Abstract
The paucity of integral membrane protein structures creates a major bioinformatics gap, whose origin is the difficulty of crystallizing these detergent-solubilized proteins. The problem is particularly formidable for hetero-oligomeric integral membrane proteins, where crystallization is impeded by the heterogeneity and instability of the protein subunits and the small lateral pressure imposed by the detergent micelle envelope that surrounds the hydrophobic domain. In studies of the hetero (eight subunit)-dimeric 220,000 molecular weight cytochrome b6f complex, derived from the thermophilic cyanobacterium, Mastigocladus laminosus, crystals of the complex in an intact state could not be obtained from highly purified delipidated complex despite exhaustive screening. Crystals of proteolyzed complex could be obtained that grew very slowly and diffracted poorly. Addition to the purified lipid-depleted complex of a small amount of synthetic nonnative lipid, dioleolyl-phosphatidylcholine, resulted in a dramatic improvement in crystallization efficiency. Large crystals of the intact complex grew overnight, whose diffraction parameters are as follows: 94% complete at 3.40 Å spacing; Rmerge = 8.8% (38.5%), space group, P6122; and unit cell parameters, a = b = 156.3 Å, c = 364.0 Å, α = β = 90o, γ = 120o. It is proposed that the methodology of augmentation of a well-defined lipid-depleted integral membrane protein complex with synthetic nonnative lipid, which can provide conformational stability to the protein complex, may be of general use in the crystallization of integral membrane proteins.
Presently, the Protein Data Bank contains ≈70 structures of integral membrane proteins (IMP), of which fewer than half have an independent fold. Thus the total number of solved IMP is currently about equal to the number of soluble protein structures deposited per week. Crystallization of IMP in a complex with detergent requires that preparations be pure and stable and that detergent(s) compatible with crystal formation be identified (1). Extensive purification of a membrane protein can lead to lipid depletion, which may be destructive to IMP (2, 3). Thus, the problem of protein purification may not center on obtaining the highest possible purity, but rather an optimum purity that does not cause depletion of the native lipid content. Two roles of IMP-bound lipids have been discussed: (i) a functional role that was inferred from structure studies on cytochrome (cyt) oxidase (4), bacteriorhodopsin (bR; ref. 5), the yeast cyt bc1 complex (6), the Escherichia coli outer membrane protein OmpX (7), and the bacterial photosynthetic reaction center (8, 9); (ii) a structural role has been proposed for the specific lipids digalactosyl-diacylglycerol and phosphatidylglycerol in crystallization of the plant light-harvesting chlorophyll–protein complex (10), and for cardiolipin bound to the bacterial photosynthetic reaction center where it forms a partial peripheral shell around the hydrophobic protein core, acting as boundary lipid at the protein–detergent interface (9). In the presence of detergent and absence of lipid, the much smaller lateral pressure exerted by the detergent micelle may result in greater conformational freedom of the side chains and backbone of the protein (11–13). Together with increased H2O accessibility and resulting lability of the IMP (12), crystallization may be frustrated. This “detergent problem” is represented in Fig. 1 a and b by the ordered arrangement of the transmembrane helices of an oligomeric IMP such as the cyt b6f complex in a lipid bilayer membrane (Fig. 1a), compared with their disordered arrangement in a detergent micelle, which is depicted by a splayed arrangement of the transmembrane α-helices (Fig. 1b). The spatial- and time-dependent disorder of the detergent micelle (2, 14), described by the heterogeneity of the micelle shown in Fig. 1b, is also an impediment to crystallization. From crystallization and diffraction studies of a hetero-oligomeric integral membrane protein, discussed below, it is proposed that addition of a small amount of pure lipid to the protein–detergent complex can stabilize the protein and the micelle.
Figure 1.
Schematic of cyt b6f complex in a membrane bilayer (a), a detergent micelle (b), and a detergent–lipid mixture (c). The 13 transmembrane helices predicted for the complex are shown, with cyt b6 (pink) and subunit IV (yellow) having four and three transmembrane spans, respectively. In b, the greater conformational freedom of the subunits of the protein complex in the detergent micelle is depicted qualitatively by an exaggerated tilting of the α-helices. The prediction of a more ordered ensemble of subunits in the presence of a low stoichiometry of lipid (≈10 per monomer) is shown. Red, lipid head group; green, detergent head group.
The lipidic “cubic phase” method was developed as one approach to the “detergent problem” (12, 13, 15). bR, other members of the bR family, the bacterial photosynthetic reaction center, and the bacterial light-harvesting complex have been crystallized by this method (13, 16). However, except for bR, the diffraction quality of the crystals formed by this method, which may not yet have been thoroughly tested with a wide range of proteins, is lower than that obtained by the conventional detergent method. The robust bR also forms well-diffracting crystals in lipid bicelles (17). The cubic phase method has not yet been shown to be applicable to the more fragile hetero-oligomeric IMP, and such tests with the recently developed lipid bicelle method have not yet been reported.
The hetero-oligomeric cyt b6f complex (plastoquinol:plastocyanin oxidoreductase) is the subject of the crystallization studies reported in the present work. The complex is an obligate dimer (18, 19) that contains three hemes per monomer [two noncovalently linked to the cyt b polypeptide (20), and one covalently to cyt f], and two non-heme Fe in the Rieske iron-sulfur protein. The complex functions in oxygenic photosynthetic membranes to transfer electrons from the photosystem II reaction center to photosystem I, and to translocate H+ in this process, thus generating a transmembrane proton electrochemical gradient that supports ATP synthesis (21). The b6f complex also mediates transmembrane activation of the light-harvesting complex protein kinase (22). The b6f complex is an example of a hetero-oligomeric IMP whose successful crystallization was not successful until the development of the methodology described herein.
Methods
Cyanobacterial Source of the Cytochrome b6f Complex.
The thermophilic cyanobacterium, Mastigocladus laminosus, was grown in 10-liter carboys at 55°C to late log phase and harvested, and thylakoid membranes were prepared by passage through a French pressure cell at 18,000 lb/in2.
The Cytochrome b6f Complex.
The complex was extracted in 30 mM Tris⋅HCl (pH 7.5), 5 mM MgCl2, 5 mM KCl, 50 mM NaCl, 1 mM EDTA (TMKNE), containing 0.2% sodium cholate and 28 mM n-octyl-β-d-glucoside at a chlorophyll concentration of 2 mg/ml.
The membrane suspension was stirred at room temperature for 20 min and centrifuged at 300,000 × g for 40 min. The supernatant was collected, and solid ammonium sulfate was added to 35% saturation. The precipitate was removed by centrifugation at 160,000 × g for 30 min, and the supernatant was loaded on a propyl-agarose column (1.5 × 10 cm) equilibrated with 35% saturated ammonium sulfate in TMKNE containing 0.05% n-undecyl-β-d-maltopyranoside (UDM). The column was washed with equilibration solution, and the cyt b6f complex was eluted with 10% saturated ammonium sulfate in TMKNE with 0.05% UDM. Fractions containing cyt b6f complex were pooled, concentrated in a Centriprep 10, loaded on a sucrose gradient (8–35%) in TMKNE, 0.05% UDM, and centrifuged at 35,000 rpm (16 h) in an SW-41 rotor (Beckman). The brown band in the middle of the gradient was collected. After purification, it was stored on ice in 50 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA. 0.05% β-d-undecyl-maltoside (Anatrace, Maumee, OH). Synthetic dioleolyl-phosphatidylcholine (Avanti Polar Lipids) was added to the complex at a final concentration of 0.1% (wt/vol) by diluting a 20% (wt/vol) stock solution in 50 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA. 0.05% β-d-undecyl-maltoside, 20% DMSO.
Crystallization.
Crystals of the intact complex were obtained by vapor diffusion at 18°C. Two microliters of concentrated protein (20 mg/ml) in 50 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 0.05% β-d-undecyl-maltoside, and 0.1% dioleolyl-phosphatidylcholine were mixed with an equal volume of reservoir solution containing 100 mM Tris⋅HCl (pH 7.5), 100 mM MgCl2, and 30% polyethylene glycol (PEG) 400. Hexagonal crystals with space group P6122 grew overnight to 0.2 × 0.15 × 0.1 mm, and reached a size of 0.4 × 0.25 × 0.15 mm after 3–4 days. For data collection, crystals were frozen in 28% PEG 400 with 5% 2,3-butanediol.
Crystallographic Analysis.
X-ray experiments were carried out at the Advanced Photon Source (APS), Argonne National Laboratory, Argonne, IL, and the Super Photon Ring 8 GeV (SPring-8), Hyogo, Japan. Diffraction data without added lipid was collected at the APS BioCARS 14-BMC beamline, and with added lipid at APS beamline SBC-19ID and 44XU. The x-ray wavelength was 0.900 Å. Diffraction data were processed with denzo/scalepack (23) and mosflm/scala (24).
Difference Spectrophotometry.
Crystals of the cyt b6f complex were dissolved in a buffer containing 30 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, and 0.05% undecyl-maltoside. Visible spectra of the cytochromes were measured on Cary 3 UV-visible spectrophotometer (25). Ferricyanide, ascorbate, and dithionite were used as oxidant and reductant, respectively. Cytochrome f and b6 spectra were measured, respectively, as the difference spectra, ascorbate minus ferricyanide, and dithionite minus ascorbate.
Results and Discussion
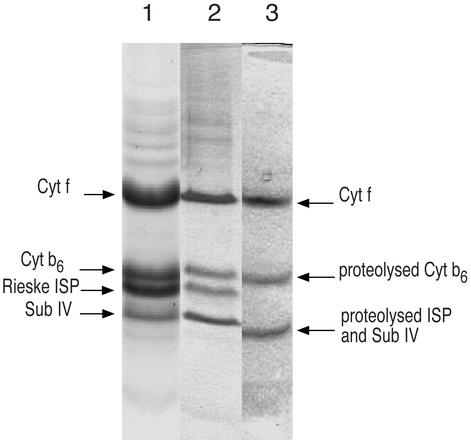
The cyt b6f complex was purified from the thermophilic cyanobacterium, M. laminosus (26, 27) in a very active form (electron transfer rate from decyl-plastoquinol to plastocyanin-ferricyanide, ≈300 electrons transferred per cyt f/s (26). The complex contains eight polypeptide subunits ranging in molecular mass from 3.3 to 32 kDa as determined by mass spectrometry (27, 28), and has a total molecular weight of 217,070 in its dimeric state. Until the development of the present methodology involving lipid addition, crystals of the cyanobacterial complex could be obtained only in a proteolyzed state, and a complete data set measured only to a spacing of 12 Å.‡ In the absence of lipid augmentation, SDS/PAGE of crystals showed significant proteolysis of three of the four large subunits (Fig. 2, lane 3). The Rieske iron-sulfur protein (ISP, petC gene product) was completely cleaved at a site (between Phe-40 and Phe-41) in the “hinge” region, leaving a 14-kDa fragment in the mother liquor. N-terminal proteolysis also occurred in subunit IV (petD gene product) between Met-22 and Gly-23, and in cyt b6 (petB) between Gly-49 and Ala-50. The use of the quinone analogue inhibitors 2,5-dibromo-6-isopropyl-3-methyl-1,4-benzoquinone, stigmatellin, or tridecyl-stigmatellin to preferentially position the ISP in a quinol-proximal conformation (30–34) did not inhibit proteolysis. Despite exhaustive screening of detergents and all definable crystallization parameters, it was not possible to obtain crystals of the b6f complex in an intact state.
Figure 2.
SDS/PAGE of purified cyt b6f complex from M. laminosus (lane 1), crystals of the intact complex obtained with lipid augmentation (lane 2), and of proteolyzed complex obtained without lipid (lane 3).
The purified complex was found to contain <0.5 molecules of monogalactosyldiacyl glycerol (MGDG) of bound lipid per monomer (data not shown). Thus, the instability of the complex might be related to depletion of bound lipid. MGDG is the major lipid (>50%) in M. laminosus (35). No other lipid was detected in the purified complex, although one equivalent each of chlorophyll a (18, 29) and β-carotene (25) is present. The substoichiometric content of lipid in the purified M. laminosus b6f complex contrasts with the five lipid molecules per monomer found in the yeast cyt bc1 complex (6), and approximately five molecules of lipid previously found in a less pure preparation of the b6f complex (26). Thus, the b6f complex was “over-purified” in the sense that it had been effectively stripped of lipid. Addition of egg phosphatidylcholine, which was used to stabilize b6f complex isolated from the green alga Chlamydomonas reinhardtii (19), did not facilitate crystallization.
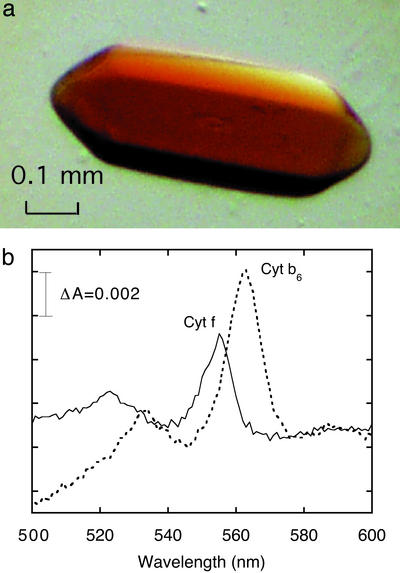
However, the consequences of addition to the purified complex of a small amount of pure homogeneous synthetic lipid were dramatic. A common commercially available lipid, dioleolyl-phosphatidylcholine (DOPC; 18:1), which contains a mono-unsaturated fatty acid chain found in M. laminosus (35), and which exists in a fluid state at room temperature, was added (0.1%, wt/vol) to the b6f complex (DOPC:cyt f ≈ 10) after purification. The addition of DOPC resulted in rapid (overnight) crystallization of intact cyt b6f complex and a great improvement in diffraction quality. SDS/PAGE of dissolved crystals showed that the four large polypeptides, cyt f, cyt b6, Rieske ISP, and subunit IV, of the complex are intact (Fig. 2, lane 2), in contrast with crystals of the lipid-depleted complex where the degraded ISP and sub IV can be seen (Fig. 2, lane 3). The color of the crystals is brownish-red (Fig. 3a), due mainly to the presence of the bound chlorophyll a molecule (18, 29), in contrast to the pink color of crystals of the cyt bc1 complex (30–33). Denaturation of the b6f complex is accompanied by loss of the noncovalently bound b-type heme and a decrease of the 2:1 ratio of heme b6:heme f. A reduced minus oxidized visible difference spectrum measured with a dissolved crystal shows the characteristic ratio of heme b6:heme f = 2:1 (Fig. 3b).
Figure 3.
(a) A crystal of the cyt b6f complex from the cyanobacterium M. laminosus. The brownish nature relative to crystals of the cyt bc1 complex (30–33) is derived from one molecule of chlorophyll a (18, 29) in the complex. (b) Difference spectra of cyt f (ascorbate minus ferricyanide) and cyt b6 (dithionite minus ascorbate) in the cyt complex obtained from crystals.
Diffraction from crystals of lipid-augmented complex, currently limited by freezing conditions, extends to 3.40 Å using synchrotron radiation from an undulator source (BL44XU, SPring-8). The space group is P6122, with unit cell dimensions of a = b = 156.3 Å, c = 364.0 Å (Table 1). A complete 3.40-Å data set has been measured (fivefold redundancy with overall Rmerge = 8.8%, 38% in the outermost shell). Crystallization conditions and crystal forms are different for the lipid-augmented and lipid-depleted preparations of the b6f complex (Table 1). It is of interest that the phosphatidylcholine head group of the lipid used for augmentation is not native to the M. laminosus cyanobacterium (35). Preliminary data indicate that the phosphatidylglycerol lipid head group, which is native to M. laminosus, can substitute for phosphatidylcholine.
Table 1.
Crystallographic parameters from diffraction of crystals of cytochrome b6f complex from M. laminosus with (A) and without (B) lipid
| Crystal data | A | B |
|---|---|---|
| Space group | P6122 | P222* |
| a, Å | 156.3 | 125 |
| b, Å | 156.3 | 143 |
| c, Å | 364.0 | 185 |
| Completeness (%) | 94.1 (79.8) | 75.6 (78.9) |
| Rmerge | 0.088 (0.385) | 0.046 (0.200) |
| I/σ | 11.2 (2.6) | 6.2 (2.2) |
| Resolution, Å | 30–3.40 | 76.7–12.0 |
| No. of measured reflections | 151,834 | 10,592 |
| No. of unique reflections | 38,824 | 906 |
Insufficient spots to distinguish systematic absences.
Structures of several oligomeric IMP have shown that IMP crystallized in detergent must be regarded as protein–detergent–lipid complexes (2). Retention of a proper lipid composition is essential for stability of the complex. One approach to the problem of optimizing the lipid:protein ratio is to retain a native or optimum lipid composition in the purified preparation. The achievement of the correct level of purity is not a simple task. Alternatively, we propose a “bare-bones” approach in which IMP are prepared to a high level of purity and stripped of lipid. The stripped preparation is then reconstituted by addition of purified synthetic lipid to the complex at a defined stoichiometry. The optimum lipid fatty acid chain is likely to depend on the acyl chain composition of the original membrane. For the cyt complex, nearly all lipids, except for the β-carotene and chlorophyll a, were initially stripped from the preparation, and reconstitution was achieved by addition of ≈10 lipid molecules per monomer unit.
The use of the nonnative lipid head-group, phosphatidylcholine, to catalyze crystallization suggests that the augmented lipid is not bound specifically within the protein complex, but more likely in the protein–detergent boundary layer, where it has an essential structural role. Such a location of one cardiolipin molecule in the bacterial photosynthetic reaction center was suggested from a structure study of the reaction center (9). We propose that the added lipid forms a partial annulus around the protein complex. If the effective diameter of the complex is 100 Å, then the stoichiometry of ten lipids added per b6f monomer corresponds to a lipid annulus whose density on the protein surface is approximately one-third of a complete monolayer on each side of the b6f complex (Fig. 1C). The function of the lipid would be (i) to increase the lateral pressure on the complex and thus to restrict the conformational freedom of the protein in the detergent micelle, as depicted in Fig. 1C, and (ii) possibly to limit the heterogeneity of the micelle population. Thus, the conceptual rationale for the improved crystallization efficiency provided by the “lipid augmentation” methodology may be somewhat similar to that of the lipidic cubic phase approach (12, 13). Application of the former methodology to the hetero-oligomeric cyt b6f complex has yielded promising results.
Acknowledgments
We thank P. Chitnis and R. Nechushtai for supplying a strain of M. laminosus, W. Minor for helpful discussions and for generously sharing beam time, R. Alkire and F. Rotella for assistance on the SBC-19 beam line, and J. T. Bolin, S. Ferguson-Miller, P. Fromme, A. Friedman, and J. Whitelegge for important discussions. These studies have been supported by National Institutes of Health Grant GM-32383 (to W.A.C.), a fellowship from the Japanese Ministry of Science and Education (to G.K.), and Department of Energy Grant W31-109-ENG-38 9 (SBC-19 beam line). Early studies on the crystallization of cyt b6f were supported by fellowships to W.A.C. from the J. S. Guggenheim Foundation and A. von Humboldt Stiftung, and hospitality from H. Michel and the Max Planck Institute for Biophysics (Frankfurt).
Abbreviations
- bR
bacteriorhodopsin
- cyt
cytochrome
- IMP
integral membrane protein
- ISP
iron-sulfur protein
Footnotes
Zhang, H., Huang, D., Sainz, G., Chaudhuri, B. N., Smith, J. L. & Cramer, W. A., Eighth International Congress on the Crystallization of Biological Macromolecules, May 14–19, 2000, Sandestin, FL, p. 210.
References
- 1.Michel H. In: Crystallization of Membrane Proteins. Michel H, editor. Boca Raton, FL: CRC; 1991. pp. 73–88. [Google Scholar]
- 2.Garavito R M, Ferguson-Miller S. J Biol Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]
- 3.Gouaux E, White S H. Curr Opin Struct Biol. 2001;11:393–396. doi: 10.1016/s0959-440x(00)00222-0. [DOI] [PubMed] [Google Scholar]
- 4.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itono K, Nakashima R, Yaono R, Yoshikawa S. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 5.Essen L-O, Siegert R, Lehmann W D, Oesterhelt D. Proc Natl Acad Sci USA. 1998;95:11673–11678. doi: 10.1073/pnas.95.20.11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lange C, Nett J H, Trumpower B L, Hunte C. EMBO J. 2001;20:6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez C, Hilty C, Wider G, Wuethrich K. Proc Natl Acad Sci USA. 2002;99:13533–13537. doi: 10.1073/pnas.212515099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones M R, Fyfe P K, Roszak A W, Isaacs N W, Cogdell R J. Biochim Biophys Acta. 2002;1565:206–214. doi: 10.1016/s0005-2736(02)00570-9. [DOI] [PubMed] [Google Scholar]
- 9.Camara-Artigas A, Brune D, Allen J P. Proc Natl Acad Sci USA. 2002;99:11055–11060. doi: 10.1073/pnas.162368399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nussberger S, Doerr K, Wang D N, Kuehlbrandt W. J Mol Biol. 1993;234:347–356. doi: 10.1006/jmbi.1993.1591. [DOI] [PubMed] [Google Scholar]
- 11.Cantor R S. Biophys J. 1999;76:2625–2639. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landau E M, Rosenbusch J P. Proc Natl Acad Sci USA. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pebay-Peyroula E, Rosenbusch J P. Curr Opin Struct Biol. 2001;11:427–432. doi: 10.1016/s0959-440x(00)00228-1. [DOI] [PubMed] [Google Scholar]
- 14.Zulauf M. In: Crystallization of Membrane Proteins. Michel H, editor. Boca Raton, FL: CRC; 1991. pp. 53–72. [Google Scholar]
- 15.Rummel G, Hardmeyer A, Widmer C, Chiu M, Nollert P, Locher K P, Pedruzzi I I. J Struct Biol. 1998;121:82–91. doi: 10.1006/jsbi.1997.3952. [DOI] [PubMed] [Google Scholar]
- 16.Luecke H, Schobert B, Richter H T, Cartailler J P, Lanyi J K. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 17.Faham S, Bowie J U. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 18.Huang D, Everly R M, Cheng R H, Heymann J B, Schägger H, Sled V, Ohnishi T, Baker T S, Cramer W A. Biochemistry. 1994;33:4401–4409. doi: 10.1021/bi00180a038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierre Y, Breyton C, Kramer D, Popot J L. J Biol Chem. 1995;270:29342–29349. doi: 10.1074/jbc.270.49.29342. [DOI] [PubMed] [Google Scholar]
- 20.Widger W R, Cramer W A, Herrmann R G, Trebst A. Proc Natl Acad Sci USA. 1984;81:674–678. doi: 10.1073/pnas.81.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cramer W A, Soriano G M, Ponomarev M, Huang D, Zhang H, Martinez S E, Smith J L. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:477–508. doi: 10.1146/annurev.arplant.47.1.477. [DOI] [PubMed] [Google Scholar]
- 22.Vener A, van Kan P J, Rich P R, Ohad I I, Andersson B. Proc Natl Acad Sci USA. 1997;94:1585–1590. doi: 10.1073/pnas.94.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project, Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 25.Zhang H, Huang D, Cramer W A. J Biol Chem. 1999;274:1581–1587. doi: 10.1074/jbc.274.3.1581. [DOI] [PubMed] [Google Scholar]
- 26.Huang D, Zhang H, Soriano G M, Dahms T E S, Krahn J M, Smith J L, Cramer W A. In: Photosynthesis: Mechanisms and Effects. Garab G, editor. Vol. 3. Dordrecht, The Netherlands: Kluwer; 1999. pp. 1577–1580. [Google Scholar]
- 27.Zhang H, Whitelegge J P, Cramer W A. J Biol Chem. 2001;276:38159–38165. doi: 10.1074/jbc.M105454200. [DOI] [PubMed] [Google Scholar]
- 28.Whitelegge J P, Zhang H, Taylor R, Cramer W A. Mol Cell Proteomics. 2002;1:816–827. doi: 10.1074/mcp.m200045-mcp200. [DOI] [PubMed] [Google Scholar]
- 29.Pierre Y, Breyton C, Lemoine Y, Robert B, Vernotte C, Popot J-L. J Biol Chem. 1997;272:21901–21908. doi: 10.1074/jbc.272.35.21901. [DOI] [PubMed] [Google Scholar]
- 30.Kim H, Xia D, Yu C A, Xia J Z, Kachurin A M, Zhang L, Yu L, Deisenhofer J. Proc Natl Acad Sci USA. 1998;95:8026–8033. doi: 10.1073/pnas.95.14.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Huang L, Shulmeister V M, Chi Y I, Kim K K, Hung L W, Crofts A R, Berry E A, Kim S H. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 32.Iwata S, Lee J W, Okada K, Lee J K, Iwata M, Rasmussen B, Link T A, Ramaswamy S, Jap B K. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 33.Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure. 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 34.Schoepp B, Brugna M, Riedel A, Nitschke W, Kramer D M. FEBS Lett. 1999;450:245–250. doi: 10.1016/s0014-5793(99)00511-6. [DOI] [PubMed] [Google Scholar]
- 35.Murata N, Wada H, Gombos Z. Plant Cell Physiol. 1992;33:933–941. [Google Scholar]