 4A). The similarity of the potencies between 5-Helix and 5-Helix-PE (IC50 values of 24 and 18 nM, respectively) suggests that the added bulk of the recombinant toxin does not restrict ability of the 5-Helix moiety to access the gp41 C-peptide region during membrane fusion.
Figure 4.
Inhibition of pseudoviral and viral infection. (A) U87-CD4+-CXCR4+ cells were infected with pNL43-Luc-R−E− pseudovirions encapsidated with either HIV-1 EnvHXB2 (filled symbols) or MuLV Env (open symbols) for 48 h in the presence of various concentrations of 5-Helix or 5-Helix-PE. Luciferase activity was measured and normalized to 100% for no added protein. (B) CEM cells infected with HIV-1NL4-3 were incubated with 5-Helix or 5-Helix-PE for 5–6 days before p24 antigen levels in the viral supernatants were measured by ELISA. The data are normalized to 100% for no added protein and represent the mean ± SEM of three experiments.
Inhibition of HIV-1 Infection.
With its cytotoxic properties and antifusion activity, 5-Helix-PE has two potential mechanisms to inhibit live HIV-1 infection in cell culture. We tested the ability of 5-Helix-PE to inhibit spreading infection by titrating the recombinant toxin into CEM cell cultures in which 25–50% of the cells were infected with HIV-1NL4–3. Consistent with our previous observations, 5-Helix-PE potently inhibits viral production (as measured by the amount of viral antigen p24 in cell-free medium), displaying an IC50 value of 2 nM. By contrast, the pure entry inhibitor 5-Helix poorly blocked spreading HIV-1 infection under these conditions [IC50 value > 40 nM; although 5-Helix inhibits initial HIV-1NL4-3 infection of CEM cells with an IC50 value of 9 nM (M.J.R., unpublished results), it cannot block the production of new HIV-1 from previously infected cells]. These data indicate that the cytotoxic properties of 5-Helix-PE confer the predominant antiviral effect for this recombinant protein. They also show that the gp41 C-peptide region is exposed in sufficient quantities on the surface of HIV-1-infected cells to allow effective targeting of a therapeutic molecule.
Crossreactivity with Env from Diverse HIV-1 Strains.
Although the residues in the gp41 C-peptide region that interact with 5-Helix are highly conserved across divergent HIV-1 strains, it was not known whether these C-peptide regions would be accessible outside the context of membrane fusion. Indeed, epitope exposure on Env has been shown to be highly variable, especially between laboratory-adapted strains compared with primary isolates (5). We used flow cytometry and cell fusion assays of HeLa cells infected with vaccinia virus recombinants to test the ability of 5-Helix-PE to interact with Env from a diverse set of laboratory-adapted and primary HIV-1 strains with different geographic origins and coreceptor utilizations. Although this system cannot be used to determine absolute binding efficiencies because of variations in Env expression levels, it can be used to determine the relative binding capabilities of different proteins.
Flow-cytometry analysis showed that 5-Helix-PE is capable of binding to the surface of cells expressing Env from five different clades (B, D, E, G, and H) that use both CXCR4 and CCR5 for viral entry (Fig. 5A). Of note, six of eight Envs tested are derived from primary isolate strains. For comparison, we show similar binding studies for the well characterized, potent immunotoxin 3B3-PE, which targets the CD4-binding site of gp120 (27). Although 3B3-PE bound significantly better than 5-Helix-PE to Env from B clade viruses (the source of both the 5-Helix sequence and the antigen used to generate the 3B3 antibody), this trend was not universal. The two toxins bound about equally well to a clade D Env, and 5-Helix-PE actually was superior for the more distantly related Envs from clades E, G, and H.
We characterized the biological activity of 5-Helix-PE against six of the eight Env glycoproteins by using a cell–cell fusion assay (the Envs from clades G and H virus showed no fusion activity). This assay is designed specifically to test the cytotoxic activity of recombinant toxins, rather than their antifusion activity, as shown by the reduced inhibitory potency of unmodified 5-Helix protein and a 3B3 immunotoxin containing a point mutation that destroys its enzymatic activity (23). 5-Helix-PE potently inhibited cell–cell fusion for all of the tested constructs (Fig. 5B). Moreover, its pattern of inhibition compared with that of 3B3-PE closely matched the pattern of binding observed in the fluorescence-activated cell sorter (FACS) analysis. Again, although 3B3-PE was significantly better at inhibiting cells expressing Env from the homologous clade B virus, 5-Helix-PE displayed similar or more effective inhibition of cells expressing Env from the heterologous clade D and E viruses.
Discussion
We report that the C-terminal region of the HIV-1 gp41 ectodomain (and gp160 precursor molecule) appears to be partially exposed and vulnerable to an antiviral agent before the receptor-mediated conformational changes that initiate membrane fusion. Both precipitation and cellular cytotoxicity experiments demonstrated that the interaction of 5-Helix with gp41 was enhanced significantly by, but not strictly dependent on, CD4. While this manuscript was under review, Koshiba and Chan (29) reported qualitatively similar results by using an epitope-tagged version of 5-Helix; however, binding to gp41 and gp160 in their experiments was more strongly induced by CD4, perhaps because of differences in the assay procedures or expression conditions. These studies do not determine whether 5-Helix interacts with the native conformation of Env or, rather, some misfolded conformation of gp41 and gp160 on the cell surface. Nevertheless, the C-terminal region of the gp41 ectodomain appears to be sufficiently exposed (in some context) on the cellular surface to form a helical structure that can bind 5-Helix.
The interaction of 5-Helix with Env+ cells in the absence of sCD4 is consistent with a similar interaction observed for 2F5, a neutralizing mAb that recognizes a peptide sequence found at the C terminus of the gp41 C-peptide region (30, 31). By contrast, the interaction of agents, such as C-peptides, that target the gp41 N-terminal region appears to strictly require conformational changes initiated by the gp120/CD4 interaction (11). Indeed, our C-peptide–toxin construct C34-PE shows no preference in killing Env+ or Env− cells, even in the presence of sCD4. The global enhancement of C34-PE cytotoxicity (compared with that of 5-Helix-PE and 3B3-PE) likely stems from the propensity of C34 to form an amphipathic helix and interact nonspecifically with cell membranes (32), an effect that possibly masks any increased binding in the presence of sCD4.
As a potential therapeutic targeting moiety, 5-Helix has several useful features. First, it interacts with highly conserved residues involved in a critical conformational change necessary for HIV-1 entry. We found that 5-Helix-PE was active against a wide variety of laboratory-adapted and primary isolates, including several from Africa, the continent with the highest levels of HIV-1 infection worldwide. By contrast, most anti-Env antibodies are directed against nonessential sequences and display a narrower range of reactivity, often limited to the very isolate to which the antibodies were raised (2, 5). Even the broadly neutralizing immunotoxin 3B3-PE, which recognizes the conserved CD4-binding domain of gp120, reacts most potently with the clade B Env from which the 3B3 antibody was derived.
A second advantage of using 5-Helix as a targeting moiety is that it is likely to be highly specific for HIV-1-infected cells. 5-Helix is a stable protein that presents a complex, virally derived interaction surface designed to recognize a specific viral epitope exposed on the cell surface. By contrast, C-peptides, with their propensity for nonspecific interactions and the inaccessibility of their binding sites, are unlikely to be good targeting moieties. Further, CD4, which also has been used as a targeting moiety for anti-HIV-1 recombinant toxins, is a normal cellular receptor that binds to other cellular components, including the widely expressed MHC molecules. It also has many basic residues, resulting in positively charged surface patches that may bind nonspecifically to uninfected cells. These features may account for the unacceptable level of nonspecific toxicity observed in the unsuccessful clinical trails of CD4-PE (33, 34).
Our findings could be extended in several useful ways. To overcome potential problems because of immunogenicity, 5-Helix-PE could be modified chemically with polyethylene glycol, or the PE portion could be replaced by a smaller and less immunogenic cytotoxic agent such as a membrane-destabilizing peptide, methotrexate, or radioisotope. Recombinant toxins might be designed with 5-Helix-like targeting moieties based on the trimer-of-hairpins structures of other enveloped viruses such as Ebola, influenza, and respiratory syncytial virus (10). Even for HIV-1, 5-Helix-PE could, in principle, be altered to recognize any Env glycoprotein of known sequence with high affinity and specificity. It might even be possible to construct a recombinant toxin specific for the latent virus present in a particular infected individual. Such a molecule, administered together with agents that induce the expression of dormant integrated provirus (e.g., cytokines or activators of protein kinase C), might help to reduce or possibly eliminate latent reservoirs of HIV-1 (35). Preliminary ex vivo experiments show that 5-Helix-PE indeed can block the deoxyphorbol ester-mediated induction of latent HIV-1 replication in CD8-depleted peripheral blood mononuclear cells from an HIV-1-infected individual receiving antiretroviral therapy.
Finally, the ability of 5-Helix-PE to inhibit viral entry as effectively as 5-Helix alone suggests that the gp41 C-peptide region is accessible to proteins of substantial size. In fact, at 65 kDa, 5-Helix-PE is actually bigger than an antibody Fab fragment. Our results lend further evidence that C-peptides constrained in their helical conformation might be useful in the development of an HIV-1 vaccine.
Acknowledgments
We thank Dr. Peter Kim, in whose lab at Massachusetts Institute of Technology the pull-down experiments were begun; Stella Hu and Louise McHugh for technical assistance; and Peter Kwong, Claude Klee, Charles Brenner, and Michael Kay for critically reviewing the manuscript. This work was supported partially by National Institutes of Health Grant R01 GM66682 (to M.J.R.).
Abbreviations
- HIV-1
HIV type 1
- Env
envelope
- sCD4
soluble CD4
- PE
Pseudomonas exotoxin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Moore J P, Stevenson M. Nat Rev Mol Cell Biol. 2000;1:40–49. doi: 10.1038/35036060. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt R, Sodroski J. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 3.Freed E O, Martin M A. In: Field's Virology. Howley P M, editor. Philadelphia: Lippincott; 2001. pp. 1971–2041. [Google Scholar]
- 4.Biscone M, Pierson T, Doms R. Curr Opin Pharmacol. 2002;2:529. doi: 10.1016/s1471-4892(02)00200-x. [DOI] [PubMed] [Google Scholar]
- 5.Burton D R. Proc Natl Acad Sci USA. 1997;94:10018–10023. doi: 10.1073/pnas.94.19.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanna S L, Yang C, Owen S M, Lal R B. AIDS. 2002;16:1603–1608. doi: 10.1097/00002030-200208160-00005. [DOI] [PubMed] [Google Scholar]
- 7.Chan D C, Chutkowski C T, Kim P S. Proc Natl Acad Sci USA. 1998;95:15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan K, Liu J, Wang J, Shen S, Lu M. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 10.Eckert D M, Kim P S. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 11.Furuta R A, Wild C T, Weng Y, Weiss C D. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 12.Melikyan G B, Markosyan R M, Hemmati H, Delmedico M K, Lambert D M, Cohen F S. J Cell Biol. 2000;151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang S, Lin K, Strick N, Neurath A R. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 15.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, et al. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 16.Root M J, Kay M S, Kim P S. Science. 2001;291:884–888. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- 17.Eckert D M, Kim P S. Proc Natl Acad Sci USA. 2001;98:11187–11192. doi: 10.1073/pnas.201392898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis J M, Bewley C A, Clore G M. J Biol Chem. 2001;276:29485–29489. doi: 10.1074/jbc.C100317200. [DOI] [PubMed] [Google Scholar]
- 19.Rowell J F, Stanhope P E, Siliciano R F. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 20.Ohno H, Aguilar R C, Fournier M C, Hennecke S, Cosson P, Bonifacino J S. Virology. 1997;238:305–315. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- 21.Reiter Y, Pastan I. Clin Cancer Res. 1996;2:245–252. [PubMed] [Google Scholar]
- 22.Newton D L, Nicholls P J, Rybak S M, Youle R J. J Biol Chem. 1994;269:26739–26745. [PubMed] [Google Scholar]
- 23.McHugh L, Hu S, Lee B K, Santora K, Kennedy P E, Berger E A, Pastan I, Hamer D H. J Biol Chem. 2002;277:34383–34390. doi: 10.1074/jbc.M205456200. [DOI] [PubMed] [Google Scholar]
- 24.Pitts T W, Bohanon M J, Leach M F, McQuade T J, Marschke C K, Merritt J A, Wierenga W, Nicholas J A. AIDS Res Hum Retroviruses. 1991;7:741–750. doi: 10.1089/aid.1991.7.741. [DOI] [PubMed] [Google Scholar]
- 25.Connor R I, Chen B K, Choe S, Landau N R. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 26.Weng Y, Weiss C D. J Virol. 1998;72:9676–9682. doi: 10.1128/jvi.72.12.9676-9682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bera T K, Kennedy P E, Berger E A, Barbas C F, III, Pastan I. Mol Med. 1998;4:384–391. [PMC free article] [PubMed] [Google Scholar]
- 28.Ashorn P, Berger E A, Moss B. Methods Enzymol. 1993;221:12–18. doi: 10.1016/0076-6879(93)21004-r. [DOI] [PubMed] [Google Scholar]
- 29.Koshiba T, Chan D C. J Biol Chem. 2003;278:7573–7579. doi: 10.1074/jbc.M211154200. [DOI] [PubMed] [Google Scholar]
- 30.Jiang S, Lin K, Lu M. J Virol. 1998;72:10213–10217. doi: 10.1128/jvi.72.12.10213-10217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sattentau Q J, Zolla-Pazner S, Poignard P. Virology. 1995;206:713–717. doi: 10.1016/s0042-6822(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 32.Kliger Y, Peisajovich S G, Blumenthal R, Shai Y. J Mol Biol. 2000;301:905–914. doi: 10.1006/jmbi.2000.4004. [DOI] [PubMed] [Google Scholar]
- 33.Ramachandran R V, Katzenstein D A, Wood R, Batts D H, Merigan T C. J Infect Dis. 1994;170:1009–1013. doi: 10.1093/infdis/170.4.1009. [DOI] [PubMed] [Google Scholar]
- 34.Davey R T, Jr, Boenning C M, Herpin B R, Batts D H, Metcalf J A, Wathen L, Cox S R, Polis M A, Kovacs J A, Falloon J, et al. J Infect Dis. 1994;170:1180–1188. doi: 10.1093/infdis/170.5.1180. [DOI] [PubMed] [Google Scholar]
- 35.Berger E A, Moss B, Pastan I. Proc Natl Acad Sci USA. 1998;95:11511–11513. doi: 10.1073/pnas.95.20.11511. [DOI] [PMC free article] [PubMed] [Google Scholar]