Abstract
Many surface proteins of pathogenic gram-positive bacteria are linked to the cell wall envelope by a mechanism requiring a C-terminal sorting signal with an LPXTG motif. Surface proteins of Streptococcus pneumoniae harbor another motif, YSIRK-G/S, which is positioned within signal peptides. The signal peptides of some, but not all, of the 20 surface proteins of Staphylococcus aureus carry a YSIRK-G/S motif, whereas those of surface proteins of Listeria monocytogenes and Bacillus anthracis do not. To determine whether the YSIRK-G/S motif is required for the secretion or cell wall anchoring of surface proteins, we analyzed variants of staphylococcal protein A, an immunoglobulin binding protein with an LPXTG sorting signal. Deletion of the YSIR sequence or replacement of G or S significantly reduced the rate of signal peptide processing of protein A precursors. In contrast, cell wall anchoring or the functional display of protein A was not affected. The fusion of cell wall sorting signals to reporter proteins bearing N-terminal signal peptides with or without the YSIRK-G/S motif resulted in hybrid proteins that were anchored in a manner similar to that of wild-type protein A. The requirement of the YSIRK-G/S motif for efficient secretion implies the existence of a specialized mode of substrate recognition by the secretion pathway of gram-positive cocci. It seems, however, that this mechanism is not essential for surface protein anchoring to the cell wall envelope.
Signal peptide-bearing precursor proteins are initiated into the secretory pathway and translocated across the plasma membranes of bacterial cells (3, 5, 46). All signal peptides comprise a string of 13 to 20 hydrophobic amino acids, which are necessary and sufficient for the recognition and transport of precursor proteins by the secretion machinery (1, 6, 17). Two modes of precursor translocation have been described. During posttranslational translocation, cytoplasmic chaperones, for example, Escherichia coli SecB (24), bind newly synthesized precursors, which are subsequently initiated into the secretion pathway (42, 43). SecA, an ATPase that binds signal peptide-bearing precursors (16, 37), pushes polypeptides through the membrane translocon (11). The translocon can be viewed as a channel-forming membrane protein complex and is composed of SecY, SecE, and SecG (13, 18, 47). SecD, SecF, and YajC represent other components of the secretion machinery that are required for in vivo secretion but are dispensable for in vitro translocation of precursors; the precise function of these factors is still unknown (10, 12, 40). Signal peptides initiate some precursor proteins into the signal recognition particle (SRP)-mediated cotranslational translocation pathway (68). Binding of the SRP to nascent polypeptides leads to the binding of ribosome-SRP complexes first to the SRP receptor and then to ribosomes docking on the translocon (4, 69). In this manner, translation and translocation of a portion of nascent polypeptides seem coupled as the ribosomes extrude polypeptides into the translocon channel.
Although all signal peptide-bearing proteins are by default translocated across the plasma membrane, the subsequent fate of precursors can be modified by the presence or absence of specific cleavage sites for signal peptidases (7). Type I signal peptides comprise a cleavage site for signal (leader) peptidase, and the mature polypeptides are released from the membrane (8). Type II signal peptides are the substrate for covalent modification with thioether-linked diacylglycerol (15). After cleavage by type II signal peptidases (59), the resulting lipoproteins can traffic to the plasma (inner) or outer membranes of gram-negative bacteria (72). Prepilin signal peptides are cleaved by prepilin signal peptidases (36), enzymes that remove an N-terminal sequence tag from signal peptides, which is followed by methylation of the amino group of phenylalanine at the N termini of mature pilins (55). Prepilin signal peptidases use signal peptide-bearing precursors and S-adenosylmethionine as substrates and act in the bacterial cytoplasm, i.e., prior to translocation (36). The mature polypeptides retain their signal peptide function for subsequent translocation by the Sec pathway (55). Translocated pilins are assembled into pilus structures that penetrate through a protein pore in the outer membrane onto the bacterial surface. Although it has been speculated that pilins may be translocated by the Sec pathway at designated locations, the presumed assembly sites of pili, all available experimental evidence supports the notion that signal peptide-bearing precursors are transported by Sec machineries without consideration for the final destination of mature polypeptides (56). In this view of protein trafficking, all modification and targeting steps involving translocated polypeptides occur subsequently to and independently of the activation of the Sec machinery (41).
Rosenstein and Götz first noticed the presence of a signal peptide motif in staphylococcal lipases (44). The same motif, YSIRK-G/S, was also identified during genome sequence analysis of Streptococcus pneumoniae and was found within signal peptides of proteins bearing C-terminal cell wall sorting signals with an LPXTG motif (58). Those authors proposed that the signal peptide motif may be required for the anchoring of surface proteins to the cell wall envelope (58). Although this mechanism has been studied only in Staphylococcus aureus and in a few other microbes, it is assumed that all gram-positive bacteria anchor surface proteins bearing C-terminal sorting signals by a universal process involving five steps (30, 33). Precursor proteins are initiated into the secretory pathway by their N-terminal signal peptides and translocated, and their signal peptides are cleaved (step 1) (51). The C-terminal sorting signal first retains polypeptides within the secretory pathway (step 2) (50) and then allows cleavage of the peptide bond between the threonine (T) and the glycine (G) of a conserved LPXTG motif (step 3) (32). The carboxyl group of threonine is subsequently amide linked to the amino group of the pentaglycine crossbridge within lipid II precursor molecules (step 4) (39, 49). The sorting intermediate of surface protein linked to lipid II is incorporated into the cell wall via the transpeptidation and transglycosylation reactions of peptidoglycan synthesis (step 5), thereby tethering the C termini of surface proteins to the cell wall envelope (34, 60, 62). Thus, in contrast to signal peptides, which act in the bacterial cytoplasm and prior to translocation (3), sorting signals function immediately after, but not during, the translocation of polypeptides. If a specific involvement of signal peptides in surface protein anchoring is assumed, one could entertain a model in which signal peptides with YSIRK-G/S motifs initiate proteins into a dedicated secretion pathway.
It is shown here that the YSIRK-G/S motif plays a role in the efficiency of the secretion of protein A, a cell wall-anchored surface protein of S. aureus. The YSIRK-G/S motif is, however, dispensable for the cell wall anchoring of surface proteins. Several models are discussed to account for the existence of two classes of signal peptides in gram-positive cocci.
MATERIALS AND METHODS
Bacterial strains.
S. aureus strains RN4220 (wild-type protein A) (23) and OS2 (spa::ermC) (51) were used as hosts for plasmid constructs and for protein-targeting experiments. E. coli DH5α was used for recombinant-DNA experiments (14).
Plasmids and mutagenesis.
Plasmids pSPA and pSPA1-519 were used as templates for generating mutations of the YSIRK-G/S motif (51). Each mutagenesis was carried out with a QuickChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's recommendations. To delete the YSIR sequence, the oligonucleotides spa-dSSM-1 (GAAAAAGAAAAACATTAAACTAGGTGTAGGTATTGC) and spa-dSSM-2 (GCAATACCTACACCTAGTTTAATGTTTTTCTTTTTC) were used as primers for DNA polymerization. Alanine substitutions for glycine and serine residues were carried out with the oligonucleotides G/A-spa-F (TTCGTAAACTAGGTGTAGCTATTGCATCTGTAACTTT) and G/A-spa-R (AAAGTTACAGATGCAATAGCTACACCTAGTTTACGAA) and with the oligonucleotides S/A-spa-F (TAGGTGTAGGTATTGCAGCTGTAACTTTAGGTACATT) and S/A-spa-R (AATGTACCTAAAGTTACAGCTGCAATACCTACACCTA) as primers for DNA polymerization, respectively. To replace both the glycine and serine residues with alanine, GS/AA-spa-F (TAAACTAGGTGTAGCTATTGCAGCTGTAACTTTAGGTACA) and GS/AA-spa-R (TGTACCTAAAGTTACAGCTGCAATAGCTACACCTAGTTTA) were used as primers. All mutations were confirmed by DNA sequence analysis and electroporated into S. aureus OS2. pSEB and pSEB-CWS have previously been reported (50). To construct the pSPASP-SEB plasmid, an seb fragment lacking the coding sequence for the N-terminal signal peptide was amplified from chromosomal DNA of S. aureus S6 by PCR amplification with the primers SEB-2 (AAGGATCCAGATCCTAAACCAGATGAGT) and SEB-3 (AAATATGAAGAGTTAGTAATTAAGGATCCTT). The resulting product was cut with BamHI and then ligated with pSPA digested with BclI and BamHI. To generate pSPASP-SEB-CWS, the seb gene was amplified with the primers SEB-2 and SEB-4 (TGAAGTTTATCTTACGACAAAGAAAGCTTTT) and cut with BamHI and HindIII. The spa fragment containing the sorting signal was cut from pSPA by HindIII/BamHI digestion. The two DNA fragments were mixed and ligated with pSPA digested with BclI and BamHI.
Cell fractionation, immunoblotting, and pulse-chase analysis.
Cells were grown in tryptic soy broth (TSB) to log phase (optical density at 600 nm [OD600], 0.5 to 1.0), and an aliquot corresponding to 1 ml of cells with an OD600 of 1 was collected by centrifugation. The cells were washed twice with TSB prewarmed to 37°C and suspended in 0.5 ml of the prewarmed TSB. After incubation at 37°C for 15 min, the cells were collected by centrifugation and 450 μl of the supernatant was removed (medium fraction). The cell pellet was suspended in 0.5 ml of TSM (50 mM Tris HCl [pH 7.5], 0.5 M sucrose, 10 mM MgCl2) containing 0.1 mg of lysostaphin per ml and incubated at 37°C for 15 min. The sample was centrifuged, and 450 μl of the supernatant was removed (cell wall fraction). The pellet was suspended in 500 μl of lysis buffer (50 mM Tris HCl [pH 7.5], 0.15 M sodium chloride) and subjected to repeated freezing and thawing in a dry ice-ethanol bath, and the cell lysate was centrifuged in an Optima Max Ultracentrifuge (Beckman) for 35 min at 100,000 × g for 35 min. The supernatant (450 μl) was removed (cytoplasmic fraction), and the pellet was suspended in 500 μl of 0.1 M Tris-HCl (pH 7.5)-0.8% sodium dodecyl sulfate (SDS) (membrane fraction). Subsequently, an aliquot (5 μl) of each fraction was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) followed by immunoblotting analysis. For immunoblotting of protein A, the monoclonal antibody clone SPA-27 (Sigma) and peroxidase-conjugated goat immunoglobulin G (IgG) (Pierce) were used as primary and secondary antibodies, respectively. To visualize staphylococcal enterotoxin B (SEB) fusion proteins, anti-SEB antibody (Sigma), and goat anti-rabbit IgG antibody (Cell Signaling Tech) were used. Pulse-labeling experiments with [35S]methionine were performed as previously described (51). Immunoprecipitated protein A species were separated by SDS-PAGE, and radioactive signals were quantified by PhosphorImager analysis. The acquired data for P1 and P2 precursor species as well as mature protein A were analyzed with ImageQuant 1.1 software (Molecular Dynamics) and plotted against sampling times. Regression curves were calculated and used for precursor half-life predictions.
Immunofluorescence microscopy.
Cells were grown in TSB to mid-log phase (OD600, 0.5), and 1 ml of the culture was centrifuged for 5 min at 13,000 × g. The bacterial sediment was washed with 1 ml of phosphate-buffered saline (PBS; 10 mM sodium phosphate [pH 7.2], 0.15 M sodium chloride) and suspended in 1 ml of PBS. An aliquot of the cell suspension (100 μl) was mixed with 20 μl of 5% bovine serum albumin in PBS and incubated at room temperature for 15 min. Then 5 μl of Cy3-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) was added, and the mixture was incubated at room temperature for 1 h in the dark. After the bacteria were washed with 200 μl of PBS, the cells were suspended in 100 μl of PBS and viewed with an Olympus AX-70 fluorescence microscope and the images were captured with a charge-coupled-device camera.
FACS analysis.
During fluorescence-activated cell sorter (FACS) analysis, solutions were filtered through 0.22-μm-pore-size filters and reactions were carried out at room temperature. Staphylococci were grown to mid-log phase, collected by centrifugation for 5 min at 13,000 × g, washed twice with 1 ml of PBS, and then suspended in 1 ml of PBS. An aliquot of the cell suspension (100 μl) was mixed with 20 μl of 5% bovine serum albumin in PBS and incubated for 15 min. Then 4.8 μl of Cy5-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) was added and the mixture was incubated for 1 h in the dark. To stain bacterial cells, 120 μl of Live/Dead BacLight reagent (Molecular Probes) was added and the solution was incubated for 15 min. Cells were washed with 200 μl of PBS, suspended in 1 ml of PBS, and then analyzed with an LSR flow cytometer (BD Biosciences), with recordings being taken at wavelengths of 488 and 633 nm.
RESULTS
YSIRK-G/S motif in signal peptides of surface proteins from gram-positive bacteria.
To determine whether the YSIRK motif is present within signal peptides of surface proteins from gram-positive bacteria, we inspected the predicted amino acid sequences of surface proteins deposited in GenBank or genes identified by BLAST homology searches in which the LPXTG motif sorting signals were used as queries (29, 33). YSIRK-G/S motifs were found in surface proteins of streptococci (S. pneumoniae, Streptococcus pyogenes, Streptococcus gordonii, Streptococcus agalactiae, and Enterococcus faecalis) and staphylococci but were absent from surface proteins of listeriae, bacilli (Bacillus anthracis and Bacillus halodurans), clostridia, corynebacteria, Streptomyces spp., and Actinomyces spp. (data not shown). Table 1 shows a list of YSIRK-G/S motifs found within the predicted signal peptides of staphylococcal open reading frames. Some, but not all, of the surface proteins bearing a C-terminal sorting signal with an LPXTG motif harbor the YSIRK-G/S motif. Furthermore, YSIRK-G/S was found in four predicted precursor proteins that are likely secreted into the extracellular medium (Table 1).
TABLE 1.
Staphylococcal surface proteins and secreted proteins carrying the YSIRK-G/S motif
| Genea | LPXTGb motif | YSIRK-G/Sc motif | Sequence | Reference |
|---|---|---|---|---|
| clfA | + | + | HAIRKKSIGVAS | 31 |
| clfB | + | + | YSIRRFTVGTTS | 35 |
| fnbA | + | + | YGIRKHKLGAAS | 52 |
| fnbB | + | + | YGIRKHKLGAAS | 21 |
| mrp | + | + | FSIRKFNVGIFS | 70 |
| pls | + | + | YSIRRFTVGTAS | 30 |
| sdrC | + | + | FSIRKYSVGTAS | 22 |
| sdrD | + | + | FSIRKYTVGTAS | 22 |
| sdrE | + | + | FSIRKYTVGTAS | 22 |
| spa | + | + | YSIRKLGVGIAS | 64 |
| sasC | + | + | YSIRKYKVGIFS | 30 |
| sasE | + | + | SAMKKITMGTAS | 30 |
| sasG | + | + | YSIRKFTVGTAS | 30 |
| sasI | + | + | YSIRKSTLGVAS | 30 |
| sasJ | + | + | YSIRKSSLGVAS | 30 |
| cna | + | − | 38 | |
| sasA | + | − | 30 | |
| sasF | + | − | 30 | |
| sasH | + | − | 30 | |
| sasK | + | − | 30 | |
| sasL | + | − | 30 | |
| sasM | + | − | 30 | |
| geh1 | − | + | YSIRKYSIGVVS | 44 |
| geh2 | − | + | YSIRKFSVGASS | 44 |
| embB | − | + | FSIRKYTVGTFS | 25 |
| lytN | − | + | YSIRKVSIGILS | 25 |
Sas denotes S. aureus surface protein (29).
Sortase cleavage site. L, leucine; P, proline; X, any amino acid; T, threonine; G, glycine. Sortase cleaves between T and G.
(F/Y)SIRKXXXGXXS. F, phenylalanine; Y, tyrosine; S, serine; I, isoleucine; R, arginine; K, lysine; X, any amino acid; G, glycine.
Signal peptide mutants of protein A.
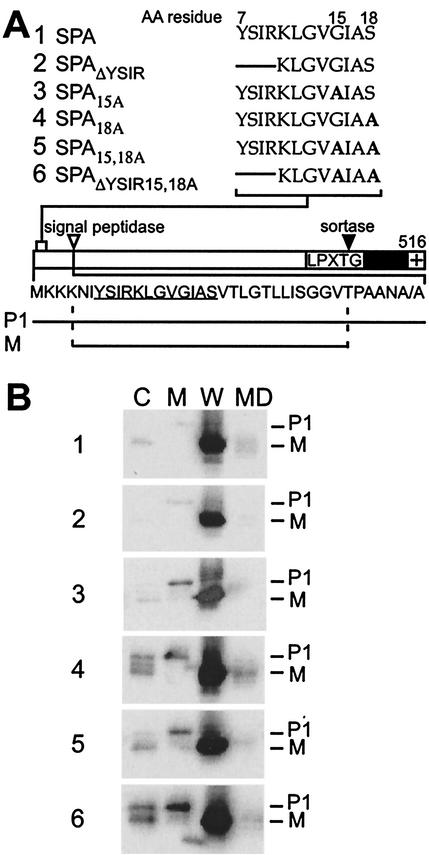
The YSIRK-G/S motif of staphylococcal protein A, an immunoglobulin binding surface protein (19), is located at amino acid position 7 of the precursor species (64). Spacers of 3 and 6 amino acids separate the lysine residue (K) of the YSIRK-G/S motif from the conserved glycine (G) and serine (S) residues, respectively (Fig. 1A). To analyze the function of the signal peptide motif, we employed S. aureus strain OS2 with an insertion of the ermC resistance gene upstream of the protein A coding sequence (51). The defect in the protein A expression of strain OS2 is complemented by plasmid-encoded wild-type protein A (SPA) and by various mutant alleles that are summarized in Fig. 1A. SPAΔYSIR carries a deletion of the YSIR sequence; the lysine residue (K) of the YSIRK-G/S motif was not deleted, as we feared that the removal of two positively charged residues might have a general impact on signal peptide function (67). The variants SPA15A and SPA18A harbor single amino acid substitutions replacing glycine 15 and serine 18 with alanine, respectively. SPA15,18A carries both alanine substitutions, whereas SPAΔYSIR/15,18A combines the two amino acid substitutions with a deletion of the YSIR sequence.
FIG. 1.
Cellular locations of protein A mutants. (A) The diagram displays wild-type protein A (SPA) with the N-terminal signal peptide, the LPXTG motif, the C-terminal hydrophobic domain (black bar), and the charged tail (+). The amino acid sequence of the signal peptide is shown below the diagram, in which the YSIRK-G/S sequence is underlined. The mutations in the YSIRK-G/S motif are shown above the diagram. The lines in the sequences indicate a deletion of amino acids (AA), while boldfaced A's indicate alanine substitutions. The precursor (P1) and mature species of protein A (M) are indicated below the diagram. (B) Cell fractionation of S. aureus OS2 expressing protein A mutants. Bacteria were grown to mid-log phase, and the cultures were fractionated into medium (MD), cell wall (W), membrane (M), and cytoplasmic (C) compartments. TCA-precipitated samples were subjected to SDS-10% PAGE, and protein A was detected by immunoblotting with monoclonal antibody.
To analyze the expression of various protein A alleles and the subcellular locations of their protein products, cultures of staphylococci harboring various plasmids were fractionated into the medium, cell wall envelope, membrane, and cytoplasmic compartments (51). Proteins were suspended in sample buffer and analyzed by SDS-PAGE and immunoblotting. We chose immunoblotting over immunoprecipitation of pulse-labeled protein, as the former technique permits analysis of the steady-state distribution of precursors and mature products in the cell, whereas the latter reveals only the final destination of the mature product. As reported previously, wild-type protein A is located in the cell wall compartment (51, 53). Signal peptide processing of protein A occurs very rapidly (see below), since P1 precursors, which bear an N-terminal signal peptide and a C-terminal sorting signal, cannot be detected by immunoblotting of membrane fractions. Deletion of the YSIR sequence did not affect the subcellular location of mutant protein A compared to that of wild-type SPA. Replacement of serine 18 with alanine, either alone or in combination with a glycine 15 replacement and YSIR deletion, affected the processing of signal peptides, as some of SPA18A appeared in the membrane and cytoplasmic compartments and migrated as a P1 precursor. Further, the cytoplasmic compartment of staphylococci expressing protein A variants with replacements of serine 18 revealed a small amount of immunoreactive species that migrated faster than P1 and more slowly than the mature, cell wall-anchored product. This observation is consistent with the hypothesis that some P1 precursor of SPA18A may be degraded in the cytoplasm without initiation into the secretory pathway.
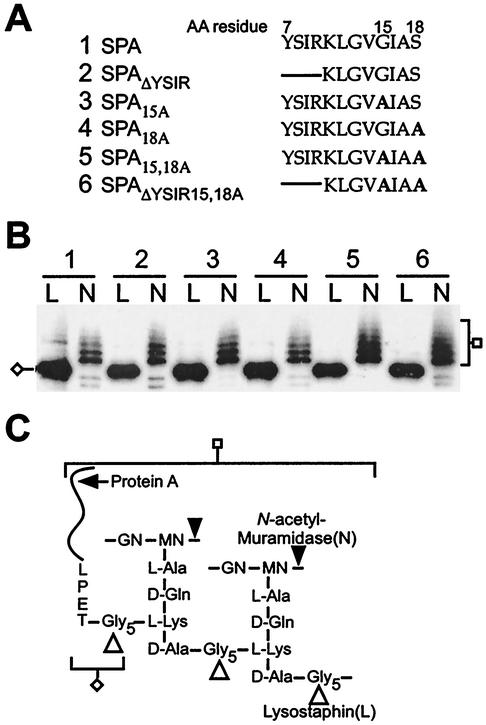
Cell wall anchoring of protein A signal peptide mutants.
To solubilize anchored surface proteins, the cell wall envelope of S. aureus must be cleaved with enzymes that attack the peptidoglycan macromolecule (51). Lysostaphin (Fig. 2) is a glycyl-glycine endopeptidase that cleaves the pentaglycine crossbridge (48), the anchoring point of surface proteins bearing sorting signals with an LPXTG motif (49). Mutanolysin, an N-acetylmuramidase (Fig. 2) (71), cleaves the glycan strands and is known to release a spectrum of protein A molecules with linked peptidoglycan fragments of various sizes (34). To assess the cell wall anchoring of protein A variants to the peptidoglycan, the cell wall envelope of S. aureus OS2 strains expressing various protein A mutants was isolated and digested with lysostaphin or muramidase and analyzed by SDS-PAGE and immunoblotting. Removal of the YSIR sequence or replacement of glycine 15 or serine 18 did not affect the cell wall anchoring of protein A, as all variants migrated upon SDS-PAGE with the characteristic lysostaphin and muramidase pattern observed for wild-type protein A (Fig. 2B).
FIG. 2.
Anchoring of protein A mutants to the cell wall. (A) YSIRK-G/S motif and mutant sequences of protein A and its variants. The lines in the sequences indicate a deletion of amino acids (AA), while boldfaced A's represent alanine substitutions. (B) S. aureus OS2 cells expressing protein A mutants were treated with either lysostaphin (L) or N-acetylmuramidase (N). The digested cell wall components were subjected to SDS-10% PAGE, and protein A mutants were detected by immunoblotting with monoclonal antibody. Cleavage products of specific enzymes are indicated. (C) Structure of the cell wall of S. aureus and cleavage sites for lysostaphin and N-acetylmuramidase. Protein A anchored to the peptidoglycan is drawn as a curved line, and the LPET sequence is tethered to the pentaglycine cell wall crossbridge (cleaved and anchored LPXTG motif).
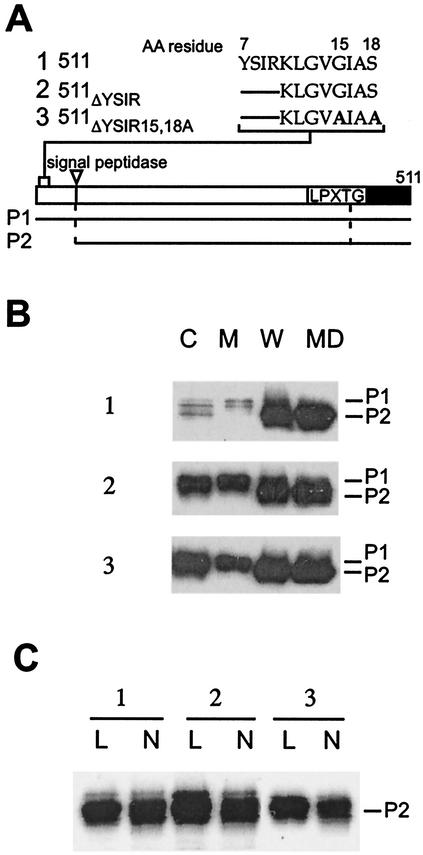
Signal peptide mutants of protein A truncated at the C terminus.
Deletion of the C-terminal charged tail of the sorting signal abolishes the cell wall anchoring of mutant protein A (51). The charged tail is thought to act as a signal for retaining the P2 precursors of surface proteins (signal peptide cleaved, sorting signal present) within the secretory pathway (50). This mechanism is a prerequisite for LPXTG substrate recognition by sortase (SrtA) (27, 28), the transpeptidase that anchors surface proteins to the peptidoglycan precursor lipid II (61). In fact, SPA511 (hereinafter referred to as 511), the variant lacking the C-terminal charged tail, cannot advance in the sorting pathway and is secreted as a P2 precursor species (51). Mutations in the signal peptide of 511 therefore provide an experimental advantage over full-length protein A mutants, as one can determine signal peptide precursor processing as a measure of protein secretion without considering its further processing by the cell wall-anchoring pathway. 511ΔYSIR carries a deletion of the YSIR sequence in truncated protein A (Fig. 3). 511ΔYSIR/15,18A combines the amino acid replacements of glycine and serine with a deletion of the YSIR sequence. Staphylococcal cultures expressing protein A variants were fractionated into the medium, cell wall envelope, membrane, and cytoplasmic compartments and analyzed by immunoblotting.
FIG. 3.
YSIRK-G/S motif mutants of C-terminally truncated protein A. (A) Diagram of the truncated protein A and sequences of YSIRK-G/S motif mutants. The diagram shows the cleavage site for signal peptidase as well as the LPXTG motif and the C-terminal hydrophobic domain (black bar). The P1 and P2 precursors (cleavage products of signal peptidase) are shown below the diagram. AA, amino acid. (B) S. aureus OS2 cells expressing protein A variants were grown to mid-log phase, and cultures were fractionated into medium (MD), cell wall (W), membrane (M), and cytoplasmic (C) compartments. The samples were subjected to SDS-10% PAGE, and protein A was detected by immunoblotting with monoclonal antibody. The migration of P1 and P2 precursors is indicated. (C) S. aureus OS2 cells expressing protein A mutants were treated with either lysostaphin (L) or N-acetylmuramidase (N). The digested cell wall components were subjected to SDS-10% PAGE, and protein A was detected by immunoblotting with monoclonal antibody.
Signal peptide processing of the protein A variant 511 occurs very rapidly, as only small amounts of the P1 precursor were detected during immunoblotting of membrane fractions. Equal amounts of 511 were detected in the cell wall and extracellular medium fractions (Fig. 3AB). Cell wall anchoring of 511 did not occur, however, as cell wall treatment with lysostaphin or muramidase did not affect the migration of 511 upon SDS-PAGE (Fig. 3C). Deletion of the YSIR sequence caused an increased amount of 511ΔYSIR to appear as the P1 precursor in the membrane and cytoplasmic compartments (Fig. 3B). Unlike 511, which bears the wild-type signal peptide, the P1 precursor of 511ΔYSIR/15,18A could be detected by immunoblotting. Thus, although the intrabacterial concentration of 511ΔYSIR/15,18A was not significantly increased over that of 511, the P1 precursor of the signal peptide mutant accumulated in the cytoplasm and membrane fractions.
Signal peptide processing and secretion of protein A mutants.
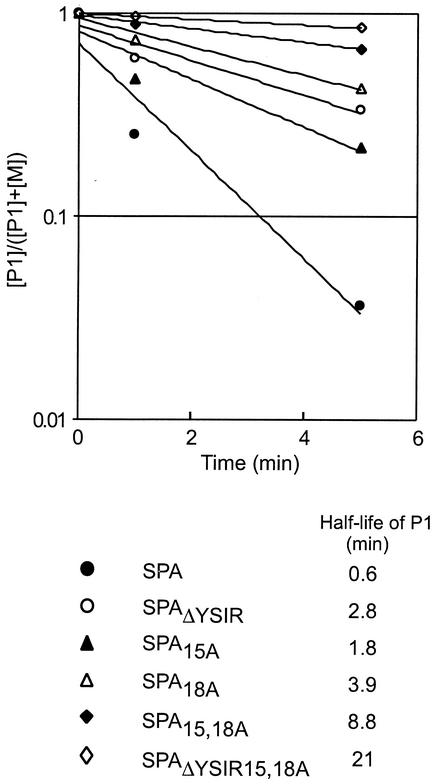
Bacterial type I and II signal peptidases are positioned on the trans side of the plasma membrane. Thus, signal peptide cleavage is a measure for the translocation of precursor proteins by the Sec machinery. We wondered whether the YSIRK-G/S motif is required for the efficient secretion of protein A and measured the rate of signal peptide processing. S. aureus OS2 carrying plasmids that encode protein A variants was suspended in minimal medium, and newly synthesized polypeptides were pulse-labeled by the addition of [35S]methionine. One minute after the addition of radioactive amino acid, an excess of nonradioactive methionine was added to quench all further incorporation of [35S]methionine. At timed intervals during or after the pulse, signal peptide processing and secretion were stopped by trichloroacetic acid (TCA) precipitation. Staphylococci were digested with lysostaphin, and protein A molecules were analyzed by immunoprecipitation followed by SDS-PAGE and PhosphorImager analysis (Fig. 4).
FIG. 4.
Precursor processing of YSIRK-G/S mutants of protein A. S. aureus cultures were pulse-labeled with [35S]methionine, and labeling was quenched with nonradioactive methionine. Aliquots of the culture were precipitated with TCA to stop protein processing at 0, 1, or 5 min after the conclusion of the pulse. Staphylococci were digested with lysostaphin, and proteins were again precipitated with TCA. Proteins were solubilized in hot SDS, immunoprecipitated with polyclonal antibody, and subjected to SDS-10% PAGE and PhosphorImager analysis. The logarithmic ratio of the concentration of the P1 precursor ([P1]) to the sum of the P1 precursor and mature protein A ([P1] + [M]) was calculated and plotted against time. The half-life of each P1 species was calculated by using regression curves.
The P1 precursor of wild-type protein A was rapidly processed (half-life, 0.6 min) to generate the mature anchored polypeptide lacking both the N-terminal signal peptide and the C-terminal sorting signal (Fig. 4). We failed to detect the P2 intermediate (signal peptide cleaved and sorting signal present) of protein A (data not shown). During our previous work, the P2 precursor of the cell wall sorting pathway could be detected only in SEB-cell wall sorting signal (CWS) hybrids and not in protein A (50). It appears that signal peptide processing of protein A may be the rate-limiting step of the sorting pathway, whereas the sortase-catalyzed cleavage of sorting signals does not influence the rate of anchoring. Replacement of glycine 15 with alanine showed the least effect on secretion or signal peptide processing (half-life, 1.8 min). In contrast, the mutant carrying deletions of YSIR and alanine substitutions at glycine 15 and serine 18 displayed the most significant reduction (half-life, 21 min) in the rate of protein A secretion and signal peptide processing. All other mutants also showed significantly delayed precursor processing (half-life, 2.8 to 8.8 min).
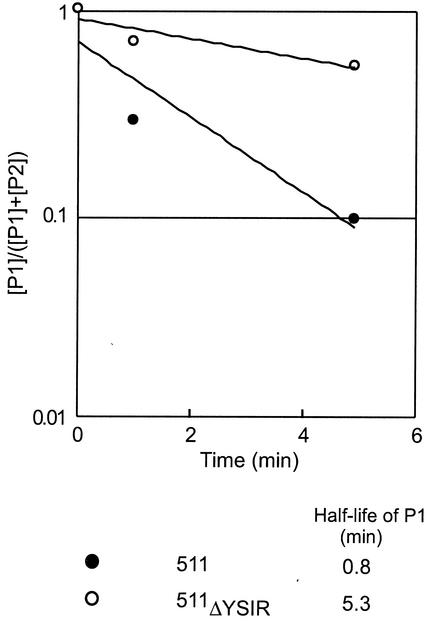
We wished to measure signal peptide processing and secretion of the C-terminally truncated protein A (511) and its variants, as these mutants are not linked to the cell wall envelope (Fig. 5). Thus, cleavage of the P1 precursor is a direct measure of signal peptide processing and secretion, without consideration of cell wall sorting. The signal peptide of protein A 511 is rapidly processed (half-life, 0.8 min). Deletion of the YSIR sequence caused a significant reduction in the processing of P1 precursors (half-life, 5.3 min). These results are consistent with the hypothesis that the YSIRK-G/S motif plays a role in the efficient secretion of protein A.
FIG. 5.
Precursor processing of a YSIRK-G/S mutant of C-terminally truncated protein A. S. aureus was pulse-labeled with [35S]methionine, and the labeling was quenched with nonradioactive methionine. Aliquots of the culture were precipitated with TCA to stop protein processing 0, 1, or 5 min after the pulse. Staphylococci were digested with lysostaphin, and proteins were again precipitated with TCA. Proteins were solubilized in hot SDS, immunoprecipitated with polyclonal antibody, and subjected to SDS-10% PAGE and PhosphorImager analysis. Because C-terminally truncated protein A is not subject to sortase-mediated cleavage and anchoring at the LPXTG motif, signal peptide processing generates only the P2 precursor. The logarithmic ratio of the concentration of the P1 precursor ([P1]) to the sum of the P1 precursor and the P2 precursor of protein A ([P1] + [P2]) was calculated and plotted against time. The half-life of each P1 species was calculated by using regression curves.
Surface display and function of protein A mutants.
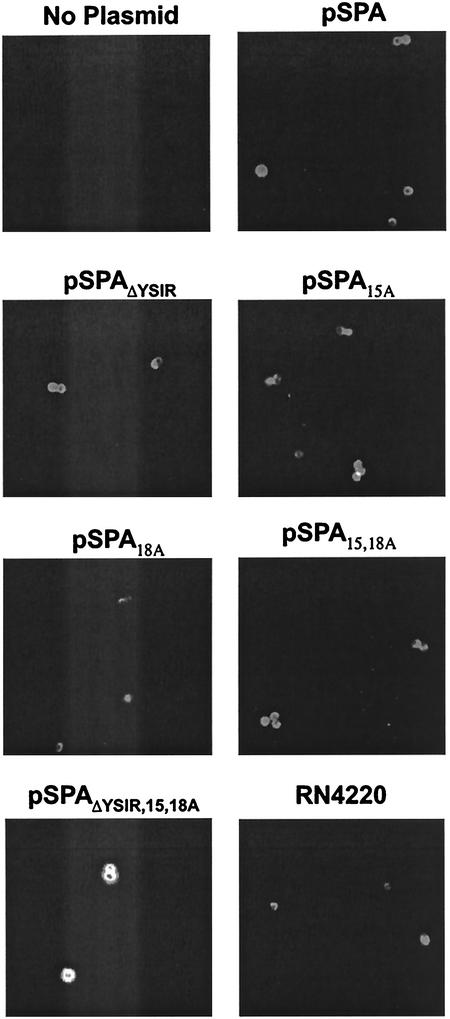
Our experimental scheme to determine the role of signal peptide motifs sought to measure surface protein display and function. Previous work revealed that protein A is distributed uniformly over the bacterial surface, presumably involving a mechanism of peptidoglycan growth beginning at defined assembly sites and spreading laterally over the protoplast surface (33). If so, is the YSIRK-G/S motif required for the uniform distribution of protein A on the bacterial surface? Furthermore, are cell wall protein A molecules folded and functional? To answer these questions, we measured the nonimmune binding of protein A to the C terminus of Cy3-labeled immunoglobulins. Binding was detected in a fluorescence microscopy experiment (Fig. 6). Wild-type S. aureus bound Cy3-labeled IgG and coated the entire bacterial surface with the protein A ligand. In contrast, S. aureus OS2 (spa::ermC) failed to bind Cy3-labeled IgG. Transformation of S. aureus OS2 with pSPA encoding wild-type protein A and plasmid selection with chloramphenicol restored the ability of the mutant strain to bind immunoglobulin in a manner similar to that of strain RN4220 (Fig. 6). Transformation of S. aureus OS2 with plasmid encoding protein A signal peptide variants also restored the binding of Cy3-labeled IgG to staphylococci (Fig. 6).
FIG. 6.
Surface display of protein A mutants as examined by immunofluorescence microscopy. Cells were grown to mid-log phase, and surface-displayed protein A was stained with Cy5-conjugated goat IgG. Samples were viewed by fluorescence microscopy, and images were captured with a charge-coupled-device camera. The S. aureus RN4220 strain (spa+) was used as a positive control. Note that all plasmids are transformed into strain OS2 (spa::ermC), which does not express protein A.
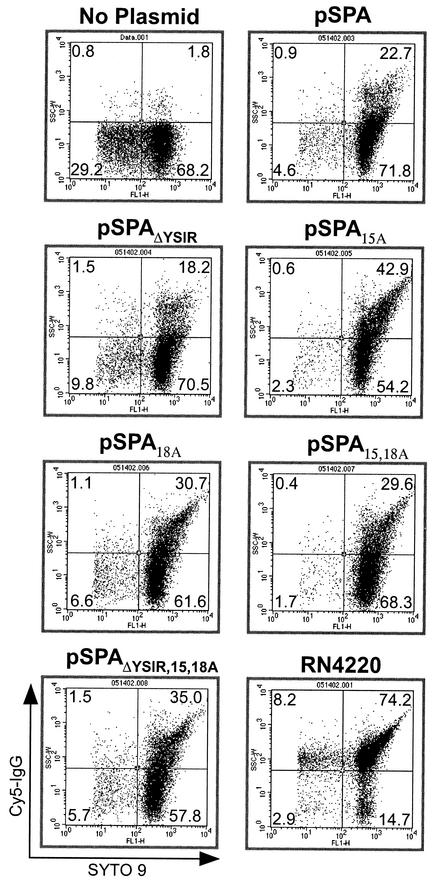
Fluorescence microscopy of staphylococci provides an assay for protein A ligand binding; however, only a limited number of bacterial cells are assessed. To determine whether signal peptide mutations caused an effect on surface protein function in larger populations of cells, a FACS experiment was performed. Staphylococci were incubated with Cy5-labeled IgG, and nucleic acids were stained with a mixture of SYTO 9, a green fluorescent dye that stains all bacteria, and propidium iodide, a red fluorescent dye that penetrates only bacteria with damaged membranes. In this condition, only cells with intact membranes are stained green. Binding of Cy5-labeled IgG to protein A was measured by plotting the number of green fluorescent cells that also emit the Cy5 signal (Fig. 7). Wild-type cultures harbor a large population of double-stained bacteria, whereas S. aureus OS2 cultures contained very few double-positive cells. S. aureus OS2 expressing either wild-type protein A or signal peptide variants displayed similar increases in the surface display of protein A molecules that are functional in binding Cy5-labeled IgG. Thus, the signal peptide mutations described here do not affect the surface display and functional folding of protein A.
FIG. 7.
Measurements of the surface display of protein A in staphylococcal populations by FACS analysis. Staphylococci were grown to mid-log phase, stained with Cy5-conjugated goat IgG and SYTO 9, and analyzed with a FACS. The S. aureus RN4220 strain (spa+) was used as positive control, while the OS2 strain (spa::ermC) (no plasmid) was used as a negative control. Note that all plasmids are transformed into strain OS2 (spa::ermC), which does not express protein A.
If protein A signal peptide mutations affect the rate of precursor processing, one would expect this to cause downstream defects, either in the amount of cell wall-anchored surface protein or in the functional assembly of surface receptors. As the immunofluorescence experiments revealed no defect in the functional assembly of protein A, we wondered whether the total amount of anchored surface protein was reduced in variants with signal peptide mutations. To test this, we measured the overall concentration of wild-type protein A (SPA) and a signal peptide mutant (SPAΔYSIR) in staphylococcal cells or purified cell walls by calibrated immunoblotting experiments. Compared to the amount of the wild-type species, the total amounts of SPAΔYSIR in whole cells (56%) and in isolated cell wall compartments (31%) were reduced. In contrast, the rate of polypeptide synthesis, assessed by immunoprecipitating radiolabeled protein A after a 1-min pulse with [35S]methionine followed by an immediate quenching with TCA, was increased for the SPAΔYSIR mutant (121%) compared to that of wild-type protein A. Together, these results suggest that the deletion of the YSIR sequence caused not only a reduction in protein A signal peptide processing but also a reduction in the amount of anchored polypeptide.
Secretion and cell wall anchoring of enterotoxin B.
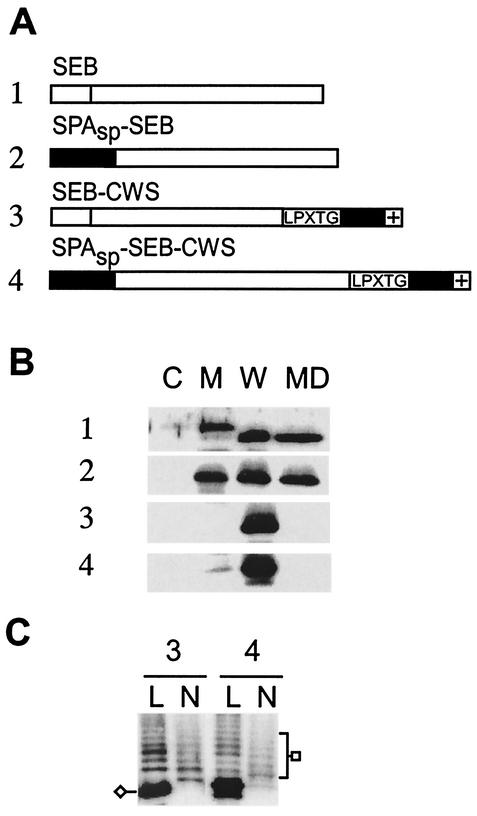
SEB is synthesized as a signal peptide-bearing precursor without the YSIRK-G/S motif, and the mature polypeptide is secreted across the plasma membrane into the extracellular medium (63). We wondered what would be the effect of introducing the YSIRK-G/S motif in SEB and replaced its signal peptide with that of protein A (SPASP-SEB). Expression and secretion of SEB was measured by immunoblotting. As expected, SEB was secreted into the extracellular medium (63) (Fig. 8B, blot 1). A small but significant amount of the SEB precursor was located in the membrane fraction. SPASP-SEB was also secreted into the extracellular medium. However, in contrast to that of SEB, the signal peptide motif-bearing precursor of SPASP-SEB could not be detected in the membrane fraction and only mature protein was found (Fig. 8B, lane M of blot 2). These data are consistent with the notion that the YSIRK-G/S motif containing the SPASP initiates SEB more efficiently into the secretory pathway. Fusions of the LPXTG motif containing the sorting signal to the C terminus of SPASP-SEB or SEB led to the cell wall targeting of hybrid polypeptides. Both SPASP-SEB-CWS and SEB-CWS were covalently linked to peptidoglycan, as cell wall digestion with muramidase released the characteristic spectrum of immunoreactive SEB species (Fig. 8C).
FIG. 8.
Cell wall sorting of enterotoxin B fusions. (A) The diagram displays SEB and SPASP-SEB fusion proteins. The signal peptide of SEB does not harbor the YSIRK-G/S motif and is represented by a white block. SPASP does encompass the YSIRK-G/S motif and is shown as a gray block. The LPXTG motif, the C-terminal hydrophobic domain (black bar), and the charged tail (+) of SPA are indicated in the drawing. (B) Cellular locations of the SPA-SEB fusion proteins. Cells were grown to mid-log phase and fractionated into medium (MD), cell wall (W), membrane (M), and cytoplasm (C) compartments. The samples were subjected to SDS-12% PAGE and analyzed by immunoblotting with polyclonal anti-SEB antibody. (C) Cells at mid-log phase were treated with either lysostaphin (L) or N-acetylmuramidase (N) in the presence of 0.5 M sucrose. The digested cell wall components were collected, subjected to SDS-12% PAGE, and then analyzed by immunoblotting with anti-SEB antibody.
DISCUSSION
In this study, we examined the function of the recently identified YSIRK-G/S signal peptide motif using staphylococcal protein A as a model system. Both deletion of the YSIR sequence and/or replacements of the G and S residues significantly reduced signal peptide processing and the secretion of protein A. In contrast, mutational changes in the YSIRK-G/S motif did not affect the cell wall anchoring or the functional assembly of protein A.
The hypothesis that signal peptides contain information other than that for the default initiation into the secretory pathway has been previously examined. Unlike eukaryotic cells, where the SRP pathway is responsible for the initiation of all polypeptides into the secretory pathway, bacteria such as E. coli use the SRP pathway for the initiation of a set of inner membrane proteins (65). Presecretory proteins, i.e., precursors destined for secretion into the periplasm or the outer membrane, use a SecB or DnaK chaperone-mediated initiation mechanism (9). If signal peptides contain other information, how does the SRP distinguish between the two classes of proteins? Photochemical protein cross-linking and in vitro translocation experiments suggest that trigger factor, a ribosome-associated peptidyl-prolyl isomerase (54), binds to presecretory proteins and prevents their association with the SRP (2). Lee and Bernstein reported that the hydrophobic properties of signal peptides determine the initiation of membrane proteins into the SRP pathway (26). The first transmembrane segment is significantly more hydrophobic than signal peptides of presecretory proteins and, as determined by the reciprocal replacement of these elements, dictates the requirement for the chaperone or SRP-mediated pathway (26). Both models predict the existence of different classes of signal peptides with discrete signals (trigger factor or SRP binding). The YSIRK-G/S motif can be viewed as a signal that plays a role in the efficient secretion of protein A but that is not absolutely essential for precursor protein translocation.
Why would bacterial cells distinguish between different classes of signal peptides when all substrates for protein translocation are moved through the same SecYEG pore? The recent characterization of YidC provides one example to answer to this question, as YidC is required for membrane protein insertion but is dispensable for the translocation of presecretory proteins (45). Thus, E. coli SRP and YidC can be viewed as modifiers of the secretory pathway and are dedicated to the translocation of a subset of substrates (57, 66). If this is true, do precursor proteins with YSIRK-G/S motifs represent a subset of secretion substrates that require dedicated factors for efficient translocation? Some gram-positive bacteria, for example, streptococci, listeriae, and staphylococci, harbor secA and secY paralog genes in the chromosome (30). While secA1 and secY1 are essential for staphylococcal growth, secA2 and secY2 are not (20). Preliminary results suggest that S. aureus cells do not require secA2 and secY2 for the efficient secretion of protein A or other surface proteins (T. Bae and O Schneewind, unpublished information). Thus, we suspect that hitherto unknown factors act on the YSIRK-G/S motif of signal peptides in streptococci and staphylococci.
Acknowledgments
We thank members of our laboratory for critical reading of the manuscript.
This work was supported by NIAID-NIH grants AI38897 and AI52474 to O.S.
REFERENCES
- 1.Bassford, P. J., Jr., T. J. Silhavy, and J. Beckwith. 1979. Use of gene fusion to study secretion of maltose-binding protein into Escherichia coli periplasm. J. Bacteriol. 139:19-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, K., L.-F. Wu, J. Brunner, and M. Müller. 2000. Discrimination of SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 19:134-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, S. A., M. N. Hall, and T. J. Silhavy. 1985. Genetic analysis of protein export in Escherichia coli K12. Annu. Rev. Biochem. 54:101-134. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, H. D., M. A. Poritz, K. Strub, P. J. Hoben, S. Brenner, and P. Walter. 1989. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature 340:482-486. [DOI] [PubMed] [Google Scholar]
- 5.Blobel, G. 1980. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA 77:1496-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blobel, G., and B. Dobberstein. 1975. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 67:835-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, C. N., G. Blobel, and P. Model. 1978. Detection of prokaryotic signal peptidase in an Escherichia coli membrane fraction: endoproteolytic cleavage of nascent f1 pre-coat protein. Proc. Natl. Acad. Sci. USA 75:361-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalbey, R. E., and W. Wickner. 1985. Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coli plasma membrane. J. Biol. Chem. 260:15925-15931. [PubMed] [Google Scholar]
- 9.de Gier, J.-W., and J. Luirink. 2001. Biogenesis of inner membrane proteins in Escherichia coli. Mol. Microbiol. 40:314-322. [DOI] [PubMed] [Google Scholar]
- 10.Duong, F., and W. Wickner. 1997. The SecDFYajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 16:4871-4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Economou, A., J. A. Pogliano, J. Beckwith, D. B. Oliver, and W. Wickner. 1995. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83:1171-1181. [DOI] [PubMed] [Google Scholar]
- 12.Gardel, C., S. Benson, J. Hunt, S. Michaelis, and J. Beckwith. 1987. secD, a new gene involved in protein export in Escherichia coli. J. Bacteriol. 169:1286-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanada, M., K. I. Nishiyama, S. Mizushima, and H. Tokuda. 1994. Reconstitution of an efficient protein translocation machinery comprising SecA and the three membrane proteins, SecY, SecE and SecG (p12). J. Biol. Chem. 269:23625-23631. [PubMed] [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-572. [DOI] [PubMed] [Google Scholar]
- 15.Hantke, K., and V. Braun. 1973. Covalent binding of lipid to protein. Eur. J. Biochem. 34:284-296. [DOI] [PubMed] [Google Scholar]
- 16.Hartl, F. U., S. Lecker, E. Schiebel, J. P. Hendrick, and W. Wickner. 1990. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell 63:269-279. [DOI] [PubMed] [Google Scholar]
- 17.Inouye, S., S. Wang, J. Sekizawa, S. Halegoua, and M. Inouye. 1977. Amino acid sequence for the peptide extension on the prolipoprotein of the Escherichia coli outer membrane. Proc. Natl. Acad. Sci. USA 74:1004-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, K., M. Wittekind, M. Nomura, K. Shiba, T. Yura, A. Miura, and H. Nashimoto. 1983. A temperature-sensitive mutant of E. coli exhibiting slow processing of exported proteins. Cell 32:789-797. [DOI] [PubMed] [Google Scholar]
- 19.Jensen, K. 1958. A normally occurring staphylococcus antibody in human serum. Acta Pathol. Microbiol. Scand. 44:421-428. [Google Scholar]
- 20.Ji, Y., B. Zhang, S. F. Van, Horn, P. Warren, G. Woodnutt, M. K. Burnham, and M. Rosenberg. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266-2269. [DOI] [PubMed] [Google Scholar]
- 21.Jönsson, K., C. Signäs, H. P. Müller, and M. Lindberg. 1991. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur. J. Biochem. 202:1041-1048. [DOI] [PubMed] [Google Scholar]
- 22.Josefsson, E., K. W. McCrea, D. Ní Eidhin, D. O'Connell, J. Cox, M. Höök, and T. J. Foster. 1998. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144:3387-3395. [DOI] [PubMed] [Google Scholar]
- 23.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 24.Kumamoto, C. A. 1989. Escherichia coli SecB protein associates with exported protein precursors in vivo. Proc. Natl. Acad. Sci. USA 86:5320-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mitsutani-Ui, N. Kobayashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 26.Lee, H. C., and H. D. Bernstein. 2001. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad. Sci. USA 98:3471-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazmanian, S. K., G. Liu, E. R. Jensen, E. Lenoy, and O. Schneewind. 2000. Staphylococcus aureus mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA 97:5510-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 29.Mazmanian, S. K., and O. Schneewind. 2001. Cell wall anchored surface proteins and lipoproteins of gram-positive bacteria. In A. Sonenshine, R. Losick, and J. Hoch (ed.), Bacillus subtilis and other gram-positive bacteria, 2nd ed., in press. ASM Press, Washington, D.C.
- 30.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalyzed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 31.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 32.Navarre, W. W., and O. Schneewind. 1994. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol. Microbiol. 14:115-121. [DOI] [PubMed] [Google Scholar]
- 33.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and the mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarre, W. W., H. Ton-That, K. F. Faull, and O. Schneewind. 1998. Anchor structure of staphylococcal surface proteins. II. COOH-terminal structure of muramidase and amidase-solubilized surface protein. J. Biol. Chem. 273:29135-29142. [DOI] [PubMed] [Google Scholar]
- 35.Ní Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Höök, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 36.Nunn, D. N., and S. Lory. 1991. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. USA 88:3281-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver, D. B., and J. Beckwith. 1981. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell 25:765-772. [DOI] [PubMed] [Google Scholar]
- 38.Patti, J. M., H. Jonsson, B. Guss, L. M. Switalski, K. Wiberg, M. Lindberg, and M. Höök. 1992. Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J. Biol. Chem. 267:4766-4772. [PubMed] [Google Scholar]
- 39.Perry, A. M., H. Ton-That, S. K. Mazmanian, and O. Schneewind. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J. Biol. Chem. 277:16241-16248. [DOI] [PubMed] [Google Scholar]
- 40.Pogliano, K. J., and J. Beckwith. 1993. The Cs sec mutants of Escherichia coli reflect the cold sensitivity of protein export itself. J. Bacteriol. 133:763-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Randall, L. L. 1992. Peptide binding by chaperone SecB: implications for recognition of non-native structure. Science 257:241-245. [DOI] [PubMed] [Google Scholar]
- 43.Randall, L. L. 1983. Translocation of domains of nascent periplasmic proteins across the cytoplasmic membrane is independent of elongation. Cell 33:231-240. [DOI] [PubMed] [Google Scholar]
- 44.Rosenstein, R., and F. Götz. 2000. Staphylococcal lipases: biochemical and molecular characterization. Biochimie 82:1005-1014. [DOI] [PubMed] [Google Scholar]
- 45.Samuelson, J. C., M. Chen, F. Jiang, I. Moller, M. Wiedmann, A. Kuhn, G. J. Phillips, and R. E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637-641. [DOI] [PubMed] [Google Scholar]
- 46.Schatz, P. J., and J. Beckwith. 1990. Genetic analysis of protein export in Escherichia coli. Annu. Rev. Genet. 24:215-248. [DOI] [PubMed] [Google Scholar]
- 47.Schatz, P. J., P. D. Riggs, A. Jacq, M. J. Fath, and J. Beckwith. 1989. The secE gene encodes an integral membrane protein required for protein export in Escherichia coli. Genes Dev. 3:1035-1044. [DOI] [PubMed] [Google Scholar]
- 48.Schindler, C. A., and V. T. Schuhardt. 1964. Lysostaphin: a new bacteriolytic agent for the staphylococcus. Proc. Natl. Acad. Sci. USA 51:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103-106. [DOI] [PubMed] [Google Scholar]
- 50.Schneewind, O., D. Mihaylova-Petkov, and P. Model. 1993. Cell wall sorting signals in surface protein of Gram-positive bacteria. EMBO J. 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 52.Signas, C., G. Raucci, K. Jonsson, P.-E. Lindgren, G. M. Anantharamaiah, M. Höök, and M. Lindberg. 1989. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc. Natl. Acad. Sci. USA 86:699-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sjöquist, J., J. Movitz, I.-B. Johansson, and H. Hjelm. 1972. Localization of protein A in the bacteria. Eur. J. Biochem. 30:190-194. [DOI] [PubMed] [Google Scholar]
- 54.Stoller, G., K. P. Rucknagel, K. H. Nierhaus, F. X. Schmid, G. Fischer, and J. U. Rahfeld. 1995. A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J. 14:4939-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strom, M. S., and S. Lory. 1992. Kinetics and sequence specificity of processing prepilin by PilD, the type IV leader peptidase of Pseudomonas aeruginosa. J. Bacteriol. 174:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 57.Stuart, R. A., and W. Neupert. 2000. Making membranes in bacteria. Nature 406:575-577. [DOI] [PubMed] [Google Scholar]
- 58.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, et al. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 59.Tokunaga, M., J. M. Loranger, and H. C. Wu. 1984. Prolipoprotein modification and processing enzymes in Escherichia coli. J. Biol. Chem. 259:3825-3830. [PubMed] [Google Scholar]
- 60.Ton-That, H., K. F. Faull, and O. Schneewind. 1997. Anchor structure of staphylococcal surface proteins. I. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J. Biol. Chem. 272:22285-22292. [DOI] [PubMed] [Google Scholar]
- 61.Ton-That, H., G. Liu, S. K. Mazmanian, K. F. Faull, and O. Schneewind. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 96:12424-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ton-That, H., and O. Schneewind. 1999. Anchor structure of staphylococcal surface proteins. IV. Inhibitors of the cell wall sorting reaction. J. Biol. Chem. 274:24316-24320. [DOI] [PubMed] [Google Scholar]
- 63.Tweten, R. K., and J. J. Iandolo. 1983. Transport and processing of staphylococcal enterotoxin B. J. Bacteriol. 153:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uhlén, M., B. Guss, B. Nilsson, S. Gatenbeck, L. Philipson, and M. Lindberg. 1984. Complete sequence of the staphylococcal gene encoding protein A. J. Biol. Chem. 259:1695-1702. [PubMed] [Google Scholar]
- 65.Ulbrandt, N. D., J. A. Newitt, and H. D. Bernstein. 1997. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88:187-196. [DOI] [PubMed] [Google Scholar]
- 66.Urbanus, M. L., L. Froderberg, D. Drew, P. Bjorck, J.-W. L. de Gier, J. Brunner, B. Oudega, and J. Luirink. 2002. Targeting, insertion and localization of Escherichia coli YidC. J. Biol. Chem. 277:12718-12723. [DOI] [PubMed] [Google Scholar]
- 67.von Heijne, G. 1983. Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem. 133:17-21. [DOI] [PubMed] [Google Scholar]
- 68.Walter, P., and G. Blobel. 1981. Translocation of proteins across the endoplasmic reticulum. III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 91:557-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walter, P., R. Gilmore, and G. Blobel. 1984. Protein translocation across the endoplasmic reticulum. Cell 38:5-8. [DOI] [PubMed] [Google Scholar]
- 70.Wu, S. W., and H. de Lencastre. 1999. Mrp—a new auxiliary gene essential for optimal expression of methicillin resistance in Staphylococcus aureus. Microb. Drug Res. 5:9-18. [DOI] [PubMed] [Google Scholar]
- 71.Yokogawa, K., S. Kawata, S. Nishimura, Y. Ikeda, and Y. Yoshimura. 1974. Mutanolysin, bacteriolytic agent for cariogenic streptococci: partial purification and properties. Antimicrob. Agents Chemother. 6:156-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yokota, N., T. Kuroda, S. Matsuyama, and H. Tokuda. 1999. Characterization of the LolA-LolB system as the general lipoprotein localization mechanism of Escherichia coli. J. Biol. Chem. 274:30995-30999. [DOI] [PubMed] [Google Scholar]