Abstract
It has long been suspected that a double Holliday junction (dHJ) could be resolved by a topoisomerase partnered with a helicase by convergent branch migration of the HJs. Genetic analysis of yeast TOP3 and SGS1 has lent considerable evidence to the notion that the protein products of these genes are involved in just such a process, although biochemical analysis of the metabolism of a dHJ has been hindered by the lack of a substrate that adequately replicates the endogenous structure. We have synthesized a dHJ substrate that recapitulates many of the features of an endogenous dHJ and represents a much earlier intermediate in the resolution pathway. Here, we show that Drosophila topoisomerase IIIα (Topo IIIα) and Blm (a homolog of Sgs1) are capable of resolving this substrate to non-cross-over products and that this activity is stimulated by replication protein A (RPA). We investigated the ability of other Drosophila topoisomerases to perform this reaction in concert with Blm and RPA and discovered that this resolution activity is unique to Topo IIIα. Examination of the mechanism of resolution reveals that Topo IIIα, Blm, and RPA resolve this substrate by convergent migration of the two HJs toward each other, collapsing the dHJ. This mechanism stands in contrast to classic resolvase activities that use a structure-specific endonuclease to cleave the HJs.
Keywords: recombination, repair, DNA strand exchange, topological constraint
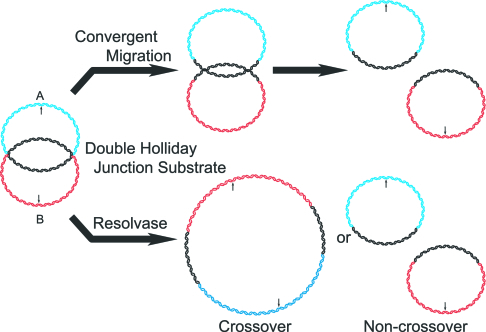
There are primarily two proposed pathways for the processing of the double Holliday junction (dHJ) intermediate of the double-strand break repair model. Structure-specific endonucleases (resolvases) have been identified that can cleave HJs, with the orientation of the cleavage events dictating whether the resolution results in gene conversion events either with (cross-overs, COs) or without (non-COs, NCOs) the exchange of sequences flanking the original break site (1–3). This model of resolution is represented by the lower pathway in Fig. 1. In a second potential pathway, the dHJ is resolved by the convergent branch migration of the two HJs by a topoisomerase and helicase (4, 5). This mode of resolution would result exclusively in NCO products and is represented by the upper pathway of Fig. 1. Elegant genetic studies in Saccharomyces cerevisiae have pointed toward topoisomerase III (Topo III) and Sgs1, a recQ family helicase, as a topoisomerase/helicase pair that may play this role in vivo (6).
Fig. 1.
Pathways of resolution of the DHJS. In the upper pathway, the linking number between circles A and B is continuously reduced by the convergent branch migration of the HJs by a topoisomerase/helicase complex until the two circles are completely unlinked to yield NCO products. In the lower pathway, a structure-specific endonuclease can cut the two HJs, with the orientation of the two cuts relative to each other determining whether CO or NCO products will be produced. The arrows indicate the location of BamHI restriction sites on the molecules. Digestion of either the CO or NCO products by BamHI will result in two linear molecules that are 416 and 465 bp in length.
In higher eukaryotes, there are two isoforms of Topo III, α and β, and multiple recQ helicases. Of the five recQ helicases in humans, BLM, WRN, and RecQ4 all have been implicated in syndromes involving genetic instability and susceptibility to a wide range of cancers (6). In Drosophila, there are three recQ helicases: Blm, RecQ4, and RecQ5. Flies mutant for DmBlm (Drosophila melanogaster Blm) show elevated mitotic recombination, nondisjunction, and chromosome loss and are nearly sterile (7–9), recapitulating many of the features of BLM deficiency in humans. Drosophila Blm has been genetically implicated in synthesis-dependent strand annealing and nonhomologous end-joining repair pathways in response to an induced double-strand break (10–12). These data are not inconsistent with a potential role for Blm in homologous recombination, because SGS1 has also been shown to function outside of homologous recombination in roles that are independent of TOP3 (13). There are no known disorders associated with mutations in Topo IIIα or β, although deletion of Topo IIIα is lethal in mice and Drosophila, and deletion of Topo IIIβ results in adult mice with reduced lifespan, reduced fertility, and an increased incidence of tumors (14–17).
A biochemical analysis of human BLM (hBLM) indicates that it may be ideally suited to migrate HJs. In addition to being a 3′–5′ helicase, characterization of hBLM revealed that it can bind to and migrate oligonucleotide-based synthetic HJs (18, 19). In addition, hBLM also possesses ssDNA annealing activity, as do most of the other characterized human recQ helicases (20–23). With both helicase and annealing activities, hBLM can perform strand exchange reactions, although the mechanism of this strand exchange reaction is certainly different from the one performed by rad51. This strand exchange activity is particularly interesting in the context of HJ migration; one could envision the migration of a HJ as a pair of strand exchange reactions.
With human Topo IIIα (hTopo IIIα), hBLM can resolve a small synthetic dHJ (24). Wu and Hickson (24) showed that these proteins resolve a synthetic DNA structure termed a double CO molecule (DCM) (25). The DCM used in this study is constructed from two oligonucleotides that are annealed and ligated to form a dHJ separated by 14 bp, with short duplexes outside the junctions terminated by hairpin structures. This substrate is resolved only in the presence of both hTopo IIIα and hBLM to form products that are analogous to NCOs. Although these results are consistent with convergent migration of the HJs, one cannot exclude other mechanisms of resolution. Because the HJs in the DCM are necessarily immobile, any migration of a HJ will generate unpaired DNA regions: a preferred substrate of type IA topoisomerases. Because of the short distance between the HJs, the two ssDNA circles that compose the substrate possess a linking number of two, making resolution by random strand passage events feasible. To better understand whether and how Topo III and recQ helicases are capable of resolving a dHJ, a dHJ-containing substrate is needed that better mimics the endogenous intermediate.
We have recently developed a method to synthesize a substrate that consists of two small dsDNA circles conjoined by two HJs (26). Because this Double Holliday Junction Substrate (DHJS) is circular in nature, it recapitulates the topological constraint that is inherent to endogenous dHJs. The DHJS contains a pair of HJs separated by 165 bp of homologous sequence, allowing the junctions to migrate (in the presence of a helicase and topoisomerase) without the obligatory generation of unpaired ssDNA (Fig. 1). In this study, we investigated the ability and requirements of DmTopo IIIα and DmBlm to resolve this substrate and the mechanism of that resolution.
Results
Topo IIIα and Blm Resolve the DHJS.
The resolution of the DHJS could proceed by two different pathways, as illustrated in Fig. 1. In previous work, we demonstrated that T7 endonuclease I (T7 endo I), a structure-specific endonuclease, could resolve the DHJS by the resolvase pathway (26). This reaction produced both CO products (dimeric circle), which have reduced electrophoretic mobility relative to the DHJS, and NCO products (two monomeric circles), which have increased electrophoretic mobility. In contrast, the convergent migration pathway of resolution should produce only NCO products. We endeavored to determine whether Drosophila Topo IIIα and Blm could also resolve the DHJS and, if so, by what pathway this resolution occurs.
The expression and purification of Drosophila Topo IIIα from a baculovirus system is described in ref. 17, and the same system was used to express and purify Drosophila Blm (Fig. 8, which is published as supporting information on the PNAS web site), whose cDNA had been isolated and cloned from an embryonic cDNA library. When the DHJS was incubated with either Topo IIIα or Blm, no resolution of the DHJS could be detected (Fig. 2, lanes 2 and 3). However, when the two proteins were both added to a reaction, some resolution products were formed that comigrated with the A and B circles, markers for NCO products (Fig. 2, lane 4 vs. lanes 7 and 8).
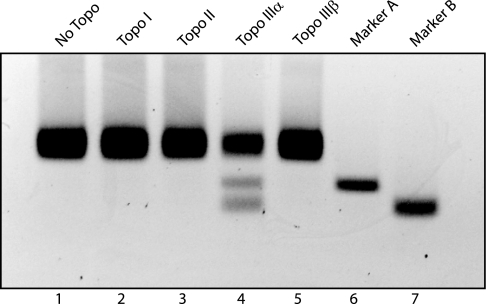
Fig. 2.
Topo IIIα and Blm can resolve the DHJS. There was no detectable activity on the substrate with either Topo IIIα alone (lane 2) or Blm alone (lane 3). When both proteins are added to the reaction, resolved DNA circles can be detected (lane 4). This activity depends on the activity of the Topo IIIα, because Topo IIIα with a mutated active-site tyrosine (mut) cannot support resolution (lane 5); it also depends on the helicase activity of Blm, because omission of ATP from the reaction buffer also abolishes activity (lane 6). Markers A and B are for the resolved products (lanes 7 and 8).
We next wanted to determine whether the enzymatic activity of both proteins is required for resolution or whether one or the other is simply required for correct localization/binding to the substrate. The active-site tyrosine of Topo IIIα was determined by homology to be amino acid 356 and was subsequently mutated to phenylalanine. The mutant enzyme, Topo IIIα-Y356F, was expressed and purified as the wild-type enzyme (Fig. 8) and was verified to be inactive on hypernegatively supercoiled DNA as well as negatively supercoiled bubble substrate (data not shown). When this protein was substituted for the wild-type Topo IIIα, no resolution products could be detected (Fig. 2, lane 5). To determine whether Blm’s helicase activity was required for resolution, ATP was omitted from the reaction buffer, because ATP is an essential cofactor for Blm but not for Topo IIIα. When the ATP was omitted, no resolution of the DHJS could be detected (Fig. 2, lane 6). These results verify that the enzymatic activities of both proteins are required for DHJS resolution.
Replication Protein A (RPA) Stimulates Topo IIIα/Blm Resolution of the DHJS.
Although Topo IIIα and Blm were able to resolve the DHJS, the activity of these proteins on the substrate was not robust (≈15% of the substrate was resolved). To test whether any cofactors can promote the DHJS resolution activity, we looked toward RPA, a protein that has been shown to stimulate hBLM helicase activity on larger substrates (27). Human RPA (hRPA) is 29–42% identical and 43–60% similar across the three subunits to Drosophila RPA, showing the highest degree of identity and similarity within the 70-kDa subunit that physically interacts with BLM (28). When hRPA is included in the reaction, the efficiency of the resolution of DHJS is raised to nearly 50%, whereas hRPA alone has no activity on the substrate (Fig. 3, lanes 2–6). Notably, the stimulatory concentrations of RPA on the resolution reactions of Topo IIIα and Blm are very similar to those that are reported to stimulate hBLM helicase activity (27). It has been established that hBLM and hRPA physically interact, but we could not rule out that the stimulation of the resolution reaction by RPA was simply through its ability to bind ssDNA. To determine whether this possibility was the case, Topo IIIα and Blm reactions were also supplemented with two other ssDNA-binding proteins: Escherichia coli SSB and T4 gp32. The stimulation of Topo IIIα- and Blm-mediated resolution was specific to RPA; neither SSB nor gp32 showed any stimulation of the resolution of the DHJS (Fig. 3, lanes 7–9 and 11–13, respectively). These results are consistent with the previously published report by Brosh et al. (27) in which it was demonstrated that hRPA stimulated hBLM helicase activity on large (259 bp) DNA duplexes but that SSB and gp32 could not.
Fig. 3.
The activity of Topo IIIα and Blm is specifically stimulated by RPA. The amount of resolution of the DHJS increases with increasing concentrations of hRPA in the reaction (compare lane 2 with lanes 3–5), whereas hRPA alone has no activity on the substrate (lane 6). This stimulation was specific to RPA; including other ssDNA-binding proteins, E. coli SSB (lanes 7–9) or T4 gp32 (lanes 11–13), showed no stimulation of the resolution of DHJS by Topo IIIα and Blm. The supplemented proteins are at 44, 88, and 175 nM concentrations, respectively.
Resolution of the DHJS with Blm and RPA Is Specific for Topo IIIα.
Next, we examined the role of Topo IIIα in these reactions. Although we have established that inactive Topo IIIα cannot support resolution, we proceeded to determine whether other topoisomerase activities could support resolution of the DHJS. All of the topoisomerases were first verified to be active in the reaction conditions used here by their ability to relax hypernegatively supercoiled plasmid DNA (data not shown). No resolution of the DHJS is detected when Drosophila Topo I or Topo II is substituted for Topo IIIα (Fig. 4, lanes 2–4), indicating that the major type IB and type II topoisomerases cannot perform this reaction in conjunction with Blm. Drosophila Topo IIIα and Topo IIIβ are both type IA topoisomerases, and both have been demonstrated to have similar DNA relaxation activities. However, when Topo IIIβ is included in the reaction, no resolution of the DHJS is detected (Fig. 4, lane 5). Although hTopo IIIα has been shown to physically interact with hBLM, no such interaction has been demonstrated between hBLM and hTopo IIIβ. In Drosophila, we have also seen a physical interaction between Topo IIIα and Blm but have seen no evidence of such an interaction between Topo IIIβ and Blm (data not shown). When taken together, these data suggest not only that a type IA topoisomerase is required for this reaction but also that the topoisomerase must physically interact with Blm.
Fig. 4.
Of all of the Drosophila topoisomerases, only Topo IIIα supports resolution with Blm and RPA. Blm and RPA were incubated with 36 nM Topo I (lane 2), Topo II (lane 3), Topo IIIα (lane 4), or Topo IIIβ (lane 5) in resolution reactions. Only Topo IIIα produced products that comigrated with markers A and B (lanes 6 and 7, respectively).
Although we have established that Topo IIIα and Blm can resolve a DHJS and that this activity is stimulated by RPA, there are also suggestions that this resolution is indeed being carried out through the convergent migration of the HJs. As previously demonstrated (26), the resolution of this substrate by a resolvase would result in two different sets of products: monomeric circles that are analogous to NCO products, which migrate more quickly than the substrate during electrophoresis, and dimeric circles that are analogous CO products, which migrate more slowly (Fig. 1). In all of the data shown thus far, the only resolution products that are produced by Topo IIIα and Blm correspond to the NCO pathway. In addition, the resolution of the DHJS by Topo IIIα and Blm is stimulated by RPA, a protein that was identified previously as stimulating hBLM helicase activity only on long duplexed DNA substrates, presumably by increasing the helicase’s processivity (27). These data suggest that Blm helicase activity is required across the entire homologous region and not simply for a short period to create a substrate for a resolvase reaction mediated by Topo IIIα. Although these data are supportive of the convergent migration model, it is still a formal possibility that Topo IIIα is acting as a resolvase, with the activity of Blm required to orient Topo IIIα such that only NCO products are formed. The experiments presented in the following more rigorously demonstrate the mechanism of resolution by the Topo IIIα/Blm/RPA complex.
Topo IIIα/Blm/RPA Cannot Resolve a Psoralen Cross-Linked DHJS.
The requirement for the DNA to be unwound in front of the HJ and re-paired behind the HJ during convergent migration is one fundamental difference between this mechanism and a resolvase mechanism of resolution. We exploited this difference by cross-linking the DHJS with psoralen, a chemical that creates interstrand cross-links, primarily at TA sequences, once the psoralen/DNA complex is irradiated with long-wavelength UV light (29, 30). It was previously shown that after exposure to UV light in the presence of psoralen, the DHJS was cross-linked between the HJs, with the majority of the cross-links within the loxP sequence as judged by restriction digestion analysis (26).
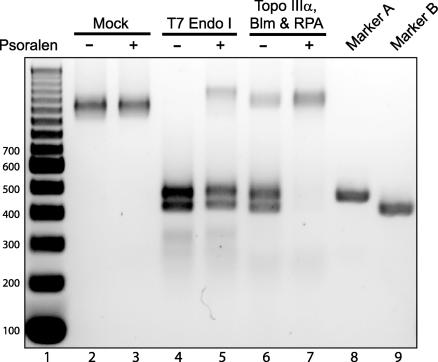
We investigated the effect of the cross-linking on T7 endo I and Topo IIIα/Blm/RPA resolution of the DHJS. To simplify the interpretation of the results, the products of these reactions were digested by BamHI before electrophoresis (Fig. 1; both the A and B circles contain a unique BamHI site in the nonhomologous sequence). This digest collapses both the CO and NCO products of the T7 endo I resolution down to two bands, 416 and 465 bp in length. The process of cross-linking could, in theory, result in nicking the DNA such that the HJs could spontaneously migrate and the substrate would resolve on its own; however, UV irradiation with or without psoralen showed no detectable resolution products (Fig. 5, lanes 2 and 3). As predicted, the cross-linking had little effect on the resolution activity of T7 endo I, with the majority of the cross-linked DHJS resolved to the expected products (Fig. 5, lanes 4 and 5). However, the presence of the interstrand cross-links in the DHJS almost completely inhibits Topo IIIα/Blm/RPA-mediated resolution of the substrate (Fig. 5, lanes 6 and 7). This result demonstrates that the mechanism of the Topo IIIα/Blm/RPA complex is distinct from that of a resolvase, and it is consistent with the resolution of this complex occurring through convergent migration of the HJs.
Fig. 5.
Psoralen cross-linking inhibits DHJS resolution by Topo IIIα/Blm/RPA. After the DHJS, with or without psoralen cross-links, was reacted in the presence of the indicated proteins, the products of the reactions were digested with BamHI. Lane 1 contains a DNA size marker, with the size of the bands denoted to the left of the gel (in base pairs). There is no resolution of the substrate in the mock reactions (lanes 2 and 3). T7 endo I can resolve the cross-linked DHJS with little inhibition of activity (lanes 4 and 5), whereas resolution by Topo IIIα/Blm/RPA is almost completely inhibited by the psoralen cross-links (lanes 6 and 7). Lanes 8 and 9 contain BamHI-digested markers A and B, respectively.
Topo IIIα/Blm/RPA Migrates the HJs in the DHJS.
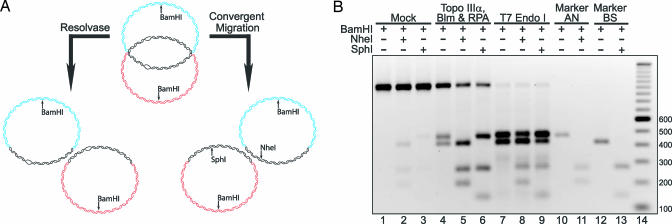
Although evidence presented thus far supports the notion that the DHJS is resolved by convergent branch migration, we wanted to generate a DHJS that could better differentiate the two mechanisms than our original substrate, which we accomplished by preparing a DHJS substrate that contains mismatches within the homologous sequences. If the dHJ undergoes convergent migration, these mismatches would be correctly paired in the resolved product to create two different restriction enzyme sites, whereas resolution products generated by a structure-specific endonuclease or strand transfer reactions would retain the mismatches (Fig. 6A). To make this substrate, two plasmids were created by replacing the NheI restriction site with the SphI restriction site in the pDHJS BN+/− plasmids to create pDHJS BS+/−. Using the two plasmids that we created and the pDHJS AN+/− plasmids, a substrate was prepared that we have termed MM-DHJS. MM-DHJS is identical to DHJS with the exception of having two mismatches: AT·AT on one homologous strand and TA·TA on the other, 35 bp from the nearest HJ. The structure of MM-DHJS is shown diagrammatically in Fig. 6A. In contrast to the DHJS, MM-DHJS is refractory to digestion by NheI and SphI (Fig. 6B, compare lane 1 with lanes 2 and 3).
Fig. 6.
DHJS resolution by Topo IIIα, Blm, and RPA occurs through migration of the HJs rather than by a resolvase mechanism. (A) A DHJS containing mismatches allows differentiation between resolvase and migration mechanisms of resolution. We created a DHJS by replacing the NheI restriction sites in the “B” vectors with a SphI site, pDHJS BS+/−. The DHJS that we made with these vectors, MM-DHJS, contains a pair of two nucleotide mismatches, which render the substrate refractory to digestion by NheI and SphI. If the substrate is resolved by a classic resolvase activity (left pathway), the mismatches will be retained in the products. If the substrate is resolved through migration of the HJs (right pathway), then the mismatches will be re-paired with their complementary sequence, and the products of the reaction will now be sensitive to digestion by NheI and SphI. (B) The products of the Topo IIIα/Blm/RPA reactions are generated by the migration of the dHJ. MM-DHJS is refractory to digestion by NheI (lane 2) or SphI (lane 3). After incubation with Topo IIIα/Blm/RPA, resolution products are detected (lane 4), and these products are sensitive to NheI (lane 5) and SphI (lane 6) digestion. In contrast, resolution of MM-DHJS by T7 endo I (lane 7) yields products that are largely refractory to digestion by NheI (lane 8) and SphI (lane 9). Lanes 10–13 contain the digested marker molecules, and lane 14 contains a DNA size marker with the size of the bands denoted to the right of the gel in base pairs.
When MM-DHJS is incubated with Topo IIIα/Blm/RPA, the substrate is resolved as its predecessor was (Fig. 6B, lane 4). When these reaction products are digested by the restriction enzymes, NheI digests the A circles to completion (Fig. 6B, lane 5), and SphI digests the B circles to completion (Fig. 6B, lane 6). In contrast, the T7 endo I-generated products remained largely refractory to digestion with NheI or SphI (Fig. 6B, lanes 8 and 9, respectively). There are products from the T7 endo I reaction that are digested by NheI or SphI, but these products most likely arise from the spontaneous migration of the second HJ after the first has been cut by T7 endo I, as observed during the characterization of the DHJS (26).
Another interesting point to take from these data is that NheI or SphI digestion diminishes the amount of unresolved material (compare the upper band in lane 4 of Fig. 6B to the upper bands in lanes 5 and 6). These data indicate that, although this material has not been fully resolved, one of the HJs has migrated past the mismatches. These molecules still contained a dHJ, but once the substrate was digested within the homologous sequence (by NheI or SphI) and the topological restraint was released (by digestion with BamHI), the dHJ could simply migrate off the ends of the DNA. So even though ≈50% of the substrate was fully resolved by Topo IIIα/Blm/RPA, ≈80% of the unresolved material had been acted upon by the complex to such a degree that the restriction sites were restored.
Discussion
In previous models of dHJ resolution, a recQ helicase had been proposed to perform convergent migration of the HJs with a topoisomerase relieving the associated superhelical stress (5). However, the ability of type IA topoisomerases to pass one ssDNA through the other makes these enzymes an ideal agent in removing the topological linkages in between dHJ (5). Recent work has shown that several type IA topoisomerases, in association with BLM, are capable of removing the last two linkages from an oligonucleotide-based substrate (24, 31). In this work, we have shown that DmTopo IIIα, in association with DmBlm, is capable of catalyzing the earlier steps of convergent branch migration of the HJs. Together, these enzymes resolved a mobile, topologically constrained dHJ containing ≈30 linkages. We have provided strong evidence that this resolution occurs through convergent migration of the HJs. This convergent migration could occur through the mobilization of both HJs or the migration of just one HJ toward the other. Showing that these proteins are capable of resolving a substrate that contains many of the features of an endogenous dHJ strengthens the convergent migration model and also confirms a role for a type IA topoisomerase in the branch migration process (Fig. 7B).
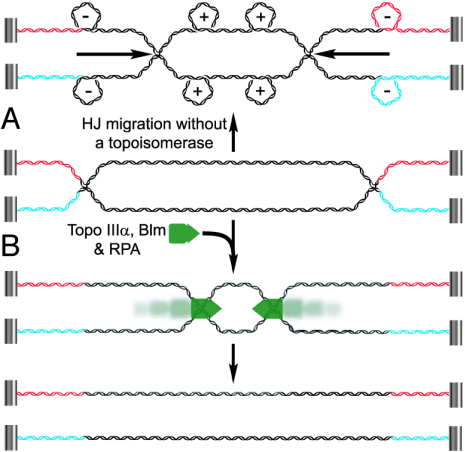
Fig. 7.
A model for Topo IIIα/Blm/RPA-mediated resolution of a dHJ. In this illustration, the ends of the DNA are attached to a hypothetically immobile structure to simulate topological constraints imposed on an endogenous dHJ. (A) If the HJs in a dHJ were to undergo convergent migration in the absence of a topoisomerase, negative writhe would be induced behind the migrating HJ, and positive writhe would accumulate in front of the HJ. (B) In the presence of Topo IIIα/Blm/RPA (represented as a green complex), the HJs undergo convergent migration until the linking number between the two DNAs is reduced to zero. Through established protein–protein interactions, it seems likely that these proteins function as a complex, and within this complex, Topo IIIα would be able to perform the strand passage events to unlink the two strands. The association between Topo IIIα and Blm could provide the directionality for the Topo IIIα strand passage events, although it is unclear at this time how directionality for Blm is established.
Although the convergent migration of a pair of HJs may seem straightforward, the process becomes complicated when one considers the reaction from a topological standpoint. As the HJs are migrated toward each other (in the absence of a topoisomerase), negative writhe accumulates behind the HJs, and positive writhe accumulates in front, much the same as would occur with an active replication fork (Fig. 7A) (32). However, during replication in eukaryotes, the relaxation of these writhes has been attributed to Topo I, a type IB topoisomerase that can relax both negative and positive supercoiling (5). Biochemical studies of Topo IIIs indicate that these enzymes only have weak to moderate relaxation activity toward negatively supercoiled DNA (17, 33–38) and can only relax positive supercoiling when a region of the DNA has been permanently denatured (17). However, during the resolution of the DHJS, Topo IIIα makes at least 30 strand passage events to separate the two DNA circles. If Topo IIIα were functioning independently of Blm, then these 30 strand passage events would need to occur in front of the migrating HJ, in a region of positive writhe. Given the biochemical characterization of Topo IIIα to date, this occurrence seems unlikely. Biochemically, Topo IIIβ is very similar to Topo IIIα (17, 38), yet it was unable to be substituted for Topo IIIα in the resolution reactions with Blm and RPA. These results, along with the fact that both Topo IIIα and RPA have been shown to physically interact with Blm, suggest that Topo IIIα, Blm, and RPA are functioning as a complex at the HJ (Fig. 7B). Complex formation could provide Topo IIIα access to ssDNA, its preferred substrate, within the HJ and allow it to perform the required strand passage events without the accumulation of positive and negative torsional stresses as the HJ migrates.
The movement of the HJ by Blm would give the strand passage events that are performed by Topo IIIα directionality, although it isn’t clear how the directionality of the entire complex is attained. Without a free DNA end to load on, the Topo IIIα/Blm/RPA complex could form on a HJ in either orientation and should function equally as well in migrating two HJs divergently and convergently. In this study, the nature of the DHJS provided refractory boundaries that likely inhibited complexes formed in a divergent orientation, but these events probably contributed to the inability of Topo IIIα/Blm/RPA to resolve all of the substrate in a reaction. It may well be that mechanisms exist in vivo to properly orient the complex (or complexes) to ensure the resolution of a dHJ.
The results presented here clearly highlight the intricacies and complexities that are involved in dHJ resolution. The reagents and approaches developed in this work may prove useful in shedding light on the mechanistic details in the process of resolving dHJs.
Materials and Methods
Purification of the Enzymes.
The purifications of Topo I (39), Topo II (40), and Topo IIIα (17) have been previously described. hRPA was a kind gift from Paul Modrich and Jochen Genschel (both of Duke University) (41).
The active-site tyrosine of Topo IIIα was predicted by homology to be at position 356, and this residue was mutated to phenylalanine by using standard procedures. Topo IIIα-Y356F was then expressed and purified as the wild-type protein (17).
Drosophila Blm was cloned from a Drosophila cDNA library and was overexpressed by using the Bac-to-Bac baculovirus expression system (Invitrogen) in Sf9 cells and purified by using a cleavable N-terminal GST tag and a C-terminal decahistidine tag by means of the procedures for the purification of Topo IIIα that are described in ref. 17.
Topo IIIβ was also overexpressed by using the baculovirus expression system in Sf9 cells and purified by using a cleavable N-terminal GST tag and a hydroxylapatite column (Fig. 8). The purification procedure is identical to that for Topo IIIα until the protein has been digested off the glutathione resin (17). After this point, Topo IIIβ is loaded onto an equilibrated hydroxylapatite column and washed with 50 mM NaPO4, pH 7.0/150 mM NaCl/10% glycerol/0.02% Triton X-100/5 mM 2-mercaptoethanol/0.1 μM 4-(2-aminoethyl)-benzenesulfonyl fluoride/80 nM aprotinin/45 μM leupeptin/3.6 μM bestatin/1.5 μM pepstatin A/1.4 μM E-64 (Sigma-Aldrich). Topo IIIβ is then eluted with the previous buffer at 400 mM NaPO4 (pH 7.0) and dialyzed as for Topo IIIα (17).
Synthesis of the DNA Substrates and Marker Molecules.
The synthesis of the DHJS, marker molecules, and the psoralen cross-linked DHJS is described in ref. 26. The MM-DHJS (DHJS with two dinucleotide mismatches) was created by replacing the portion of the homologous region containing the NheI restriction site with an identical sequence except for a TA-to-AT mutation that converts the NheI site to a SphI site. This fragment was cloned into pDHJS BN+ and pDHJS BN− (containing the NheI site) to create pDHJS BS+ and pDHJS BS− (containing the SphI site). These BS vectors were used in place of the BN vectors to synthesize MM-DHJS. The diagrammatical structure of MM-DHJS is shown in Fig. 6.
DHJS Resolution Reactions.
Resolution reactions using T7 endo I are described in ref. 26. Topo IIIα and Blm resolution reactions were carried out in 2.5 ng/μl DHJS/40 mM Tris, pH 7.5/0.1 mM EDTA/4 mM MgCl2/50 mM sodium acetate/1 mg/ml BSA/1 mM DTT/1 mM ATP unless otherwise indicated. Where included, standard protein concentrations were 36 nM Topo IIIα, 36 nM Blm, and 175 nM RPA. Reactions were set up on ice and then transferred to, and incubated at, 37°C for 1 h. Reactions were then stopped and processed as in the T7 endonuclease resolution reactions described above.
Experiments using other topoisomerases were handled in the manner described above except for Topo II. For this topoisomerase, the reaction was stopped by the addition of 10 mM EDTA and 500 mM NaCl (final concentration) and incubated for an additional 10 min at 37°C to minimize double-strand breaks. This sample was then treated with SDS and proteinase K and processed as described above.
Supplementary Material
Acknowledgments
We thank Drs. Jochen Genschel and Paul Modrich for hRPA and members of our laboratory and colleagues for stimulating discussions. This work was supported by National Institutes of Health Grant GM29006.
Abbreviations
- Topo III
topoisomerase III
- dHJ
double Holliday junction
- DHJS
Double Holliday Junction Substrate
- RPA
replication protein A
- CO
cross-over
- NCO
non-CO
- hBLM
human BLM
- hRPA
human RPA
- hTopo
human Topo
- T7 endo I
T7 endonuclease I.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Heyer W. D. Curr. Biol. 2004;14:R56–R58. doi: 10.1016/j.cub.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 2.Heyer W. D., Ehmsen K. T., Solinger J. A. Trends Biochem. Sci. 2003;28:548–557. doi: 10.1016/j.tibs.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Symington L. S., Holloman W. K. Science. 2004;303:184–185. doi: 10.1126/science.1093959. [DOI] [PubMed] [Google Scholar]
- 4.Nasmyth K. A. Annu. Rev. Genet. 1982;16:439–500. doi: 10.1146/annurev.ge.16.120182.002255. [DOI] [PubMed] [Google Scholar]
- 5.Wang J. C. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 6.Bachrati C. Z., Hickson I. D. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beall E. L., Rio D. C. Genes Dev. 1996;10:921–933. doi: 10.1101/gad.10.8.921. [DOI] [PubMed] [Google Scholar]
- 8.Boyd J. B., Golino M. D., Shaw K. E., Osgood C. J., Green M. M. Genetics. 1981;97:607–623. doi: 10.1093/genetics/97.3-4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kusano K., Johnson-Schlitz D. M., Engels W. R. Science. 2001;291:2600–2602. doi: 10.1126/science.291.5513.2600. [DOI] [PubMed] [Google Scholar]
- 10.Adams M. D., McVey M., Sekelsky J. J. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 11.McVey M., Larocque J. R., Adams M. D., Sekelsky J. J. Proc. Natl. Acad. Sci. USA. 2004;101:15694–15699. doi: 10.1073/pnas.0406157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Min B., Weinert B. T., Rio D. C. Proc. Natl. Acad. Sci. USA. 2004;101:8906–8911. doi: 10.1073/pnas.0403000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watt P. M., Hickson I. D., Borts R. H., Louis E. J. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwan K. Y., Moens P. B., Wang J. C. Proc. Natl. Acad. Sci. USA. 2003;100:2526–2531. doi: 10.1073/pnas.0437998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwan K. Y., Wang J. C. Proc. Natl. Acad. Sci. USA. 2001;98:5717–5721. doi: 10.1073/pnas.101132498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W., Wang J. C. Proc. Natl. Acad. Sci. USA. 1998;95:1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plank J. L., Chu S. H., Pohlhaus J. R., Wilson-Sali T., Hsieh T. S. J. Biol. Chem. 2005;280:3564–3573. doi: 10.1074/jbc.M411337200. [DOI] [PubMed] [Google Scholar]
- 18.Mohaghegh P., Karow J. K., Brosh R. M., Jr., Bohr V. A., Hickson I. D. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Brabant A. J., Ye T., Sanz M., German I. J., Ellis N. A., Holloman W. K. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- 20.Cheok C. F., Wu L., Garcia P. L., Janscak P., Hickson I. D. Nucleic Acids Res. 2005;33:3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia P. L., Liu Y., Jiricny J., West S. C., Janscak P. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macris M. A., Krejci L., Bussen W., Shimamoto A., Sung P. DNA Repair (Amsterdam) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Sharma S., Sommers J. A., Choudhary S., Faulkner J. K., Cui S., Andreoli L., Muzzolini L., Vindigni A., Brosh R. M., Jr. J. Biol. Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 24.Wu L., Hickson I. D. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 25.Fu T. J., Tse-Dinh Y. C., Seeman N. C. J. Mol. Biol. 1994;236:91–105. doi: 10.1006/jmbi.1994.1121. [DOI] [PubMed] [Google Scholar]
- 26.Plank J. L., Hsieh T. S. J. Biol. Chem. 2006;281:17510–17516. doi: 10.1074/jbc.M602933200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brosh R. M., Jr., Li J. L., Kenny M. K., Karow J. K., Cooper M. P., Kureekattil R. P., Hickson I. D., Bohr V. A. J. Biol. Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 28.Doherty K. M., Sommers J. A., Gray M. D., Lee J. W., von Kobbe C., Thoma N. H., Kureekattil R. P., Kenny M. K., Brosh R. M., Jr. J. Biol. Chem. 2005;280:29494–29505. doi: 10.1074/jbc.M500653200. [DOI] [PubMed] [Google Scholar]
- 29.Hearst J. E. Annu. Rev. Biophys. Bioeng. 1981;10:69–86. doi: 10.1146/annurev.bb.10.060181.000441. [DOI] [PubMed] [Google Scholar]
- 30.Zhen W. P., Buchardt O., Nielsen H., Nielsen P. E. Biochemistry. 1986;25:6598–6603. doi: 10.1021/bi00369a039. [DOI] [PubMed] [Google Scholar]
- 31.Wu L., Bachrati C. Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G. W., Hickson I. D. Proc. Natl. Acad. Sci. USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu L. F., Wang J. C. Proc. Natl. Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goulaouic H., Roulon T., Flamand O., Grondard L., Lavelle F., Riou J. F. Nucleic Acids Res. 1999;27:2443–2450. doi: 10.1093/nar/27.12.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hotoda N., Hanai R. J. Biochem. (Tokyo) 2000;127:1109–1113. doi: 10.1093/oxfordjournals.jbchem.a022705. [DOI] [PubMed] [Google Scholar]
- 35.Kim R. A., Wang J. C. J. Biol. Chem. 1992;267:17178–17185. [PubMed] [Google Scholar]
- 36.Kim Y. C., Lee J., Koo H. S. Nucleic Acids Res. 2000;28:2012–2017. doi: 10.1093/nar/28.9.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srivenugopal K. S., Lockshon D., Morris D. R. Biochemistry. 1984;23:1899–1906. doi: 10.1021/bi00304a002. [DOI] [PubMed] [Google Scholar]
- 38.Wilson T. M., Chen A. D., Hsieh T. J. Biol. Chem. 2000;275:1533–1540. doi: 10.1074/jbc.275.3.1533. [DOI] [PubMed] [Google Scholar]
- 39.Shaiu W. L., Hsieh T. S. Mol. Cell. Biol. 1998;18:4358–4367. doi: 10.1128/mcb.18.7.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh T. Methods Enzymol. 1983;100:161–170. doi: 10.1016/0076-6879(83)00052-x. [DOI] [PubMed] [Google Scholar]
- 41.Genschel J., Modrich P. Mol. Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.