Abstract
The 1,995,275-bp genome of Coxiella burnetii, Nine Mile phase I RSA493, a highly virulent zoonotic pathogen and category B bioterrorism agent, was sequenced by the random shotgun method. This bacterium is an obligate intracellular acidophile that is highly adapted for life within the eukaryotic phagolysosome. Genome analysis revealed many genes with potential roles in adhesion, invasion, intracellular trafficking, host-cell modulation, and detoxification. A previously uncharacterized 13-member family of ankyrin repeat-containing proteins is implicated in the pathogenesis of this organism. Although the lifestyle and parasitic strategies of C. burnetii resemble that of Rickettsiae and Chlamydiae, their genome architectures differ considerably in terms of presence of mobile elements, extent of genome reduction, metabolic capabilities, and transporter profiles. The presence of 83 pseudogenes displays an ongoing process of gene degradation. Unlike other obligate intracellular bacteria, 32 insertion sequences are found dispersed in the chromosome, indicating some plasticity in the C. burnetii genome. These analyses suggest that the obligate intracellular lifestyle of C. burnetii may be a relatively recent innovation.
Coxiella burnetii, the etiological agent of “Q fever,” is a category B bioterrorism agent that is highly infective to both humans and livestock (1). In humans, the disease manifests as an acute flu-like illness, with a hallmark debilitating headache and cyclic fever (1). Instances of chronic disease are characterized by endocarditis (1). A correlation has been suggested between C. burnetii infections and onset of atherosclerosis, chronic-fatigue syndrome, and other cerebrovascular incidents (2, 3). In livestock, abortion epidemics have been reported in endemic regions resulting in severe economic impacts (4, 5).
C. burnetii is an obligate intracellular, Gram-negative acidophile that replicates to high numbers, albeit with an estimated slow doubling time (12–20 h) (6), within the phagolysosome (PL) of the eukaryotic phagocyte (7). It is highly adapted to thrive within this compartment where low pH (≈4.5), hydrolytic enzymes, oxygen and nitrogen radicals, and other stresses typically result in microbial killing. C. burnetii is capable of survival outside the host for extended periods of time; high-level resistance to UV radiation, heat, desiccation, pressure [>50,000 psi (1 psi = 6.89 kPa)], and osmotic and oxidative stress has been demonstrated (8). Some of this resistance has been attributed to its biphasic (asynchronous) developmental cycle involving distinct morphological variants (9). A tick host (Dermacentor andersonii) has been implicated in transmission between various vertebrate hosts (10). Ease of aerosol dissemination, environmental persistence, and high infectivity (ID50 = 1–10) (11) make C. burnetii a serious threat for military personnel and civilians. This agent has already been weaponized and mass-produced under various biological warfare programs (12). Because the lifestyle of the pathogen causes it to be largely intractable to study by using contemporary molecular-genetic methods, the genome sequence contributes significantly to our understanding of the biology and pathogenesis of C. burnetii, additionally serving as a comparative paradigm to study the evolution of obligate intracellular parasitism.
Methods
Sequencing.
The genome of C. burnetii Nine Mile phase I RSA493, a tick isolate identified in 1935 (10), was sequenced and assembled by using the random shotgun method (13). Circular topography of the chromosome was confirmed by examining clone coverage in the region where a break was predicted (14) and validated by GC skew.
Coding Sequence (CDS) Prediction and Gene Identification.
ORFs likely to encode proteins (CDSs) were predicted by glimmer (15). All predicted proteins larger than 30 aa were searched against a nonredundant protein database (13). Frameshifts and point mutations were detected and corrected where appropriate. Remaining frameshifts and point mutations are considered to be authentic and were annotated as “authentic frameshift” or “authentic point mutation.” Protein membrane-spanning domains were identified by TOPPRED (16, 17). Putative signal peptides were identified with SIGNALP (17). The 5′ regions of each CDS were inspected to define initiation codons using homologies, position of ribosomal binding sites, and transcriptional terminators. Two sets of hidden Markov models were used to determine CDS membership in families and superfamilies: PFAM 5.5 (18) and TIGRFAM (19). PFAM 5.5 hidden Markov models were also used with a constraint of a minimum of two hits to find repeated domains within proteins and mask them. Domain-based paralogous families were built by performing all-versus-all searches on the remaining protein sequences. Base pair one was assigned within the putative origin of replication determined by colocalization of genes (dnaA, dnaN, recF, and gyrA) often found near the origin in prokaryotic genomes and GC nucleotide skew (G − C/G + C) analysis (20). All CDSs were searched with FASTA3 against all CDSs from the complete genomes, and matches with a FASTA3 P value of 10−5 were considered significant.
Trinucleotide Composition.
Distribution of all 64 trinucleotides (3-mers) for each chromosome was determined, and the 3-mer distribution in 2,000-bp windows that overlapped by half their length (1,000 bp) across the genome was computed. For each window, we computed the χ2 statistic on the difference between its 3-mer content and that of the whole chromosome. A large value for χ2 indicates that the 3-mer composition in this window is different from the rest of the chromosome. Probability values for this analysis are based on assumptions that the DNA composition is relatively uniform throughout the genome and that 3-mer composition is independent. Because these assumptions may be incorrect, we prefer to interpret high χ2 values as indicators of regions on the chromosome that appear unusual and demand further scrutiny.
Results and Discussion
General Features of the Genome.
General features of the 1,995,275-bp chromosome and 37,393-bp previously sequenced QpH1 plasmid (21) of C. burnetii Nine Mile phase I are summarized in Table 1. Although previously suspected to be linear (14), genome sequencing and analysis suggest a circular topology for the chromosome (see Methods). The genome is predicted to encode 2,134 CDSs, of which 719 (33.7%) are hypothetical, i.e., have no significant matches to other sequenced genes, which is a high proportion given the large number of γ-proteobacterial genome sequences currently available.
Table 1.
General features of the C. burnetii genome
| Chromosome | QpH1 | |
|---|---|---|
| Size, bp | 1,995,275 | 37,393 |
| G + C content, % | 42.6 | 39.3 |
| Protein-coding genes | ||
| No. similar to known proteins | 1,022 | 11 |
| No. similar to proteins of unknown function* | 179 | 5 |
| No. of conserved hypotheticals† | 200 | 1 |
| No. of hypotheticals‡ | 693 | 23 |
| Total | 2,094 | 40 |
| Average ORF size, bp | 849 | 736 |
| Coding, % | 89.1 | 78.8 |
| Stable RNAs | ||
| rRNA | 3 | 0 |
| tRNA | 42 | 0 |
Unknown function and significant sequence similarity to a named protein for which no function is currently attributed.
Conserved hypothetical protein with sequence similarity to a translation of another ORF; however, no experimental evidence for protein expression exists.
Hypothetical protein with no significant similarity to any other sequenced protein.
Although Coxiella was historically considered “Rickettsia-like,” 16S rRNA gene sequence analysis (22) and genome analysis (based on shared proteins across genomes and phylogenetic analysis of a set of 20 highly conserved proteins) indicate that it is a γ-proteobacteria (order Legionellales) and thus is distant from the α-proteobacterial Rickettsia group. Coxiella is also distant from any other lineage within the γ subgroup, its closest relationship is with Legionella pneumophila, a facultative intracellular human pathogen, and Rickettsiella grylli, an intracellular arthropod pathogen (23).
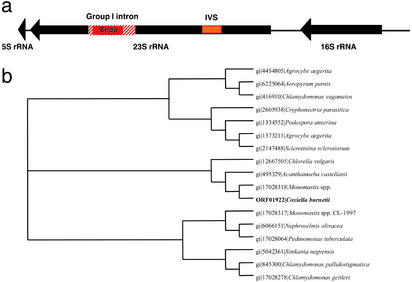
Various intervening elements have been identified within physiologically vital genes of C. burnetii. A putative excisable intein is located in the C-terminal region of the replicative DNA helicase (CBU0868), and the sole 23S rRNA gene is interrupted both by an intervening sequence (24) and a group I self-splicing intron (Fig. 1a). The mechanism of excision of these two elements in the 23S rRNA is very different; the intervening sequence is excised from the RNA without exon ligation, resulting in a fragmented but functional rRNA (24), whereas the intron is presumed to splice out precisely, resulting in no further fragmentation. The intron encodes a LAGLIDADG homing endonuclease that may play a role in intron mobility (25). This endonuclease is most similar to rRNA-associated intron endonucleases found in organelle genomes of single-celled eukaryotes (Fig. 1b); however, members of this endonuclease family are not found in most other prokaryotic genomes or nuclear genomes of eukaryotes. Although it is possible that a common ancestor of C. burnetii (γ-proteobacteria), mitochondria (α-proteobacteria), and plastids (cyanobacteria) encoded this endonuclease and this gene was subsequently lost in numerous other lineages, it is more likely that some of these lineages acquired the gene by lateral gene transfer. Because the endonuclease is associated with a mobile element, its lateral transfer seems likely, but the exact mechanism for the transfer (i.e., what the donor and recipient lineages were) is not possible to infer with the available data.
Figure 1.
(a) Schematic representation of the 5S-23S-16S components of the rRNA operon in C. burnetii. The intervening sequences (IVSs) located in the 23S rRNA gene are indicated. (b) Phylogenetic clustering of the C. burnetii group I intron endonuclease with other sequenced endonucleases. Amino acid sequences were aligned by using CLUSTALW, and a neighbor-joining tree was generated from the alignment by using the PAUP program. All nodes had bootstrap values >85%. The C. burnetii endonuclease clusters with intron-associated endonucleases from the mitochondrial genomes of Monomastix spp., Acanthamoeba castellani, and other single-celled eukaryote mitochondrial and chloroplast genomes.
Comparative Analysis of Obligate Intracellular Pathogens.
Obligate intracellular pathogens (i.e., Rickettsia, Chlamydia, and Mycobacterium leprae) appear to share characteristics such as reduced metabolic and transport capabilities and lack of mobile elements. Although C. burnetii ostensibly shares similarities in lifestyle and parasitic strategies with these bacteria, the genome differs considerably, particularly in terms of presence of mobile elements, extent of genome reduction, metabolic capabilities, and transporter profiles. However, C. burnetii does occupy a different intracellular niche compared with these other bacteria, and substrates or nutrients in the PL may be quite different from those encountered in other compartments.
Insertion sequences (ISs).
Genomes of other obligate intracellular bacteria have few to no IS elements (26–29), presumably because of limited opportunities for gene transfer. By contrast, C. burnetii possesses 29 IS elements; there are 21 copies of a unique IS110-related isotype, IS1111 (30), 5 IS30 and 3 ISAs1 family elements, and 3 degenerate transposase genes of unknown lineage. The transposase genes are >99% identical in DNA sequence within each group, suggesting either recent introduction and spread of each IS element or homogenization by concerted evolution as occurs with rRNA genes (31).
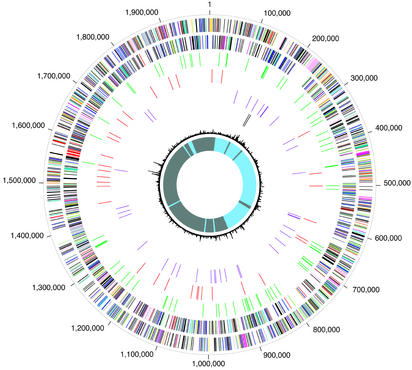
In C. burnetii the IS elements are dispersed around the chromosome (but not found on the plasmid), and there is no localized clustering. Although there is little evidence of recently acquired DNA in the genome (Fig. 2; see Methods), in one case identical elements flank groups of genes in a manner reminiscent of “pathogenicity islands” (32). This potential island is flanked by IS1111 elements (CBU1218 and CBU1186), found adjacent to tRNA-serine genes. The region includes a putative multidrug transporter protein (CBU1208), a sterol reductase (CBU1206), and numerous hypothetical proteins.
Figure 2.
Circular representation of the C. burnetii RSA493 overall genome structure. The outer scale designates coordinates in base pairs. The first circle shows predicted coding regions on the plus strand color-coded by role categories: violet, amino acid biosynthesis; light blue, biosynthesis of cofactors, prosthetic groups, and carriers; light green, cell envelope; red, cellular processes; brown, central intermediary metabolism; yellow, DNA metabolism; light gray, energy metabolism; magenta, fatty acid and phospholipid metabolism; pink, protein synthesis and fate; orange, purines, pyrimidines, nucleosides, and nucleotides; olive, regulatory functions and signal transduction; dark green, transcription; teal, transport and binding proteins; gray, unknown function; salmon, other categories; blue, hypothetical proteins. The second circle shows predicted coding regions on the minus strand color-coded by role categories: third circle, pseudogenes in green; fourth circle, IS elements in red; fifth circle, tRNA genes in violet; sixth circle, rRNA genes in dark brown; seventh circle, trinucleotide composition in black; eighth circle, GC-skew curve in blue (positive residues) and gray (negative residues).
Genome reduction in C. burnetii: A work in progress?
Genome analysis identified 83 “pseudogenes” (genes disrupted by one or more authentic frameshifts and/or point mutations or, in some cases, truncations) suggesting that some genome reduction is underway. Reductive evolution has been found to be common in obligate intracellular bacteria. During genome reduction, genes that once had an important function accumulate mutations and eventually disappear either because they are no longer under strong selective pressures (e.g., if the niche of the organism has changed) or because of population-level phenomena that lead to an inability to maintain genes under weak selection (e.g., Muller's ratchet) (33). Because many of the pseudogenes are disrupted by a single frameshift, it would seem that they are of recent origin [as suggested for Salmonella typhimurium (34)]. Additionally, a much higher percentage of the C. burnetii genome is coding (89.1%) than for species known to have undergone large amounts of genome reduction (≈76% in both Rickettsia prowazekii and M. leprae), suggesting that either the reduction process has just begun in C. burnetii or it may not be as severe in this species as in some others.
Pseudogenes are overrepresented in specific role categories (e.g., toxin production and resistance, transport and binding, conserved hypothetical, and unknown function) suggesting elimination of functions no longer relevant to the lifestyle. For example, components of a competence system (for uptake of extracellular DNA), ComA (CBU0855), ComF (CBU0464), and ComM (CBU2022), possess one or more frameshifts; suggesting C. burnetii may not be naturally competent, although putative ComE (CBU0532) and ComL (CBU0758) genes are intact. Several homologues of Bacillus spp. peptide synthetases (for nonribosomal synthesis of antimicrobial lantibiotics) are highly degenerate (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). Frameshifts are also detected in predicted β-lactamases that confer resistance to β-lactams (Table 2). The C. burnetii Nine Mile isolate is not described as being naturally resistant to antibiotics; in fact this strain is considered to be highly susceptible to various antibiotics relative to some chronic disease-causing isolates (35).
Transport and Metabolism.
The transporter content of C. burnetii seems to largely reflect its intracellular lifestyle within the hostile environment of the PL. Four predicted sodium ion/proton exchangers (CBU1590, CBU1259, CBU0459, and CBU1582–CBU1588) probably play an important role in pH homeostasis and survival within the acidic PL. Three predicted mechanosensitive ion channels and three transporters for osmoprotectants might provide resistance to osmotic stress. More than a quarter of the transporters in C. burnetii are predicted drug-efflux systems, a higher proportion than seen in other proteobacteria (Fig. 5, which is published as supporting information on the PNAS web site). One possible role of these predicted drug-efflux pumps is to provide resistance to host-produced antimicrobial defensins within the PL. Alternately, they may have had a role in secondary metabolite secretion, because several polyketide biosynthesis genes are encoded on the C. burnetii genome; however, many of them are pseudogenes (Table 2).
C. burnetii possesses greater transport capabilities for organic nutrients than Chlamydia and Rickettsia; however, these capabilities are limited compared with those of free-living bacteria. Only two sugar transport systems were identified: proton-driven systems for xylose (CBU0347) and glucose (CBU0265). Histidine phosphocarrier protein (HPr) and enzyme I of the sugar phosphotransferase system are present, as is HPr serine kinase, but no phosphotransferase system transporters were detected; thus these Hpr proteins presumably play only a regulatory role. The presence of 15 transporters for amino acids and 3 for peptides suggest that these may be the dominant carbon sources for C. burnetii, reflecting their abundance within the PL. Biochemical studies of C. burnetii have focused on glutamate utilization (36), and genome analysis allowed identification of a probable glutamate uptake transporter (CBU2020). Unlike rickettsial and chlamydial species, C. burnetii does not possess any ATP/ADP exchangers that enable these organisms to function as energy parasites by scavenging ATP from their host (37), confirming previous experimental observations (38). There are six transporters of unknown specificity belonging to a previously uncharacterized subfamily of the major facilitator superfamily, which seem to have undergone amplification in the C. burnetii lineage.
C. burnetii has greater biosynthetic capabilities in contrast to obligate intracellular bacteria that have reduced metabolic capability as a result of increased dependence on host-derived substrates (39). C. burnetii possesses enzymes for glycolysis, the Entner–Doudoroff pathway, the electron transport chain, the Embden–Meyerhof–Parnas pathway, gluconeogenesis, the pentose phosphate pathway, and the tricarboxylic acid (TCA) cycle. The glyoxylate bypass is absent, probably because C. burnetii is largely reliant on exogenously acquired amino acids and does not need to recharge the TCA cycle by synthesizing intermediates for amino acid synthesis. Additionally, exogenous amino acids can also be degraded to TCA intermediates when needed. Pathways for synthesis of purine and pyrimidines, fatty acids and phospholipids, and cofactors (biotin, ubiquinone, and folic acid) are also intact. Most components of a mevalonate pathway (CBU0607–CBU0609) for the synthesis of isopentyl diphosphate (IPP) or isoprenoids (except a HMG-CoA synthase) were identified. The glyceraldehyde-3-phosphate (GAP) pyruvate pathway for IPP synthesis seen in most Gram-negative bacteria is absent, which suggests that C. burnetii may synthesize IPP solely via a mevalonate pathway characteristic of Gram-positive bacteria and Archaea. The anabolic capabilities of C. burnetii are not comprehensive; it is predicted to be an auxotroph for 11 amino acids. Some of these biosynthetic pathways are absent in their entirety, whereas others are missing key enzymatic members. The presence of amino acid transporters probably compensates for these deficiencies, enabling acquisition of amino acids from the host.
Reductive convergent evolution of Coxiella, Chlamydia spp., and Rickettsia spp. is suggested by shared absences of enzymatic steps for tryptophan and lysine biosynthesis. Similar to Rickettsia and Chlamydia, C. burnetii encodes all steps for the lysine biosynthesis pathway except the terminal step involving diaminopimelate decarboxylase (CBU1030 is degraded), suggesting that diaminopimelate synthesis is the primary function and that C. burnetii is a lysine auxotroph. The tryptophan biosynthesis operon is unusual; TrpA, -C, and -E (CBU1156, CBU1154, and CBU1152) are intact, phosphoribosyl anthranilate isomerase (TrpF) and tryptophan synthase (TrpB) are fused (CBU1155), and anthranilate phosphoribosyl transferase (TrpDG) has multiple frameshifts (CBU1153). This disrupts the initial two steps of the pathway (from chorsimate to phosphoribosyl anthranilate); however, the presence of subsequent/terminal steps suggests that tryptophan can be synthesized from intermediates taken up from the host as seen in Chlamydiae (40, 41). Chlamydial acquisition of host-derived intermediates (released from protective IFN-γ-induced tryptophan degradation) and subsequent recycling to tryptophan is considered an adaptive virulence strategy (40).
Genome analysis reveals pathways for the utilization of few sugars (glucose, galactose, and xylose) and glycerol (however, no transporter for glycerol uptake is identified). Contents of the parasitophorous vacuole are undefined; however, the PL is typically known to contain many enzymes for degradation of polysaccharides (42), and therefore free sugars may be plentiful in this environment. Given the ability of C. burnetii to persist in the extracellular environment for prolonged periods, the absence of pathways for synthesis of storage compounds such as glycogen, trehalose, or polyhydroxybutyrate is unexpected. Although genes for synthesis of lipid A and 2-keto-3-deoxyoctulosonic acid components of the lipopolysaccharide are scattered, genes for the synthesis of lipopolysaccharide core and outer repeating units of the O-antigen are clustered into two regions (at coordinates 612,249–642,009 and 779,513–806,121). These regions contain a large number of genes pertaining to nucleotide sugar metabolism (e.g., dehydratases, dehydrogenases, epimerases, and reductases) involved in polymerization of sugars in the O-antigen chains. Also found in the region are candidates (CBU0691 and CBU0683) for synthesizing two unusual sugars present in the Coxiella lipopolysaccharide [6-deoxy-3-C-methylgulose or virenose and 3-C-(hydroxymethyl)-lyxose] (43) by methylation of pyranose and furanose nucleotide sugars, similar to pathways for macrolide and other antibiotic synthesis.
C. burnetii encodes a number of genes that are similar to eukaryotic genes and have no matches to any sequenced prokaryotic genes, for example, a eukaryotic fatty acid desaturase (CBU0920) homologue that may play a role in synthesis of diverse saturated and unsaturated fatty acids and two sterol reductases (CBU1206 and CBU1158) involved in cholesterol synthesis in eukaryotes. The absence of other related enzymes suggests that de novo synthesis of cholesterol is unlikely. It is more likely that these sterol reductases play a role in conversion of metabolites taken up from the host and possibly even contribute to the tissue- or organotropisms (placenta and reproductive tissue) attributed to this organism (1, 44). A correlation between hormonal levels of pregnant animals and increased C. burnetii infection has also been suggested (45, 46).
Pathogenesis.
Adhesion and invasion are critical early events that determine successful colonization. Although C. burnetii lacks genes encoding typical structures for adhesion (e.g., pili and other adhesins), it possesses 13 ankyrin repeat-containing proteins that may be implicated in host-cell attachment (Table 3, which is published as supporting information on the PNAS web site). Ankyrins are spectrin-binding proteins that mediate interaction of the membrane skeleton with the plasma membrane (47). However, ankyrin domains have also been identified in some transcriptional regulators that may play a role in altering host-cell gene expression as suggested for Ehrlichia spp. (48). Although ankyrin domains are not unique to C. burnetii, the presence of 13 proteins with this domain is higher than in any other sequenced genome. Although bacterial ligands and host-cell receptors that mediate internalization have not been identified previously, entry via αvβ3 integrin or CR3 complement receptor has been suggested (49). Genome analysis identified several CDSs with the integrin-binding RGD motif and signal sequence that may be candidates for facilitating internalization (Table 3). Additionally, C. burnetii contains homologues of L. pneumophila EnhA (CBU1122 and CBU1138), EnhB (CBU1137 and CBU0053), and EnhC (CBU1136) genes that have been implicated in an enhanced cell-entry phenotype (50) and may interact with the host; signal peptides are predicted for all. Cytoskeletal rearrangements have been suggested to play a critical role in internalization of C. burnetii (51). These rearrangements may be mediated by a tyrosine-phosphorylated GTPase regulator (CBU0884) analogous to BipA of enteropathogenic Escherichia coli that triggers cytoskeletal rearrangements (52).
Modification of host-cell proteins by intracellular pathogens is an important virulence strategy. A putative eukaryotic-type protein kinase (CBU1379) and an acid phosphatase (CBU0335) may play a role in modulation of host response. Phosphorylation of host proteins by C. burnetii acid phosphatase activity results in diminished oxidative burst after phagocytosis (53).
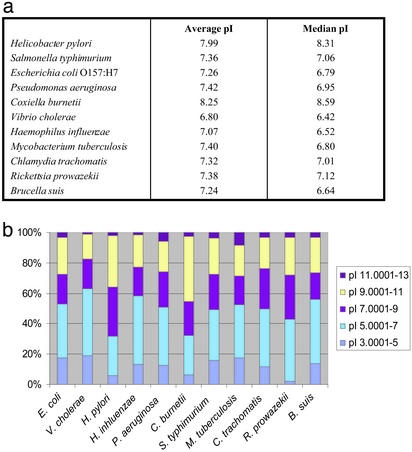
Although other intracellular pathogens typically evade phagolysosomal fusion, C. burnetii targets this prohibitive niche where it is under considerable oxidative and osmotic stress (8). A number of genes associated with stress-response or vacuole detoxification systems are present (Table 3). Protection against low pH may be conferred by basic proteins that could serve as a proton “sink” to buffer against the excess protons that may enter the bacterial cell from the acidic environment of the PL (54). C. burnetii encodes an unusually high number of basic proteins; the average pI value for all predicted proteins is 8.25, which is higher than for most other sequenced genomes, with the exception of Helicobacter pylori, which resides in the low-pH environment of the gastric mucosa (Fig. 3a). Furthermore, ≈45% of C. burnetii proteins were found to have a pI value of ≥9, in striking contrast to the proteome of other γ-proteobacteria and intracellular pathogens (Fig. 3b).
Figure 3.
(a) Average and median pI values of all predicted proteins for C. burnetii and a subset of sequenced γ-proteobacteria and intracellular bacterial genomes. (b) Percentage of total proteins in given pI ranges for C. burnetii and various sequenced bacterial genomes.
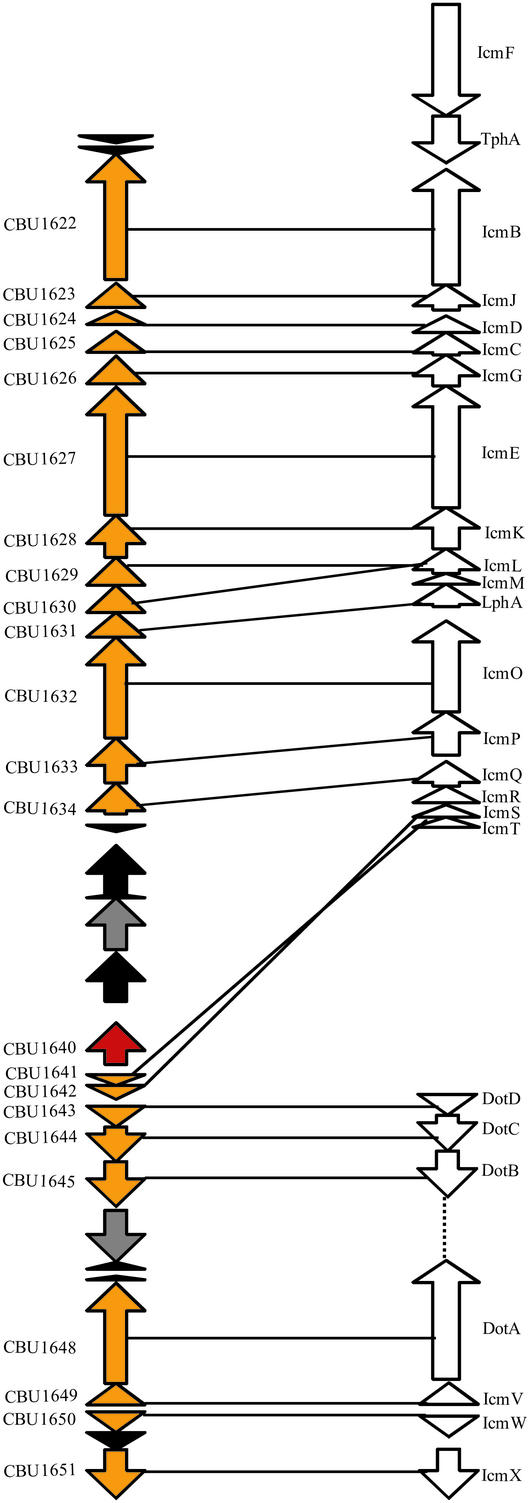
Active secretion of proteins into host cells by intracellular pathogens is often implicated in intracellular trafficking and virulence (55). Genome analysis revealed components of types I- II, and IV secretion systems (Table 3). The type IV secretion system is related to that of L. pneumophila, of which three components (IcmKST) were identified previously (56). Analysis of this region reveals a number of inversions, insertions, and deletions in C. burnetii relative to the gene organization in L. pneumophila (Fig. 4). The IS1111 transposase located in this cluster may be responsible for part of the rearrangements. An IcmF homologue (CBU0320) is located elsewhere and possesses a point mutation. No homologues of IcmM and IcmR or of the few described type IV effectors are seen; however, the hypothetical genes identified within this cluster or flanking it may be candidates.
Figure 4.
Linear representation of the locus encoding a putative type IV secretion system in C. burnetii with relation to L. pneumophila icm-dot genes. Genes are shown as arrowheads and colored by predicted function: orange, putative type IV secretion system components; black, hypothetical protein; gray, conserved hypothetical protein; red, IS1111 element transposase. Black lines connect genes with best matches to L. pneumophila genes (shown in white) at a P-value cutoff of <10−5. Both CBU1629 and CBU1630 show significant similarity to IcmL; however, they are not identical to each other.
Conclusion
Because the lifestyle of C. burnetii causes it to be largely refractory to study with contemporary molecular-genetic methods, the genome sequence has contributed significantly to our understanding of the biology and pathogenesis of this organism. Previously uncharacterized genes implicated in adhesion, invasion, intracellular trafficking, host modulation, detoxification, and other virulence-related functions have been identified. The relatively high proportion of hypothetical genes may be involved in functions relevant to the unique developmental cycle and lifestyle of C. burnetii. Genome analysis has revealed the presence of numerous mobile elements and pseudogenes, indicating plasticity and ongoing genome reduction. These observations and comparisons to genomes of other sequenced obligate intracellular bacteria suggest that C. burnetii may have transitioned relatively recently into its current lifestyle.
Supplementary Material
Acknowledgments
We thank O. White, S. Salzberg, M. Heaney, M. Covarrubias, S. Hodge, R. Karamchedu, B. Lee, S. Lo, M. Holmes, and V. Sapiro for informatics, database, and software support, and The Institute for Genomic Research faculty and sequencing core for advice and assistance. This work was supported by the Defense Advanced Research Projects Agency and National Institutes of Health, National Institute of Allergy and Infectious Disease Grant 1U01AI49034-01.
Abbreviations
- PL
phagolysosome
- CDS
coding sequence
- IS
insertion sequence
Footnotes
References
- 1.Maurin M, Raoult D. Clin Microbiol Rev. 1999;12:518–553. doi: 10.1128/cmr.12.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wildman M J, Smith E G, Groves J, Beattie J M, Caul E O, Ayres J G. Q J Med. 2002;95:527–538. doi: 10.1093/qjmed/95.8.527. [DOI] [PubMed] [Google Scholar]
- 3.Lovey P Y, Morabia A, Bleed D, Peter O, Dupuis G, Petite J. BMJ. 1999;319:284–286. doi: 10.1136/bmj.319.7205.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanford S E, Josephson G K, MacDonald A. Can Vet J. 1994;35:376–378. [PMC free article] [PubMed] [Google Scholar]
- 5.Zeman D H, Kirkbride C A, Leslie-Steen P, Duimstra J R. J Vet Diagn Invest. 1989;1:178–180. doi: 10.1177/104063878900100218. [DOI] [PubMed] [Google Scholar]
- 6.Zamboni D S, Mortara R A, Freymuller E, Rabinovitch M. Microbes Infect. 2002;4:591–598. doi: 10.1016/s1286-4579(02)01577-0. [DOI] [PubMed] [Google Scholar]
- 7.Hackstadt T, Williams J C. Proc Natl Acad Sci USA. 1981;78:3240–3244. doi: 10.1073/pnas.78.5.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinzen R A, Samuel J E. In: The Procaryotes: An Evolving Electronic Database for the Microbiological Community. Dworkin M, editor. New York: Springer; 2001. [Google Scholar]
- 9.Heinzen R A, Hackstadt T, Samuel J E. Trends Microbiol. 1999;7:149–154. doi: 10.1016/s0966-842x(99)01475-4. [DOI] [PubMed] [Google Scholar]
- 10.Davis G E, Cox H R. Public Health Rep. 1938;53:2259–2282. [Google Scholar]
- 11.Tigertt W D, Benenson A S, Gochenour W S. Bacteriol Rev. 1961;25:285–293. doi: 10.1128/br.25.3.285-293.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Regis E. The Biology of Doom: The History of America's Secret Germ Warfare Project. New York: Holt; 1999. [Google Scholar]
- 13.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, et al. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willems H, Jager C, Baljer G. J Bacteriol. 1998;180:3816–3822. doi: 10.1128/jb.180.15.3816-3822.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delcher A L, Harmon D, Kasif S, White O, Salzberg S L. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claros M G, von Heijne G. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haft D H, Loftus B J, Richardson D L, Yang F, Eisen J A, Paulsen I T, White O. Nucleic Acids Res. 2001;29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobry J R. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 21.Thiele D, Willems H, Haas M, Krauss H. Eur J Epidemiol. 1994;10:413–420. doi: 10.1007/BF01719665. [DOI] [PubMed] [Google Scholar]
- 22.Weisburg W G, Dobson M E, Samuel J E, Dasch G A, Mallavia L P, Baca O, Mandelco L, Sechrest J E, Weiss E, Woese C R. J Bacteriol. 1989;171:4202–4206. doi: 10.1128/jb.171.8.4202-4206.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roux V, Bergoin M, Lamaze N, Raoult D. Int J Syst Bacteriol. 1997;47:1255–1257. doi: 10.1099/00207713-47-4-1255. [DOI] [PubMed] [Google Scholar]
- 24.Afseth G, Mo Y Y, Mallavia L P. J Bacteriol. 1995;177:2946–2949. doi: 10.1128/jb.177.10.2946-2949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belfort M, Roberts R J. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 27.Ogata H, Audic S, Renesto-Audiffren P, Fournier P E, Barbe V, Samson D, Roux V, Cossart P, Weissenbach J, Claverie J M, Raoult D. Science. 2001;293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
- 28.Cole S T, Eiglmeier K, Parkhill J, James K D, Thomson N R, Wheeler P R, Honore N, Garnier T, Churcher C, Harris D, et al. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 29.Tamas I, Klasson L, Canback B, Naslund A K, Eriksson A S, Wernegreen J J, Sandstrom J P, Moran N A, Andersson S G. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 30.Hoover T A, Vodkin M H, Williams J C. J Bacteriol. 1992;174:5540–5548. doi: 10.1128/jb.174.17.5540-5548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao D. J Mol Evol. 2000;51:305–317. doi: 10.1007/s002390010093. [DOI] [PubMed] [Google Scholar]
- 32.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 33.Haigh J. Theor Popul Biol. 1978;14:251–267. doi: 10.1016/0040-5809(78)90027-8. [DOI] [PubMed] [Google Scholar]
- 34.McClelland M, Sanderson K E, Spieth J, Clifton S W, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 35.Yeaman M R, Baca O G. Ann NY Acad Sci. 1990;590:297–305. doi: 10.1111/j.1749-6632.1990.tb42236.x. [DOI] [PubMed] [Google Scholar]
- 36.Hackstadt T, Williams J C. J Bacteriol. 1983;154:598–603. doi: 10.1128/jb.154.2.598-603.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zomorodipour A, Andersson S G. FEBS Lett. 1999;452:11–15. doi: 10.1016/s0014-5793(99)00563-3. [DOI] [PubMed] [Google Scholar]
- 38.Miller J D, Thompson H A. Microbiology. 2002;148:2393–2403. doi: 10.1099/00221287-148-8-2393. [DOI] [PubMed] [Google Scholar]
- 39.Andersson S G, Kurland C G. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- 40. Xie, G., Bonner, C. A. & Jensen, R. A. (2002) Genome Biol.3, research0051.1–0051.17. [DOI] [PMC free article] [PubMed]
- 41.Fehlner-Gardiner C, Roshick C, Carlson J H, Hughes S, Belland R J, Caldwell H D, McClarty G. J Biol Chem. 2002;277:26893–26903. doi: 10.1074/jbc.M203937200. [DOI] [PubMed] [Google Scholar]
- 42.Barrett A J. Biochem Soc Trans. 1984;12:899–902. doi: 10.1042/bst0120899. [DOI] [PubMed] [Google Scholar]
- 43.Schramek S, Radziejewska-Lebrecht J, Mayer H. Eur J Biochem. 1985;148:455–461. doi: 10.1111/j.1432-1033.1985.tb08861.x. [DOI] [PubMed] [Google Scholar]
- 44.Baumgartner W, Bachmann S. Infect Immun. 1992;60:5232–5241. doi: 10.1128/iai.60.12.5232-5241.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biberstein E L, Behymer D E, Bushnell R, Crenshaw G, Riemann H P, Franti C E. Am J Vet Res. 1974;35:1577–1582. [PubMed] [Google Scholar]
- 46.Rose M, Wemheuer W, Schmidt F W. Dtsch Tierarztl Wochenschr. 1994;101:484–486. [PubMed] [Google Scholar]
- 47.Batrukova M A, Betin V L, Rubtsov A M, Lopina O D. Biochemistry (Moscow) 2000;65:395–408. [PubMed] [Google Scholar]
- 48.Caturegli P, Asanovich K M, Walls J J, Bakken J S, Madigan J E, Popov V L, Dumler J S. Infect Immun. 2000;68:5277–5283. doi: 10.1128/iai.68.9.5277-5283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Capo C, Lindberg F P, Meconi S, Zaffran Y, Tardei G, Brown E J, Raoult D, Mege J L. J Immunol. 1999;163:6078–6085. [PubMed] [Google Scholar]
- 50.Cirillo S L, Lum J, Cirillo J D. Microbiology. 2000;146:1345–1359. doi: 10.1099/00221287-146-6-1345. [DOI] [PubMed] [Google Scholar]
- 51.Meconi S, Jacomo V, Boquet P, Raoult D, Mege J L, Capo C. Infect Immun. 1998;66:5527–5533. doi: 10.1128/iai.66.11.5527-5533.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farris M, Grant A, Richardson T B, O'Connor C D. Mol Microbiol. 1998;28:265–279. doi: 10.1046/j.1365-2958.1998.00793.x. [DOI] [PubMed] [Google Scholar]
- 53.Baca O G, Roman M J, Glew R H, Christner R F, Buhler J E, Aragon A S. Infect Immun. 1993;61:4232–4239. doi: 10.1128/iai.61.10.4232-4239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seshadri R, Samuel J E. Infect Immun. 2001;69:4874–4883. doi: 10.1128/IAI.69.8.4874-4883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cianciotto N P. Int J Med Microbiol. 2001;291:331–343. doi: 10.1078/1438-4221-00139. [DOI] [PubMed] [Google Scholar]
- 56.Segal G, Shuman H A. Mol Microbiol. 1999;33:669–670. doi: 10.1046/j.1365-2958.1999.01511.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.