Abstract
We report the molecular identification of a sialic acid-independent host–parasite interaction in the Plasmodium falciparum malaria parasite invasion of RBCs. Two nonglycosylated exofacial regions of human band 3 in the RBC membrane were identified as a crucial host receptor binding the C-terminal processing products of merozoite surface protein 1 (MSP1). Peptides derived from the receptor region of band 3 inhibited the invasion of RBCs by P. falciparum. A major segment of the band 3 receptor (5ABC) bound to native MSP142 and blocked the interaction of native MSP142 with intact RBCs in vitro. Recombinant MSP119 (the C-terminal domain of MSP142) bound to 5ABC as well as RBCs. The binding of both native MSP142 and recombinant MSP119 was not affected by the neuraminidase treatment of RBCs, but sensitive to chymotrypsin treatment. In addition, recombinant MSP138 showed similar interactions with the band 3 receptor and RBCs, although the interaction was relatively weak. These findings suggest that the chymotrypsin-sensitive MSP1–band 3 interaction plays a role in a sialic acid-independent invasion pathway and reveal the function of MSP1 in the Plasmodium invasion of RBCs.
RBC invasion by Plasmodium falciparum is thought to proceed via two distinct pathways: sialic acid-dependent and -independent pathways (1, 2). The former pathway is primarily influenced by the interaction of the parasite ligand, EBA-175 (175-kDa erythrocyte binding antigen) with the host sialoglycoprotein, glycophorin A (GPA) (3, 4). However, a successful invasion of human En(a−) RBCs lacking GPA and MkMk RBCs lacking GPA and glycophorin B by the sialic acid-dependent Camp strain of P. falciparum, albeit with markedly reduced efficiency (5, 6), indicates that GPA alone is not sufficient as the host receptor. Further, disrupting the C terminus of EBA-175 by gene targeting switched the sialic acid-dependent W2-mef strain to a sialic acid-independent line (7). A similar disruption of EBA-175 in the sialic acid-independent Dd2/neuraminidase (Nm) clone of P. falciparum had no effect on RBC invasion (8). These lines of evidence indicate that the invasion pathway using the EBA-175 and GPA interaction is dispensable. Despite the extensive efforts made to uncover the sialic acid-independent invasion pathway, its molecular identity has remained elusive.
Previous observations have suggested a possible involvement of erythrocyte band 3 in the malaria parasite invasion of RBCs. The circumstantial evidence is as follows: A mAb binding to rhesus RBCs blocked Plasmodium knowlesi invasion and immunoprecipitated band 3 from rhesus erythrocyte ghosts solubilized in Triton X-100 (9). Human erythrocyte membrane fraction enriched in band 3 and incorporated into liposomes inhibited P. falciparum invasion of human RBCs (10). A mAb against extracellular epitopes of band 3 inhibited P. falciparum invasion of human RBCs (11). Metabolically radiolabeled P. falciparum proteins associated with band 3-enriched RBC membrane components (12). Erythroglycan, a carbohydrate component of the band 3 protein, was suggested to be involved in the invasion process (13), but evidence opposing this view has been reported (14). Proteolytic degradation of band 3 by a serine protease during RBC invasion by P. falciparum as well as the rodent malaria parasite Plasmodium chabaudi has been suggested (15, 16).
P. falciparum merozoite surface protein 1 (MSP1) is a major membrane protein attached to the merozoite surface at its C terminus through a glycosylphosphatidylinositol anchor (17). During schizogony or soon after merozoite release, full-length MSP1 is processed to give proteolytic products referred to as MSP183, MSP130, MSP138, and MSP142, which together form a noncovalent complex on the merozoite surface (18). Subsequently, MSP142 undergoes secondary processing to give MSP119 (the C-terminal domain of MSP142), which is carried into the newly invaded RBCs while other MSP1 fragments are shed off by an unknown mechanism (19). Sequence diversity analyses have identified highly conserved blocks in P. falciparum MSP183, MSP138, and MSP119 sequences (20, 21). Evidence has suggested that MSP1 is an essential malaria protein having a conserved role in the parasite invasion of RBCs in P. falciparum and P. chabaudi (22, 23). However, the molecular function of MSP1 during invasion has been unclear.
In this study, we describe the identification of a sialic acid-independent host–parasite interaction involved in the P. falciparum invasion of RBCs. We show that two nonglycosylated extracellular regions of band 3 function as a crucial host receptor. Major parasite ligands binding to the band 3 receptor are identified as MSP142 and MSP119. Our findings provide important insights into the development of a blood-stage malaria vaccine.
Materials and Methods
Synthetic Band 3 Peptides.
Human band 3 peptides were synthesized with an N-terminal biotin tag and purified to homogeneity by HPLC. Peptides were solubilized in pertinent assay buffer with minimal DMSO (≤1%) if necessary. Peptide 6C was not soluble under these conditions and could not be used in the study. The peptides and their corresponding amino acid stretches are as follows: 1, 424–435; 2, 477–491; 3A, 538–557; 3B, 551–570; 4A, 623–642; 4B, 634–653; 4C, 644–663; 5A, 720–739; 5B, 731–750; 5C, 742–761; 6A, 807–826; 6B, 823–842; and 6C, 838–857.
Binding of MSP1 and 5ABC/5BC in Solution.
Native MSP1 binding.
The P. falciparum (3D7) protein extract (200 μl) or culture supernatant (250 μl) obtained at the segmented schizont/free merozoite stage (Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org) was incubated with GST-5ABC beads (40 μl, 50% slurry) overnight at 4°C. After centrifugation, beads were washed with PBS (two times), and proteins associated with beads were analyzed by SDS/PAGE followed by Western blotting using P. falciparum MSP1 T9/94 rabbit antiserum (MRA-75, Malaria Research and Reference Reagent Resource Center, American Type Culture Collection) or mAb 5.2, a P. falciparum MSP119-specific mAb raised against the native antigen (MRA-94, Malaria Research and Reference Reagent Resource Center). GST beads were used as control.
Recombinant MSP1 binding.
Binding of 32P-labeled 5ABC and 5BC to GST-MSP142 and GST-MSP138 beads (Supporting Materials and Methods), respectively, was performed as described (24). Assay conditions were as follows: 20 mM phosphate buffer (pH 7.4)/120 mM NaCl/1 mM DTT/1.0 mg/ml BSA for 3.5 h at 25°C (280 μl final volume). Concentration-dependent binding of biotinylated thioredoxin (Trx)-MSP119 (Supporting Materials and Methods) to immobilized GST-5ABC was carried out by ELISA, using neutravidin-linked horseradish peroxidase (Pierce) and TMB as substrate. Binding conditions were as follows: 5% BSA/PBS-T (Tween 20; 0.5%) (trypsin) for 3 h at 25°C.
Blot Overlay Assay.
Naturally released P. falciparum (3D7) merozoites isolated as described (25) and human RBC ghosts prepared as described (26) were subjected to SDS/PAGE. Proteins transferred onto nitrocellulose membrane were blocked overnight with 10% milk/2% BSA/0.05% TBS-T (Tween 20; 0.05%) at 4°C. After four washes in TBS-T, the blot was incubated with biotinylated band 3 peptides (each 400 μM) in TBS-T (0.025%)/10 mM phosphate buffer (pH 8.0)/60 mM KCl for 4 h at room temperature (RT). After extensive washing, the blot was incubated with neutravidin-linked horseradish peroxidase (1:21,000; Pierce) in TBS-T (0.05%) with 2% BSA for 5–6 h at RT. After washing the blot five times with TBS-T and two times with TBS, bound peptides were visualized by the enhanced chemiluminescence method (Pierce).
Binding of MSP1 to RBCs in Suspension.
MSP138 binding.
Affinity-purified GST-MSP138 was concentrated in PBS by using a 10-kDa Amicon spin column and incubated with either Nm-treated or untreated human RBCs (7-μl packed volume) in PBS (pH 7.4) for 2 h at RT (final volume 200 μl). The mixture was passed through a bed of silicon oil (300 μl) by centrifugation. The RBC pellet was washed in 1 ml of PBS and resuspended (50–60 μl) in PBS and subjected to SDS/PAGE followed by Western blotting using a GST mAb (Amersham Pharmacia). GST was used as control.
MSP119 binding.
Enzyme-treated or untreated human (7 μl) and mouse RBCs (10 μl) were incubated with 32P-labeled MSP119 in PBS as above. On passing through silicon oil as above, radioactivity associated with the RBCs (pellet) was measured by using a β-scintillation counter. Negative controls included samples with no RBCs and no 32P-labeled MSP119 (only RBCs).
Native MSP1 binding.
Enzyme-treated or untreated intact human RBCs (25 μl) were incubated with 250 μl of P. falciparum (3D7) culture supernatant for 2 h at RT. After centrifugation, the RBCs (pellet) were washed once with PBS, lysed with 5 mM phosphate buffer (pH 8.0), and washed once with PBS again. The resulting RBC ghosts were subjected to SDS/PAGE followed by Western blotting using P. falciparum MSP1 T9/94 rabbit antiserum or mAb 5.2 (MSP119-specific).
Inhibition of native MSP1 binding.
Soluble GST-5ABC (80 μl) eluted from GSH beads by using GSH was incubated with 250 μl of the culture supernatant for 2 h at RT. Untreated human RBCs (25 μl) were then added to the reaction mixture and further incubated for 2 h at RT. Resulting RBCs were analyzed as above by Western blot. GST was used as control.
Indirect Immunofluorescence Assay.
RBCs freshly collected into citrate phosphate dextrose buffer were washed, resuspended in RPMI (20% hematocrit), and fixed on glass slides with methanol (20–30 sec) after thin smear. Fixed RBCs were washed in PBS, blocked with 10% FBS in PBS (pH 7.4) for 1.5 h at 37°C, washed again (five times, 10 min each), and incubated with 1 μM GST-MSP142, 8 μM GST-MSP138, or 9 μM GST in 10% FBS for 3.5 h at 37°C. Slide samples were washed five times in PBS, incubated with goat anti-GST antibody (1:1,000; Amersham Pharmacia), washed five times again, and incubated with rabbit anti-goat FITC-conjugated antibody (1:60; Sigma). For visualizing spectrin, RBCs blocked as above were incubated with human spectrin antibody (Sigma) followed by a FITC-conjugated secondary antibody.
Results
Band 3 Ectodomain Peptides Block Merozoite Invasion.
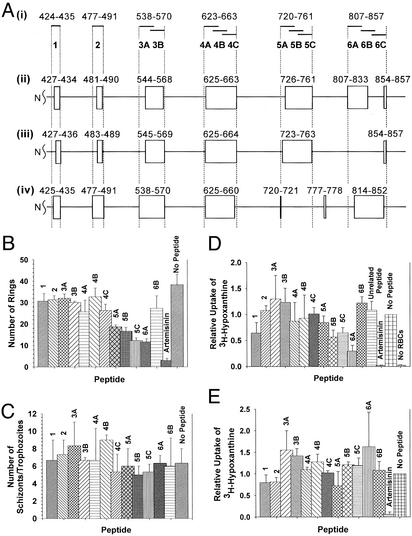
To investigate a potential role of band 3 in the P. falciparum invasion of RBCs, we prepared synthetic peptides derived from putative ectodomains of human erythrocyte band 3 based on recent topology models (refs. 27–29; Fig. 1A). These band 3 peptides were tested for their ability to inhibit the P. falciparum (3D7 strain) invasion of human RBCs in culture by using the visual counting and [3H]hypoxanthine incorporation methods (Supporting Materials and Methods). In the visual counting assay (Fig. 1B), band 3 peptides 5A (P = 0.016), 5B (P = 0.013), 5C (P = 0.006), and 6A (P = 0.006) showed significant levels of inhibition of invasion at 500 μM concentration as compared with other band 3 peptides and the control (no peptide). Inhibition by these four band 3 peptides was concentration-dependent (50-, 200-, 500-, and 1,000-μM peptides; data not shown). These peptides did not have any significant effect on retarding the growth of trophozoites and schizonts (Fig. 1C), because there was no apparent accumulation of trophozoites and schizonts as compared with the control (no peptide).
Figure 1.
Band 3 peptides inhibit the P. falciparum invasion of RBCs. (A) Human band 3 peptides. (i) Overlapping 12- to 20-aa peptides shown in solid bars were prepared according to the inclusive ectodomain boundaries based on the following three topology models: the Casey model (ii; ref. 27), the Reithmeier model (iii; ref. 28), and the Sherman model (iv; ref. 29). Boxes denote ectodomains. (B) Invasion inhibition assay by the visual counting method. The number of ring-stage parasites counted from Giemsa-stained thin smears is plotted. No Peptide, negative control. (C) Effect of peptides on intraerythrocytic parasite maturation. The total number of schizonts and trophozoites was scored from the same smears in B. (D) Invasion inhibition assay by the [3H]hypoxanthine incorporation method. Effects relative to the control sample (no peptide) are shown. Unrelated peptide was derived from RBC dematin (negative control). (E) Growth inhibition assay by the [3H]hypoxanthine incorporation method. Effects on the growth of ring-stage parasites are shown relative to the control sample (no peptide). In all assays, artemisinin (25 μM) served as a negative control for parasite development in the culture. Parasitemia was calculated as the mean of three experiments (±SE), and Student's t test was used to compare the mean.
In the [3H]hypoxanthine uptake assay (Fig. 1D) peptide 6A (P = 0.003) showed strong inhibition and peptides 5B (P = 0.031) and 5C (P = 0.023) showed moderate but significant levels of inhibition of invasion at 400 μM concentration as compared with other band 3 peptides and control (no peptide and unrelated peptide) samples. Inhibition by peptide 1 was not statistically significant (P = 0.149). IC50 values (50% inhibition of invasion) determined for peptide 5C and 6A by using the same method were 455 ± 159 μM and 279 ± 39 μM (mean ± SD), respectively (Fig. 5, which is published as supporting information on the PNAS web site). The growth inhibition study using a similar [3H]hypoxanthine uptake method (Fig. 1E) showed that none of the peptides at 400 μM had a statistically significant effect on the growth of ring-stage parasites as compared with the control sample (no peptide), although peptides 1 (P = 0.320), 2 (P = 0.140), and 5A (P = 0.448) samples had an apparent reduction in the [3H]hypoxanthine uptake. Invasion-blocking effects of peptides 5B, 5C, and 6A did not correlate with the net charge or pI of the peptides because these properties for peptides 4B, 1, and 3A, respectively, were closely similar (Table 1, which is published as supporting information on the PNAS web site). The peptides 5B, 5C, and 6A were designed by randomly dividing the putative ectodomains and, therefore, do not necessarily have the optimal composition to effect maximum inhibition. Nevertheless, our results taken together show that peptides 5B, 5C, and 6A inhibit the blood-stage development of the parasites by specifically targeting the invasion process.
Band 3 Peptides Interact with Merozoite Proteins.
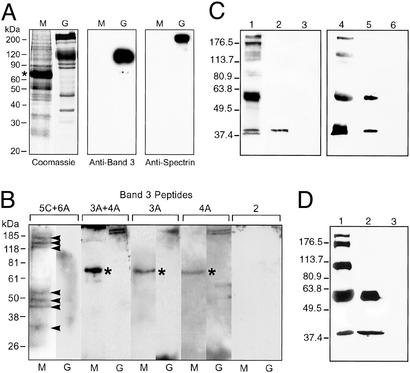
Naturally released P. falciparum merozoites (3D7) were isolated, and total merozoite proteins were separated by SDS/PAGE. The merozoite protein mixture did not have contaminating human RBC ghost proteins, as judged by the Coomassie staining of the gel and Western blotting using anti-band 3 and anti-spectrin antibodies (Fig. 2A). A 1:1 mixture of peptides 5C and 6A showed specific binding to a number of merozoite proteins in the binding assay, using a blot overlay method (Fig. 2B). These merozoite proteins had apparent molecular masses of 175, 150, 125, 52, 48, 42, and 35 kDa (shown by arrowheads). In control samples, peptides 3A, 4A, and 2 did not show significant binding to any of these merozoite proteins, although a couple of weak signals were observed in the peptide 3A + 4A sample. The peptide 5C + 6A mixture did not show specific binding to RBC ghost proteins. Our results provide evidence that human band 3, especially the regions corresponding to peptides 5C and 6A, functions as a host receptor in the P. falciparum invasion of RBCs. In fact, these findings suggest that in our earlier experiment (Fig. 1), peptides 5B, 5C, and 6A inhibited the parasite invasion of RBCs by competitively blocking one or more merozoite proteins from binding to the RBC band 3 receptor.
Figure 2.
The binding of human band 3 and native P. falciparum merozoite proteins is shown. (A) Merozoite protein separation. Total proteins in isolated merozoites (M) and human RBC ghosts (G) are shown in the Coomassie gel. Residual human serum albumin (apparent mass 67 kDa) from the culture medium is marked with an asterisk. Western blotting using anti-band 3 and anti-spectrin antibodies (Sigma) is shown. (B) Blot overlay assay. Negative control peptides 3A and 4A showed only nonspecific bindings to human serum albumin (asterisk) in merozoite samples and α/β spectrin (240/220 kDa) in ghost samples. Peptide 2 also served as negative control. (C) Western blot using P. falciparum MSP1 T9/94 antiserum. Binding of native MSP1 to 5ABC is shown. Lanes: 1, parasite lysate (PL); 2, PL + GST-5ABC beads; 3, PL + GST beads; 4, P. falciparum culture supernatant (CS); 5, CS + GST-5ABC beads; and 6, CS + GST beads. (D) Western blot using mAb 5.2. Lanes: 1, CS; 2, CS + GST-5ABC beads; and 3, CS + GST beads.
Native MSP1 Binds to Recombinant Band 3.
Among the merozoite proteins that specifically interacted with the peptide 5C + 6A mixture in the above blot overlay assay were polypeptides migrating at ≈42 and 35 kDa. We postulated that at least one of the two polypeptides could be the 42- or 38-kDa processing product of MSP1 (MSP142 and MSP138, respectively). To investigate this possibility, we prepared recombinant human band 3 peptide 5ABC (amino acids 720–761) as a GST-fusion protein and affinity-purified GST-5ABC on GSH beads (Fig. 6, which is published as supporting information on the PNAS web site). P. falciparum (3D7) proteins were extracted from the mixture of segmented schizonts and naturally released merozoites by using 0.5% Triton X-100 (Supporting Materials and Methods). The protein extract (parasite lysate) was reacted with either GST-5ABC or GST beads, and proteins associated with the beads were analyzed by Western blot (Fig. 2C) using P. falciparum MSP1 T9/94 rabbit antiserum generated against full-length MSP1. Native MSP142 bound to GST-5ABC (lane 2), but not to GST (lane 3), demonstrating a specific interaction between MSP142 and the 5ABC domain. A similar binding assay was also carried out by using the parasite culture supernatant collected at the same segmented-schizont/merozoite stage. Consistent with the above result, MSP142 bound specifically to the 5ABC domain in this assay (lane 5). In both assays, neither full-length MSP1 (≈200 kDa) nor MSP138 appeared to have interacted with 5ABC, although they were present in the parasite lysate (lane 1) and culture supernatant (lane 4). Repeating the binding assay with the parasite culture supernatant followed by a Western blot analysis using mAb 5.2 (P. falciparum MSP119-specific mAb) confirmed the binding of MSP142 to 5ABC (Fig. 2D). The 60-kDa protein binding to 5ABC (Fig. 2 C, lane 5, and D, lane 2) could be a truncated form of MSP1, because full-length MSP1 degraded into this and a few other lower-molecular-weight protein when the culture supernatant was stored for an extended period.
Characterization of MSP1–Band 3 Interaction.
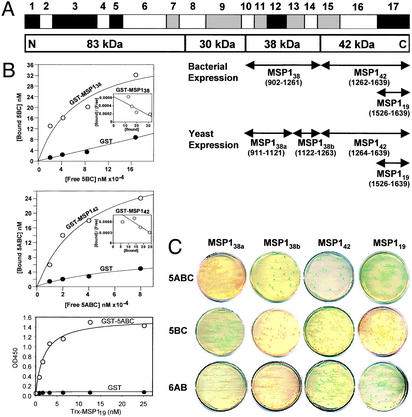
To characterize the interaction of band 3 with the processing products of MSP1, we prepared human band 3 peptide constructs 5ABC (amino acids 720–761), 5BC (amino acids 731–761), and 6AB (amino acids 807–842) and a number of P. falciparum (FCB-1 strain) MSP1 constructs shown in Fig. 3A (see Supporting Materials and Methods). GST-MSP138 expressed in bacteria was purified as a mixture of three C-terminal truncated polypeptides of 45 kDa (major) and 28–32 kDa (Fig. 7, which is published as supporting information on the PNAS web site). In the in vitro solution binding assay, 32P-labeled 5ABC and 5BC, respectively, bound to GST-MSP142 (Kd = 36 μM) and GST-MSP138 (Kd = 67 μM), whereas neither showed significant binding to GST alone (Fig. 3B). Further, MSP119 fused to the Trx domain bound strongly to GST-5ABC (Kd = 2.1 nM) in ELISA (Fig. 3B Bottom), whereas the Trx and GST control samples did not interact with GST-5ABC and Trx-MSP119, respectively. These results demonstrate that the MSP1 domains interact specifically with the band 3 peptides. The 6AB peptide expressed in bacteria was poorly soluble and could not be used in the study.
Figure 3.
The band 3–MSP1 interaction is shown. (A) P. falciparum MSP1. Conserved (black), semiconserved (gray), and polymorphic (white) regions are shown based on previous sequence diversity analyses (20, 21). Various MSP1 constructs derived from the FCB-1 strain were prepared for solution binding and yeast two-hybrid assays as illustrated. (B) Solution binding assay. 32P-labeled 5ABC (10, 20, 40, and 80 μM) and 5BC (21, 42, 84, and 168 μM) bound to GST-MSP142 (Top) and GST-MSP138 (Middle), respectively, on beads in a concentration-dependent manner. Biotinylated Trx-MSP119 (0.78, 1.56, 3.13, 6.25, 12.5, and 25.0 nM) bound to immobilized GST-5ABC in ELISA when GST alone did not (Bottom). Trx did not interact with GST-5ABC (not shown). (C) Yeast two-hybrid assay. The protein interaction was analyzed by a cotransformation method in yeast AH109 cells using SD/−Leu/−Trp selection plates. Activation of the MEL1 reporter gene on specific binding of the MSP1 domain to the band 3 peptide gave positive blue colonies when using the α-galactosidase assay.
In the in vivo yeast two-hybrid assay, band 3 peptides (5ABC, 5BC, and 6AB) and MSP1 domains (MSP138a, MSP138b, MSP142, and MSP119 as shown in Fig. 3A) were coexpressed as a fusion to the GAL4 DNA-binding domain and GAL4 activation domain, respectively (Matchmaker Two-Hybrid System 3, CLONTECH). MSP138 was divided into MSP138a and MSP138b in view of the C-terminal truncated MSP138 (45-kDa GST-fusion protein) used in our solution binding assay. Peptide 5ABC interacted with MSP142, MSP119, and MSP138b, 5BC interacted only with MSP138a, and 6AB interacted only with MSP142 and MSP119 (Fig. 3C). The expression of peptide 6AB as a soluble DNA-binding domain fusion protein in yeast was confirmed by Western blot (data not shown). Consistent with the solution binding results above, the interaction of band 3 peptides with MSP138a and MSP138b was relatively weak (Table 2, which is published as supporting information on the PNAS web site). No interaction was observed in other two-hybrid samples. Specific interactions observed between band 3 and MSP1 are remarkably similar in the solution binding and yeast two-hybrid methods. These results implicate that a key function of MSP142 and/or MSP119 is to interact with the host receptor band 3 during the merozoite invasion of RBCs. It is possible that MSP138 is a ligand binding to band 3 with relatively weak affinity.
Sialic Acid-Independent Binding of MSP1 to RBCs.
Earlier studies implicated that native P. falciparum MSP1 (full length) binds to RBCs in a sialic acid-dependent manner (30, 31). In contrast, it has been shown recently that a recombinant segment of MSP138 (p115MSP-1) binds to both wild-type and GPA-deficient En(a−) human RBCs (32). Further, short peptides derived from MSP183, MSP138, and MSP142 bound to sialic acid-depleted RBCs with relatively high affinity (33). To elucidate RBC binding properties of the MSP1 processing products, we carried out the binding assays shown in Fig. 4.
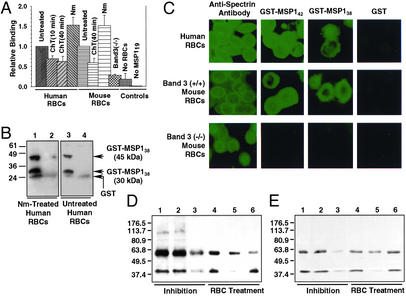
Figure 4.
The binding of P. falciparum MSP1 to RBCs is shown. (A) Relative binding of 32P-labeled MSP119 to various RBCs in suspension. Means (±SE) were plotted relative to the control (untreated wild type) and compared using Student's t test. Assays were repeated three to five times. Normalization: human RBC control, 1.0 = 9,387 counts per minute (cpm); mouse RBC control, 1.0 = 17,787 cpm. (B) Binding of MSP138 to RBCs. Anti-GST Western blotting shows that GST-MSP138 binds to Nm-treated (lane 1) and untreated (lane 3) human RBCs. GST was used as a control (lanes 2 and 4). (C) Indirect immunofluorescence assay. The binding of GST-MSP142 and GST-MSP138 is shown by using anti-GST antibody and FITC-conjugated secondary antibody. GST was used as a control. Anti-spectrin antibody staining confirmed that all RBC types were morphologically normal. (D) Western blotting using P. falciparum MSP1 T9/94 rabbit antiserum. The binding of native P. falciparum MSP1 to human RBCs is shown. Lanes: 1, culture supernatant (CS) + untreated RBCs; 2, CS + GST + RBCs; 3, CS + GST-5ABC + RBCs; 4, CS + RBCs treated with T (1.0 mg/ml); 5, CS + RBCs treated with ChT (0.5 mg/ml); and 6, CS + RBCs treated with Nm (3 milliunits/ml). (E) Western blot analysis using mAb 5.2 confirms the 42-kDa protein shown in D is MSP142. The lanes are identical to those in D.
In the first assay, human and mouse RBCs treated with either Nm or α-chymotrypsin (ChT) (Supporting Materials and Methods and Fig. 8, which are published as supporting information on the PNAS web site) were reacted with 32P-labeled MSP119 (Fig. 7, lane 10) and sedimented through a bed of silicon oil as described (32). The radioactivity associated with the RBC pellet was analyzed quantitatively. Human RBCs treated with ChT for 10 min and 40 min and Nm showed a 30% reduction (P = 0.010), a 37% reduction (P = 0.040), and a 52% increase (P = 0.054), respectively, in their ability to bind MSP119 as compared with untreated RBCs (Fig. 4A). In mouse RBC samples, ChT-treated RBCs (40 min) showed 41% reduction (P = 0.005) and Nm-treated RBCs showed 51% increase (P = 0.086) as compared with untreated RBCs. These results show that P. falciparum MSP119 binds to RBCs in a sialic acid-independent manner.
The reduced binding of MSP119 in ChT-treated RBC samples in our assays is directly correlated with the well characterized proteolysis of band 3 (34) and GPA (15) peptide backbones on the ChT-treated RBC surface (Fig. 8). Previously, it was shown that the ChT treatment of human RBCs resulted in a 36–94% reduction in RBC invasion by P. falciparum, depending on the concentration of ChT (35–37). Band 3 (−/−) mouse RBCs lacking both band 3 and GPA in the RBC membrane (38, 39) showed a 72% reduction (P = 0.277 × 10−6) in binding 32P-labeled MSP119 as compared with untreated wild-type mouse RBCs (Fig. 4A). However, because the background radioactivity in the negative control sample (no RBCs) was ≈18% of the positive control (untreated wild-type mouse RBCs), the actual radioactivity associated with the band 3 (−/−) mouse RBCs was a mere 10% above the background. Thus, band 3 (−/−) mouse RBCs completely lacking both band 3 and GPA from the plasma membrane bound MSP119 at a relatively insignificant level.
We then examined whether the binding of MSP138 to RBCs is sialic acid-independent. Wild-type and Nm-treated human RBCs were each incubated with purified GST-MSP138 (C-terminal truncated forms; Supporting Materials and Methods) and subjected to SDS/PAGE followed by Western blotting using anti-GST antibody. Truncated GST-MSP138 bound to both Nm-treated and untreated intact RBCs in suspension (Fig. 4B). GST alone did not bind to either type of RBCs. These results demonstrate that MSP138 specifically interacted with the extracellular component of human RBCs in sialic acid-independent manner. This is consistent with our finding that peptide 5BC, representing a nonglycosylated ectodomain of band 3, bound to MSP138 in solution and to MSP138a in the yeast two-hybrid system.
Band 3 Is Essential for the Binding of MSP1 to RBCs.
In the indirect immunofluorescence assay, using fixed RBCs, GST-MSP142 (1 μM final concentration) and GST-MSP138 (truncated forms, 8 μM) bound to human as well as mouse wild-type RBCs, whereas GST (9 μM) alone did not, demonstrating that the observed binding was specific to the MSP142 and MSP138 domain, respectively (Fig. 4C). It is noteworthy that an 8-fold higher concentration of GST-MSP138 as compared with GST-MSP142 was necessary to achieve a comparable level of immunofluorescence on the RBC surface by using the anti-GST antibody. This observation suggests a relatively weak interaction of MSP138 with RBCs as compared with MSP142. In the band 3 (−/−) mouse RBC sample, on the other hand, neither GST-MSP142, GST-MSP138, nor GST showed binding. Because truncated MSP138 interacted with sialic acid-depleted RBCs (Fig. 4B) and p115MSP-1 (>60 aa overlap with our 45-kDa GST-MSP138) derived from MSP138 bound to GPA-deficient human RBCs (32), our immunofluorescence assay results suggest that band 3 is involved in the binding of MSP138 to the RBC surface. In this context, peptides 5ABC and 5BC, representing a putative ectoplasmic region (amino acids 720–761) of human band 3, specifically bound to MSP138, MSP138a, and MSP138b in our binding assays (Fig. 3). In this region of band 3 there is 98% sequence identity between mouse and human proteins.
The binding of recombinant MSP142 in our immunofluorescence assay is consistent with the binding of its C-terminal domain MSP119 to the wild-type, enzyme-treated, and band 3 (−/−) mouse RBCs in suspension (Fig. 4A). To investigate the essential role of band 3 in binding MSP142 to the RBC surface, we carried out solution binding studies using native MSP142 in the P. falciparum (3D7) culture supernatant and wild-type and enzyme-treated human RBCs. Both P. falciparum MSP1 T9/94 rabbit antiserum (Fig. 4D) and mAb 5.2 (Fig. 4E) were used to analyze the binding interaction by Western blot. Native MSP142 indeed bound to untreated wild-type human RBCs in solution (Fig. 4 D and E, lane 1). Consistent with the binding assay using 5ABC beads (Fig. 2C, lanes 4–6), neither full-length MSP1 nor the MSP138 processing product showed apparent binding to human RBCs, but the 60-kDa protein (presumably, truncated MSP1) bound to RBCs. When GST-5ABC was added to this reaction mixture, the binding of MSP142 to RBCs was substantially reduced (Fig. 4 D and E, lane 3). An equivalent amount of GST added to the reaction mixture did not affect the binding of MSP142 (lane 2). These results demonstrate that the 5ABC domain specifically blocks the binding of native MSP142 to RBCs. When the binding assay was carried out using enzyme-treated human RBCs, native MSP142 bound to T-treated (Fig. 4 D and E, lane 4), Nm-treated (lane 6), and untreated (lane 1) RBCs at a comparable level. However, the binding of native MSP142 was nearly abolished in the ChT-treated RBC sample (Fig. 4 D and E, lane 5). On treatment of intact RBCs with T, we have confirmed that GPA is completely removed, but band 3 is undigested from the extracellular surface (Fig. 8), as reported (40, 41). These results demonstrate that band 3 is essential for binding native MSP142 to RBCs by a ChT-sensitive, T-insensitive, Nm-insensitive (sialic acid-independent) mechanism and that 5ABC is a crucial region of the host band 3 receptor in the P. falciparum invasion of RBCs.
Discussion
We describe a host–parasite interaction in the P. falciparum invasion of human RBCs that involves two nonglycosylated ectoplasmic regions of band 3 and processing products of MSP1. The core region of the band 3 receptor includes amino acid residues 720–761 (peptides 5A, 5B, 5C, or 5ABC) and 807–826 (peptide 6A). The native ligand interacting with the band 3 receptor has been identified as MSP142. Apparently, recombinant MSP119 and MSP138 also bound to this receptor, although MSP138 consistently showed relatively weak binding to band 3 peptides and RBCs. Recombinant MSP119 interacted with 5ABC with the Kd of 2.1 nM, which was four orders of magnitude lower than the MSP142–5ABC interaction. These results suggest that a major band 3 binding site is located within the 19-kDa domain of MSP1. It was shown earlier that disrupting the 19-kDa C-terminal region of the MSP1 gene is lethal to blood-stage P. falciparum (22), indicating that the processing products MSP142 and/or MSP119 play a critical role in RBC invasion. The binding of native MSP142 as well as recombinant MSP142, MSP119, and MSP138 to intact RBCs is directly correlated with the peptide backbone of band 3 on the RBC surface. These findings taken together demonstrate the essential role of band 3 as the host receptor mediating the sialic acid-independent interaction of P. falciparum MSP1 and RBCs. Further, our study reveals the function of MSP1 in the P. falciparum invasion of RBCs.
The finding that the band 3 receptor interacts with MSP142 and MSP138 suggests that MSP1–band 3 interactions begin before MSP1 processing products are shed from the merozoite surface. A recent study has reported that the C terminus of Plasmodium yoelii MSP138 interacts with the N terminus of P. yoelii MSP142, suggesting a binding mechanism between the two MSP1 processing products (42). On secondary processing of MSP142 and shedding of non-covalently associated MSP1 fragments, the merozoite–band 3 interaction might be retained through MSP119 anchored to the merozoite surface. It is possible that the band 3 binding site in MSP142 is located within its highly conserved C-terminal domain, MSP119 (block 17). Our recombinant MSP138a (yeast) also has a conserved region (block 12), and the C-terminal truncated MSP138 protein (bacteria) is believed to contain most of this conserved region (Fig. 3A).
Evidence from P. falciparum field isolates indicates that the parasites commonly use alternate invasion pathways that do not depend on sialic acid residues of GPA (43). The extracellular region of RBC band 3 is sensitive to ChT but insensitive to T and Nm. Whether the MSP1–band 3 interaction plays a primary role in a dominant sialic acid-independent invasion pathway across P. falciparum lines remains to be seen. Association between band 3 and GPA in the human RBC membrane has been shown (44, 45). In this regard, an RBC invasion mechanism in which the parasites use band 3 and GPA in a cooperative or complementary manner could be possible. On the other hand, it remains possible that the MSP1–band 3 interaction could be responsible for the initial attachment of merozoites to RBCs in the circulation. Presumably, the initial attachment is a critical event requiring both the selectivity of the interaction and efficiency of the process at the cellular level. Band 3 and MSP1 appear to satisfy the intricate requirements of the initial attachment process at the molecular level: they are the most abundant protein on the RBC and merozoite surface, respectively, highly conserved throughout the respective species, especially at the binding interface, and yet interact with a remarkable specificity.
Band 3 peptides 5C, 6A, and 5ABC interacted with a number of merozoite proteins in our binding assays, which leaves open the possibility that ligands other than MSP1 could also be accommodated by this crucial region of band 3, perhaps at different stages of the invasion process. A recent study has suggested that P. falciparum acidic–basic repeat antigen (ABRA) located on the merozoite surface interacts with RBCs through band 3 (46). On the other hand, multiple parasite ligands binding the band 3 receptor might exist to promote redundancy that ensures the selectivity and/or efficiency of the initial attachment process. Future studies should pursue these points of interest and may provide an ensemble of novel targets for a multivalent malaria vaccine.
Supplementary Material
Acknowledgments
We thank John B. Dame (University of Florida, Gainesville) for the P. falciparum (FCB-1 strain) cDNA library and the Malaria Research and Reference Reagent Resource Center (MR4), National Institute of Allergy and Infectious Diseases, for MSP1 antibodies (MRA-75 from Anthony A. Holder and MRA-94 from David C. Kaslow). This work was supported by National Institutes of Health Grants HL 60961 and HL 60755 and the Tufts University E. P. Charlton Award.
Abbreviations
- ChT
α-chymotrypsin
- GPA
glycophorin A
- MSP1
merozoite surface protein 1
- Nm
neuraminidase
- T
trypsin
- Trx
thioredoxin
Footnotes
References
- 1.Mitchell G H, Hadley T J, McGinniss M H, Klotz F W, Miller L H. Blood. 1986;67:1519–1521. [PubMed] [Google Scholar]
- 2.Dolan S A, Miller L H, Wellems T E. J Clin Invest. 1990;86:618–624. doi: 10.1172/JCI114753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orlandi P A, Klotz F W, Haynes J D. J Cell Biol. 1992;116:901–909. doi: 10.1083/jcb.116.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sim B K, Chitnis C E, Wasniowska K, Hadley T J, Miller L H. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 5.Miller L H, Haynes J D, McAuliffe F M, Shiroishi T, Durocher J R, McGinniss M H. J Exp Med. 1977;146:277–281. doi: 10.1084/jem.146.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadley T J, Klotz F W, Pasvol G, Haynes J D, McGinniss M H, Okubo Y, Miller L H. J Clin Invest. 1987;80:1190–1193. doi: 10.1172/JCI113178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reed M B, Caruana S R, Batchelor A H, Thompson J K, Crabb B S, Cowman A F. Proc Natl Acad Sci USA. 2000;97:7509–7514. doi: 10.1073/pnas.97.13.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaneko O, Fidock D A, Schwartz O M, Miller L H. Mol Biochem Parasitol. 2000;110:135–146. doi: 10.1016/s0166-6851(00)00263-2. [DOI] [PubMed] [Google Scholar]
- 9.Miller L H, Hudson D, Rener J, Taylor D, Hadley T J, Zilberstein D. J Clin Invest. 1983;72:1357–1364. doi: 10.1172/JCI111092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okoye V C, Bennett V. Science. 1985;227:169–171. doi: 10.1126/science.3880920. [DOI] [PubMed] [Google Scholar]
- 11.Clough B, Paulitschke M, Nash G B, Bayley P M, Anstee D J, Wilson R J, Pasvol G, Gratzer W B. Mol Biochem Parasitol. 1995;69:19–27. doi: 10.1016/0166-6851(94)00185-p. [DOI] [PubMed] [Google Scholar]
- 12.Jones G L, Edmundson H M. Biochim Biophys Acta. 1991;1097:71–76. doi: 10.1016/0925-4439(91)90026-6. [DOI] [PubMed] [Google Scholar]
- 13.Friedman M J, Fukuda M, Laine R A. Science. 1985;228:75–77. doi: 10.1126/science.3883494. [DOI] [PubMed] [Google Scholar]
- 14.Dhume S T, Adams-Burton C R, Shumak K H, Laine R A. Glycobiology. 1994;4:903–908. doi: 10.1093/glycob/4.6.903. [DOI] [PubMed] [Google Scholar]
- 15.Roggwiller E, Betoulle M E, Blisnick T, Braun Breton C. Mol Biochem Parasitol. 1996;82:13–24. doi: 10.1016/0166-6851(96)02714-4. [DOI] [PubMed] [Google Scholar]
- 16.McPherson R A, Donald D R, Sawyer W H, Tilley L. Mol Biochem Parasitol. 1993;62:233–242. doi: 10.1016/0166-6851(93)90112-b. [DOI] [PubMed] [Google Scholar]
- 17.Gerold P, Schofield L, Blackman M J, Holder A A, Schwarz R T. Mol Biochem Parasitol. 1996;75:131–143. doi: 10.1016/0166-6851(95)02518-9. [DOI] [PubMed] [Google Scholar]
- 18.Holder A A, Blackman M J, Burghaus P A, Chappel J A, Ling I T, McCallum-Deighton N, Shai S. Mem Inst Oswaldo Cruz. 1992;3:37–42. doi: 10.1590/s0074-02761992000700004. [DOI] [PubMed] [Google Scholar]
- 19.Blackman M J, Heidrich H G, Donachie S, McBride J S, Holder A A. J Exp Med. 1990;172:379–382. doi: 10.1084/jem.172.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanabe K, Mackay M, Goman M, Scaife J G. J Mol Biol. 1987;195:273–287. doi: 10.1016/0022-2836(87)90649-8. [DOI] [PubMed] [Google Scholar]
- 21.Miller L H, Roberts T, Shahabuddin M, McCutchan T F. Mol Biochem Parasitol. 1993;59:1–14. doi: 10.1016/0166-6851(93)90002-f. [DOI] [PubMed] [Google Scholar]
- 22.O'Donnell R A, Saul A, Cowman A F, Crabb B S. Nat Med. 2000;6:91–95. doi: 10.1038/71595. [DOI] [PubMed] [Google Scholar]
- 23.O'Donnell R A, de Koning-Ward T F, Burt R A, Bockarie M, Reeder J C, Cowman A F, Crabb B S. J Exp Med. 2001;193:1403–1412. doi: 10.1084/jem.193.12.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh S S, Voigt S, Fisher D, Yi S J, LeRoy P J, Derick L H, Liu S C, Chishti A H. Mol Biochem Parasitol. 2000;108:237–247. doi: 10.1016/s0166-6851(00)00227-9. [DOI] [PubMed] [Google Scholar]
- 25.Mrema J E, Langreth S G, Jost R C, Rieckmann K H, Heidrich H G. Exp Parasitol. 1982;54:285–295. doi: 10.1016/0014-4894(82)90037-6. [DOI] [PubMed] [Google Scholar]
- 26.Dodge J T, Mitchell C, Hanahan D J. Arch Biochem Biophys. 1963;100:119–130. doi: 10.1016/0003-9861(63)90042-0. [DOI] [PubMed] [Google Scholar]
- 27.Fujinaga J, Tang X B, Casey J R. J Biol Chem. 1999;274:6626–6633. doi: 10.1074/jbc.274.10.6626. [DOI] [PubMed] [Google Scholar]
- 28.Popov M, Tam L Y, Li J, Reithmeier R A. J Biol Chem. 1997;272:18325–18332. doi: 10.1074/jbc.272.29.18325. [DOI] [PubMed] [Google Scholar]
- 29.Crandall I, Sherman I W. Parasitology. 1994;108:257–267. doi: 10.1017/s0031182000076101. [DOI] [PubMed] [Google Scholar]
- 30.Perkins M E, Rocco L J. J Immunol. 1988;141:3190–3196. [PubMed] [Google Scholar]
- 31.Su S, Sanadi A R, Ifon E, Davidson E A. J Immunol. 1993;151:2309–2317. [PubMed] [Google Scholar]
- 32.Nikodem D, Davidson E. Mol Biochem Parasitol. 2000;108:79–91. doi: 10.1016/s0166-6851(00)00206-1. [DOI] [PubMed] [Google Scholar]
- 33.Urquiza M, Rodriguez L E, Suarez J E, Guzman F, Ocampo M, Curtidor H, Segura C, Trujillo E, Patarroyo M E. Parasite Immunol (Oxf) 1996;18:515–526. doi: 10.1046/j.1365-3024.1996.d01-15.x. [DOI] [PubMed] [Google Scholar]
- 34.Steck T L, Koziarz J J, Singh M K, Reddy G, Kohler H. Biochemistry. 1978;17:1216–1222. doi: 10.1021/bi00600a013. [DOI] [PubMed] [Google Scholar]
- 35.Perkins M. J Cell Biol. 1981;90:563–567. doi: 10.1083/jcb.90.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Facer C A. Trans R Soc Trop Med Hyg. 1983;77:524–530. doi: 10.1016/0035-9203(83)90130-x. [DOI] [PubMed] [Google Scholar]
- 37.Facer C A. Bull Soc Pathol Exot Ses Fil. 1983;76:463–469. [PubMed] [Google Scholar]
- 38.Southgate C D, Chishti A H, Mitchell B, Yi S J, Palek J. Nat Genet. 1996;14:227–230. doi: 10.1038/ng1096-227. [DOI] [PubMed] [Google Scholar]
- 39.Hassoun H, Hanada T, Lutchman M, Sahr K E, Palek J, Hanspal M, Chishti A H. Blood. 1998;91:2146–2151. [PubMed] [Google Scholar]
- 40.Steck T L, Fairbanks G, Wallach D F. Biochemistry. 1971;10:2617–2624. doi: 10.1021/bi00789a031. [DOI] [PubMed] [Google Scholar]
- 41.Furthmayr H. J Supramol Struct. 1978;9:79–95. doi: 10.1002/jss.400090109. [DOI] [PubMed] [Google Scholar]
- 42.Daly T M, Long C A, Bergman L W. Mol Biochem Parasitol. 2001;117:27–35. doi: 10.1016/s0166-6851(01)00329-2. [DOI] [PubMed] [Google Scholar]
- 43.Okoyeh J N, Pillai C R, Chitnis C E. Infect Immun. 1999;67:5784–5791. doi: 10.1128/iai.67.11.5784-5791.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nigg E A, Bron C, Girardet M, Cherry R J. Biochemistry. 1980;19:1887–1893. doi: 10.1021/bi00550a024. [DOI] [PubMed] [Google Scholar]
- 45.Auffray I, Marfatia S, de Jong K, Lee G, Huang C H, Paszty C, Tanner M J, Mohandas N, Chasis J A. Blood. 2001;97:2872–2878. doi: 10.1182/blood.v97.9.2872. [DOI] [PubMed] [Google Scholar]
- 46.Kushwaha A, Perween A, Mukund S, Majumdar S, Bhardwaj D, Chowdhury N R, Chauhan V S. Mol Biochem Parasitol. 2002;122:45–54. doi: 10.1016/s0166-6851(02)00077-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.