Abstract
Cytosine methylation at CpG dinucleotides contributes to the epigenetic maintenance of gene silencing. Dynamic reprogramming of DNA methylation patterns is believed to play a key role during development and differentiation in vertebrates. The mechanisms of DNA demethylation remain unclear and controversial. Here, we present a detailed characterization of the demethylation of an endogenous gene in cultured cells. This demethylation is triggered in a regulatory region by a transcriptional activator, the glucocorticoid receptor. We show that DNA demethylation is an active process, occurring independently of DNA replication, and in a distributive manner without concerted demethylation of cytosines on both strands. We demonstrate that the DNA backbone is cleaved 3′ to the methyl cytidine during demethylation, and we suggest that a DNA repair pathway may therefore be involved in this demethylation.
Keywords: DNA repair, epigenetics, transcription, chromatin, glucocorticoids
In vertebrates, DNA methylation occurs at the carbon-5 position of cytosine in the dinucleotide CpG. This DNA modification is involved in stabilizing the silent state of genes, either locally by preventing regulators from binding to their target, or by recruiting specific proteins that favor the formation of silent chromatin (1, 2). DNA methylation participates in the control of several aspects of mammalian development, including X chromosome inactivation, parental imprinting, and tissue-specific expression of genes (1, 2). Alterations in the methylation patterns of a number of genes are believed to be involved in both the initiation and the progression of cancer (3, 4).
The propagation of the methylated state through DNA replication is ensured by the action of maintenance and de novo DNA methyltransferases (DNMTs) (1, 5, 6). DNA methylation is subject to regulated reprogramming, in particular during development (1, 7, 8). Genome-wide waves of demethylation and remethylation are observed during gametogenesis and after fertilization. Localized DNA demethylation occurs later at specific genes, mostly when and where differentiation requires their activation. Although maintenance and de novo methylation are relatively well understood, the mechanisms of DNA demethylation are still unclear and controversial (1, 6, 9, 10). Two distinct mechanisms have been shown to act during the genome-wide demethylation occurring after fertilization in mouse: A passive mechanism resulting from the absence of maintenance methylation after replication is responsible for the demethylation of the maternal genome (11), whereas an active mechanism of unknown nature appears responsible for the demethylation of the paternal genome (7). The mechanisms of local gene-specific demethylation are also unclear (9). Both passive, active, and a combination of passive and active demethylation have been proposed to occur in the various local events that were analyzed (12–15).
Several attempts have been made to characterize the mechanism of DNA demethylation in vitro (reviewed in ref. 9). Three classes of mechanisms have been proposed: direct removal of the methyl moiety from the base, excision of the methylated base, or excision of the methylated nucleotide. These pathways were supported by the identification of corresponding enzymatic activities (16), but these demethylases have failed to gain widespread acceptance because of the lack of reproducibility of the data and to the caveats in the experimental approaches used (17, 18). It is thus essential to firmly establish the biochemical pathways of MeCpG demethylation occurring in live cells.
To study the mechanisms of local targeted DNA demethylation in living cells, we have investigated an enhancer-specific demethylation event at an endogenous gene, triggered by a transcriptional activator during development. We analyzed the demethylation occurring at a glucocorticoid-responsive unit (GRU) located 2.5 kb upstream of the transcription start site of the tyrosine aminotransferase (Tat) gene upon activation by the glucocorticoid receptor. This event takes place in the liver before birth and is involved in the memorization of the first stimulation of the gene by glucocorticoids (19). We demonstrate that demethylation of cytosines at this gene under physiological conditions results from an active mechanism that involves the creation of nicks in the DNA 3′ to the methylcytidine. This mechanism would be consistent with the involvement of a demethylase initiating a base or nucleotide excision repair.
Results
Demethylation of the Tat Gene Enhancer Does Not Require Passage Through S Phase.
DNA demethylation of the Tat enhancer can be triggered ex vivo by glucocorticoid treatment of either cultured embryonic day 15 (E15) fetal hepatocytes or a rat hepatoma cell line (H4IIEC3). The glucocorticoid receptor induces a chromatin-remodeling event over 450 bp encompassing the four CpGs that are demethylated (20). This chromatin remodeling is rapid (15 min–1 h) and reversible after hormone withdrawal. In contrast, DNA demethylation is slow and persists in nonstimulated cells (19).
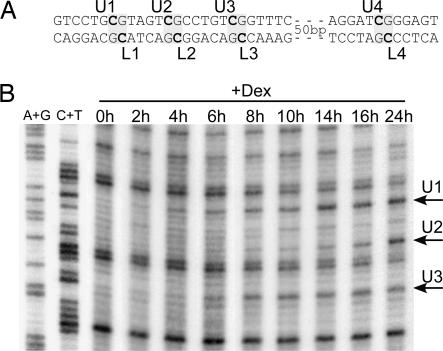
Three of the four MeCpGs that are demethylated are clustered within a 15-bp region (Fig. 1A). Features of the demethylation kinetics of these MeCpGs are best seen by ligation-mediated PCR (LM-PCR) analysis of genomic DNA treated with hydrazine and piperidine under conditions where DNA is cleaved only at unmethylated cytosines (Fig. 1B). In hepatoma cells, the three Cs were all demethylated slowly. Demethylation was detectable after a few hours of stimulation and proceeded steadily over a 24-h time period. The relatively slow kinetics of methylation loss seems consistent with a passive mechanism, but the different rates observed for neighboring Cs on the same strand (compare U2 with U1 and U3 on Fig. 1) suggests that demethylation is not the mere consequence of the absence of maintenance methylation after DNA replication. Thus, the slow kinetics could reflect a more subtle connection between DNA replication and demethylation or another requirement for a passage through a specific phase of the cell cycle.
Fig. 1.
Kinetics of glucocorticoid-induced DNA demethylation at the −2.5 Tat GRU in hepatoma cells. (A) The four MeCpGs within the Tat GRU (−2,425 to −2,336) that are demethylated upon glucocorticoid treatment are numbered from 1 to 4 (upper strand, U1 to U4; lower strand, L1 to L4). (B) Demethylation kinetics of the three neighboring Cs of the upper strand. Rat hepatoma cells were grown with 10−7 M dexamethasone (+Dex) for the indicated time. The corresponding genomic DNA was treated with hydrazine and piperidine and analyzed by LM-PCR (refs. 19 and 40; Fig. 7). Genomic DNA cleaved at A+G and C+T was analyzed simultaneously.
Arresting cells at particular phases of the cell cycle by means of drugs and/or specific culture conditions was not well suited for the investigation of a putative role of DNA replication in DNA demethylation, because the activation of the Tat gene by the glucocorticoid receptor was affected by these treatments, independently of the DNA methylation status of the GRU (data not shown and ref. 21). In growing cells, however, the Tat gene can be activated throughout the cell cycle except during mitosis (21). Cells released from a treatment inducing a cell cycle arrest resume growth synchronously and regain their capacity to respond normally to glucocorticoids (data not shown). They are therefore well suited for the study of the demethylation mechanism of the Tat gene without any interference caused by the continued presence of the drug. To determine rigorously whether passage through a specific phase of the cell cycle was important for demethylation, we thus compared asynchronously and synchronously growing cells. Because LM-PCR analysis of hydrazine-piperidine-treated DNA is not exactly quantitative, the demethylation rate was quantified precisely using MethylQuant, a novel real-time PCR approach for quantifying the methylation status of a single cytosine (22). Bisulfite-treated genomic DNA is PCR-amplified, and the methylation status of a specific cytosine is quantified by real-time PCR by using a primer whose 3′ end discriminates for the methylation status of the cytosine of interest. Fig. 2 shows the quantification of the methylation status of U3 that has an intermediate demethylation rate as was shown in Fig. 1. Within an asynchronous population not stimulated by glucocorticoids, the level of unmethylation for each position is 2–10%, because of the presence of unmethylated cytosines scattered among Tat GRU molecules (vide infra). The proportion of unmethylated U3 cytosines did not change for the first 6 h of glucocorticoid treatment, and, after this lag, it steadily increased in the next 18 h to reach 85%. The rapid demethylation of 70% of the cytosines that occurred between 6 and 24 h argues against a purely passive demethylation mechanism, because the cells had a doubling time of ≈24 h in the conditions used here, and thus went through at most a single replication event during this period.
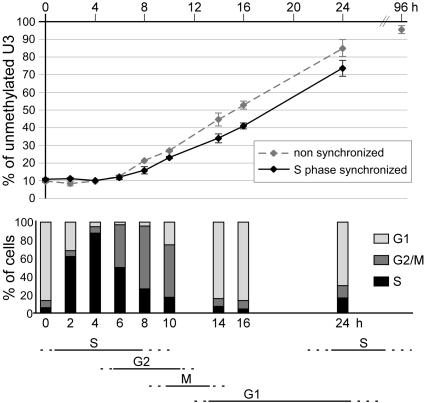
Fig. 2.
Demethylation of the −2.5 Tat GRU is not restricted to a particular phase of the cell cycle. Hepatoma cells were synchronized in early S phase in two steps. They were first presynchronized in G1 by isoleucine and serum deprivation and then released from this block in the presence of aphidicolin. They were then released from the early S block in the presence of 10−7 M dexamethasone for the indicated time (0 h, arrested cells). The demethylation kinetics in nonsynchronized cells was analyzed in parallel. (Upper) The proportion of unmethylated U3 during glucocorticoid treatment as determined by using the MethylQuant assay (22). (Lower) The proportion of cells in the various phases of the cell cycle in the synchronized culture (determined by using propidium iodide staining and flow cytometry) and the deduced time frames of the phases of the cell cycle.
To determine whether passive demethylation might nevertheless be involved, in combination with active demethylation (i.e., if there was active demethylation of hemimethylated cytosines produced upon replication), we analyzed whether demethylation occurs preferentially during S phase. Cells were synchronized at the beginning of S phase by aphidicolin treatment and then released from the block in the presence of glucocorticoids. A 6-h time lag was found again before demethylation was observed (Fig. 2), and, consequently, no detectable loss of methylation occurred during S phase, revealing that maintenance methylation occurred normally at this stage. Demethylation was detected at 8 h when the majority of the cells had completed S phase and proceeded thereafter steadily between 8 and 24 h, during G2 and the following G1, before most of the cells had initiated their next S phase. Thus, there was no evidence for a preferential demethylation of hemimethylated CpGs resulting from replication: Demethylation began with an active mechanism on fully methylated CpGs. The rate of demethylation was similar in nonsynchronized and synchronized cells, the latter being slightly retarded for the start, probably because of the passage through mitosis. We also observed that demethylation could occur during S phase, because the demethylation rate was not slowed down by the entry in S phase in cell populations synchronized and treated with glucocorticoid in G1 (data not shown). Quantitative analyses of demethylation at positions 1 and 2, which were demethylated faster or slower than U3, also revealed that demethylation can occur outside the S phase (vide infra and data not shown). This conclusion is also supported by the observation that demethylation of these three Cs occurred in aphidicolin-treated cells even though the drug decreased the efficiency of both glucocorticoid induction and DNA demethylation (data not shown). In conclusion, demethylation appears to occur at all phases of the cell cycle permissive for Tat gene activation, with no preferential phase, and is performed mainly by an active mechanism.
Distributive Nature of Cytosine Demethylation in the Tat GRU.
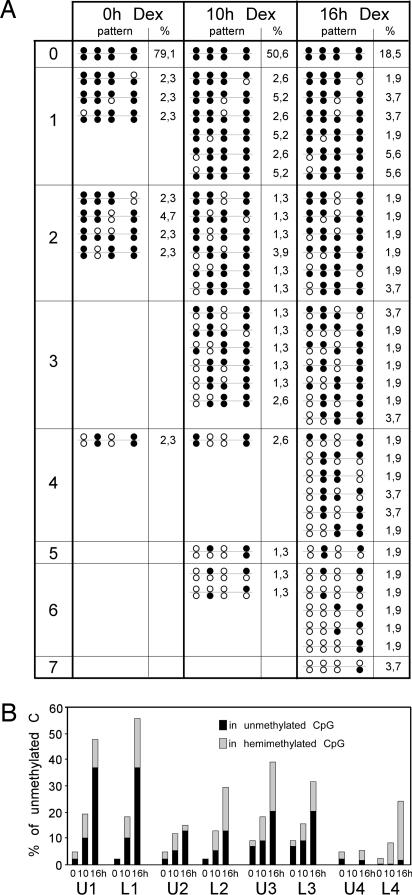
We next assessed whether demethylation of the four MeCpGs occurred through a processive or a distributive mechanism in living cells, i.e., whether the various Cs of individual Tat gene molecules, in particular 2 Cs belonging to the same CpG, are demethylated in a concerted fashion. To this end, we used hairpin-bisulfite PCR that allows simultaneous analysis of the two strands of a DNA molecule (5). A hairpin linker, targeted and ligated to restriction enzyme-cleaved genomic DNA, prevents the strand separation that occurs upon subsequent bisulfite conversion and PCR amplification. Single PCR molecules corresponding to the two linked strands are then cloned and sequenced. The method was applied to DNA samples prepared from the synchronized cells undergoing demethylation. We focused on the cells that had not yet undergone demethylation (0 h time point) and on the cells that were undergoing active demethylation after the end of S and during G1 (10 and 16 h time points). The various methylation patterns of the four CpGs of the Tat GRU, and their frequencies, show that during the demethylation process there is no bimodal distribution of fully demethylated and fully methylated molecule (Fig. 3). The Cs were demethylated progressively in a rather stochastic manner, resulting in an increase of frequency of molecules with multiple unmethylated positions with the duration of glucocorticoid stimulation. The various Cs were not demethylated at the same rate, even though similar rates were observed for Cs belonging to the same CpG (Fig. 3B). CpG#1 was demethylated the fastest, whereas CpG#4 was demethylated at the slowest pace. For most positions, extensive demethylation occurred between 10 and 16 h, when most of the cells were outside S phase, further demonstrating the replication-independent nature of this demethylation event.
Fig. 3.
Distribution of the unmethylated cytosines during demethylation of the −2.5 Tat GRU. Methylation patterns of individual double-stranded molecules were assessed by using the hairpin-bisulfite PCR method (5). DNA from the synchronized cells described in Fig. 2 was analyzed 0, 10, and 16 h after release from the aphidicolin block in the presence of dexamethasone (Dex). (A) Frequencies of the various methylation patterns obtained during demethylation of the −2.5 Tat GRU. The patterns are represented by a succession of circles symbolizing the methylation status of cytosines U1/L1, U2/L2, U3/L3, U4/L4: filled, methylated; open, unmethylated. The patterns are organized in subgroups according to the number of unmethylated Cs as indicated on the left. The frequency of each pattern is indicated (%). Because the total numbers of clones analyzed were 43, 77, and 54 for the dexamethasone treatments of 0, 10, and 16 h, respectively, the frequencies corresponding to one event are 2.3, 1.3, and 1.85%, respectively. (B) Time course of the demethylation of each individual cytosine. The percentages of unmethylated cytosines, belonging to either hemimethylated or fully demethylated CpGs as indicated, are plotted for the three analyzed times of dexamethasone induction.
When the frequencies of demethylated Cs found at each position in either hemimethylated or fully demethylated CpGs are compared, there appears to be a bias in favor of fully demethylated CpG except for CpG#4 where hardly any demethylation could be observed at U4 even when demethylation of L4 was above 20%. (Fig. 3B). This bias in favor of fully demethylated CpG could have been because of a preferential demethylation of hemimethylated CpG. Statistical analyses revealed, however, that it mainly reflected the presence of a population of slowly demethylating cells (Fig. 6, which is published as supporting information on the PNAS web site), presumably those refractory to induction during mitosis (21). There was only a slight preferential demethylation of a MeC facing a demethylated C and no preferential demethylation of neighboring Cs. Overall, demethylation appeared to proceed in a distributive manner, essentially with independent demethylation of Cs.
Cytosine Demethylation Is Associated with Cleavage of the DNA Backbone.
In cultured E15 fetal hepatocytes, glucocorticoid-dependent DNA demethylation of the Tat enhancer occurred as in hepatoma cells but with distinctive features: Rapid demethylation was observed in the first 5 h after hormonal activation and the various Cs were demethylated at similar rates (Fig. 4A). After 5 h, however, demethylation did not progress any further because the demethylation levels reached 10 and 24 h after glucocorticoid addition were similar. Presumably, only a subpopulation of the Tat gene enhancer responded to glucocorticoids, because the fraction of chromatin that became accessible to restriction enzyme cleavage was ≈3- to 4-fold lower in E15 fetal hepatocytes than in hepatoma cells (compare Fig. 7 in ref. 19 with Fig. 1 in ref. 20). Thus, demethylation occurred rapidly in the fraction of responsive enhancer, which prompted us to test whether this rapidity would make fetal hepatocytes a valuable system to reveal the properties of the active demethylation observed in hepatoma cells.
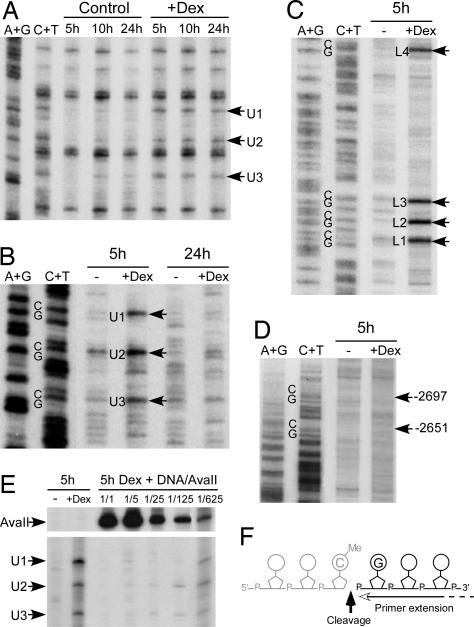
Fig. 4.
DNA is cleaved 3′ to the methylated cytosines during glucocorticoid-induced demethylation. Hepatocytes isolated from E15 fetuses were cultured for the indicated time (hours) without (control or −) or with (+Dex) 10−7 M dexamethasone. (A) Kinetics of glucocorticoid-induced DNA demethylation in hepatocytes analyzed by LM-PCR by using genomic DNA treated with hydrazine-piperidine as described in Fig. 1B. (B) Native genomic DNA was analyzed by LM-PCR to visualize strand breaks in the upper strand of the −2.5 Tat GRU. The arrows indicate the bands revealing cleavage at MeCpG. (C) Analysis of the lower strand as in B. (D) Analysis of the upper strand in an upstream region where there are two MeCpGs that are not demethylated. The arrows indicate the location of the corresponding Cs. (E) Competitive PCR analysis of the abundance of the glucocorticoid-induced cleavage products. A fixed amount of genomic DNA from fetal hepatocytes treated for 5 h with dexamethasone was diluted with varying amounts of rat genomic DNA digested with AvaII that cleaves just upstream the −2.5 Tat GRU and analyzed by LM-PCR. The relative amount of the competing liver DNA corresponded to either the same amount (1/1) or serial 5-fold dilutions (1/5 to 1/625). The regions of the gel showing the bands corresponding to the AvaII cleavages and the dexamethasone-induced cleavages at the MeCpG are shown. (F) Representation of the location of the strand break detected relative to the CpG dinucleotide. Note that LM-PCR allows only analysis of the fragment downstream the break and thus that the location and nature of the 3′ end of the upstream fragment (represented in gray) cannot be assessed.
As some of the cytosine demethylase activities that have been previously proposed involve base or nucleotide excision, we wished to assess whether glucocorticoid treatment might induce DNA backbone cleavage within the Tat GRU when it underwent demethylation. To this end, we used the same LM-PCR procedure that was used to analyze hydrazine-piperidine-cleaved genomic DNA except that we performed it directly on the genomic DNA, without any prior treatment. In LM-PCR, a DNA linker is ligated to free 5′ P ends available within the region of interest without prior knowledge of their distribution (Fig. 7, which is published as supporting information on the PNAS web site). PCR amplification is then performed with the linker and a gene-specific primer, and, after a few further rounds of PCR amplification with a labeled nested gene-specific primer, the amplified products are detected on a gel (23). PCR amplification generally reaches a plateau in conditions allowing detection of minor cleavage products, and the same overall plateau is reached for different initial numbers of molecules. Thus, specific cleavage products show up if they have a rather discrete distribution and if they are more abundant than the background cleavages that occur during DNA preparation, which give rise to products distributed throughout the amplified region (23). In this way, we could detect glucocorticoid-induced cleavage products within the Tat GRU when it was undergoing demethylation, i.e., after 5 h of dexamethasone treatment (Fig. 4 B and C). The corresponding bands were seen on both strands at specific positions above a background of widely distributed bands. These bands were not visible after a 24-h dexamethasone treatment when demethylation was completed (Fig. 4B) but could already be detected 3 h after hormone addition (data not shown). No cleavages above background were detected at CpGs that were not demethylated upon glucocorticoid treatment, such as those in the vicinity of the Tat GRU (Fig. 4D).
To estimate the proportion of gene copies that were giving rise to the cleavage products detected, we performed competitive LM-PCR by mixing a fixed amount of genomic DNA from fetal hepatocytes treated for 5 h with dexamethasone with varying amounts of restriction enzyme digested rat genomic DNA and then analyzing the mixture by LM-PCR (Fig. 4E). The genomic DNA competed for the detection of the three cleavage products up to the 1/625th dilution, showing that about one gene copy of a thousand presented a specific cleavage. This result is consistent with a short half-life intermediate produced during the demethylation process. Registration with the genomic sequencing ladders revealed that the glucocorticoid-induced strand breaks affected every MeCpG undergoing demethylation and were located 3′ to the methyl cytidine, yielding fragments whose 5′ end corresponds to the guanosine of the dinucleotide CpG (Fig. 4F). Phosphorylation of these 5′ ends is required for linker ligation allowing LM-PCR amplification, and thus the strand breaks detected here had a 5′ P end. Other types of demethylation intermediates, if they exist, would have escaped the detection. These breaks were detected on both strands, but they were distributed on different DNA molecules because they were not detected with linker ligation strategies that allow analysis of double-stranded breaks with either blunt or staggered ends (ref. 24 and data not shown). This result is consistent with the distributive nature of demethylation. We have not detected glucocorticoid-induced specific strand breaks above background in hepatoma cells despite the evidence for an active demethylation in these cells. As the demethylation occurring in hepatoma cells was slower than in fetal hepatocytes, it is likely that there were fewer demethylation events per unit of time, and thus the number of demethylation intermediates produced could have been too low to show up against background cleavages. Alternatively, a faster repair of the cleavage products in the hepatoma cells could also have precluded their detection. In conclusion, during demethylation of cytosines, the DNA backbone was cleaved 3′ to the modified nucleotide.
Discussion
DNA Demethylation Mechanism.
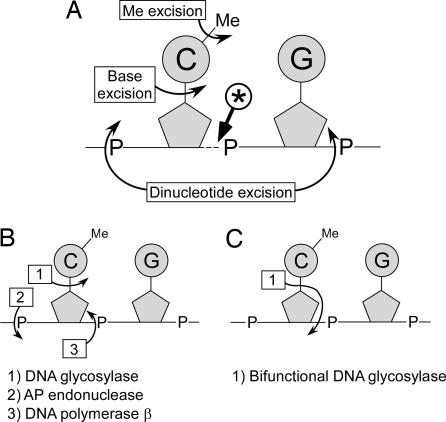
The demethylation event that we have described here differs from those that have previously been described in vertebrates based on biochemical analyses. The cleavage product 3′ to the methyl cytidine is not compatible with an activity directly removing the methyl group and leaving the base intact (ref. 16; see Fig. 5A). It indicates the involvement of a mechanism producing DNA strand breaks, and suggests that the methylated base is processed by a base lesion repair pathway. Nucleotide excision repair of the MeC would be compatible with the cleavage observed, but there is no evidence for such a mechanism, because the only demethylating nuclease activity reported so far excises the dinucleotide MeCpG (ref. 25; see Fig. 5A). Base excision repair initiated by a MeC glycosylase could also lead to the 3′ cleavage (Fig. 5 B and C). Bifunctional glycosylase/lyases couple base excision (DNA glycosylase activity) with 3′ phosphodiester bond breakage (DNA nicking activity) leaving a 5′ P end 3′ to the excised methyl cytidine, i.e., the type of ends we have detected with LM-PCR (Fig. 5C). Monofunctional enzymes do not have this nicking activity, but a transient 5′ P end 3′ to the MeC is also produced during the short-patch repair of the abasic lesion (Fig. 5B). Hydrolysis of abasic sites before the ligation step of LM-PCR, which may occur during DNA purification and denaturation before LM-PCR first extension step, would also give 5′ P ends 3′ to the MeC. Thus, both mono- and bifunctional DNA glycosylases are compatible with the cleavage products detected.
Fig. 5.
Scheme depicting the possible DNA demethylation pathways. (A) Location of the cleavages expected for the published putative demethylation mechanisms. The curved arrows indicate the cleavages expected for the methyl excision pathway (16), the base excision pathway (26), and the dinucleotide excision pathway (25). The cleavage observed herein is indicated by the arrow labeled with an asterisk. (B) The short patch repair pathway involving a monofunctional DNA glycosylase that could generate the cleavage product observed (41). (C) The action of a bifunctional DNA glycosylase/AP lyase that could generate the cleavage product observed (41). The opening of the sugar ring that normally occurs has not been represented.
Putative DNA Demethylases.
Two monofunctional glycosylases involved in thymine excision in T:G base pairs, TDG and MBD4, have been proposed to be active on MeC (26, 27), but these in vitro data have not been reproduced in other studies (28, 29) and could have been due to deamination of some of the MeC within the substrates used, which would have created unappreciated G/T mismatches (9). A demethylation pathway involving deamination of the MeC by cytidine deaminases followed by DNA repair of the T:G mismatch has also been suggested (30). The involvement of TDG in DNA demethylation was investigated in cultured cells. In one study, overexpression of TDG led to DNA demethylation of a retinoic acid receptor (RAR) target gene, but, surprisingly, it was observed in the absence of RAR activation (31). In a second study, inhibition of TDG by antisense oligonucleotide interfered with the transient demethylation of repetitive sequences that was observed upon myoblast differentiation (32). TDG, however, has a coactivator activity that is independent from its glycosylase activity, and it participates with the coactivator p300/CBP in the transactivation of various target genes (33, 34). Thus, the effect exerted by TDG on DNA demethylation might have been indirect and resulted from its transcriptional activation properties. MBD4−/− mice do not show developmental defects suggestive of perturbed DNA demethylation but rather show an increase in the frequency of C→T transitions at CpGs consistent with a role in DNA repair (35). Hence, in the absence of solid and reproducible biochemical evidence, or of well controlled genetic experiments showing a glycosylase-dependent involvement of TDG or MBD4 in DNA demethylation, it is not clear whether these enzymes are good candidates for this demethylation process. Furthermore, TDG and MBD4 have been proposed to act preferentially on hemimethylated CpGs (26, 27), but we observed that active DNA demethylation occurred on symmetrically methylated DNA and did not show a significant preference for generating symmetrically demethylated CpGs. In plants, two DNA glycosylase-lyases, ROS1 and DEMETER, antagonize the repression exerted by DNA methylation (36, 37). DEMETER has just been shown to remove MeC in vivo and in vitro (37) and, in contrast to TDG, its DNA glycosylase activity is required for activation of gene expression (38). Our results suggest that similar activities are also involved in DNA demethylation in vertebrates and revive the need to clarify the connections between DNA repair and demethylation. These activities could be targeted to specific regulatory sequences either directly through interaction with one of the transcription factors binding these sequences or indirectly through transcription factor-induced chromatin remodeling (9).
Prevention of Double-Stranded Breaks During DNA Demethylation.
DNA demethylation through a mechanism inducing strand cleavage raises a question about the risk of appearance of double-stranded breaks, because multiple modified bases are often found in proximity. Previous characterizations of demethylation events taking place in cells suggested that a coupling of replication-dependent and active demethylation (14), or a slow two-step active mechanism producing hemimethylated sites first (12), could prevent the generation of these detrimental DNA damages. We have not found evidence of such a two-step mechanism. Demethylation of MeCpGs occurred on both strands independently of replication by an active mechanism acting on fully methylated CpGs, and the various Cs were not first altogether hemimethylated before being fully demethylated in a clear two-step manner. The two-step demethylation observed previously could be the consequence of the use of transgenes as demethylation substrates. Indeed, they may not undergo the same regulation of methylation patterns as the endogenous gene that we studied in its natural chromatin context, especially as regards chromatin organization dependent modifications. Tat GRU demethylation seems rather slow and not processive, and this feature might minimize the risk of double-stranded breaks. But does such a mechanism perform the demethylation of the paternal genome occurring in the mouse egg after fertilization (7), which is rapid, replication-independent, and affects numerous fully methylated CpGs? A mechanism introducing a large number of DNA strand breaks would bear the risk of damaging genome integrity, and, indeed, we obtained no evidence of extensive cytidine incorporation at this developmental stage. This observation raises the possibility that there are several active mechanisms of cytosine demethylation operating at different stages in development.
Materials and Methods
Cell Synchronization.
We used a subclone of the rat hepatoma cell line (H4IIEC3). Cells were presynchronized in G1 by a 30-h incubation in Coon’s modified Ham-F12 medium without isoleucine in the presence of 0.5% FCS. They were then synchronized as follows: The medium was replaced by isoleucine-containing medium supplemented with 5% FCS and 5 μg/ml aphidicolin, and cells were cultured for 18 h. Cells were then released from the S phase block in aphidicolin-free medium supplemented with 10−7 M dexamethasone. Cells were subsequently collected at regular intervals by trypsinization, and genomic DNA was purified using the Wizard Genomic DNA purification kit (Promega). For cytometry analysis, cells were fixed in ice-cooled 70% ethanol, washed with PBS, and incubated at 37°C for 30 min in 500 μl of PBS containing RNase (100 μg/ml) and propidium iodide (50 μg/ml). Analyses of at least 10,000 cells were performed with a Coulter Elite-ESP flow cytometer (Beckman Coulter) using a 15 mW air-cooled argon-ion laser tuned at 488 nm, and fluorescence was measured through a 620-nm band-pass filter. Cell doublets were eliminated on the basis of DNA peak vs. DNA area signals, and cell cycle quantification was performed by using the multicycle software (Phoenix Flow Systems, San Diego).
Methylation Analysis.
Hydrazine-piperidine-LM-PCR analysis was performed as described (39). Briefly, after chemical treatment, genomic DNA was denatured, and a gene-specific primer was extended to create blunt ends suitable for linker ligation allowing subsequent LM-PCR amplification (Fig. 7).
Quantification of the methylation level of individual cytosines was done by using MethylQuant (22). Genomic DNA was treated with sodium bisulfite, and the converted upper strand was PCR-amplified as described. Real-time PCR quantification of the total amplified product and of the subpopulation corresponding to the specific methylation status of the cytosine of interest was performed by using, respectively, the nondiscriminative (ND) and the discriminative (D) primer sets designed for the analysis of the U3 cytosine (22).
Haipin-bisulfite PCR was adapted to the −2.5 Tat GRU from the protocol described (5). The two strands of the GRU were ligated together by using a hairpin linker (5′P-AAAGAGCGATGCGTTCGAGCATCGCT) that is compatible with the ends resulting from cleavage at the −2,302 BsmAI site. Bisulfite conversion was carried out as described (39), except that the incubation was performed in a PCR machine with slow ramp temperatures: 3× (98°C for 1 s, 50°C for 5 min); 3× (98°C for 1 s, 50°C for 10 min); 3× (98°C for 1 s, 50°C for 20 min); 3× (98°C for 1 s, 50°C for 40 min). PCR amplification of the −2.5 Tat GRU was performed in trimethyl ammonium chloride-containing buffer (39) by using the following primers: TTTGTTGTATAGGATGTTTTAGT and CCAAAATTTACCAATCTCTACTA. Amplified DNA was cloned in pGEM-T Easy vector (Promega).
DNA Strand Cleavage Analysis.
Rat fetal (E15) hepatocytes were isolated and cultured as described (19), except that the nonadhering hematopoietic cells were removed 16 h after plating. Genomic DNA was analyzed by LM-PCR as described (24, 39). The LM-PCR was performed to map the 5′ P ends of DNA molecules that were cleaved on at least one strand during the demethylation process as follows: DNA was denatured, and a gene-specific primer was extended to create blunt-ends allowing linker ligation (Fig. 7).
Supplementary Material
Acknowledgments
We thank M. C. Gendron for the flow cytometry analysis; C. Brossas for technical support; M. Georges for the injection of fluorescent dCTP in fertilized mouse embryo; W. Bickmore, E. Heard, C. Darzacq, and D. Higuet for helpful discussions and critical reading of the manuscript; and A. Kropfinger for corrections of the English language. This work was supported in part by the Centre National de la Recherche Scientifique (CNRS) and grants from the Association de Recherche Sur le Cancer and the Ligue Nationale Contre le Cancer (to T.G.). C.K. was supported by fellowships from CNRS and the Ligue Nationale Contre le Cancer.
Abbreviations
- En
embryonic day n
- GRU
glucocorticoid-responsive unit
- LM-PCR
ligation-mediated PCR
- Tat
tyrosine aminotransferase
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Meehan R. R. Semin. Cell Dev. Biol. 2003;14:53–65. doi: 10.1016/s1084-9521(02)00137-4. [DOI] [PubMed] [Google Scholar]
- 3.Jones P. A., Baylin S. B. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 4.Laird P. W. Nat. Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 5.Laird C. D., Pleasant N. D., Clark A. D., Sneeden J. L., Hassan K. M., Manley N. C., Vary J. C., Jr, Morgan T., Hansen R. S., Stoger R. Proc. Natl. Acad. Sci. USA. 2004;101:204–209. doi: 10.1073/pnas.2536758100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z. X., Riggs A. D. Biochem. Cell Biol. 2005;83:438–448. doi: 10.1139/o05-138. [DOI] [PubMed] [Google Scholar]
- 7.Reik W., Dean W., Walter J. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 8.Simonsson S., Gurdon J. Nat. Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- 9.Kress C., Thomassin H., Grange T. FEBS Lett. 2001;494:135–140. doi: 10.1016/s0014-5793(01)02328-6. [DOI] [PubMed] [Google Scholar]
- 10.Bird A. Nat. Immunol. 2003;4:208–209. doi: 10.1038/ni0303-208. [DOI] [PubMed] [Google Scholar]
- 11.Rougier N., Bourc’his D., Gomes D. M., Niveleau A., Plachot M., Paldi A., Viegas-Pequignot E. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paroush Z., Keshet I., Yisraeli J., Cedar H. Cell. 1990;63:1229–1237. doi: 10.1016/0092-8674(90)90418-e. [DOI] [PubMed] [Google Scholar]
- 13.Matsuo K., Silke J., Georgiev O., Marti P., Giovannini N., Rungger D. EMBO J. 1998;17:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh C. L. Mol. Cell. Biol. 1999;19:46–56. doi: 10.1128/mcb.19.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruniquel D., Schwartz R. H. Nat. Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharya S. K., Ramchandani S., Cervoni N., Szyf M. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 17.Wolffe A. P., Jones P. L., Wade P. A. Proc. Natl. Acad. Sci. USA. 1999;96:5894–5896. doi: 10.1073/pnas.96.11.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng H. H., Zhang Y., Hendrich B., Johnson C. A., Turner B. M., Erdjument-Bromage H., Tempst P., Reinberg D., Bird A. Nat. Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 19.Thomassin H., Flavin M., Espinas M. L., Grange T. EMBO J. 2001;20:1974–1983. doi: 10.1093/emboj/20.8.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flavin M., Cappabianca L., Kress C., Thomassin H., Grange T. Mol. Cell. Biol. 2004;24:7891–7901. doi: 10.1128/MCB.24.18.7891-7901.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abel G. A., Wochnik G. M., Ruegg J., Rouyer A., Holsboer F., Rein T. Mol. Endocrinol. 2002;16:1352–1366. doi: 10.1210/mend.16.6.0842. [DOI] [PubMed] [Google Scholar]
- 22.Thomassin H., Kress C., Grange T. Nucleic Acids Res. 2004;32:e168. doi: 10.1093/nar/gnh166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grange T., Bertrand E., Rigaud G., Espinás M. L., Fromont-Racine M., Roux J., Pictet R. Methods. 1997;11:151–163. doi: 10.1006/meth.1996.0401. [DOI] [PubMed] [Google Scholar]
- 24.Cappabianca L., Thomassin H., Pictet R., Grange T. In: Chromatin Protocols. Becker P. B., editor. Vol. 119. Totowa, NJ: Humana; 1999. pp. 427–442. [Google Scholar]
- 25.Weiss A., Keshet I., Razin A., Cedar H. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 26.Zhu B., Zheng Y., Hess D., Angliker H., Schwarz S., Siegmann M., Thiry S., Jost J. P. Proc. Natl. Acad. Sci. USA. 2000;97:5135–5139. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu B., Zheng Y., Angliker H., Schwarz S., Thiry S., Siegmann M., Jost J. P. Nucleic Acids Res. 2000;28:4157–4165. doi: 10.1093/nar/28.21.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrich B., Hardeland U., Ng H. H., Jiricny J., Bird A. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 29.Hardeland U., Bentele M., Jiricny J., Schar P. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan H. D., Dean W., Coker H. A., Reik W., Petersen-Mahrt S. K. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 31.Zhu B., Benjamin D., Zheng Y., Angliker H., Thiry S., Siegmann M., Jost J. P. Proc. Natl. Acad. Sci. USA. 2001;98:5031–5036. doi: 10.1073/pnas.091097298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jost J. P., Oakeley E. J., Zhu B., Benjamin D., Thiry S., Siegmann M., Jost Y. C. Nucleic Acids Res. 2001;29:4452–4461. doi: 10.1093/nar/29.21.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tini M., Benecke A., Um S. J., Torchia J., Evans R. M., Chambon P. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 34.Chen D., Lucey M. J., Phoenix F., Lopez-Garcia J., Hart S. M., Losson R., Buluwela L., Coombes R. C., Chambon P., Schar P., Ali S. J. Biol. Chem. 2003;278:38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- 35.Millar C. B., Guy J., Sansom O. J., Selfridge J., MacDougall E., Hendrich B., Keightley P. D., Bishop S. M., Clarke A. R., Bird A. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- 36.Gong Z., Morales-Ruiz T., Ariza R. R., Roldan-Arjona T., David L., Zhu J. K. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 37.Gehring M., Huh J. H., Hsieh T. F., Penterman J., Choi Y., Harada J. J., Goldberg R. B., Fischer R. L. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi Y., Harada J. J., Goldberg R. B., Fischer R. L. Proc. Natl. Acad. Sci. USA. 2004;101:7481–7486. doi: 10.1073/pnas.0402328101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomassin H., Oakeley E. J., Grange T. Methods. 1999;19:465–475. doi: 10.1006/meth.1999.0883. [DOI] [PubMed] [Google Scholar]
- 40.Rigaud G., Roux J., Pictet R., Grange T. Cell. 1991;67:977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- 41.Scharer O. D., Jiricny J. BioEssays. 2001;23:270–281. doi: 10.1002/1521-1878(200103)23:3<270::AID-BIES1037>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.