Abstract
Defining the hardwiring of transcription factors to their cognate genomic binding sites is essential for our understanding of biological processes. We used scanning chromatin immunoprecipitation to identify in vivo binding regions (E boxes) for c-Myc in three target genes as a model system. Along with other c-Myc target genes that have been validated by chromatin immunoprecipitation, we used the publicly available genomic sequences to determine whether experimentally derived in vivo binding sites might be predictable from nonexonic sequence conservation across species. Our studies revealed two classes of target genomic binding sites. Although the majority of target genes studied [class I: B23 (NPM1), CAD, CDK4, cyclin D2, ID2, LDH-A, MNT, PTMa, ODC, NM23B, nucleolin, prohibitin, SHMT1, and SHMT2] demonstrate significant sequence conservation of the E boxes and flanking regions, several genes (cyclin B1, JPO1, and PRDX3) belong to a second class (class II) that does not display sequence conservation at and around the site of c-Myc binding. On the basis of our model, we propose a strategy for predicting transcription factor binding sites using phylogenetic sequence comparisons, which will select potential class I target genes among the many emerging candidates from DNA-microarray studies for experimental validation by chromatin immunoprecipitation.
Keywords: chromatin immunoprecipitation‖DNA binding‖phylogenetic footprinting
Genomic regulatory systems, which consist of transcription factors interacting with gene regulatory modules to produce specific gene-expression levels, provide the molecular basis for the regulation of cell growth and development (1). Hence, the identification of authentic transcription factor genomic binding sites is fundamental for an understanding of biological processes including cancer development (2). The emergence of DNA-microarray technologies has greatly expanded the number of transcription factor candidate target genes that require further experimental validation. The technologies for the identification of transcription factor binding sites such as electrophoretic mobility-shift assays (EMSAs) have been performed in vitro primarily with naked DNA templates without the consideration of chromatin structure and the accessibility of transcription factors to their binding sites in vivo (2). Other approaches such as promoter-reporter transfection assays are also fraught with potential artifacts that do not reflect in situ binding of transcription factors to endogenous genes (2). The development of scanning chromatin immunoprecipitation (SChIP) to identify in vivo target binding sites along with the availability of human, rat, and mouse genomic sequences provide a unique opportunity to assess different approaches to the identification of bona fide transcription factor binding sites (3). In particular, mammalian transcription factor binding sites determined by ChIP had not been correlated with binding sites predicted from comparative genomic sequence analysis.
As a model system, we chose the oncogenic transcription factor c-Myc, which is a helix–loop–helix factor that binds DNA with its obligate partner Max (4–7). The consensus core binding sites of the Myc–Max heterodimer have been identified previously as 5′-CA(C/T)GTG-3′ or variations thereof (8). We chose three established c-Myc targets, nucleophosmin (B23 or NPM1) (3), peroxiredoxin 3 (PRDX3) (9), and JPO1 (10), to investigate c-Myc genomic binding sequences by SChIP and EMSA. B23 or nucleophosmin is a nuclear protein that participates in ribosomal biogenesis. PRDX3 is a mitochondrial peroxiredoxin that is necessary for c-Myc-mediated transformation of Rat1a fibroblasts. JPO1 is a nuclear protein with limited cellular transforming activity but without a known biochemical function.
In the studies we report here, we observed that binding in vivo as determined by SChIP correlates with in vitro DNA-binding assays. However, sequences capable of binding c-Myc in vitro are not necessarily bound in vivo, suggesting that the endogenous chromatin structure plays a role in regulating c-Myc binding. We studied the phylogenetic conservation of the c-Myc binding sequences of the three genes that we studied and 14 other genes that have been identified as c-Myc target genes and confirmed by ChIP. We used the publicly available genomic sequences to determine whether experimentally identified in vivo binding sites might be predictable from sequence conservation across species, an approach termed phylogenetic footprinting. Our studies reveal two classes of target DNA-binding sites. Class I contains a high level of nonexonic sequence conservation between organisms for transcription factor binding sequences, whereas class II sites display significant divergence of sequence between man and mouse. We propose a strategy to identify potential class I c-Myc target binding sites using phylogenetic comparisons, which will be effective for the validation of a large number of c-Myc target genes that have begun to emerge (www.myc-cancer-gene.org).
Experimental Procedures
EMSA.
The purified c-Myc and Max heterodimeric proteins were a gift from S. Nair and S. Burley (Rockefeller University, New York). The proteins are histidine-tagged truncated forms of the human c-Myc protein (amino acids 353–434) and human Max (amino acids 22–103) with the addition of amino acids GGCD at the C terminus (11). One 38-bp and one 46-bp oligonucleotide (some 36 and 44) were designed from the human genomic sequences (NT_023132 for B23, NT_008902 for PRDX3, and NT_005332 for JPO1) with the E box situated in the middle of the oligonucleotide. Fifty nanograms of double-stranded oligonucleotide was labeled with [γ-32P]ATP by using T4 polynucleotide kinase (NEB, Beverly, MA) following manufacturer protocol. Labeled oligonucleotides were purified by using the QiaQuick kit (Qiagen, Valencia, CA). Approximately 0.5 ng of probe (5 × 105 to 1 × 106 cpm) was used per reaction. Reactions were carried out in a 20-μl volume at 20°C for 20 min in 50 mM KCl/10 mM Tris, pH 8.0/5% glycerol/1 mM EDTA/1 mM DTT/50 ng of poly(dI-dC) (Amersham Pharmacia) as described (12). The entire reaction was loaded on a 6% polyacrylamide gel and run at 20°C and 30 mA for 2 h. Quantitation of gels was performed by using a PhosphorImager and IMAGEQUANT software (Molecular Dynamics), and the percentage bound was calculated from the ratio of counts in the shifted complex to the total counts in the lane. For competition experiments, 100 ng of Myc–Max were incubated with wild-type E-box probe and either cold wild-type (E box) or mutant (no E box) oligonucleotide. The sequences of one strand of the oligonucleotides used to show Myc–Max binding were E-box oligonucleotide (5′-tgccttaagtctagtatcacgtgcagatcgctacaacgac-3′) and no E-box oligonucleotide (5′-tgccttaagtctagtatgcagctcagatcgctacaacgac-3′).
ChIP.
The primary human fibroblasts 2091 (American Type Culture Collection) was used for all ChIP analyses (13). Cells (1 × 106) were plated on 15-cm dishes, incubated for 24 h, and then starved for 24 h in 0.1% serum-containing medium. After 0 or 2 h of serum stimulation, cells were exposed to formaldehyde, and chromatin was precipitated as described (14). The rabbit polyclonal anti-c-Myc sc-764 antibody (Santa Cruz Biotechnology) was used to precipitate chromatin from 2 × 107 cells. Immunoprecipitated samples were suspended in 30 μl of TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). The total input represents the supernatant from the no-antibody control and was suspended in 100 μl of TE buffer and diluted an additional 10-fold. Mock sample was treated similarly to other samples with all solutions but contained no chromatin. SChIP was performed as described (3).
PCR.
The sequences of primer pairs used for PCR are listed in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. PCR was performed with 1 μl of sample DNA or 1 ng of total input DNA, 0.5 mM each primer, 2.5 mM MgCl2, 0.4 mM each dNTP, 1× Taq buffer (Invitrogen), and 1.25 units of Platinum Taq (Invitrogen) for 35 cycles. PCR products were analyzed on a 1.2% agarose gel, and bands were visualized by staining with ethidium bromide (3). Quantitation was performed by using LABWORKS image-analysis software (Ultraviolet Products). Relative amounts of each region were calculated by normalizing the 2-h ChIP sample to the quantity in the total input. For real-time PCR, a SYBR green reagents kit was used (Applied Biosystems) (3). Known quantities of total input were used to generate a standard curve for determining nanogram equivalents for each sample. All amplifications were carried out in the linear range.
Phylogenetic Comparisons.
The sequences were downloaded from the NCBI web site. The sequences analyzed were limited to 2 kb upstream of the transcriptional start site and to the first exon, first intron, and second exon of the target gene. The transcriptional start site and intron–exon boundaries were determined from annotation of the NCBI sequences or by direct comparison of mRNA and genomic DNA. To find putative c-Myc binding sites, sequences were searched by using the nucleic acid motifs feature of OMIGA software (Oxford Molecular Limited, Oxford, U.K.). The search parameters were user-defined as the canonical or noncanonical c-Myc binding sites. To find homologous regions, sequences of human and mouse or rat orthologs were compared by using the dot-plot feature of OMIGA software. The occurrence of putative c-Myc binding sites in regions of homology were determined by examining alignments produced from the dot-plot analysis.
Results
Identification of E Boxes in B23, PRDX3, and JPO1 Sequences.
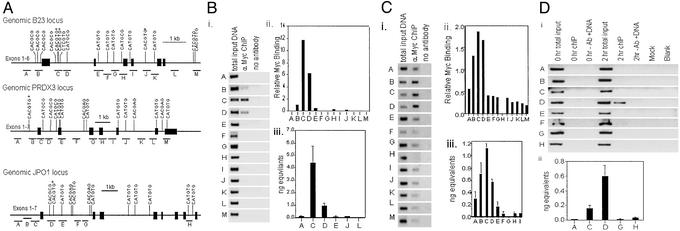
The genomic sequences for human B23, PRDX3, and JPO1 were analyzed for the presence of canonical and noncanonical E boxes. The sequence of a canonical E box is 5′-CACGTG-3′, and the sequence of the noncanonical E boxes that c-Myc binds are 5′-CATGTG-3′, 5′-CACGCG-3′, 5′-CATGCG-3′, 5′-CACGAG-3′, and 5′-CACGTTG-3′. Fig. 1A shows the position of the E boxes found in the three genes. Identification of these putative c-Myc binding sites facilitates the investigation of in vitro and in vivo binding to these sequences.
Figure 1.
(A) Location of putative c-Myc binding sites. (Top) Genomic organization of the B23 promoter and the first six exons are shown. (Middle) Genomic organization of the PRDX3 promoter and the first seven exons are shown. (Bottom) Genomic organization of JPO1 with the first eight exons shown. Exons are represented by black rectangles. The position and sequence of potential E boxes are indicated. The location of regions amplified for ChIP analysis are indicated by the lines under the gene and labeled with the letters. Drawings are to scale with 1 kb equal to the bar shown. Canonical E boxes are indicated by an asterisk. (B) Localization of c-Myc in vivo binding sites within B23 by SChIP. (i) Ethidium bromide-stained agarose gel of PCR products. PCR was carried out on total-input chromatin, chromatin precipitated with anti-c-Myc antibody, chromatin precipitated without antibody, or a water control (mock). All the chromatin samples were from cells that had been starved and then serum-stimulated for 2 h. (ii) Quantitation of the agarose gels. The relative amount of c-Myc binding was calculated by dividing the signal from the 2-h anti-c-Myc PCR products by the total input signal. (iii) Absolute quantitation of 2-h ChIP samples by using SYBR green and real-time PCR for a subset of regions as labeled. The quantity of product was calculated by using a standard curve generated with a range of total-input DNA concentrations and the same set of primers and conditions. Values represent the average of three replicates, and error bars indicate the standard deviation. (C) c-Myc in vivo binding sites within the PRDX3 locus. (i) Ethidium bromide-stained agarose gels of PCR products. PCRs are as described for B. (ii) Relative amount of c-Myc binding from agarose gels. (iii) Graph of absolute quantitation of 2-h ChIP by real-time PCR for a subset of regions as labeled. (D) c-Myc in vivo binding sites within the JPO1 locus. (i) Ethidium bromide-stained agarose gels of PCR products. (ii) Graph of absolute quantitation of 2-h ChIP by real-time PCR for a subset of regions as labeled.
SChIP Localizes Regions Bound in Vivo.
We used SChIP to evaluate the potential c-Myc binding sites of the B23, PRDX3, and JPO1 loci (3). This technique involves designing a series of primer pairs throughout a gene locus for use in ChIP experiments. We previously established the effectiveness of this technique in a system in which addition of serum to serum-starved 2091 primary human fibroblasts induces c-Myc to a high level 2 h after stimulation. c-Myc targets have also been shown to be induced after the expression of c-Myc in this system (9). Chromatin from the unstimulated cells (0-h time point) was used as a control for background binding, and chromatin obtained 2 h after stimulation was used to immunoprecipitate c-Myc target genes.
The human B23 locus was scanned previously by ChIP (3), and we now report an extended scan of the B23 locus including a region that contained a canonical E box in intron four (Fig. 1B, region J). Only two adjacent regions in intron one give strong signals for c-Myc binding in the ChIP assay (regions C and D). The peak of c-Myc binding occurs in region C, which contains two adjacent canonical E boxes, whereas the intron-four E box (region J) displayed no c-Myc binding. We also extended the scanning of PRDX3 to include four more regions between those published previously (9). A peak of c-Myc binding activity was again seen (Fig. 1C). Unlike the case for B23, the peak of Myc binding was broader and contained three regions encompassing the promoter and first intron (regions B–D). The B region contains a canonical E box, and the C and D regions contain noncanonical E boxes. None of the other regions were bound by c-Myc to significant levels in vivo. We also subjected the Myc target JPO1 to SChIP analysis and found that region D (Fig. 1D) is bound by Myc. This region contains a canonical Myc E box. Using SChIP we have localized the in vivo binding regions for c-Myc in three of its target genes.
c-Myc Binds B23 and PRDX3 E Boxes in Vitro.
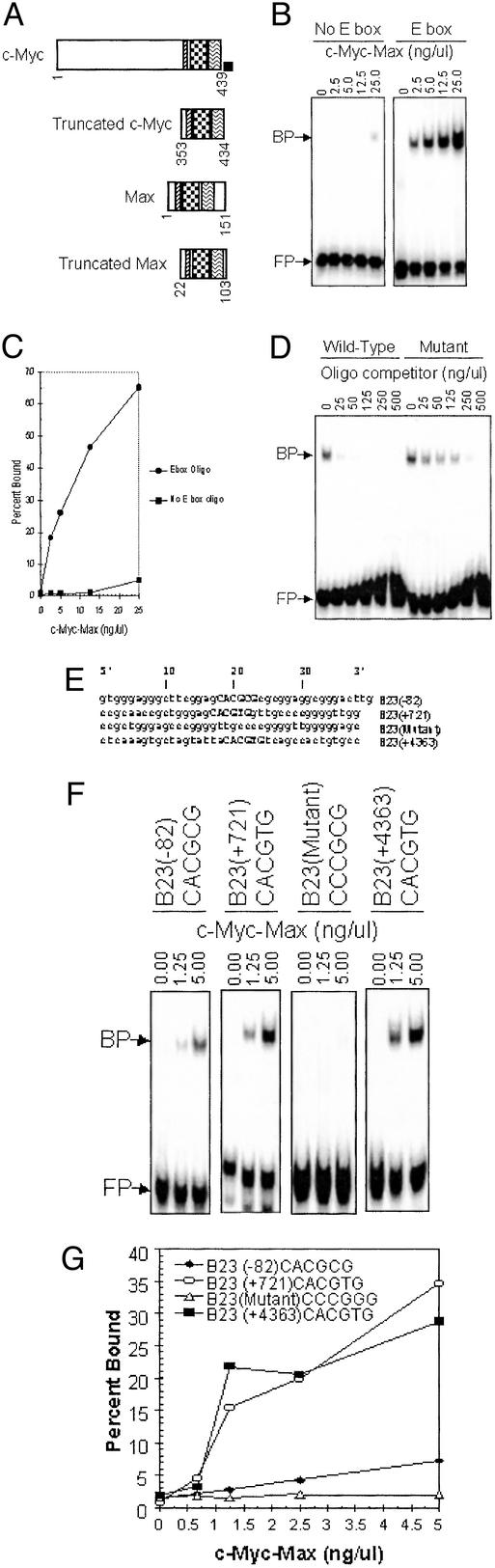
We sought to determine whether c-Myc binding in vivo as determined by SChIP correlates with its ability to bind target sequences in vitro. We designed EMSA probes with the putative E box in the middle of a 40-bp double-stranded oligonucleotide. We used purified truncated c-Myc and Max heterodimers that contain the basic helix–loop–helix leucine-zipper part of these proteins (Fig. 2A). Fig. 2B demonstrates the specificity of the EMSA, showing that an oligonucleotide with a canonical E box (5′-CACGTG-3′) is shifted by c-Myc–Max heterodimers, whereas an oligonucleotide with a mutated E box (5′-CCCGGG-3′) is not. The EMSA experiment was quantitated as the percentage of probe bound (Fig. 2C). The specificity of binding was demonstrated further by competition with unlabeled, cold oligonucleotide containing an E box that competes much better than an unlabeled oligonucleotide containing a mutated E box (Fig. 2D).
Figure 2.
c-Myc in vitro EMSA. (A) Diagram of the c-Myc and Max proteins. Full-length wild-type proteins are shown for comparison to truncated and histidine-tagged versions purified for use in the in vitro system. The numbers below each diagram indicate the amino acids present in each protein. (B) EMSA experiment with 40-bp oligonucleotides and the indicated concentrations of c-Myc–Max. FP, free probe not bound by c-Myc–Max heterodimers; BP, bound probe consisting of the complex of the probe and c-Myc–Max heterodimer. (Left) An oligonucleotide that does not contain an E box. (Right) Performed with an oligonucleotide containing a canonical E box. (C) An EMSA experiment was quantitated, and the percentage of bound probe is plotted as a function of c-Myc–Max concentrations. ●, oligonucleotide containing a canonical E box; ■, mutant oligonucleotide with no E box. (D) An EMSA competition experiment is pictured. A constant amount of c-Myc–Max and labeled probe was incubated with increasing amounts of the cold competing oligonucleotide indicated. (E) Diagram showing the sequence of the oligonucleotides used for EMSA. Capital letters indicate the location of the E boxes. The numbers associated with each oligonucleotide indicate the position of the E box relative to the B23 transcriptional start site. B23(−82) is the first E box that occurs upstream of B23. Note that in the B23(+721) oligonucleotide, the E box 5 nt downstream have been mutated such that this oligonucleotide can be used for comparison to other oligonucleotides containing one putative E box. B23(+721) is one of the canonical E boxes that occurs in intron one of the B23 locus. B23(+4363) is the first E box that occurs within intron four of the B23 gene (see Fig. 1 Top). (F) Relative binding of c-Myc–Max to the oligonucleotides indicated. (G) Graph of percentage of each oligonucleotide bound by increasing amounts of c-Myc–Max proteins. ○, B23(+721) oligonucleotide; ■, B23(+4363) oligonucleotide; ⋄, an oligonucleotide with no E boxes; ♦, data from the upstream oligonucleotide, B23(−82).
We next determined whether putative c-Myc binding sites identified on the basis of consensus sequences could be bound by c-Myc–Max specifically in EMSA. The c-Myc Max protein bound the first E box (5′-CACGCG-3′; Fig. 1A, region B of B23) upstream of the B23 promoter although weakly as compared with its binding to an oligonucleotide with a canonical (5′CACGTG-3′; Fig. 1A, region C of B23) intron-one E box (Fig. 2 E–G). In contrast to the noncanonical upstream E box, c-Myc–Max heterodimers bound the canonical E box (5′-CACGTG-3′) from intron four of B23 to high levels comparable to the canonical E box in the first intron of B23 (Fig. 2 E–G). As shown earlier, however, the intron-four E box (Fig. 1A, region J of B23) was not bound by c-Myc in vivo. This specific instance illustrates that in vitro DNA binding does not correlate with in vivo binding (Table 2, which is published as supporting information on the PNAS web site).
We further evaluated some of the putative E boxes located in the PRDX3 locus. We studied the canonical E box upstream of the transcriptional start site, the three noncanonical E boxes in the first intron, and a noncanonical E box in exon two. The upstream canonical E box (Fig. 1A, region B or PRDX3) bound with the highest affinity, whereas the first two E boxes in intron one (Fig. 1A, regions C and D of PRDX3) bound with similar but lower affinity. The third E box in intron one was bound slightly less well, and the E box in exon two (Fig. 1A, region E of PRDX3) was barely bound above background of an oligonucleotide with a mutant E box (Fig. 4 A–C, which is published as supporting information on the PNAS web site). In the case of PRDX3, in vivo binding of c-Myc corresponds to the in vitro binding activities (Table 2).
Phylogenetic Comparison of c-Myc Target Genes.
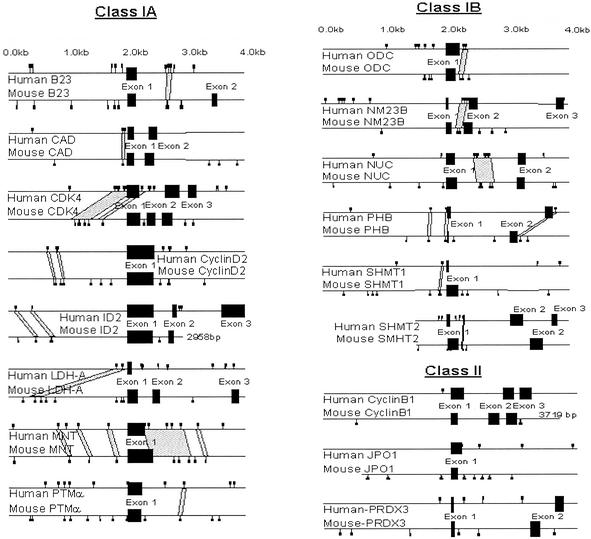
Given the limitation of in vitro DNA-binding assays for the identification of transcription factor binding sites, we used genomic sequences to determine whether experimentally identified in vivo binding sites might be predictable from phylogenetic sequence comparisons. We selected the B23, PRDX3, and JPO1 as three well characterized Myc targets and 14 additional genes that have been shown to bind c-Myc directly through ChIP assays. These other genes include the multifunctional carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase (CAD) (14), cyclin-dependent kinase 4 (CDK4) (15, 16), cyclin B1 (CCNB1) (16), cyclin D2 (CCND2) (17), inhibitor of differentiation (ID2) (18), lactate dehydrogenase A (LDH-A) (3, 19), the Max-binding protein MNT (16), NM23B (20), nucleolin (20), ornithine decarboxylase (ODC) (3, 20), prohibitin (PHB) (16), prothymosin α (PTMA) (20), cytoplasmic serine hydroxymethyltransferase (SHMT1) (21), and mitochondrial serine hydroxymethyltransferase (SHMT2) (21). We examined these genes for putative E boxes within the first 2 kb upstream of the transcriptional start site and sequences containing the first exon, first intron, and second exon. We then determined whether these human E boxes are phylogenetically conserved in the mouse and in some cases in the rat.
We identified two classes of c-Myc target genes. We found that the majority of these c-Myc targets belong to class I target genes that have E boxes that are evolutionarily conserved (Fig. 3). In class I (B23, CAD, CDK4, cyclin D2, ID2, LDH-A, MNT, PTMa, ODC, NM23B, nucleolin, prohibitin, SHMT1, and SHMT2), the E boxes occur in nonexonic regions of DNA that are identically conserved among the species for longer than 12 bp (Fig. 5, which is published as supporting information on the PNAS web site). In some cases the region of high sequence identity extends over several hundred base pairs. With the criteria of a 30-bp window and a minimum of 80% sequence identity, which is the average level of identity between human and murine exonic sequences, we identified 8 of 14 class I genes termed class IA (Fig. 3 and Fig. 5). The class IB genes contain conserved islands that are detectable with a window of 30 bp and a minimum of 65% sequence identity. In class II (cyclin B1, JPO1, and PRDX3), there is no region of homology at or flanking the genomic regions that c-Myc bound in ChIP studies.
Figure 3.
Phylogenetic comparisons of c-Myc target genes. The diagrams represent the human (top of each pair) and mouse (bottom of each pair) target genomic sequences with exons depicted by black boxes. Small squares depict locations of canonical and noncanonical Myc E boxes, and the gray areas represent regions of high identity between human and murine nonexonic sequences. Note that class II genes display no nonexonic areas of high sequence homology.
Discussion
The emergence of complete genome sequences from a variety of species provides an opportunity for sequence analysis to contribute to the identification of phylogenetically conserved, physiologically relevant transcription factor binding sites. We have chosen the c-Myc oncogenic transcription factor and its target genes to determine the extent to which evolutionarily conserved sequences could predict bona fide binding sites. Although our analysis is limited only to a small number of c-Myc target genes, specific examples illustrate that in vitro DNA binding does not necessarily predict in vivo binding of c-Myc to its binding site or E box. In the case of B23, in vitro DNA-binding assays demonstrate similar binding of c-Myc–Max to intron-one or -four E boxes, but SChIP revealed that c-Myc is only bound to intron-one E boxes in vivo. It is intriguing to note that sequence analysis reveals extensive phylogenetic conservation of the B23 intron-one tandem E boxes (Fig. 5), but the human intron-four E box is not conserved in rat or mouse. Furthermore, we found that the in vivo binding of c-Myc as determined by ChIP correlates well with the majority of c-Myc target genes that contain phylogenetically conserved E boxes and flanking sequences. It is notable that with all target genes studied here, the regions of c-Myc binding determined by ChIP are all within several hundred base pairs of the conserved E boxes. Although sequence conservation can lead to the accurate identification of authentic transcription factor binding sites, there is a fraction of target sites that would be missed through the use of sequence analysis alone. We surmised that the lack of binding sequence conservation in the class II c-Myc target genes may arise from the drift of the c-Myc binding sites through evolution.
The drift of transcription factor binding sites has been suggested to result from mutations of the relatively short nucleotide sequences that define a binding site (22). Through phylogenetic comparisons of 51 different promoter sequences, it was observed that 40% of the human transcription factor binding sites are not conserved in rodents. Both the small size of transcription factor binding sites and the degeneracy of the consensus binding sites may allow for the emergence of new binding sites through nucleotide substitutions. In particular, because many different nucleotide combinations satisfy the DNA-binding requirements of a transcription factor, new sites may emerge and relax the constraint on previously required sites. Loss of the previously essential sites in the class II genes then could be replaced by the emergence of new sites.
In contrast to the class II genes, the class I genes display conservation in flanking sequences well beyond the E boxes in nonexonic regions. These islands of highly conserved sequences are likely to contain other transcriptional regulatory sequences that, together with the E boxes, constitute specific transcriptional cis-regulatory modules or regulons. The nature of c-Myc-responsive regulons needs further studies, although in most cases that we studied the preferred c-Myc binding site is the E box (5′-CACGTG-3′). Of the 14 class I genes that we studied, 7 contain conserved regions in promoter regions, and the other 7 have conserved sequences in intron one. The occurrence of intronic conserved c-Myc binding regions is compatible with the distribution of regulatory cis elements close to promoter regions, extending well into the 5′ end of first introns (23). In particular, it is intriguing to note that the frequency of CpG islands is higher than expected at the 5′ ends of first introns, indicating that transcriptional regulatory elements extend into the first intron. The fact that c-Myc binding to 5′-CACGTG-3′ is inhibited by methylation of cytosine in the central dinucleotide CpG suggests that methylation of E boxes may regulate the responsiveness of c-Myc target genes (24).
With the development of technologies for the identification of differentially expressed genes, the number of putative c-Myc target genes are now totaling ≈550 genes (www.myc-cancer-gene.org) (25–29). On the basis of our studies and the findings of others, we propose a strategy to predict c-Myc binding sites in the large number of emerging putative c-Myc target genes. Phylogenetic comparison through available tools such as VISTA (30), PIPMAKER (31), rVISTA (32), and TRAFAC (33) or through dot-plot analysis used in our study will identify nonexonic islands of highly conserved sequences. Our use of dot-plot analysis is confirmed for several genes available through the TRAFAC server including B23 (NPM1) and LDH-A. With these computational tools, candidate c-Myc target genes, which have not been validated by ChIP assays, could be analyzed for nonexonic conserved sequences that contain canonical 5′-CACGTG-3′ E boxes. In particular, sequences 2 kb upstream and downstream of putative transcriptional start sites would be subject to phylogenetic comparisons. Candidate binding sites then could be verified experimentally by SChIP analysis as reported previously. With the availability of the Myc target gene database (www.myc-cancer-gene.org), which prioritizes candidate target genes according to the level of experimental evidence, we selected 12 putative target genes that have not been validated by ChIP. Of these, phylogenetic comparisons reveal four target genes with phylogenetically conserved c-Myc binding sites, three of which have been validated experimentally by ChIP (K.I.Z. and C.V.D., unpublished data).
In conclusion, we found that phylogenetic sequence conservation readily identifies class I target gene binding sites that are likely to be validated by ChIP. However, the lack of these conserved regions in class II type targets will require experimental approaches such as SChIP to identify bona fide binding sites.
Supplementary Material
Acknowledgments
We thank members of the Dang laboratory, Greg Kato, Linda Lee, and Dan Wechsler for comments. This work was supported in part by National Institutes of Health Grants RO1 CA57341, T32HL07525, and T32GM07819.
Abbreviations
- EMSA
electrophoretic mobility-shift assay
- ChIP
chromatin immunoprecipitation
- SChIP
scanning ChIP
References
- 1.Davidson E H, Rast J P, Oliveri P, Ransick A, Calestani C, Yuh C H, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 2.Pennacchio L A, Rubin E M. Nat Rev Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- 3.Zeller K I, Haggerty T J, Barrett J F, Guo Q, Wonsey D R, Dang C V. J Biol Chem. 2001;276:48285–48291. doi: 10.1074/jbc.M108506200. [DOI] [PubMed] [Google Scholar]
- 4.Eisenman R N. Genes Dev. 2001;15:2023–2030. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- 5.Grandori C, Cowley S M, James L P, Eisenman R N. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 6.Pelengaris S, Khan M, Evan G. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 7.Cole M D, McMahon S B. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 8.Dang C V. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wonsey D R, Zeller K I, Dang C V. Proc Natl Acad Sci USA. 2002;99:6649–6654. doi: 10.1073/pnas.102523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prescott J E, Osthus R C, Lee L A, Lewis B C, Shim H, Barrett J F, Guo Q, Hawkins A L, Griffin C A, Dang C V. J Biol Chem. 2001;276:48276–48284. doi: 10.1074/jbc.M107357200. [DOI] [PubMed] [Google Scholar]
- 11.Ferre-D'Amare A R, Prendergast G C, Ziff E B, Burley S K. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 12.Kato G J, Lee W M, Chen L L, Dang C V. Genes Dev. 1992;6:81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- 13.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 14.Boyd K E, Wells J, Gutman J, Bartley S M, Farnham P J. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett J F, Obaya A J, O'Connell B C, Mateyak M K, Tam W, Kohlhuber F, Dang C V, et al. Proc Natl Acad Sci USA. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menssen A, Hermeking H. Proc Natl Acad Sci USA. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Luscher B. Genes Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 19.Shim H, Dolde C, Lewis B C, Wu C S, Dang G, Jungmann R A, Dalla-Favera R, Dang C V. Proc Natl Acad Sci USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank S R, Schroeder M, Fernandez P, Taubert S, Amati B. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikiforov M A, Chandriani S, O'Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy J M, Cole M D. Mol Cell Biol. 2002;22:5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dermitzakis E T, Clark A G. Mol Biol Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- 23.Majewski J, Ott J. Genome Res. 2002;12:1827–1836. doi: 10.1101/gr.606402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prendergast G C, Ziff E B. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 25.Coller H A, Grandori C, Tamayo P, Colbert T, Lander E S, Eisenman R N, Golub T R. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Q M, Malek R L, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee N H, Dang C V, Liu E T. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- 27.Neiman P E, Ruddell A, Jasoni C, Loring G, Thomas S J, Brandvold K A, Lee R, Burnside J, Delrow J. Proc Natl Acad Sci USA. 2001;98:6378–6383. doi: 10.1073/pnas.111144898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuldiner O, Benvenisty N. Oncogene. 2001;20:4984–4994. doi: 10.1038/sj.onc.1204459. [DOI] [PubMed] [Google Scholar]
- 29.Watson J D, Oster S K, Shago M, Khosravi F, Penn L Z. J Biol Chem. 2002;277:36921–36930. doi: 10.1074/jbc.M201493200. [DOI] [PubMed] [Google Scholar]
- 30.Mayor C, Brudno M, Schwartz J R, Poliakov A, Rubin E M, Frazer K A, Pachter L S, Dubchak I. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz S, Zhang Z, Frazer K A, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loots G G, Ovcharenko I, Pachter L, Dubchak I, Rubin E M. Genome Res. 2002;12:832–839. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jegga A G, Sherwood S P, Carman J W, Pinski A T, Phillips J L, Pestian J P, Aronow B J. Genome Res. 2002;12:1408–1417. doi: 10.1101/gr.255002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.