Abstract
Reelin is synthesized and secreted into extracellular matrix by cortical γ-aminobutyric acid (GABA)ergic interneurons and binds with high affinity to the extracellular domain of integrin receptors expressed in dendritic shaft and spine postsynaptic densities (DSPSD) of pyramidal neurons. In heterozygous reeler mice, reelin bound to DSPSD, and the expression of Arc (activity-regulated cytoskeletal protein) is lower than in wild-type mice. We studied the effect of reelin on Arc and total protein synthesis in synaptoneurosomes (SNSs) prepared from mouse neocortex. Recombinant full-length mouse reelin displaces the high affinity (KD = 60 fM) binding of [125I]echistatin (a competitive integrin receptor antagonist) to integrin receptors with a Ki of 22 pM and with a Hill slope close to 1. Echistatin (50–100 nM) competitively antagonizes and abates reelin binding. The addition of reelin (2–40 pM) to SNSs enhances the incorporation of [35S]methionine into Arc and other rapidly translated proteins in a concentration-dependent manner. This incorporation is virtually abolished by 50–100 nM echistatin or by 5–10 nM rapamycin, a blocker of the mammalian target of rapamycin kinase. We conclude that reelin binds with high affinity to integrin receptors expressed in SNSs and thereby activates Arc protein synthesis.
In mammalian cortex, reelin is selectively synthesized by γ-aminobutyric acid (GABA)ergic interneurons, which constitutively secrete this protein in the extracellular matrix (ECM) very likely in proximity of dendritic spine postsynaptic densities (DSPSD) of pyramidal neurons (1–3). Immunoelectron microscopy shows that reelin preferentially localizes around the extracellular domain of integrin receptors highly expressed in DSPSD of cortical pyramidal neurons (1, 3). In the embryonic rodent brain, reelin not only binds to the very low density lipoprotein and apoliprotein E2 receptors (4) but also to α3,β1 integrin receptors with high affinity (5) thereby activating a signal transduction pathway that induces the adapter function of the DAB1 protein (4–6). It is therefore possible that in the adult mammalian brain integrin receptors initiate a signal transduction cascade between ECM reelin and pyramidal neuron DSPSD.
In rat hippocampal slices, integrin receptors may contribute to synapse maturation (7) or to long-term potentiation (LTP) consolidation by transducing ECM signals, leading to a cytoskeleton reorganization via the activation of extrasomal mRNA translation (8–10). This view is supported by inhibition of LTP consolidation by the addition of function-blocking antibodies to α5/β1 or α3/β1 integrin receptors or by integrin receptor antagonists including an RGD (Arg-Gly-Asp) motif (9, 10). Hence, an integrin-mediated signal transduction may be critical to the stabilization of the LTP-induced plasticity at central excitatory synapses.
The application of reelin to adult rat hippocampal slices enhances LTP induction (11). Moreover, in heterozygous reeler mice (HRM) that contain reduced reelin gene dosage, cortical neuropil and dendritic spine density are reduced (12). These considerations suggest that reelin, acting at integrin receptors, could stabilize DSPSD, providing a molecular scaffold for the assembly of cytoskeletal proteins that facilitate dendrite resident mRNA translation and provide the increased protein synthesis required for LTP consolidation and memory trace formation. This hypothesis is consistent with the deficit in memory acquisition and the increased dizocilpine amnestic action characteristic of HRM (13, ‡).
The synthesis of activity-regulated cytoskeletal protein (Arc) is encoded by dendritic resident mRNAs. This rapidly inducible protein (14) functions as an immediate early gene product that very likely is involved in spine skeleton formation during LTP stabilization (15). The reduction of Arc synthesis elicited by antisense oligonucleotides curtails LTP duration and the associated long-term spatial memory consolidation without affecting task acquisition or short-term performance (16). In frontal cortex and hippocampus of reeler and heterozygous reeler mice, Arc expression is curtailed.§ To determine whether reelin can modulate dendritic resident Arc mRNA translation, we studied whether reelin can change Arc and total protein synthesis in synaptoneurosomes (SNSs). Our results are consistent with the view that recombinant reelin acting at integrin receptors activates Arc biosynthesis in a manner inhibited by echistatin, a competitive antagonist at integrin receptors (10, 17), and rapamycin, a blocker of the mammalian target of rapamycin (mTOR) kinase (18).
Materials and Methods
Preparation of SNSs.
SNSs were prepared according to the flotation-sedimentation density gradient centrifugation method described by Jones and Matus (19). Briefly, mouse forebrains were homogenized in 10% sucrose. The crude mitochondrial pellet fraction (P2) was brought to 34% sucrose by the addition of 48% sucrose. A sucrose upper phase (28.5%) was overlaid on the P2 phase, and a small volume of 10% sucrose was overlaid onto the upper phase to obtain a gradient of 36 ml. The gradients were centrifuged at 60,000 × g for 110 min in a Surespin 630 Sorvall rotor, and three fractions were separated. The middle fraction (fraction 2) contained purified SNSs, which were then treated with 0.01% Triton X-100 for 30 min and washed three times with buffer to remove native adhering reelin. SNS membranes were prepared by resuspending SNSs in 5 mM Tris⋅HCl buffer (pH 8.1) at 0°C for 30 min, and sonicating for 15 s.
Label-Fracture.
SNSs were fixed with a solution of cold 4% paraformaldehyde in PBS for 1 h and washed in PBS, osmicated, embedded in EPON812, and processed for electron microscopy. Other fixed SNSs were incubated with an anti-α3 integrin receptor subunit antiserum diluted at 1:250 (20 h at 4°C and 2-h incubation at 22°C). Samples were then incubated for 2 h with a secondary antibody conjugated with colloidal gold particles 10 nm in size. Immunolabeled SNS samples were then cryoprotected by 30 min incubation in a solution of 30% glycerol, frozen in Freon 22 cooled by liquid nitrogen, fractured, and replicated in a Balzers 400D freeze-fracture unit. The replicas were cleaned in distilled water (for at least 6 h) before being transferred onto formvar-coated grids.
Western Blot Analysis.
Western blot immunostaining was performed with G10 anti-reelin (1:5,000; a generous gift of A. M. Goffinet, University of Namur, Brussels), anti-integrin α3 and β1 (1:250; Chemicon), anti-synaptophysin (1:500; Upstate Biotechnology, Lake Placid, NY), anti-Arc (1:1,000, Santa Cruz Biotechnology), anti-PSD95 (1:1,000, Upstate Biotechnology), and anti-β-actin (Sigma) antibodies, followed by a peroxidase-conjugated secondary antibody. The signals were visualized by chemiluminescent detection system (Amersham Pharmacia Biotech). For the autoradiography of radioactive blots, membranes were exposed to Storage Phosphor Screens (Molecular Dynamics) for 2–4 days. Signals were visualized by using a STORM 840 Optical Scanner (Amersham) and quantified by using imagequant software (Molecular Dynamics).
Arc mRNA Expression in SNSs and Polyribosomes.
To separate polysomes from the cytosolic fraction, SNSs were lysed with 0.25% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) containing 1 mg/ml heparin. The lysate was spun 30 s in a microcentrifuge, and the supernatant was overlayered 1 M sucrose buffer (10 mM Tris, pH 7.6/1 mM potassium acetate/1.5 mM MgCl2/2 mM DTT/0.5 mg/ml heparin), and centrifuged for 11 min at 400,000 × g. The polyribosomal pellet was resuspended in 300 μl of diethyl pyrocarbonate water, and total RNA was extracted with an RNeasy Mini Kit (Qiagen, Valencia, CA). Arc mRNA was amplified by RT-PCR with forward primer, ATGGGCAACCAAGCCCAGCGT, and reverse primer, AGTGTCTGGTACAGGTCCCG [330-bp fragment from 821 to 1151 of the Arc cDNA sequence (U19866, GenBank)]. After agarose gel electrophoresis, the amplified signals were visualized by ethidium bromide staining.
Expression and Purification of Recombinant Reelin.
Mouse recombinant reelin was expressed in 293T cells by using a full-length reelin cDNA clone (generously provided by G. D'Arcangelo, St. Jude Children's Hospital, Memphis, TN). The serum-free DMEM was collected for 2 days. After lyophilization, the medium was purified through a Superdex-200 (Pharmacia) column (1 × 20 cm; separation range, 10–600 kDa) equilibrated with 150 mM NaCl solution. Reelin was detected in the column eluate (1-ml fractions) by dot immunoblot with G10 anti-reelin antibodies. The reelin-containing fractions were pooled and dialyzed against the buffers used for binding or protein synthesis studies. The final reelin concentration was estimated by dot blot by using reelin H fusion protein as a reference standard (20). Recombinant reelin analyzed by immunoblotting after SDS/PAGE separation is composed mainly of a high molecular mass form (≈400 kDa) and by trace amounts of the 320- and 180-kDa proteins.
Receptor Binding Assay.
The binding of [125I]echistatin to integrin receptors expressed by SNS membranes was performed as described by Kumar et al. (17). SNS membranes were diluted in buffer containing 50 mM Tris, pH 7.4, 100 mM NaCl, 2 mM CaCl2, 1 mM MgCl2, 1 mM MnCl2, and 1% BSA. Triplicate samples (200 μl/0.2 mg of protein) were incubated with different concentrations of [125I]echistatin [2,000 Ci/mmol (1 Ci = 37 GBq), Amersham Pharmacia] in a final volume of 300 μl for 2 h at room temperature. At the end of the incubation period, the mixture was filtered on Millipore filter paper that had been pretreated with 0.3% polyethylenimine (Sigma). Nonspecific binding was measured in the presence of a 200-fold molar excess of echistatin (Sigma) and was subtracted from the total binding to yield specific binding. Protein amounts were determined by the Bradford method (21). The dissociation constant, KD, the number of binding sites, Bmax, and the inhibition constant, Ki, were calculated for each experiment by a computer-assisted curve-fitting program based on ebda and ligand (22).
[35S]Methionine Incorporation into SNS Proteins.
SNSs were resuspended in a minimum volume (1–2 ml) of cold buffer containing 125 mM NaCl, 2 mM potassium acetate, 100 mM sucrose, 50 mM Hepes, pH 7.5, 0.2 mg/ml heparin, 0.05 mg/ml cycloheximide, 2 mM DTT, 2 mM CaCl2, ATP (3.5 mM), and [35S]methionine (30 μCi/30 pmol/ml, NEN) (23). The sample was then diluted in the same buffer to reach a protein content of 6–8 mg/ml. Triplicate 0.5-ml aliquots were incubated at 37°C, for the time and treatment indicated in Results. The reaction was terminated by adding 0.5 ml cold 10% trichloroacetic acid (TCA) and filtering on glass microfiber membranes, washing three times with 5 ml of 5% TCA.
To study [35S]methionine incorporation into Arc, the SNSs were lysed with buffer [1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and a complete mixture of protease inhibitors (Sigma)]. The lysate was incubated overnight at 4°C with anti-Arc (1:500) antibodies. The immunocomplex was captured by adding 50 μl of Protein G agarose bead slurry. The agarose beads were then collected by microcentrifugation and washed three times with PBS. The immunocomplex was eluted by using glycine buffer, pH 2.2, and incorporated [35S]methionine was measured in a liquid scintillation counter.
Results
Characteristics of SNSs.
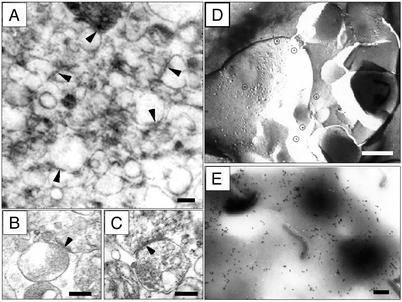
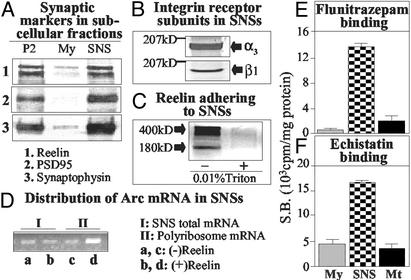
SNS fractions from mouse neocortex were studied electron-microscopically (Fig. 1) and biochemically (Fig. 2). The presence of presynaptic axon-terminals is indicated by the visualization of synaptic endings containing vesicles (Fig. 1 A, B, and C) and the enrichment in synaptophysin immunoreactivity (Fig. 2A). The electron microscopic characterization of the postsynaptic components included: (i) the visualization of postsynaptic densities and postsynaptic particles (polysomes?) (Fig. 1 B and C), (ii) the detection of α3 integrin receptor subunit immunoreactivity in the label fracture replicas of the SNS (Fig. 1 D and E); (iii) enrichment of PSD95 (Fig. 2A), (iv) the expression of α3,β1 integrin receptor subunits (Fig. 2B), (v) the enrichment of [3H]flunitrazepam binding to GABAA receptors (Fig. 2E), and (vi) enrichment of [125I]echistatin binding to integrin receptors (Fig. 2F). Electron microscopy studies of the SNS preparation failed to detect nuclear and myelin contaminations. Also, the glial cell contamination was low, as less than 5% of the total cyclic 2′,3′ esterase activity present in the homogenate before fractionation was expressed in the SNS fraction. The SNS fraction also contained mRNAs. Fig. 2D shows that Arc mRNA is compartmentalized in the cytosolic and translocates in the polyribosomal fraction after reelin addition.
Figure 1.
Electron microscopy of SNSs. (A–C) Presynaptic endings containing vesicles and postsynaptic structures (arrows) with postsynaptic densities and scattered polysomes. (Scale bar = 0.5 μm.) (B and C) Larger magnification of SNSs with or without 0.01% Triton X-100 treatment, respectively. (Scale bar = 0.5 μm.) (D) Label-fracture replica showing the ImmunoGold (Aurion, Wageningen, The Netherlands) labeling of the α3 integrin receptor subunit (circles) in a postsynaptic membrane, E-face. (Calibration bar = 0.4 μm.) (E) Electron micrograph showing the ImmunoGold labeling of the α3 integrin subunit in unfractured SNSs. The round black areas observed in the micrograph correspond to presynaptic terminals to which remain attached postsynaptic membranes showing heavy labeling for the α3 integrin subunit (black dots). (Calibration bar = 0.4 μm.)
Figure 2.
Biochemical characteristics of SNSs. (A) Western immunoblot with antibodies specific for reelin, 400- and 320-kDa (A1); postsynaptic density protein, 95-kDa (A2); and synaptophysin, 35-kDa proteins (A3). (B) Western immunoblot with α3 or β1 integrin receptor subunit-specific antibodies in 0.01% Triton X-100-treated SNSs. (C) Western immunoblot with reelin-specific (G10) antibody of SNSs before and after treatment with 0.01% Triton X-100. (D) RT-PCR of Arc mRNA in SNSs incubated without (lanes a and c) or with (lanes b and d) reelin (30 pM/45 min at 22°C and 30 min at 37°C). Total SNS (I) and polyribosome (II) Arc mRNA. (E) S.B. of [3H]flunitrazepam (2 nM) to SNS, myelin (My), and mitochondria (Mt) fractions. Each bar is the mean ± SE of triplicate assays. (F) S.B. of [125I]echistatin (50 fM) to the samples described in E. Each bar is the mean ± SE of triplicate assays.
Binding of Echistatin and Reelin to SNS Membranes.
Reelin-like immunoreactive proteins (mostly 400-kDa, but including a relative small amount of 180-kDa, and only traces of 320-kDa) adhere to SNS (Fig. 2 A1 and C). These reelin-like immunoreactive proteins, unlike the integrin receptor subunits, can be easily removed from SNSs with a 0.01% Triton X-100 treatment (Fig. 2C).
The binding characteristics of echistatin or reelin to integrin receptors were studied in synaptic membranes obtained from 0.01% Triton X-100-treated SNSs.
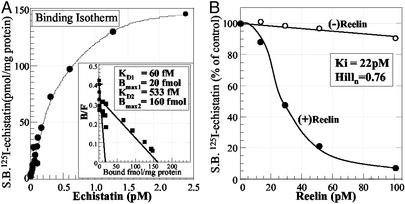
Fig. 3A shows that [125I]echistatin binds to two apparent integrin recognition sites: one with a very high affinity (KD ≈ 60 fM) and another with a nine times lower affinity (KD ≈ 533 fM). The binding of [125I]echistatin is almost completely displaced (85 ± 10% SE of control binding, n = 3) by 5 μM GRGDSP, a polypeptide with specific integrin receptor antagonist activity but not by GRADSP, a polypeptide devoid of integrin receptor binding (8–10). Recombinant mouse reelin displaces the high-affinity [125I]echistatin binding with a Ki of 22 pM and with a Hill slope close to the unit, suggestive of a competitive binding interaction between [125I]echistatin and reelin (Fig. 3B). Under our binding conditions, reelin displaces [125I]echistatin by the same extent in the presence or absence of 1 mM PMSF, an inhibitor of the possible serine-protease activity associated with recombinant reelin (24). Reelin does not influence the binding of flunitrazepam [specific binding of [3H]flunitrazepam (2 nM) with 100 pM reelin is decreased by only 8 ± 0.05% SE, n = 3, when compared with control binding without reelin).
Figure 3.
Competition between echistatin and reelin binding to integrin receptors. (A) S.B. [125I]echistatin to SNS membranes treated with 0.01% Triton X-100 to remove native reelin. (Inset) Scatchard analysis of the data. (B) Recombinant reelin dose-dependent displacement of [125I]echistatin binding to integrin receptors. Culture medium from mock transfected cells [(−)reelin] was used as control. Each data point is the average of triplicate measurements. Experiments were repeated three to five times, yielding similar results.
Increased [35S]Methionine Incorporation into Proteins Elicited by Reelin Is Blocked by Echistatin and Rapamycin.
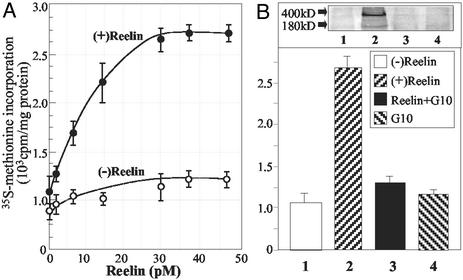
When SNSs treated with 0.01% Triton X-100 were incubated at 37°C with [35S]methionine, they incorporated the labeled amino acid into proteins with a linear rate during the first 45 to 60 min of incubation. This incorporation was blocked if the SNSs were incubated at 0°C or if the samples were treated with cycloheximide (0.05 mg/ml) but not with chloramphenicol (100 μg/ml), an inhibitor of mitochondrial protein synthesis (25, 26). When SNSs were incubated at 22°C with reelin, they bound reelin; equilibrium was reached after ≈1 h of incubation. Reelin (2 to 40 pM) enhanced the incorporation of [35S]methionine into proteins in a concentration-dependent manner, reaching a plateau (≈200% stimulation) at 30 pM (Fig. 4A). This stimulation was prevented by reelin immunoprecipitation with anti-reelin G-10 antibody (Fig. 4B).
Figure 4.
Reelin stimulates [35S]methionine incorporation into SNS proteins. (A) Dose–response curve of reelin stimulation of [35S]methionine incorporation into proteins. SNSs treated with 0.01% Triton X-100 were preincubated with recombinant reelin [(+)reelin)] or culture medium of mock transfected cells [(−)reelin] (45 min at 22°C before adding [35S]methionine and additional 30 min at 37°C after methionine addition). Each value is the mean ± SE of triplicate assays. (B) An aliquot of reelin (30 pmol in 10 μl) was incubated with equal volume of G10 antibody (10 mg protein/ml) at 4°C overnight. After absorption with Protein-G Sepharose, an aliquot of the supernatant, diluted to obtain a final reelin concentration of 30 pM, was applied to SNSs. Western immunoblot (Inset) of reelin content in supernatants preabsorbed with or without G10 antibody. Each value is the mean ± SE of triplicate assays.
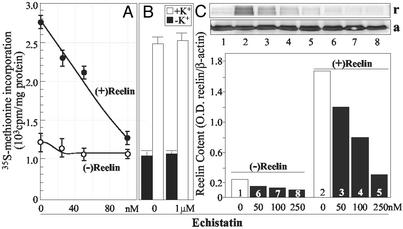
Reelin-mediated stimulation of protein synthesis was abated by echistatin (Fig. 5A) at concentrations that reduce the binding of reelin to SNSs (see Fig. 5C). These echistatin concentrations had a small inhibitory effect on the basal protein synthesis (Fig. 5A) but failed to inhibit the protein synthesis stimulated by 40 mM K+ (Fig. 5B). The incorporation of [35S]methionine into proteins elicited by reelin and high K+ was not modified by addition of 1 mM PMSF.
Figure 5.
Echistatin blocks reelin but not high K+ ion-induced [35S]methionine incorporation into proteins. (A) Echistatin dose dependently antagonizes the action of reelin. (B) Echistatin (1 μM) failed to abolish K+ (40 mM)-induced [35S]methionine incorporation into SNS proteins. (C) Echistatin displaces reelin bound to SNSs. Bars represent immunofluorescence intensity ratio between reelin (r) and β-actin (a). The SNSs were first incubated with or without recombinant reelin (30 pM) for 45 min at room temperature, then echistatin was added to the mixture and incubated for another 30 min at 37°C. After two washes, reelin bound to SNS membranes was measured by Western immunoblot (Inset) with G-10 anti-reelin antibody and referred to β-actin immunoreactivity. Each bar is the average of determinations made in triplicate. Open bars, non-echistatin treatment; filled bar, echistatin treatment. Lane 1, non-treatment; lane 2, reelin; lanes 3–5, reelin plus echistatin; lanes 6–8, echistatin.
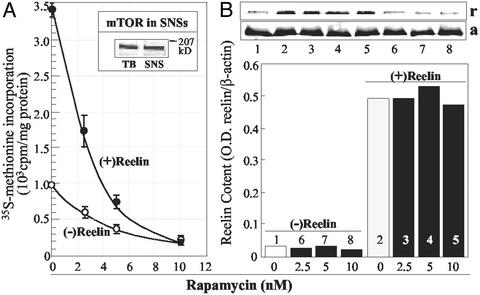
Because SNSs express mTOR (Fig. 6A Insert), a kinase operative in the translation of resident mRNAs (18, 27), we studied whether rapamycin, a blocker of the mTOR-signaling pathway, reduces the protein synthesis stimulated by reelin. As shown in Fig. 6A, rapamycin reduces the incorporation of [35S]methionine into proteins in a concentration-dependent manner without affecting the binding of reelin to SNSs (Fig. 6B).
Figure 6.
Rapamycin blocks reelin-induced [35S]methionine incorporation into proteins. (A) Rapamycin dose dependently inhibits reelin (30 pM) stimulation of [35S]methionine incorporation into SNS proteins. Each value is the mean ± SE of triplicate assays. (Inset) Western immunoblot with mTOR-specific antibody. TB, total brain. (B) Western immunoblot of reelin bound to SNSs after incubation with or without reelin in the presence or absence of different doses of rapamycin. Bars represent immunofluorescence intensity ratio between reelin (r) and β-actin (a). Lane 1, non-treatment; lane 2, reelin; lanes 3–5, reelin plus rapamycin; lanes 6–8, only rapamycin.
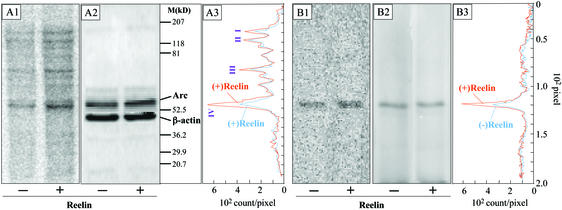
Fig. 7A shows the SDS/PAGE profile of TCA precipitated [35S]methionine-labeled proteins from control and reelin-treated SNSs. The intensity of the radioactivity incorporated within the protein bands (Fig. 7A1) and the densitometric measurements (Fig. 7A3) indicated that, under basal conditions, the extent of 35S incorporation into proteins of 150, 120, 80, and 55 kDa was smaller than that occurring in preparations stimulated by reelin. Reelin stimulated the incorporation of [35S]methionine into the 55-kDa protein band more efficiently than in the 150-, 120-, and 80-kDa protein bands (Fig. 7 A1 and A3). The radioactive band corresponding to the 55-kDa protein has an electrophoretic mobility identical to that of Arc immunoreactivity (Fig. 7A2). When the incorporation of [35S]methionine into Arc was measured after Arc immunoprecipitation, only the band (55 kDa) immunopositive for Arc associates with a radioactive peak (Fig. 7 B1, B2, and B3). The specific activity of [35S]methionine incorporated into Arc under basal condition was smaller than that of [35S]methionine incorporation after reelin stimulation. However, Arc steady-state protein levels were not increased significantly.
Figure 7.
Reelin enhances [35S]methionine incorporation into selected proteins. (A) SNSs preincubated for 45 min at room temperature without or with reelin (30 pM) were incubated for 30 min at 37°C with [35S]methionine, and proteins were precipitated with 10% TCA. Equal amounts of proteins (10 μg) from (−)- or (+)Reelin-stimulated samples were subjected to SDS/7% PAGE and Western blotting. (A1) Western blot autoradiography of unstimulated (−) and reelin-stimulated (+) SNS samples, respectively. (A2) Western immunoblot of A1 with Arc and β-actin antibodies. No apparent differences in the amount of β-actin or Arc immunoreactivity were observed. (A3) Phosphor screen peaks (I, 150 kDa; II, 120 kDa; III, 80 kDa; and IV, 55 kDa) of the radioactive bands of A1. The areas under the peaks were corrected for the amount of β-actin measured in A2 by fluorescence intensity [i.e., (−)reelin = 2.7; (+)reelin 2.7 pixel 106]. The ratios of the area under peak I, II, III, and IV between (+)- and (−)reelin after β-actin correction were 1.7, 1.1, 1.2, and 2.2, respectively. Peak IV radioactive band corresponds to the A2 band of Arc immunoreactivity. (B) SNSs incubated as described in A were lysed and then were subjected to immunoprecipitation with anti-Arc antibody (see Materials and Methods) before SDS/7% PAGE and Western blot. The data show that reelin increases the incorporation of [35S]methionine into Arc. (B1) Western blot autoradiography showing a single radioactive band of 55 kDa in unstimulated (−) and reelin-stimulated (+) SNS samples. (B2) Western immunoblot of B1 with Arc-specific antibody, indicating the radioactive band in B1 has an electrophoretic position identical to that of Arc. (B3) Phosphor screen peaks of 55-kDa radioactive bands of B1 in (−)reelin and (+)reelin SNS-stimulated samples. The areas under the peaks were corrected for the amount of Arc measured in B2 by fluorescence intensity [i.e., (−)reelin = 3.2; (+)reelin 3.0 pixel 106]. The ratios of the area under the 55-kDa peaks between (+)- and (−)reelin was 1.55. Experiments were repeated three times, yielding similar results.
Reelin Enhances [35S]Methionine Incorporation into Arc.
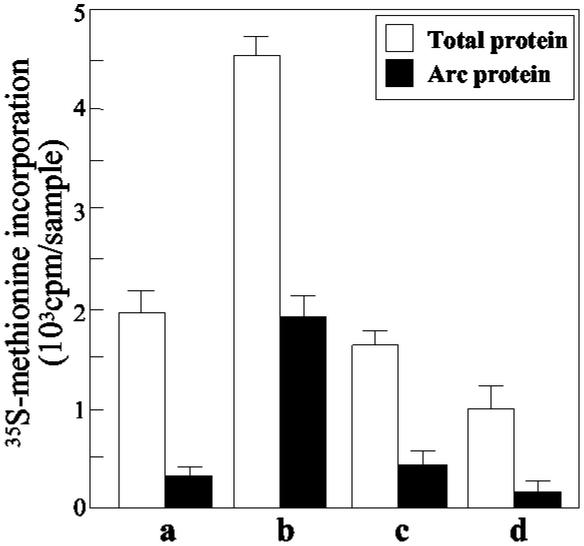
Arc mRNA is expressed in SNS preparations, and, although the total amount of Arc mRNA is not affected by reelin, after this treatment, there was a significant shift of Arc mRNA from the cytosolic to the polyribosomal fraction (Fig. 2D). The [35S]methionine incorporation into Arc was measured after Arc immunoprecipitation. As shown in Fig. 8, under basal conditions Arc [35S]methionine incorporation accounted for ≈10% of the total incorporation of this amino acid whereas, in the presence of reelin (30 pM) ≈40% of the radioactive amino acid incorporated into total proteins was immunoprecipitated with the Arc antibody. Despite this increased synthesis, no change of steady state Arc protein levels was measured in SNSs when the fluorescent intensity of the Arc immunoreactive band was related to the intensity of the β-actin band (Arc/β-actin ratio: basal = 0.13 ± 0.01; reelin stimulated = 0.12 ± 0.015; n = 3, nonsignificant). The reelin-induced increased [35S]methionine incorporated into Arc as well as into other proteins was prevented by preincubation of SNSs with rapamycin (Fig. 8).
Figure 8.
Reelin selectively stimulates [35S]methionine incorporation into Arc: inhibition by rapamycin. Open bars represent [35S]methionine incorporated into total sample (2 mg of protein per sample). Filled bars represent [35S]methionine incorporated into Arc protein (immunoprecipitated with Arc polyclonal-specific antibody, see Materials and Methods). SNSs were treated with none, basal condition (a), reelin (30 pM) (b), reelin (30 pM) and rapamycin (5 nM) (c), and rapamycin only (5 nM) (d). Each bar is the mean ± SE of triplicate assays.
The incorporation of [35S]methionine into Arc can be stimulated by brain-derived neurotrophic factor [BDNF; control = 730 ± 84; BDNF (35 ng/ml) = 1,208 ± 104 cpm per sample; P < 0.05, n = 3]. However, this stimulation was not blocked by echistatin [BDNF (35 ng/ml) + echistatin (200 nM) = 1,248 ± 79 cpm per sample, n = 3] in a dose that blocks reelin-induced [35S]methionine incorporation into Arc.
Discussion
To the best of our knowledge, the results of this study provide the first direct evidence that integrin receptors expressed in DSPSD mediate the reelin-induced facilitation of Arc mRNA translation and very likely also that of other dendritic and/or spine resident mRNAs. This reelin activation of extrasomal protein synthesis was studied in an SNS preparation devoid of endogenous reelin immunoreactivity but enriched in Arc mRNA, mTOR protein kinase, and integrin receptors located on DSPSD.
We have characterized the properties of reelin adhesion to integrin receptors by studying the displacement of echistatin bound to SNS membranes (Fig. 3B). Previous studies have shown that [125I]echistatin binds with different affinities to recombinant RGD-dependent integrin receptor subtypes (i.e., α3β1, α5β1, α8β1, and ανβ1, etc.; ref. 28). Because α3,α5,α8,αν and β1,β5,β8 subunits are found in brain (9, 10), it is not surprising that binding of echistatin to SNS membranes shows positive cooperativity (see Fig. 3A), suggesting that a heterogeneous population of integrin receptors susceptible to RGD agonist activity is expressed in the SNS membrane preparations.
Reelin in pM concentrations seems to compete (Hill number close to 1) with the high affinity binding of [125I]echistatin to SNS membranes. The endogenous protease activity that is present in the recombinant reelin does not seem to be a factor in the displacement of [125I]echistatin binding because reelin action was unabated by addition of 1 mM PMSF. Moreover, the specificity of reelin binding to integrin receptors is supported by the complete absence of reelin binding to other specific high-affinity recognition sites located in DSPSD (see, for example, flunitrazepam binding).
The subtype(s) of integrin receptors that bind reelin were not identified directly in the present experiments; however, their presence is inferred by the expression of α3β1 subunits in spine postsynaptic densities (Fig. 1 D and E and Fig. 2B). The α3β1 integrin receptor subtype displays high affinity for echistatin and has been considered a putative brain receptor for ECM proteins such as reelin (5). Hence, one can speculate that the α3β1 integrin receptor may be a major high-affinity reelin receptor candidate operative in mediating reelin action at SNSs.
Several lines of evidence suggest that binding of ECM proteins to integrin receptors very likely underlies, in a synapse-specific manner, the morphological dendritic spine changes that are associated with consolidation of synaptic strength after repeated synaptic stimulation (29). The structural and functional changes at synapses sustained by an interaction of integrin receptors located at DSPSD with ECM proteins are likely mediated by dendritic activation of focal adhesion kinase as indicated by increases in tyrosine phosphorylation, activation of the src-family kinase fin, and mRNA translation (29). To gain insight into the consequences that the binding of ECM reelin to integrin receptors may have on local protein synthesis at dendrites and dendritic spines, we studied the incorporation of [35S]methionine into newly synthesized proteins in cortical SNSs. Our studies demonstrate that reelin stimulates the initial fast synthesis rate of Arc and of other unidentified rapidly inducible proteins and that this stimulation is blocked by echistatin concentrations that compete for reelin binding to integrin receptors expressed in SNSs.
A recent study reports that BDNF added to SNSs produces a specific increase in Arc expression (30). Our results have established that reelin, similar to BDNF, elicits a significant increase in the initial rate of Arc biosynthesis. However, this reelin activation of Arc biosynthesis can be blocked by echistatin in doses that do not block high potassium- or BDNF-induced stimulation of Arc mRNA translation. Thus, the specificity of reelin on the initial rates of Arc biosynthesis may be related to the fact that, under basal conditions, SNS protein synthesis occurs in more than one pool, but only a neuronal synaptosomal pool enriched in Arc mRNA and integrin receptors might selectively respond to reelin.
The increased incorporation of [35S]methionine into Arc does not seem to be due to a decrease of Arc degradation rate because the incorporation rate of [35S]methionine into Arc occurs in the absence of Arc accumulation. On the contrary, it is very likely that an increased rate of Arc mRNA translation is responsible for the increased incorporation of [35S]methionine into Arc because, after reelin addition, Arc mRNA is found associated with polyribosomes (Fig. 2D).
The increase of Arc and of other protein synthesis induced by reelin was blocked by rapamycin. Because rapamycin specifically inhibits the mTOR kinase signaling pathway, we suggest that this pathway may be operative in facilitating the mechanism for reelin-induced up-regulation of Arc biosynthesis specifically mediated by an activation of integrin receptors.
On reelin addition, Arc and other spine-resident mRNAs very likely acquire a polyadenylation tail that allows them to associate with polyribosomes and initiate the translation of dendritic resident mRNAs (31), which is blocked by echistatin and by rapamycin. Such a mechanism is apparently operative in the stimulation of Ca2+/calmodulin-dependent kinase II (CaMKII) translation (32, 33) but has not been yet demonstrated for Arc.
We are aware that the SNS preparation used in these experiments exhibits synaptic heterogeneity. Therefore, only some of the reelin–integrin responses observed in these in vitro experiments may be applicable to mechanisms controlling translation of dendritic resident mRNAs in vivo.
However, our data are consistent with the hypothesis that reelin–integrin interactions may be generating pivotal signal transduction events in the control of dendritic spine mRNA translation, thereby contributing to synaptic plasticity. A reelin action on synaptic plasticity in vivo can be inferred from reports that: (i) reelin in ECM is associated with integrin receptors at synapses between GABAergic or glutamatergic axon terminals and dendritic shafts and spines of pyramidal neurons in cortex (1–3), (ii) reelin application to rat hippocampal slices facilitates LTP (11), and (iii) the down-regulation of reelin expression in heterozygous reeler mice results in decreased cortical spine density (3, 12). Among dendritic resident mRNAs that are translated locally at synapses, Arc mRNA translation is rapidly induced in response to excessive electrical or sensory motor stimulation (14–16, ‡). Arc protein may bind to actin and other cytoskeletal proteins and thus may participate in synaptic remodeling and in synaptic strength stabilization. Arc was found to be decreased in the brain of reeler mice where the abundance of filopodia-like dendritic spines could be an index of a dendritic spine maturation deficit. These findings raise the possibility that reelin binding to integrin receptors at specific synapses (i.e., between GABAergic axon terminals and dendrites of glutamatergic pyramidal neurons) is a pivotal event that promotes highly selective synaptic modifications during event-related LTP plasticity associated with memory trace formation.
Acknowledgments
We thank Dr. E. S. Anton (University of North Carolina, Chapel Hill) and Dr. P. W. Vanderklish (The Scripps Research Institute, La Jolla, CA) for constructive criticisms and suggestions in the preparation of the manuscript. This work was supported by grants from the National Institutes of Health (MH062090 to E.C., MH062682 to D.R.G., MH60778 to N.R.S., and MH062188 to A.G.).
Abbreviations
- SNS
synaptoneurosomes
- ECM
extracellular matrix
- DSPSD
dendritic spine postsynaptic densities
- mTOR
mammalian target of rapamycin
- GABA
γ-aminobutyric acid
- LTP
long-term potentiation
- Arc
activity-regulated cytoskeletal protein
- TCA
trichloroacetic acid
- BDNF
brain-derived neurotrophic factor
Footnotes
Carboni, G., Liu, W. S., Pesold, C., Tueting, P., Costa, E. & Guidotti, A. (2001) Soc. Neurosci. Abstr. 27, 617.
Lacor, P. N., Rodriguez, M. A., Larson, J., Guidotti, A. & Costa, E. (2001) Soc. Neurosci. Abstr. 27, 1759.
References
- 1.Rodriguez M A, Pesold C, Liu W S, Kriho V, Guidotti A, Pappas G D, Costa E. Proc Natl Acad Sci USA. 2000;97:3550–3555. doi: 10.1073/pnas.050589797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pappas G D, Kriho V, Pesold C. J Neurocytol. 2001;30:413–425. doi: 10.1023/a:1015017710332. [DOI] [PubMed] [Google Scholar]
- 3.Costa E, Davis J, Grayson D R, Guidotti A, Pappas G D, Pesold C. Neurobiol Dis. 2000;8:723–742. doi: 10.1006/nbdi.2001.0436. [DOI] [PubMed] [Google Scholar]
- 4.Rice D S, Curran T. Annu Rev Neurosci. 2001;24:1005–1039. doi: 10.1146/annurev.neuro.24.1.1005. [DOI] [PubMed] [Google Scholar]
- 5.Dulabon L, Olson E C, Taglienti M G, Eisenhuth S, McGrath B, Walsh C A, Kreidberg J A, Anton E S. Neuron. 2000;27:33–44. doi: 10.1016/s0896-6273(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 6.Howell B W, Herrick T M, Hildebrand J D, Zhang Y, Cooper J A. Curr Biol. 2000;10:877–885. doi: 10.1016/s0960-9822(00)00608-4. [DOI] [PubMed] [Google Scholar]
- 7.Chavis P, Westbrook G. Nature. 2001;411:317–321. doi: 10.1038/35077101. [DOI] [PubMed] [Google Scholar]
- 8.Staubli U, Chun D, Lynch G. J Neurosci. 1998;18:3460–3469. doi: 10.1523/JNEUROSCI.18-09-03460.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun D, Gall C M, Bi X, Lynch G. Neuroscience. 2001;85:815–829. doi: 10.1016/s0306-4522(01)00173-7. [DOI] [PubMed] [Google Scholar]
- 10.Kramar E A, Bernard J A, Gall C M, Lynch G. Neuroscience. 2002;110:29–39. doi: 10.1016/s0306-4522(01)00540-1. [DOI] [PubMed] [Google Scholar]
- 11.Weeber E J, Beffert U, Jones C, Christian J M, Forster E, Sweatt J D, Herz J. J Biol Chem. 2002;277:39944–39952. doi: 10.1074/jbc.M205147200. [DOI] [PubMed] [Google Scholar]
- 12.Liu W S, Pesold C, Rodriguez M A, Carboni G, Auta J, Lacor P, Larson J, Condie B, Guidotti A, Costa E. Proc Natl Acad Sci USA. 2001;98:3477–3482. doi: 10.1073/pnas.051614698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larson J, Hoffman J S, Guidotti A, Costa E. Brain Res. 2003;971:40–46. doi: 10.1016/s0006-8993(03)02353-9. [DOI] [PubMed] [Google Scholar]
- 14.Lyford G L, Yamagata K, Kaufmann W E, Barnes C A, Sanders L K, Copeland N G, Gilbert D J, Jenkins N A, Lanahan A A, Worley P F. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 15.Steward O, Schuman E M. Annu Rev Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- 16.Steward O, Worley P F. Proc Natl Acad Sci USA. 2001;98:7062–7068. doi: 10.1073/pnas.131146398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar C C, Nie H, Rogers C P, Malkowski M, Maxwell E, Catino J J, Armstrong L. J Pharmacol Exp Ther. 1997;283:843–853. [PubMed] [Google Scholar]
- 18.Sabatini D M, Lai M M, Snyder S H. Mol Neurobiol. 1997;15:223–239. doi: 10.1007/BF02740635. [DOI] [PubMed] [Google Scholar]
- 19.Jones D H, Matus A I. Biochim Biophys Acta. 1974;356:276–287. doi: 10.1016/0005-2736(74)90268-5. [DOI] [PubMed] [Google Scholar]
- 20.Lacor P N, Grayson D R, Auta J, Sugaya I, Costa E, Guidotti A. Proc Natl Acad Sci USA. 2000;97:3556–3561. doi: 10.1073/pnas.050589597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.McPherson G A. A Collection of Radioligand Analysis Programs. Cambridge, U.K.: Elsevier; 1986. [Google Scholar]
- 23.Weiler I J, Greenough W T. Mol Cell Neurosci. 1991;2:305–314. doi: 10.1016/1044-7431(91)90060-2. [DOI] [PubMed] [Google Scholar]
- 24.Quattrocchi C C, Wannenes R, Persico A M, Ciafre S A, D'Arcangelo G, Farace M G, Keller F. J Biol Chem. 2002;277:303–309. doi: 10.1074/jbc.M106996200. [DOI] [PubMed] [Google Scholar]
- 25.Polosa P L, Attardi G. J Biol Chem. 1991;266:10011–10017. [PubMed] [Google Scholar]
- 26.Rao A, Seward O. J Neurosci. 1991;11:2881–2895. doi: 10.1523/JNEUROSCI.11-09-02881.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown E J, Schreiber S L. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 28.Thibault G. Mol Pharmacol. 2000;58:1137–1145. doi: 10.1124/mol.58.5.1137. [DOI] [PubMed] [Google Scholar]
- 29.Martin K C, Kosik K S. Nat Rev Neurosci. 2002;3:813–820. doi: 10.1038/nrn942. [DOI] [PubMed] [Google Scholar]
- 30.Yin Y, Edelman G M, Vanderklish P W. Proc Natl Acad Sci USA. 2002;99:2368–2373. doi: 10.1073/pnas.042693699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter J D, Lorenz L J. Curr Opin Neurobiol. 2002;12:300–304. doi: 10.1016/s0959-4388(02)00318-5. [DOI] [PubMed] [Google Scholar]
- 32.Bagni C, Mannucci L, Dotti C G, Amaldi F. J Neurosci. 2000;20:RC76. doi: 10.1523/JNEUROSCI.20-10-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu L, Wells D, Tay J, Mendis D, Abbott M A, Barnitt A, Quinlan E, Heynen A, Fallon J R, Richter J D. Neuron. 1988;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]