Abstract
Generation of biological function by chemical methods is potentially of great importance for the understanding and targeting of physiological processes. Chemical synthesis of proteins offers the ability to alter the properties of target protein molecules in a tailor-made fashion. In the present work it is demonstrated that this methodology can be expanded to the elucidation of protein–protein interactions as exemplified by the complete chemical synthesis of the protooncogene product H-Ras as well as of the Ras-binding domain (RBD) of its effector c-Raf1. The 166-aa polypeptide chain of H-Ras was synthesized by native chemical ligation of three unprotected peptide segments. Similarly, the 81-aa RBD was prepared by ligation of two peptide segments. Both RBD and Ras displayed functional and spectroscopic properties indistinguishable from their recombinant forms as judged by CD spectroscopy and from transient kinetic measurements of the Ras–RBD interaction as well as from nucleotide replacement reactions in Ras. An unnatural amino acid bearing a nitrobenzofurazan side chain was introduced into position 91 of the RBD, providing unique fluorescence properties. The association transient of nitrobenzofurazan labeled with Ras⋅guanosine 5′-β,γ-imidotriphosphate showed a slow phase that had not been detected in earlier work by using other signals.
Eukaryotic signaling cascades are made up of intricate networks of interacting proteins. Generally, the external stimulus, which for example can be a hormone, a cytokine, or light, activates transmembrane receptors that then turn on cytoplasmic signaling molecules, resulting, in many cases, in signal transduction to the nucleus and diverse cellular responses such as cell growth, differentiation, and action (1).
A central requirement for the normal functioning of these signal transduction pathways is the numerous protein–protein interactions, which are the essence of the signal relay. Members of a well studied example of a signaling cascade are the GTPases of the Ras (rat sarcoma) superfamily, of which the Ras protein (p21ras) plays a particularly important role (2). A signal coming from either a receptor tyrosine kinase or a G protein-coupled receptor stimulates nucleotide dissociation from the inactive Ras⋅GDP, which allows subsequent generation of Ras⋅GTP. In the latter state, Ras activates downstream effector proteins (3). As long as Ras stays in its GTP-bound form, it can recruit one of its effector proteins, such as c-Raf1, to the plasma membrane, thereby activating the C-terminal threonine-serine-specific protein kinase domain of this protein. Subsequently, the downstream mitogen-activated protein kinase pathway is turned on, which finally leads to the activation of genes in the nucleus (4). Ras⋅GTP is deactivated by a low intrinsic GTPase activity or, more efficiently, by binding to GTPase-activating proteins, which increase the hydrolysis rate at saturation by up to approximately five orders of magnitude (5).
The crucial steps in these signal transfer cascades rely on the specific properties of protein–protein contacts. A paradigm of such an interacting protein pair is Ras and c-Raf1, which has been studied kinetically as well as structurally in great detail (6–8). The effector c-Raf1, originally identified in humans, is 648 aa long and contains a domain [residues 51–131, named Ras-binding domain (RBD)] specific for Ras⋅GTP binding (7) but not for Ras⋅GDP binding (6, 9, 10). The importance of this interaction is exemplified by the fact that ≈30% of all human solid cancers have mutations in Ras that accelerate GDP release or slow down GTP hydrolysis, thereby continuously activating the downstream kinases (3, 11). This central role of the Ras family and its signaling partners for cellular processes and in tumors has made these proteins prime targets not only for mechanistic studies but also for medicinal purposes, in particular for the development of anticancer drugs (12).
An in-depth analysis of the functional properties of Ras would not only help to understand the principles of protein–protein signal transfer and the intrinsic mechanism of GTP hydrolysis and GDP/GTP exchange but would also facilitate the design of potent inhibitors that could be applied in cancer treatment. These aims can be approached by chemical means, e.g., by the complete chemical synthesis of Ras and RBD, because a chemically synthesized protein–protein pair enables versatile preparation of analogs with tailor-made properties for probing the molecular basis of function. In this work the total chemical synthesis of Ras as well as RBD is described. The purified and renatured proteins are functionally indistinguishable from their counterparts prepared by heterologous expression.
Materials and Methods
Materials.
Tert-butyloxycarbonyl (Boc)-protected amino acids were purchased from Peptides International. 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) was obtained from Spectrum (Gardena, CA). The Boc-aminoacyl-OCH2-phenylacetamidomethyl resins and N,N-diisopropylethylamine were from Applied Biosystems. Trifluoroacetic acid was from Halocarbon Products (Hackensack, NJ). Hydrogen fluoride was purchased from Matheson. Nα-Boc-l-diamino-propionic acid was from Bachem. All other chemicals were obtained from Fluka at the highest purity available.
Synthesis of N2-Boc-N3-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-l-2,3-diamino-propionic Acid (Dap).
4-Chloro-7-nitrobenz-2-oxa-1,3-diazol was coupled with N2-Boc-Dap in ethanol/water at 1:1 in the presence of 1.2 eq of triethylamine for 6 h at room temperature. The resulting mixture was purified by flash-silica gel chromatography by using dichloromethane/methanol at 9:1 (13).
Peptide Synthesis.
Solid-phase peptide synthesis (14) was performed manually or on a custom-modified 430A peptide synthesizer from Applied Biosystems by using Boc chemistry, in situ neutralization, and HBTU-activation protocols (15). The unnatural fluorescent amino acid Nα-Boc-l-Nβ-nitrobenzofurazan (NBD)-2,3-diamino-propionic acid was coupled manually by using the above-mentioned neutralization and activation protocol. RBD was synthesized from two unprotected peptide segments, RBD-(51–95)αCOSR + RBD-(Cys-96–131), and Ras was synthesized from three segments, Ras-(1–50)αCOSR, Ras-[Cys(Acm)51–117]αCOSR, and Ras-(Cys-118–166). Both N-terminal peptide segments and the middle segment Ras-(51–117) were synthesized on a resin that generates a C-terminal thioester after HF cleavage (16). The thiol group of the N-terminal cysteine residue of Ras-(51–117) was protected by the acetamidomethyl (Acm) group, which can be removed by treatment with 1.5 eq of Hg(OAc)2 per thiol group in a buffer containing 2 M urea at pH 3.5–4 (17). The peptides were deprotected and simultaneously cleaved from the resin by using anhydrous HF. Subsequently, they were precipitated with ether, and the crude products were dissolved in 50% aqueous acetonitrile (0.1% trifluoroacetic acid) and lyophilized. The peptide segments were purified by RP-HPLC on a C4 column from Vydac (Hesperia, CA) by using linear gradients of solution B (acetonitrile with 0.08% trifluoroacetic acid) in solution A (water with 0.1% trifluoroacetic acid). The elution of the peptide was monitored at 214 nm. Fractions were analyzed by electrospray mass spectrometry with a Perkin–Elmer/Sciex API-I or API-III quadrupole mass spectrometer, and the product-containing fractions were combined and lyophilized.
Native Chemical Ligation.
Ligation of the peptide segments {10 mg/ml for RBD [RBD-(51–95)–RBD-(96–131)] and 2.5 mg/ml for Ras [Ras-(51–117)–Ras-(118–166)]} was carried out in 6 M guanidinium hydrochloride/300 mM phosphate, pH 7/1% thiophenol for 14–48 h and produced the corresponding full-length polypeptide chains (18, 19). The ligation of Ras-(1–50) with Ras-(51–166) (Acm group removed from the N-terminal cysteine residue) was carried out in the presence of 1% dodecyl-β-d-maltoside for 48 h (yield: 70%). Purification and analysis was carried out as described above. The Ras protein was obtained in a purity of >95%; the electrospray mass spectrum showed a mass of 18,836 Da (calculated: 18,854 Da), indicating that an unexpected dehydration event had occurred in this last step of the chemical synthesis.
The experimental strategy for the synthesis of RBD from two peptides and introduction of the NBD label in position 91 has been described (13). The RP-HPLC chromatogram of RBD/L91NBD-Dap revealed a purity of >96%, and the corresponding electrospray mass spectrum shows a mass of 9,324 Da (calculated: 9,323 Da; data not shown). The polypeptide was obtained with a recovered yield of ≈30% based on the amount of peptide segments used (≈20 mg).
Protein Folding.
The purified polypeptide chain of RBD was folded by dissolving 1 mg of protein in 1 ml of buffer 1 (100 mM NaCl/50 mM Tris⋅HCl/5 mM MgCl2/5 mM DTT, pH 7.4). After the reaction mixture had been stirred gently overnight at 4°C, the solution was shock-frozen in liquid nitrogen and stored at −80°C. For complete folding of Ras, which was carried out in the presence of a 20× molar excess of the nonhydrolyzable GTP analog guanosine 5′-β,γ-imidotriphosphate (GppNHp), the protein was first dissolved in a buffer containing 5 M guanidine hydrochloride, 100 mM NaCl, 50 mM Tris, 5 mM MgCl2, and 5 mM DTT at pH 8, and GppNHp was added (20, 21). This solution was diluted 100-fold with the same buffer lacking guanidine and dialyzed against 600 volumes of buffer 1 for 24 h at 4°C. Buffer 1 was exchanged against fresh buffer two times further. For CD spectroscopic measurements, the samples were dialyzed against 20 mM phosphate buffer at pH 7.4. CD spectra were recorded on a Jasco J-710 spectropolarimeter at 25°C in a quartz cell with a 0.2-cm path length and a final protein concentration of 60 μM.
Stopped-Flow Measurements.
Stopped-flow measurements of the Ras–RBD interaction were carried out as described by Sydor et al. (7, 22). Briefly, reagents were mixed rapidly in a Hi-Tech (Salisbury, U.K.) stopped-flow apparatus, and fluorescence intensity was monitored in the millisecond to second time range. RBD/L91NBD-Dap and methylanthraniloyl (mant)-GDP were excited with light at 436 and 366 nm, respectively, and the fluorescence emission was monitored through filters with cutoff wavelengths of 530 and 408 nm, respectively. The experiment, designed to monitor the displacement of GppNHp from its complex with Ras by mantGDP, was carried out in the presence of 2.5 mM EDTA in buffer 1 without MgCl2.
Results and Discussion
Synthesis of Ras-(1–166).
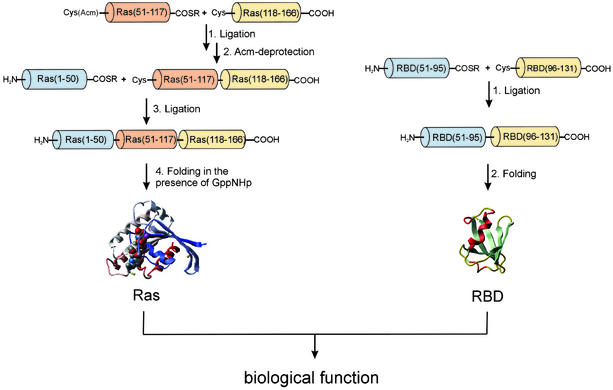
The strategy for the total chemical synthesis of Ras-(1–166) and RBD is outlined in Scheme S1. Because of the length of the polypeptide chain, it was necessary to assemble Ras from more than two fragments. Ras contains three cysteine residues at positions 51, 80, and 118 that could serve as possible ligation sites. The corresponding peptides that would have to be synthesized would be of convenient length (50, 30, 38, and 48 aa, respectively). However, the necessary purification of additional intermediate ligation products would reduce the yields considerably. Therefore, a strategy with only three segments (1–50, 51–117, and 118–166) was pursued, although this approach required synthesis of a peptide of considerable length [Ras-(51–117), i.e., 67 aa].
Scheme 1.
Strategy for the total chemical synthesis of Ras-(1–166) and RBD proteins (numbering for RBD was taken from the sequence of full-length c-Raf1).
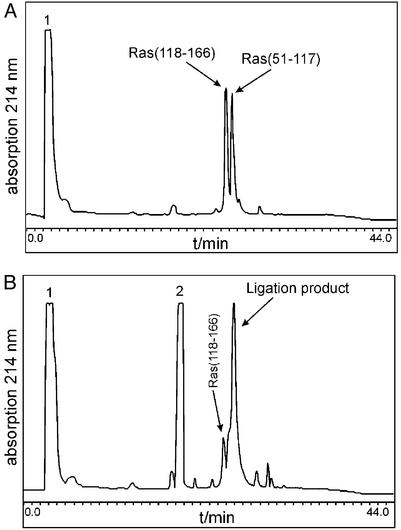
All peptide segments were prepared by stepwise solid-phase synthesis by using the in situ neutralization/HBTU-activation protocols and Boc chemistry (14). The two segments Ras-(1–50) and Ras-(51–117) were synthesized on a resin designed to generate peptide α-thioesters after HF cleavage (16). The C-terminal segment of Ras, comprising amino acids 118–166, was synthesized on a standard –OCH2–Pam resin. The second segment [Ras-(51–117)] was S-Acm-protected at its N-terminal cysteine residue to prevent cyclization (23). The first native chemical ligation of Ras-(51–117) and Ras-(118–166) was carried out at the Lys-117–Cys-118 site in 6 M guanidine hydrochloride buffer at pH 7.0. The ligation reaction was complete within 20 h (see Fig. 1), and the resulting 116-residue polypeptide was purified by RP-HPLC. The thiol-protecting Acm group on the N-terminal cysteine residue was then removed by treatment with Hg(OAc)2. An electrospray mass spectrum of the fully deprotected polypeptide Ras-(51–166) gave a mass of 13,343 ± 1.6 Da (calculated: 13,342 Da).
Figure 1.
HPLC analytical control of the ligation of Ras-(51–117) with Ras-(118–166). (A) Reaction mixture at t = 0. (B) Reaction mixture after 20 h at ambient temperature. Peak 1, guanidine; peak 2, thiophenol; detection wavelength, 214 nm.
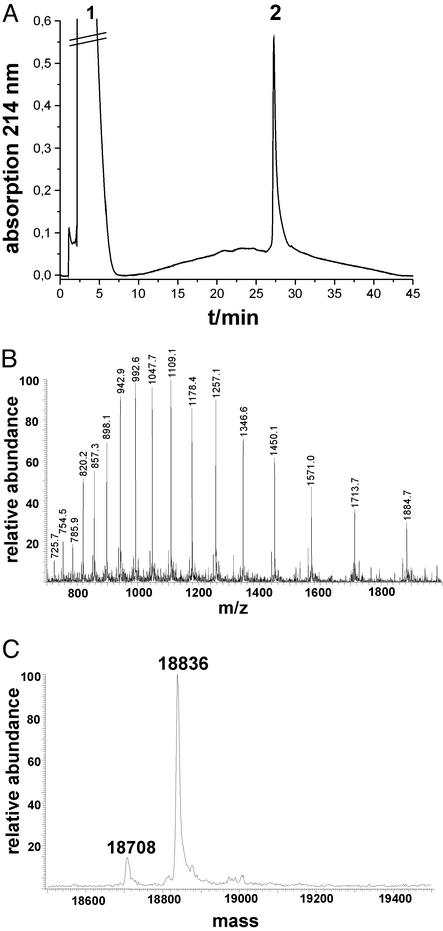
The ligation of Ras-(1–50) and Ras-(51–166) was carried out initially under the same conditions as described above. However, precipitation of peptides and incomplete coupling necessitated a modification of the experimental protocol, which included the addition of 1% dodecyl-β-maltoside to keep the polypeptides in solution and to drive the reaction to completion. Under these conditions a reaction yield of 70% was obtained after 48 h. At this point the ligation reaction was stopped, and the 166-residue polypeptide was purified by RP-HPLC. The elution profile of purified Ras-(1–166) is shown in Fig. 2 and reveals the high purity of the sample. However, this product had a mass of 18,836 Da as determined by electrospray mass spectrometry. This mass differs from the calculated mass (18,854 Da) of Ras-(1–166) by −18 ± 2 Da, suggesting that an unexpected dehydration or deamination event has occurred in the synthesis. This loss of water or ammonia must have occurred during the last ligation, because the starting peptide segments used at that point had masses identical to their calculated ones, or during the subsequent HPLC purification with acidic buffers. The site and nature of this side reaction, which seems to have no functional consequences (see below), may be the subject of further investigation.
Figure 2.
Characterization of purified Ras-(1–166). (A) HPLC analysis of purified Ras-(1–166) on an analytical C4 RP-HPLC column. (B) Electrospray ionization mass spectrometry of Ras-(1–166). (C) Deconvoluted mass spectrometry of Ras-(1–166).
Based on the amount of the longest peptide segment used in the synthesis, Ras-(1–166) was obtained with a recovered yield of ≈5%. The yield from a typical purification was ≈3 mg.
Folding and Secondary Structure.
Whereas the lyophilized RBD/L91NBD-Dap polypeptide chain could be renatured quite easily in an aqueous buffer (100 mM NaCl/50 mM Tris⋅HCl/5 mM MgCl2/5 mM DTT, pH 7.4), the linear polypeptide Ras-(1–166) was not soluble under these conditions, and Ras activity could not be detected in the supernatant. However, after addition of an excess of nucleotide (GppNHp, 20× molar excess) to the suspension of Ras-(1–166) in buffer and stirring gently at 4°C for 14 h, almost half of the polypeptide dissolved. This solution was used for further experiments. The remaining insoluble Ras was dissolved in a buffer containing 5 M guanidine hydrochloride and an excess of GppNHp. This solution was diluted and dialyzed to give fully soluble Ras⋅GppNHp in guanidine-free buffer (ref. 24; see Materials and Methods). Thus, the nucleotide GppNHp seems to function as a scaffold for folding of the synthesized Ras to form a soluble protein (25).
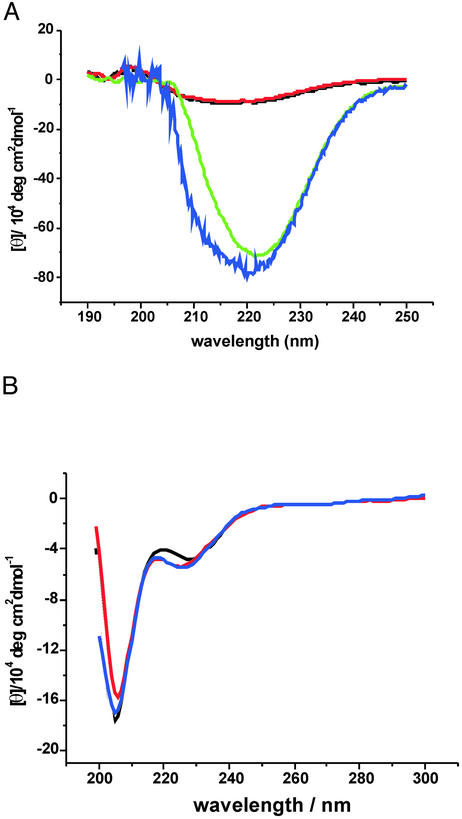
The CD spectra of chemically synthesized RBD and Ras in comparison with those of the recombinant proteins are shown in Fig. 3. The folded, chemically synthesized Ras protein shows the typical strong absorption with a maximum at 223 nm, which is indicative of α-helix/β-sheet proteins and is in good agreement with the data obtained for recombinant Ras (Fig. 3A). No folded Ras can be detected in the absence of GppNHp, confirming the result of the renaturation experiment (Fig. 3A). The CD spectra of the RBD proteins, chemically synthesized and produced by recombinant means, show a weak absorption at 225 nm, which corresponds to an nπ* transition of the peptide bond and a strong absorption band at 205 nm, typical for α-helical structures (Fig. 3B). Comparing these data with previously published spectra shows that they are almost indistinguishable, indicating that the proteins prepared by solely chemical means and by recombinant expression adopt a similar tertiary fold (9, 22). However, it does not yet imply that native functional properties are attained.
Figure 3.
CD spectra of the chemically synthesized proteins (ch) compared with the corresponding recombinant proteins (rec). (A) CD spectra of Ras-(1–166). Black, buffer; red, Ras(ch) without GppNHp; green, Ras⋅GppNHp(ch); blue, Ras(rec). (B) CD spectra of RBD-(51–131). Black, RBD(ch); blue, RBD(rec); red, RBD/L91NBD-Dap.
Characterization of the Ligand-Binding Properties of Chemically Synthesized Ras-(1–166).
Ras releases its bound nucleotide very slowly, making a measurement of the nucleotide dissociation rate constant technically difficult. However, the rate can be increased dramatically by removal of bound Mg2+ ions from the Ras⋅GTP complex. Thus, the rate of displacement of GppNHp from chemically synthesized Ras-(1–166) by mantGDP was measured in the absence of Mg2+ ions and in the presence of EDTA. As shown in Fig. 4A, by using the large increase in fluorescence intensity of mantGDP on binding to Ras, the kinetics of dissociation of GppNHp from its complex with synthetic Ras-(1–166) or its recombinant equivalent were investigated in displacement reactions. As Fig. 4A shows, essentially identical results were obtained, suggesting that an intact nucleotide-binding site is formed on folding the chemically synthesized protein. The fluorescent transients obtained were not strictly exponential because of the fact that a relatively low concentration of displacing agent was used, so that the effective rate of association of mantGDP to free Ras was not significantly faster than the dissociation rate of GppNHp. This makes the experiment especially sensitive, because the form of the curve obtained depends on all of the rate constants describing the system (i.e., dissociation and association rate constants of GppNHp and mantGDP). Thus, the high degree of similarity between the results with synthetic and recombinant proteins is a strong indicator of the integrity of the nucleotide-binding site in the synthetic protein. It also argues that the dehydration that occurred during the final ligation step does not influence the functional properties of the chemically synthesized Ras.
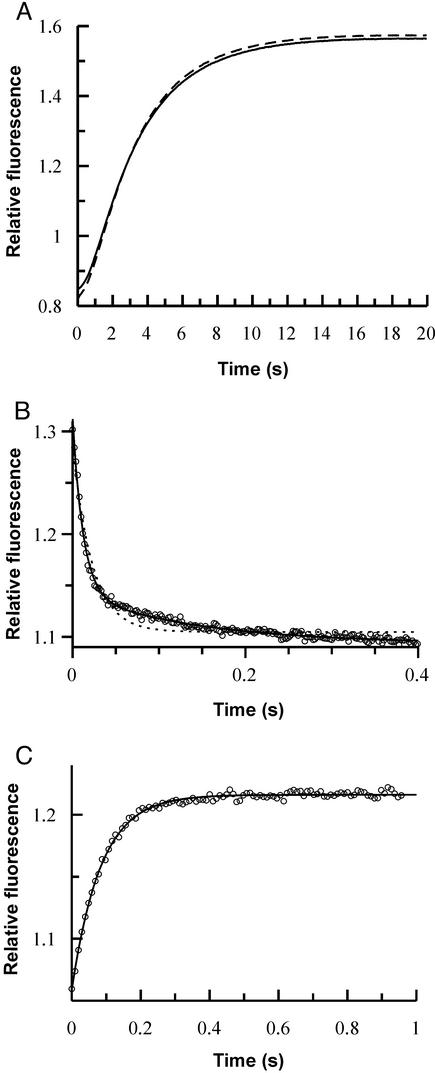
Figure 4.
(A) Fluorescence transient observed on displacement of GppNHp from Ras(reconstituted)⋅GppNHp (dashed line) or Ras(chemically synthesized)⋅GppNHp (solid line) by rapid mixing with mantGDP (1.5 μM) in a stopped-flow apparatus. The buffer used contained no Mg2+, and traces of metal ions were complexed by using 2.5 mM EDTA. Association and dissociation kinetics for the interaction of Ras and RBD were monitored by using the stopped-flow method and NBD fluorescence. (B) Biphasic association reaction of RBD/L91NBD-Dap (0.5 μM) and Ras⋅GppNHp(chemically synthesized) (4 μM). Fits are shown to monoexponential (dashed line) and biexponential (closed line) functions. Pseudo first-order rate constant for the first, concentration-dependent phase, 83 s−1; rate constant for the second phase, 6.9 s−1. (C) Displacement of RBD/L91NBD-Dap from Ras⋅GppNHp⋅RBD/L91NBD-Dap (1 μM) by RBD(reconstituted) (8 μM). Fitting to a monoexponential function (closed line) leads to a dissociation rate constant of 11 s−1.
Interaction of Chemically Synthesized Ras-(1–166) and RBD.
We have already reported that chemically synthesized and folded RBD can interact with Ras in an identical manner to recombinant RBD (13). In the present work we examined the interaction of chemically synthesized Ras-(1–166) with NBD-labeled RBD (RBD/L91NBD-Dap). Surprisingly, the association reaction, which led to a drop in NBD fluorescence, showed a biphasic time course (Fig. 4B), with a concentration-dependent first phase followed by a concentration-independent second phase. This is in contrast to the monophasic behavior seen on association of recombinant Ras⋅mantGppNHp with either recombinant or chemically synthesized RBD protein that does not contain the unnatural amino acid (NBD; Dap) (7, 22). Repeating the experiment shown in Fig. 4B with recombinant Ras showed identical biphasic behavior, demonstrating again that recombinant and chemically synthesized Ras show identical kinetic parameters for their interaction with partner molecules.
The biphasic kinetics seen on interaction of RBD/L91NBD-Dap with Ras by using NBD as the reporter group suggest a more complex binding mechanism than that deduced in our earlier work. To exclude the possibility of an artifactual result arising from two populations of the RBD/L91NBD-Dap, we performed an additional experiment making use of fluorescence resonance energy transfer between the mant group of mantGppNHp at the active site of recombinant Ras and the NBD group on association. This showed simple monophasic kinetics, suggesting a homogenous population of RBD/L91NBD-Dap (data not shown).
These results imply that, when excited directly, the NBD group provides a signal for the association reaction followed by a further signal reflecting a change in the local environment of the reporter group. This must be a step with a relatively small equilibrium constant (i.e., not very much larger than unity in the forward direction); otherwise its presence would have been detected indirectly in earlier work. Further detailed investigations are needed to clarify this point, which promises more detailed information on the mechanism of the binding reaction. For the present, the results serve to show that the ability to insert probe amino acids in a planned and controlled fashion can reveal mechanistic aspects that are not necessarily revealed by approaches that rely more heavily on the exploitation of naturally available sites of labeling.
Dissociation behavior was more straightforward, as shown in the displacement reaction of Fig. 4C. RBD/L91NBD-Dap dissociated from the Ras–RBD complex with a rate constant that was only slightly higher (11 s−1) than that determined previously for recombinant RBD and recombinant Ras (7 s−1) (7).
To estimate the affinity of the interaction between Ras and RBD/L91NBD-Dap, we used the first concentration-dependent phase of the association reaction (Fig. 4B) to calculate the association rate constant together with the dissociation rate constant obtained from the displacement reaction (Fig. 4C). This leads to KD values of 480 and 610 nM, respectively, for chemically synthesized and recombinant Ras. These values are somewhat higher than those for recombinant or chemically synthesized unmodified RBD, but they show that recombinant and chemically synthesized Ras behave similarly toward RBD (Table 1). A comparison with recombinant RBD/L91W/H, used in earlier studies (7), shows that the affinity of the interaction with Ras is in the same range as described above (KD, 380 nM; Table 1). This demonstrates that the slightly higher KD values for modified RBD proteins are due to the introduction of amino acids larger than the naturally occurring Leu at position 91.
Table 1.
Kinetic data of Ras–RBD interactions
| Reaction | kdiss/s−1 | kass/M−1⋅s−1 | KD/nM |
|---|---|---|---|
| 1. wtRBD + wtRas⋅mGppNHp | 7.4 | 4.5 × 107 | 160 |
| 2. RBD(ch) + wtRas⋅mGppNHp | 5.9 | 3.5 × 107 | 170 |
| 3. RBD/L91NBD + wtRas⋅GppNHp | 11 | 1.8 × 107 | 610 |
| 4. RBD/L91NBD + Ras⋅GppNHp(ch) | 11 | 2.3 × 107 | 480 |
| 5. wtRBD/L91W/H + wtRas⋅GppNHp* | 14 | 3.6 × 107 | 380 |
Reactions 1 and 2, kinetics were monitored by using the mant fluorescence of mGppNHp. Reactions 3 and 4, kinetics monitored by using the NBD-Dap fluorescence. Reaction 5, kinetics monitored by using the tryptophan fluorescence from ref. 7. wt, wild type; H, His tag.
Conclusions
The results described in this work demonstrate several important points that are of significance for strategies to develop proteins or protein domains with tailor-made properties. As has been shown in previous work, small functional proteins of moderate size can be prepared by chemical means and converted into a folded state with native properties. Obviously, the cellular machinery is not needed for the generation of functional properties. This is in keeping with the classic understanding that the primary sequence of the polypeptide chain determines the folding and structure of a protein and thus its properties. However, for some proteins, as exemplified by Ras in this work, folding does not occur without the assistance of its natural ligand. This might also be of relevance for the preparation of properly folded recombinant proteins, if, for example, the natural ligand is not present in the expression system used. Finally, the total chemical synthesis of an ≈27-kDa system made up of a pair of interacting proteins offers opportunities for the analysis of such important biological principles as signal transduction, hormone-receptor activation, or transmembrane signaling, as well as having potentially important medical applications.
Acknowledgments
This work is dedicated to R. B. Merrifield.
Abbreviations
- RBD
Ras-binding domain
- Boc
tert-butyloxycarbonyl
- HBTU
2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- NBD
nitrobenzofurazan (7-nitrobenz-2-oxa-1,3-diazole)
- Dap
l-2,3-diamino-propionic acid
- Acm
acetamidomethyl
- GppNHp
guanosine 5′-β,γ-imidotriphosphate
- mant
methylanthraniloyl
References
- 1.Wilson K F, Cerione R A. Biol Chem. 2000;381:357–365. doi: 10.1515/BC.2000.048. [DOI] [PubMed] [Google Scholar]
- 2.Boguski M S, McCormick F. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 3.Ahmadian M R, Zor T, Vogt D, Kabsch W, Selinger Z, Wittinghofer A, Scheffzek K. Proc Natl Acad Sci USA. 1999;96:7065–7070. doi: 10.1073/pnas.96.12.7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgering B M, Bos J L. Trends Biochem Sci. 1995;20:18–22. doi: 10.1016/s0968-0004(00)88944-6. [DOI] [PubMed] [Google Scholar]
- 5.Scheffzek K, Lautwein A, Kabsch W, Ahmadian M R, Wittinghofer A. Nature. 1996;384:591–596. doi: 10.1038/384591a0. [DOI] [PubMed] [Google Scholar]
- 6.Herrmann C, Martin G A, Wittinghofer A. J Biol Chem. 1995;270:2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- 7.Sydor J R, Engelhard M, Wittinghofer A, Goody R S, Herrmann C. Biochemistry. 1998;37:14292–14299. doi: 10.1021/bi980764f. [DOI] [PubMed] [Google Scholar]
- 8.Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. Nature. 1995;375:554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- 9.Scheffler J E, Waugh D S, Bekesi E, Kiefer S E, LoSardo J E, Neri A, Prinzo K M, Tsao K L, Wegrzynski B, Emerson S D, et al. J Biol Chem. 1994;269:22340–22346. [PubMed] [Google Scholar]
- 10.Chuang E, Barnard D, Hettich L, Zhang X F, Avruch J, Marshall M S. Mol Cell Biol. 1994;14:5318–5325. doi: 10.1128/mcb.14.8.5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbacid M. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 12.Wittinghofer A, Waldmann H. Angew Chem. 2000;39:4193–4214. doi: 10.1002/1521-3773(20001201)39:23<4192::AID-ANIE4192>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 13.Becker C F W, Hunter C L, Seidel R P, Kent S B H, Goody R S, Engelhard M. Chem Biol. 2001;8:243–252. doi: 10.1016/s1074-5521(01)00003-5. [DOI] [PubMed] [Google Scholar]
- 14.Merrifield R B. Angew Chem Int Ed Engl. 1985;97:799–810. [Google Scholar]
- 15.Schnölzer M, Alewood P, Jones A, Alewood D, Kent S B H. Int J Pept Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 16.Hojo H, Kwon Y, Kakuta Y, Tsuda S, Tanaka I, Hikichi K, Aimoto S. Bull Chem Soc Jpn. 1993;66:2700–2706. [Google Scholar]
- 17.Canne L E, Botti P, Simon R J, Chen Y J, Dennis E A, Kent S B H. J Am Chem Soc. 1999;121:8720–8727. [Google Scholar]
- 18.Dawson P E, Muir T W, Clark-Lewis I, Kent S B H. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 19.Dawson P E, Churchill M J, Ghadiri M R, Kent S B H. J Am Chem Soc. 1997;119:4325–4329. [Google Scholar]
- 20.DeLoskey R J, Van Dyk D E, Van Aken T E, Campbell-Burk S. Arch Biochem Biophys. 1994;311:72–78. doi: 10.1006/abbi.1994.1210. [DOI] [PubMed] [Google Scholar]
- 21.Campbell-Burk S L, Carpenter J W. Methods Enzymol. 1995;255:3–13. doi: 10.1016/s0076-6879(95)55003-8. [DOI] [PubMed] [Google Scholar]
- 22.Sydor J R, Herrmann C, Kent S B H, Goody R S, Engelhard M. Proc Natl Acad Sci USA. 1999;96:7865–7870. doi: 10.1073/pnas.96.14.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Tam J P. J Am Chem Soc. 1997;119:2363–2370. [Google Scholar]
- 24.Netzer W J, Hartl F U. Nature. 1997;388:343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Matthews C R. Biochemistry. 1998;37:14891–14899. doi: 10.1021/bi981116z. [DOI] [PubMed] [Google Scholar]